Summary
In eukaryotes, most RNA molecules are exported into the cytoplasm after transcription. Long noncoding RNAs (lncRNAs) reside and function primarily inside the nucleus, but nuclear localization of mRNAs has been considered rare in both animals and plants. Here we show that Arabidopsis Anaphase Promoting Complex/Cyclosome (APC/C) coactivator genes CDC20 and CCS52B (CDH1 orthologue) are co-expressed with their target cyclin B genes (CYCBs) during mitosis. CYCB transcripts can be exported and translated, whereas CDC20 and CCS52B mRNAs are confined to the nucleus at prophase and the cognate proteins are not translated until the redistribution of the mRNAs to the cytoplasm after nuclear envelope breakdown (NEBD) at prometaphase. The 5’ untranslated region (UTR) plays dual roles in CDC20 mRNA nuclear localization and translation. Mitotic accumulation of CDC20 and CCS52B transcripts enables the timely and rapid activation of APC/C, while their nuclear sequestration at prophase appears to protect cyclins from precocious degradation.
Introduction
Understanding the patterns and regulatory mechanisms of organ formation in multicellular organisms is a central aspect of developmental biology (Lander, 2011). Animal organogenesis is completed during embryonic development or, in some instances, during metamorphosis; while in plants, active division and differentiation of stem cells and their progenitors in the shoot apical meristem (SAM) and the root apical meristem (RAM) lead to continuous formation of new tissues and organs, ensuring developmental plasticity in a changing environment (Gaillochet and Lohmann, 2015; Heidstra and Sabatini, 2014; Meyerowitz, 1997; Vernoux et al., 2000). Plant cell division, as in mammalian cells, yeast and Drosophila, is triggered and maintained by the kinase complex composed of cyclin-dependent kinases (CDKs) and various cyclin subunits. Fluctuating gene expression and orderly proteolysis of cyclins, spatial positive feedback of Cdk1-cyclin B1 redistribution, combined with the antagonistic actions of Wee1 kinase and Cdc25 phosphatase, generate a robust and highly ordered mitotic process (Coudreuse and Nurse, 2010; Dewitte and Murray, 2003; De Veylder et al., 2007; Morgan, 1995; Santos et al., 2012).
Destruction of cyclins at the appropriate time in the cell cycle is mediated by APC/C, an E3 ubiquitin ligase whose catalytic activity and substrate specificity are conferred by two coactivators, CDC20 and Cdc20 homolog 1 (CDH1) (Peters, 2006; Pines, 2011; Yu, 2007). During early mitosis, phosphorylation of the APC/C subunits, such as the auto-inhibitory segment loop in APC1, exposes the binding sites of CDC20 thus facilitating CDC20 association with APC/C (Fujimitsu et al., 2016; Kraft et al., 2003; Qiao et al., 2016; Zhang et al., 2016). At prometaphase, APC/C activity is restrained by the spindle assembly checkpoint (SAC), a regulatory pathway during which unattached kinetochores generate a diffusible ‘wait anaphase’ signal that triggers the incorporation of CDC20 into a complex composed of MAD2, BUBR1 and BUB3, leading to the formation of the mitotic checkpoint complex (MCC) (Fraschini et al., 2001; Hardwick et al., 2000; Sudakin et al., 2001). Recently it has been proposed that MCC itself could function as a diffusible signal to inhibit APC/C by recognizing a second CDC20 that has already bound to and activated APC/C (Izawa and Pines, 2015). Furthermore, APC/C activity is counteracted by the F box protein early mitotic inhibitor 1 (Emi1) (Reimann et al., 2001). The multi-faceted regulation of APC/C in various organisms suggests high plasticity of APC/C activity, and also implies the existence of additional mechanisms.
Subcellular RNA localization has been implicated in multiple cellular processes by regulation of spatial gene expression (Lipshitz and Smibert, 2000). For instance, the posterior-anterior polar localization of bicoid, oskar, gurken, and nanos mRNAs in Drosophila oocytes guides proper pattern formation and embryo development (Martin and Ephrussi, 2009). Long noncoding RNAs (lncRNAs) predominantly localize to the nucleus to modulate transcription factor binding, histone modification, chromosome structures and specific nuclear body formation (Batista and Chang, 2013; Engreitz et al, 2016; Geisler and Coller, 2013; Tsai et al., 2010). While mature mRNAs are considered to reside predominantly in the cytoplasm, deep sequencing of nuclear and cytoplasmic RNA fractions from various mouse tissues identified a number of mRNAs with higher amounts in the nucleus than in the cytoplasm (Bahar Halpern et al., 2015), suggesting a potential for mRNA nuclear retention in gene expression regulation. However, nuclear localization of mRNAs or mRNA precursors and its biological relevance have rarely been documented. In Drosophila embryos, the non-polyadenylated histone mRNAs are retained in the nuclei of DNA-damaged cells, contributing to the maintenance of genome integrity (Iampietro et al., 2014). CTN-RNA, an adenosine-to-inosine (A-to-I) edited mouse-specific pre-mRNA, localizes in the nuclear paraspeckle and can be rapidly cleaved under physiologic stress to produce mCAT2 mRNA encoding a cell-surface L-arginine receptor (Prasanth et al, 2005). Apart from these examples, nuclear sequestration of non-edited mature mRNAs remains to be discovered.
Here, through a comprehensive fluorescent in situ hybridisation (FISH) analysis of mRNA distribution of core cell cycle genes in Arabidopsis stem cells, we have found that CDC20 and CDH1 orthologue CCS52B mRNAs are sequestered in the nucleus during prophase. We show that CDC20 and CCS52B transcripts accumulate to peak levels but are confined to the nucleus at prophase, and redistribute into the cytoplasm following NEBD at prometaphase. With fluorescence live cell imaging, we demonstrate that this mRNA nuclear sequestration prevents CDC20 and CCS52B protein translation, thus blocking premature APC/C activation in early mitosis. By systematic mRNA deletion and chimeric RNA localization analysis, we found that CDC20 mRNA 5’UTR confers nuclear sequestration and is also involved in protein translation. Nuclear sequestration of CDC20 and CCS52B mRNAs reveals a previously unrecognized mechanism for the tuning of APC/C activity.
Results
Systematic Analysis of mRNA Localization of Core Cell Cycle Genes in Meristematic Cells
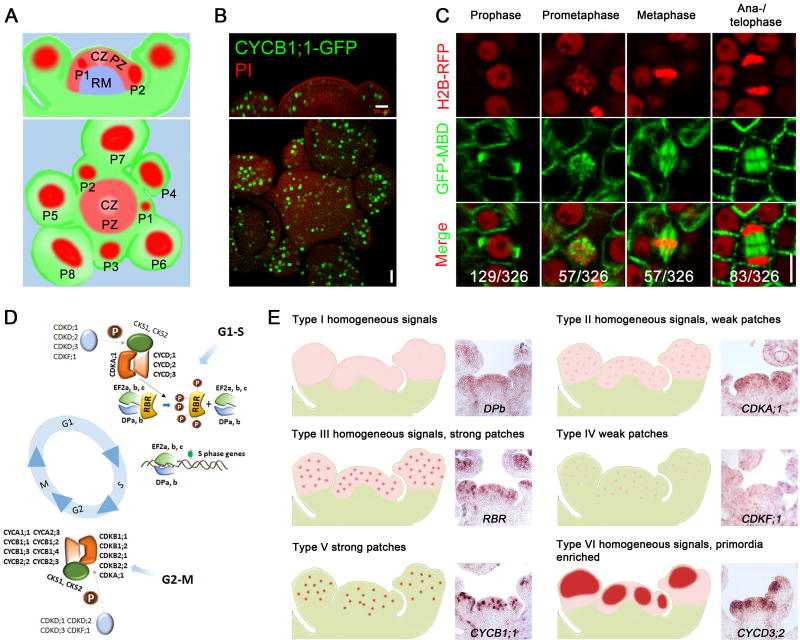
In Arabidopsis, the SAM is organized into three zones distinguished by cell division activity: the central zone (CZ) composed of slowly dividing stem cells, which is surrounded by the peripheral zone (PZ) that contains more rapidly dividing cells that give rise to primordia of leaves and flowers, and the rib meristem (RM) underlying the CZ and the PZ responsible for stem growth (Steeves and Sussex, 1989) (Figure1A). The distinct cell division activities in different SAM regions can be visualized by using a fusion of green fluorescent protein (GFP) to CyclinB1;1 (CYCB1;1-GFP), exhibiting a low number of GFP-positive cells in the slowly dividing cells of the CZ and RM, and relatively higher number in the PZ and flower primordia (Figure 1B). Using a GFP-microtubule-binding domain marker (GFP-MBD) and the nuclear reporter histone H2B fused to red fluorescent protein (H2B-RFP), we found that the microtubule and nuclear structures corresponding to different cell cycle stages could all be identified in the SAM (Figure 1C). Therefore, the SAM serves as a suitable system with which to study the control of the cell cycle in plants.
Figure 1. Expression Patterns of Core Cell Cycle Genes in Arabidopsis Meristematic Cells.
(A) A schematic representation showing the organization of the Arabidopsis inflorescence shoot apical meristem (SAM). Upper panel, side view; lower panel, top view. CZ, central zone; PZ, peripheral zone; RM, rib meristem; P, flower primordia, which form sequentially in the PZ.
(B) CYCB1;1-GFP reporter expression in wild type (WT) SAM. Scale bar, 20 µm.
(C) Expression of nuclear reporter H2B-RFP and microtubule reporter GFP-MBD in meristematic cells corresponding to different cell cycle stages. From 6 WT SAMs, 326 cells were observed to be undergoing division and the number of cells at each stage is shown. Scale bar, 5 µm.
(D) Functional modules of core cell cycle regulators in the Arabidopsis SAM.
(E) Classification of the mRNA distribution patterns of core cell cycle genes expressed in the SAM. In situ hybridisation images for representative genes in each class are shown.
CDKs, CYCs and other regulatory proteins constitute a group of core cell-cycle regulators. Multiple members in each CDK and cyclin subfamily exist in plants, suggesting a level of functional conservation but also specialized regulation of cell cycle progression in plants as compared to animals (Vandepoele et al., 2002) (Figure 1D). To explore the role of cell cycle regulatory genes in Arabidopsis SAM development, we first analysed their mRNA abundance from RNA-seq data of meristematic cells derived from dissection of enlarged clavata3 (clv3) mutant SAM (Yang et al., 2016). We focused on 130 annotated core cell-cycle regulators (Menges et al., 2005; Van Leene et al., 2010), and identified 72 genes showing detectable expression in the SAM (TPM > 10; Table S1). To investigate their expression pattern in planta, we carried out systematic RNA in situ hybridization. Using RNA probes specific to individual SAM-expressed cell cycle genes, we were able to detect the distribution of transcripts from 66 genes at single-cell resolution. In situ hybridization results are presented in Data S1. Most of the genes exhibited strong expression in the SAM compared to other tissues (e.g. stem), supporting the RNA-seq data. Based upon their expression patterns, these cell-cycle genes were classified into six groups: (i) homogeneous signal (Type I); (ii) homogeneous background signal with weak additional signal in a spotted pattern (Type II); (iii) homogeneous background signal with strong additional spots of signal (Type III); (iv) weak spots of signal in a subset of cells (Type IV); (v) only strong spots of signal in a subset of cells (Type V); and (vi) homogeneous background expression with additional strong signal in developing primordia (Type VI) (Figure 1E; Table S1). Homogeneous signals across the whole meristem indicate that the corresponding genes are expressed throughout the cell cycle; whereas patchy patterns suggest that expression correlates to specific cell cycle stages.
Most of the G1/S regulators, including CDKA;1, E2Fs (E2Fa, E2Fb and E2Fc) and DPs (DPa and DPb), displayed homogeneous expression in the shoot apex, which would maintain these meristematic cells with the capacity for active proliferation. One exception was RETINOBLASTOMA RELATED (RBR), an inhibitor of E2F and DP transcription factors, which showed a strong patchy pattern (Type III) (Figure 1E), similar to previous observations in embryonic and root meristematic cells (Wildwater et al., 2005), and implying a cell-cycle controlled regulation. Compared to G1/S genes, G2/M regulators, including plant-specific B type CDKs (CDKBs), and A and B type cyclins (CYCAs and CYCBs) were all grouped into Type V, showing a strongly patchy pattern with weak background expression (Figure 1E). RNA fluorescence in situ hybridization (RNA FISH) together with 4', 6-diamidino-2-phenylindole (DAPI) staining indicated that these genes were exclusively expressed in mitotic cells from early prophase until late anaphase (Figures S1A and S1B).
Our gene expression map data were consistent with Affymetrix microarray data of dividing Arabidopsis cell cultures (Menges et al., 2005). The mRNA distribution patterns in the shoot apex, combined with previous cell-cycle transcript in situ analysis in Arabidopsis seedlings and in the shoot/floral meristems of Antirrhinum majus (de Almeida Engler et al., 2009; Fobert et al., 1994), provide a good overview of cell cycle gene expression patterns in various plant tissues.
Mitosis-specific Expression of CDC20 and CCS52B mRNA
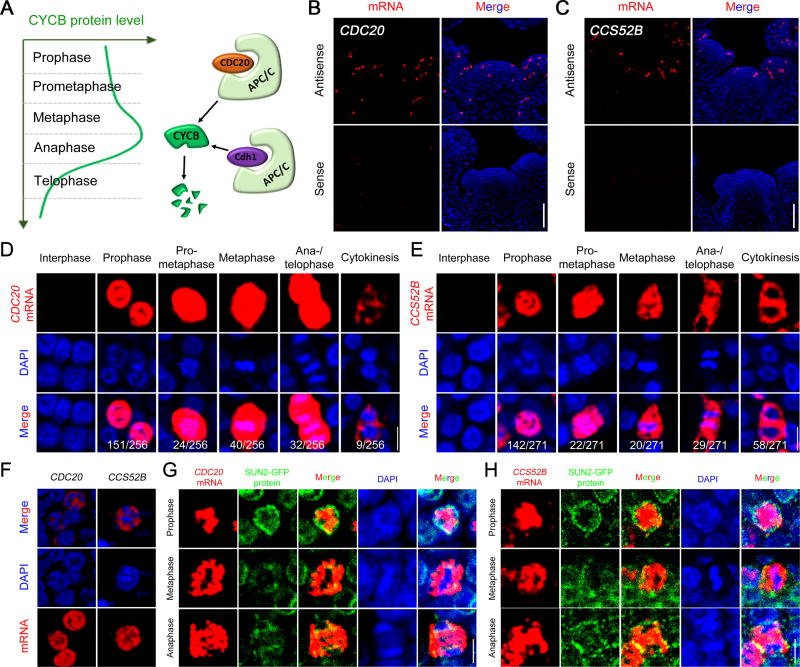
The accumulation of CYCB transcripts at M-phase (Figures S1C and S1D) would be expected to lead to a corresponding peak of CYCB proteins at this stage. Indeed, CYCB1;1-GFP fluorescence signals increased from prophase onwards, peaked at metaphase and then decreasing rapidly at anaphase, finally being undetectable in telophase cells (Figures S1E–S1G). The decline of CYCB1;1-GFP fluorescence signals could be caused by insufficient protein synthesis and/or short half-life. The rapid elimination of large amount of CYCB1 proteins may attribute to APC/C-mediated degradation (Figure 2A), a mechanism conserved among various organisms. The genes encoding Arabidopsis APC/C subunits, as well as the CDH1 orthologues CCS52A1 and CCS52A2, were all expressed homogeneously in the SAM at relatively low level (Table S1 and Data S1). By contrast, both CDC20 and CCS52B showed strong patchy patterns similar to CYCB genes (Data S1). The distinct expression patterns of A- and B-class CCS52 genes supported the predicted roles of CCS52As in regulating endoreduplication (Cebolla et al., 1999; Lammens et al., 2008; Vanstraelen et al., 2009), and CCS52B in controlling mitosis (Tarayre et al., 2004).
Figure 2. Nuclear Sequestration of CDC20 and CCS52B mRNAs in Prophase Cells.
(A) A schematic model illustrating CYCB protein dynamics during mitosis and its degradation by APC/CCDC20 and APC/CCDH1 E3 ligases.
(B and C) RNA FISH to reveal the expression patterns of CDC20 and CCS52B in the SAM. No signals were detected from the sense probes. Scale bars, 50 µm.
(D and E) Expression of CDC20 and CCS52B at different mitotic stages. Note the nuclear localization of CDC20 and CCS52B mRNAs at prophase. Scale bars, 5 µm.
(F) 3-D projection of CDC20 and CCS52B mRNAs in prophase cells. Scale bar, 5 µm.
(G and H) CDC20 and CCS52B mRNA localization with nuclear envelope reporter at different stages of mitosis. The mRNAs were detected by FISH. The nuclear envelope was revealed using GFP antibody against a nuclear envelope reporter protein, SUN2-GFP. Scale bars, 5 µm. All images, with the exception of (F), show single optical confocal sections.
See also Figure S2.
Cell-cycle controlled CDC20 and CCS52B expression was further investigated by RNA FISH. Both mRNAs accumulated exclusively in mitotic cells from prophase until cytokinesis (Figures 2B–2E). The amount of CDC20 mRNA decreased when mitosis was completed (Figure 2D), whereas a high level of CCS52B mRNA persisted until cytokinesis (Figure 2E). The extended expression of CCS52B relative to CDC20 was validated by double RNA FISH. CDC20 and CCS52B mRNAs co-expressed in early mitotic cells, but at late mitosis a population of cells were found only to express CCS52B (Figures S2A–S2C). Taken together, the enrichment of CDC20 and CCS52B transcripts, along with the constitutive expression of all APC/C components, would presumably allow for rapid APC/C activation.
CDC20 and CCS52B mRNAs Are Sequestered in the Nucleus at Prophase
Mature mRNAs are usually rapidly exported out of the nucleus (Köhler and Hurt, 2007). For example, CYCB transcripts, despite their high levels, were all found to reside in the cytoplasmic space (Figures S1A and S1C). However, when analysing the sub-cellular distribution of CDC20 and CCS52B mRNAs in prophase cells, we found that each of them is localized inside the DAPI-labelled nuclei (Figure 2F). No hybridization signals could be detected in the cytoplasm even when we increased the detection settings to saturation (data not shown). To further validate the nuclear sequestration of CDC20 and CCS52B transcripts, we examined CDC20 and CCS52B mRNA localization in mitotic cells together with a marker for the nuclear envelope. CDC20 and CCS52B mRNAs were detected by RNA FISH. The nuclear envelope was revealed by immunohistochemistry using an anti-GFP antibody in SAM sections of Arabidopsis nuclear envelope marker line SUN2-GFP (Oda and Fukuda, 2011; Varas et al, 2015). As shown in Figures 2G and 2H, both CDC20 and CCS52B mRNAs were localized inside the nucleus and were surrounded by the intact nuclear envelope in prophase cells; when cells enter metaphase and the nuclear envelope has disassembled, the transcripts were found distributed in the cytoplasm. At late telophase and cytokinesis when the nuclear envelope reforms, CDC20 and CCS52B mRNAs were excluded from the nuclei of daughter cells, suggesting that once in the cytoplasm, CDC20 and CCS52B mRNAs are not imported back or recruited into the nucleus. These cytosol-localized CDC20 and CCS52B mRNAs seem to be unstable as they could only be detected in a small group of newly divided cells. Nuclear localization of CDC20 mRNA was also detected in root apical meristem (Figures S2D and S2E) and shoot vascular cambium (Figure S2F), demonstrating that this phenomenon exists in the dividing cells of different tissues.
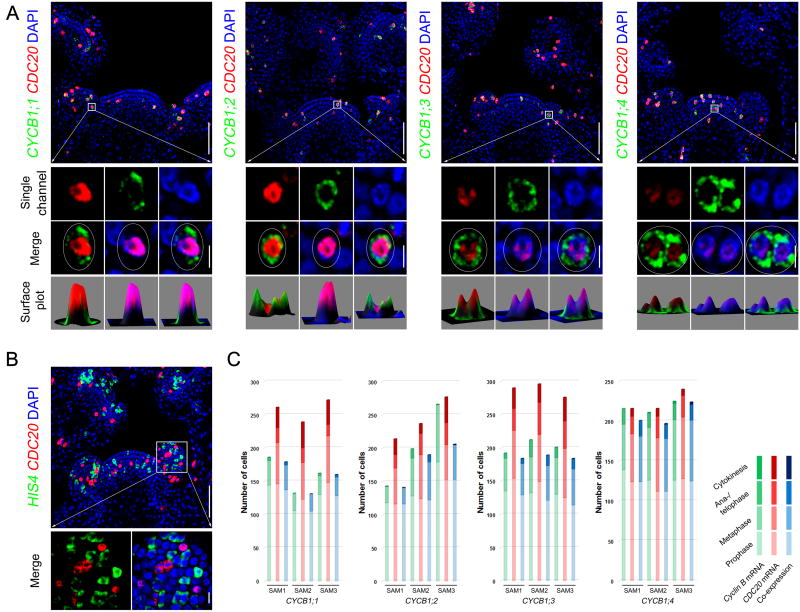
Nucleo-cytoplasmic Compartmentalization of CDC20 and CYCB mRNAs
Since both CYCBs and CDC20 transcripts could be detected at prophase, we hypothesized that they might be expressed simultaneously in the same cells, although the possibility of sequential expression could not be excluded. To clarify this, we investigated CYCBs and CDC20 expression in the same meristems by double RNA FISH. Arabidopsis wild-type meristems were hybridised with both CYCBs and CDC20 gene-specific RNA probes and the number of cells expressing different genes was quantified. CDC20 was found to largely co-express with different CYCB genes in all mitotic cells from prophase until anaphase (Figures 3A and 3C), whereas no co-expression was detected for CDC20 with the S phase marker Histone H4 (HIS4) gene (Figure 3B). In prophase cells, the localization of CDC20 and CYCB transcripts was clearly separated: CDC20 mRNA was restricted to the nuclei and surrounded by cytoplasmically localized CYCB mRNAs (Figures 3A and S3). Therefore, CYCB mRNAs can be translated, resulting in high expression of CYCB1;1-GFP in prophase cells (Figure S1E); whereas nuclear confinement of CDC20 and CCS52B transcripts might prevent protein synthesis.
Figure 3. Spatial Separation of CDC20 and CYCB mRNAs in Prophase Cells.
(A) Co-expression of CDC20 with cell cycle genes as revealed by double RNA FISH coupled with DAPI staining.
(B) CDC20 does not co-express with an S-phase expressed gene HIS4. CDC20 and cell cycle genes were detected by gene specific probes with different labelling. Scale bars in (A) and (B), SAM overview (top panels) = 50 µm; single cells (bottom panels) = 5 µm.
(C) Quantification of the number of cells that express CDC20 and CYCB genes at different mitotic stages. CYCB1 genes were mostly expressed at prophase and metaphase, and largely co-express with CDC20.
See also Figure S3.
Nuclear Sequestration of CDC20 and CCS52B mRNAs Blocks Protein Translation
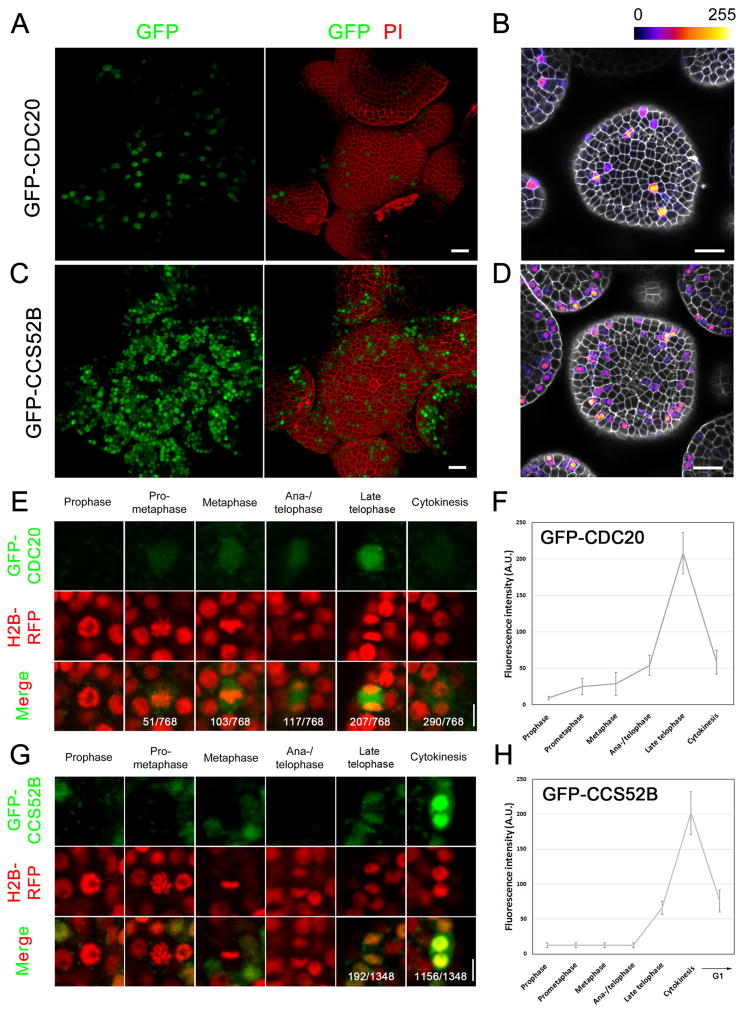
To evaluate the effect of CDC20 and CCS52B mRNA nuclear sequestration upon protein translation, we analysed the expression patterns of GFP-tagged CDC20 and CCS52B fusion proteins in living cells, an approach that has been widely used to track the dynamics of key cell cycle proteins, including CDC20 in animal cells (Nilsson et al., 2008). Genomic fragments containing the entire coding sequences of CDC20 and CCS52B were fused with GFP at the N-terminus and expressed in wild-type plants under the control of their endogenous promoters. Double RNA FISH using GFP probe and CDC20 and CCS52B gene-specific probes showed overlapping signals at different mitotic stages, suggesting that fusion of GFP coding sequence did not interfere with nuclear localization of CDC20 or CCS52B mRNAs (Figures S4A and S4B).
The meristems of pCDC20::GFP-gCDC20 and pCCS52B::GFP-gCCS52B transgenic plants were examined using confocal microscopy. GFP-CDC20 was only expressed in a small fraction of meristematic cells (Figure 4A). GFP-CCS52B protein expression could be identified in a greater proportion of SAM cells, which predominantly localized in the nucleus but also in the cytoplasm (Figure 4C). For both proteins, the expression levels varied between different cells (Figures 4B and 4D). To analyse their expression in relation to different phases of the cell cycle, we further introduced GFP-CDC20 and GFP-CCS52B into H2B-RFP plants. GFP-CDC20 fluorescence signals were detected at very low level in prometaphase cells, increased slowly at metaphase and anaphase, and reached maximal level in late telophase cells. When cytokinesis was completed, GFP-CDC20 eventually decreased and disappeared (Figures 4E and 4F). Compared to GFP-CDC20, the expression of GFP-CCS52B was much delayed, as it was not detected until cells enter late telophase. GFP-CCS52B protein expression exhibited its peak level at cytokinesis, and persisted until the next G1 stage (Figures 4G and 4H).
Figure 4. Expression Patterns of CDC20 and CCS52B Proteins during the Cell Cycle.
(A–D) GFP-CDC20 (A, B) and GFP-CCS52B (C, D) expression in the Arabidopsis SAM. The cell wall was stained with propidium iodide (PI). Expression of GFP-CDC20 and GFP-CCS52B in (B) and (D) were displayed using the Fire lookup table in ImageJ to show difference in fluorescence intensity. Scale bars, 20 µm.
(E–H) Protein dynamics of GFP-CDC20 (E) and GFP-CCS52B (G) at different stages of mitosis. The fluorescence intensity was shown in (F) and (H). Scale bars, 5 µm.
See also Figure S4.
The protein expression pattern of CDC20 beginning at prometaphase was consistent with its transcript accumulation prior to NEBD, followed by mRNA redistribution into the cytoplasm after NEBD. However, given the late appearance of CCS52B protein despite much earlier release of its RNA from the nucleus, it appears that there are additional mechanisms beyond nuclear sequestration that controls CCS52B translation, one of which could be regulation by CCS52B mRNA binding proteins as RNA-binding proteins also play crucial roles in controlling translation efficiency besides guiding RNA localization (Lipshitz and Smibert, 2000). Nevertheless, the peak accumulation of CCS52B protein at cytokinesis and subsequent stages was in line with the predicted roles of Cdh1 to degrade CDC20 and maintain a low cyclin abundance through late mitosis and G1 phases (Fang et al., 1998). After analysing a number of meristems from different transgenic lines, we were unable to detect any GFP-CDC20 or GFP-CCS52B protein expression in prophase cells, demonstrating that mRNA nuclear sequestration correlated with an absence of protein translation.
Dynamic Turnover of CDC20 and CCS52B proteins
The GFP-CDC20 and GFP-CCS52B proteins dynamics was further examined by real-time fluorescence imaging of individual cells, revealing that both proteins accumulated rapidly at late mitosis and disappeared when mitosis was completed (Figures S4C and S4D). Fluctuation in CDC20 protein levels during the cell cycle has been observed in animal cells (Fang et al., 1998; Kramer et al., 1998; Prinz et al. 1998). For CDH1, the protein level appears to remain constant throughout the cell cycle in HeLa cells (Fang et al., 1998; Kramer et al., 1998). In order to distinguish changes in gene expression from proteolytic activity, we treated SAMs with the proteasome inhibitor MG132. This treatment did not increase the protein level of GFP-CCS52B (Figure S4E), suggesting that CCS52B levels are a function of gene expression and translation. By contrast, MG132 treatment resulted in a marked increase in GFP-CDC20 fluorescence intensity in both SAM and root cells (Figure S4F), implying that CDC20 may undergo continuous synthesis and degradation. Therefore, a conserved surveillance system exists to tightly control CDC20 protein abundance in plants as in human cells (Ge et al., 2009; Izawa and Pines, 2015; Nilsson et al., 2008).
Mapping the Cis-acting Element Involved in CDC20 mRNA Nuclear Localization
To investigate how CDC20 mRNA is sequestered in the nucleus, we first tested the mechanisms proposed for known nuclear RNAs. It has been shown that mRNAs containing adenosine (A)-to-inosine (I) edited Alu inverted repeats are predominantly localized in the nucleus (Chen et al., 2008). A-to-I editing was responsible for the nuclear retention of CTN-RNA (Prasanth et al., 2005). We compared the sequences of CDC20 full-length cDNA and genomic DNA but did not find any difference, ruling out A-to-I editing in CDC20 mRNA. In addition, mRNA transcribed from CDC20 cDNA, like the genomic DNA-derived mRNA, was also confined to the nucleus (Figure S5A), suggesting that CDC20 nuclear sequestration can act upon the mature mRNA. These results indicate that the regulation of CDC20 mRNA nuclear localization was distinct from other nuclear RNAs.
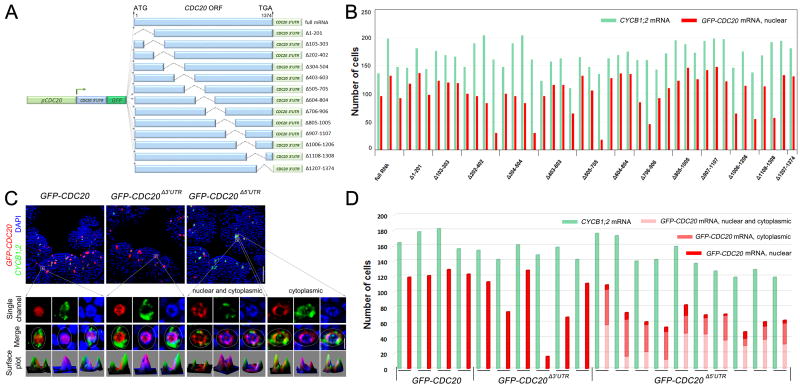
As the targeting signals of localized RNAs are usually encoded by their own sequences (Buxbaum et al., 2015), we next sought to identify the cis-acting element involved in CDC20 mRNA nuclear localization. A series of deletions spanning the entire CDC20 coding sequence, each 200 bp in length (except for Δ1207–1374) with 100 bp overlapping, were fused with GFP and expressed in wild-type plants under the control of the CDC20 promoter (Figure 5A). The localization of these truncated GFP-CDC20 chimeric RNAs was examined by RNA FISH. As cytoplasmic localization of CDC20 mRNA can be observed at late mitosis when daughter cell nuclei reform (Figure 2D), we used CYCB1;2 mRNA expression as an indicator of prophase cells. CYCB1;2 showed similar expression in these transgenic plants compared to wild-type plants (Figure 5B), suggesting that expression of these exogenous RNAs did not interfere with normal cell cycle progression. Examination of the subcellular distribution revealed that all these GFP-CDC20 truncated RNAs were all localized inside the nucleus, surrounded by the cytoplasmic CYCB1;2 mRNA (Figures 5B and S5B), indicating that deletion of a single fragment of CDC20 coding region was not sufficient to disrupt RNA nuclear localization.
Figure 5. CDC20 5’UTR Is Involved in mRNA Nuclear Localization.
(A) Schematic diagram of CDC20 mRNA deletion constructs.
(B) Quantification of the number of prophase cells expressing GFP fused CDC20 mRNAs that contain serial deletions. CYCB1;2 expression was used as a prophase marker. All GFP-CDC20 mRNAs with deletions in the CDC20 ORF were found to localize in the nucleus. Each pair of columns represents data from one meristem.
(C) Localization of GFP-CDC20 truncated mRNAs lacking CDC20 5’UTR or 3’UTR. Deletion of 5’UTR abolished GFP-CDC20 mRNA nuclear sequestration, leading to nucleocytoplasmic or mostly cytoplasmic localization. Scale bars, 50 µm for SAM overview (top panels) and 5 µm for single cells (bottom panels).
(D) Quantification of the number of prophase cells expressing full length, 3’UTR deleted, and 5’UTR deleted GFP-CDC20 mRNAs. Each pair of columns represents data from one meristem.
See also Figure S5.
We next investigated the role of UTRs in CDC20 mRNA nuclear sequestration. Chimeric mRNAs transcribed from GFP in-frame fused with CDC20 genomic fragment without the 5’UTR or 3’UTR (pCDC20::GFP-CDC20Δ5’UTR and pCDC20::GFP-CDC20Δ3’UTR) were analysed by RNA FISH. CDC20 3’UTR-truncated mRNAs showed the same nuclear localization pattern as full length GFP-CDC20 transcript. By contrast, when the 5’UTR was deleted, nuclear localization was largely reduced. In most of the prophase cells, 5’UTR-truncated GFP-CDC20 mRNAs were present either in both the nucleus and the cytoplasm, or mostly in the cytoplasm (Figures 5C, 5D and S5B), indicating that deletion of the 5’UTR abolished CDC20 mRNA nuclear sequestration.
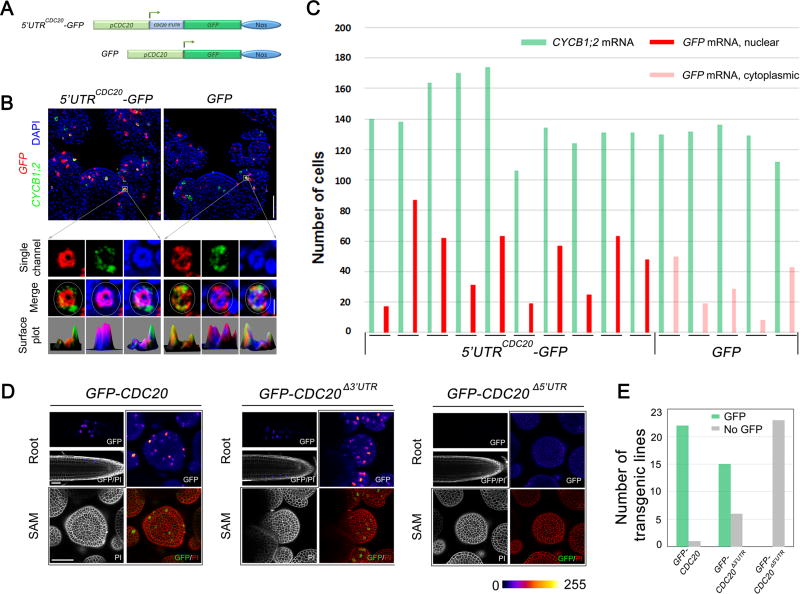
CDC20 5’UTR Is Sufficient to Confer Nuclear Sequestration
To further evaluate the function of the CDC20 5’UTR, we fused it to a GFP coding sequence (Figure 6A). This chimeric mRNA, 5’UTRCDC20-GFP, as well as GFP alone, were expressed in wild-type plants under the control of the CDC20 promoter. The number of prophase cells expressing these GFP mRNAs seem to be reduced compared to GFP fused with full length CDC20 mRNA (Figure 6C), implying that the CDC20 coding region contains one or more cis-elements contributing to transcriptional activity. Nevertheless, when expressed, 5’UTRCDC20-GFP mRNA was found to be exclusively confined to the nucleus. The control, GFP mRNA alone, was distributed in the cytoplasm similarly to CYCB1;2 mRNA (Figures 6B and S5C). The results, taken together, demonstrate that the 5’UTR was both necessary and sufficient to sequester CDC20 mRNA inside the nucleus.
Figure 6. Dual Roles of 5’UTR in CDC20 mRNA Nuclear Localization and Translation.
(A) Schematic diagram of chimeric mRNA construction in which GFP was fused with CDC20 5’UTR. GFP alone was used a control.
(B) Localization of 5’UTRCDC20-GFP and GFP mRNAs in prophase cells. Scale bars, 50 µm for SAM overview (top panels) and 5 µm for single cells (bottom panels).
(C) Quantification of the number of prophase cells expressing5’UTRCDC20-GFP and GFP mRNAs. Each pair of columns represents cell numbers from one meristem.
(D) The expression of GFP-CDC20 fusion protein in root and SAM as revealed. No GFP fluorescence could be observed in 5’UTR truncated GFP-CDC20 transgenic plants. Scale bar, 50 µm.
(E) The number of transgenic lines analysed. GFP-CDC20 expression was detected in 22/23 lines of full length GFP-CDC20 plants, 15/21 lines of 3’UTR truncated GFP-CDC20 transgenic plants, and 0/23 of 5’UTR truncated GFP-CDC20 transgenic plants.
See also Figure S5.
CDC20 5’UTR Is Required for Protein Translation
The cytoplasmic localization of GFP-CDC20Δ5’UTR mRNA in prophase cells, if translated, would be expected to interfere with proper cell cycle progression. However, we did not observe any cellular defect in chromosome alignment or segregation, and the transgenic plants grew normally. Confocal microscopy analysis revealed that in GFP-CDC20Δ3’UTR meristems, the fusion protein could be translated, showing clear GFP fluorescence in root and shoot apical meristems similar to the full length transcript (Figures 6D and 6E). However, no fluorescence could be observed in multiple independent GFP-CDC20Δ5’UTR transgenic lines, indicating that 5’UTR truncated GFP-CDC20 mRNA cannot be properly translated. These results demonstrate that the 5’UTR of CDC20 plays dual roles in mRNA nuclear localization and translation.
Discussion
To ensure the fidelity of chromosome segregation, APC/C activity needs to be precisely modulated, especially at early mitosis when APC/C targets (e.g. CYCB proteins) are playing crucial roles. Emi1 has been implicated in animals as the inhibitor of APC/C by binding to CDC20, preventing its interaction with APC/C substrates at prophase (Reimann et al., 2001). However, the role of Emi1 remains contentious as it was also shown to have little effect on APC/CCDC20 activity, and expression of a non-degradable version of Emi1 does not affect the destruction of cyclin A, cyclin B1 and securin (Di Fiore and Pines, 2007). Phosphorylation of APC/C subunits can facilitate CDC20 binding thus promoting APC/C activation (Sivakumar and Gorbsky, 2015). In mammalian cells APC/C phosphorylation is already initiated and CDC20 protein is also highly expressed at prophase (Kraft et al., 2003; Nilsson et al., 2008), which would presumably lead to APC/C activation. Therefore, it still remains obscure how APC/C activity is restrained during prophase. In plants, no Emi1 orthologue has been identified. GIG1/OSD1 and UVI4 have been suggested as the negative regulators of plant APC/C (Heyman et al., 2011; Iwata et al., 2011), but their direct effect on APC/C activity has not been determined. We found that in Arabidopsis dividing cells the mRNAs of CDC20 and CCS52B are sequestered inside the nucleus. Nuclear retention of mRNAs is expected to block their accessibility to cytoplasmic ribosomes. Consistent with this scenario, neither CDC20 nor CCS52B proteins could be detected in prophase cells. As CDC20 and CCS52B are key activators of APC/C, it seems that absence of CDC20 and CCS52B proteins at prophase due to RNA nuclear sequestration would result in very low APC/C activity, thereby allowing cyclin B function (Figure S6).
Cellular mRNA localization has been proposed as a common mechanism to control local protein abundance. A systematic study revealed that 71% of expressed mRNAs in Drosophila embryos exhibit distinct cytoplasmic distribution patterns (Lécuyer et al., 2007). Compared to the predominant distribution in the cytoplasm, nuclear localization of protein coding mRNAs has rarely been encountered. Our data demonstrate that properly processed, unedited mature mRNAs can be specifically sequestered inside the nucleus, correlating with control (absence) of protein synthesis. Nuclear sequestration of CDC20 and CCS52B mRNA, despite their high levels, prevents protein translation, but on the other hand could also generate a store of RNA molecules that can be rapidly released to the cytoplasm upon NEBD for protein synthesis, and thus efficiently activate APC/C.
RNA localization is guided by specific cis-acting elements that are mostly identified in the 3’UTR (Martin and Ephrussi, 2009). The localization signals contributing to the spatial distribution of bicoid, nanos, xcat2, β-actin mRNAs, and histone mRNAs have all been mapped to the 3’UTR (Iampietro et al., 2014; Martin and Ephrussi, 2009). However, deletion analysis revealed that the 3’UTR has no effect on CDC20 mRNA nuclear localization. By contrast, when the 5’UTR is removed, the resulting GFP-CDC20Δ5’UTR chimeric mRNA is found to distribute into the cytoplasm. Furthermore, adding the CDC20 5’UTR was sufficient to sequester GFP mRNA in the nucleus, indicating that the 5’UTR is necessary and sufficient for CDC20 mRNA nuclear sequestration. Despite being exported into the cytoplasm, the 5’UTR truncated GFP-CDC20 RNA was not detectably translated, which is consistent with the important functions of 5’UTR in ribosome recruitment and translational initiation (Hinnebusch et al., 2016). Therefore, the dual roles of the 5’UTR in CDC20 mRNA nuclear localization and translation provide a ‘belt-and-braces’ approach to avoid CDC20 protein synthesis and APC/C activation. RNA localization elements are recognized by trans-acting proteins. The RNA interactome capture method has been recently developed to identify Xist lncRNA binding proteins in human cells (Chu et al., 2015; McHugh et al., 2015; Minajigi et al., 2015). Applying this technology in plants to characterize CDC20 and CCS52B mRNA interacting protein(s) would provide further insight into the understanding of mRNA localization, translational control, and cell cycle regulation.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Elliot M. Meyerowitz (meyerowcaltech.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Arabidopsis
Arabidopsis Columbia ecotype (Col-0) was used as wild-type for the in situ hybridization analysis. The reporter lines GFP-MBD, CYCB1;1-GFP, and SUN2-GFP were described previously (Hamant et al., 2008; Oda and Fukuda, 2011; Reddy et al., 2005). Seeds were germinated on Murashige and Skoog agar plates and 7 day-old seedlings were transferred to soil. Plants were grown under long day conditions (16 h/8 h light/dark period) at 20 °C.
METHOD DETAILS
mRNA In Situ Hybridization
For RNA probe synthesis, the cDNA fragments corresponding to each cell cycle gene were amplified with gene-specific primers (Table S4), ligated into the pGEM®-T Easy vector (Promega) and verified by sequencing. The plasmids containing the cDNA fragments were then used as templates for PCR with primers T7 and SP6. The PCR products were used as templates for in vitro transcription using the DIG RNA Labeling Kit (Roche). For fluorescence in situ probes, Fluorescein-12-UTP (Roche) was used instead of Digoxigenin-11-UTP (Roche).
For sample preparation, shoot apices of Arabidopsis were harvested and fixed in FAA (3.7% formaldehyde, 5% acetic acid, 50% ethanol). The samples were embedded in wax and cut into 8-µm sections. The sections were processed by dewaxing, rehydration and dehydration, as described in (http://www.its.caltech.edu/~plantlab/protocols/insitu.pdf).
For hybridization, the sections were hybridized with gene-specific probes at 55 °C. After washing with SSC, the slices were incubated with anti-digoxigenin-AP antibody (Roche) for 2 hours at room temperature. The signals were detected by overnight colour reaction at 28 °C using NBT/BCIP (Roche). Sense-strand hybridizations, yielding no hybridization with target mRNA, are shown as controls. Images were taken using a Zeiss AxioImager M2 microscope fitted with a Zeiss Axiocam MRc colour camera and a PlanApochromat 20×/ 0.8 NA objective.
RNA Fluorescence in situ Hybridization (RNA FISH)
Samples were processed as above for in situ hybridization, except that anti-fluorescein-POD (Roche) or anti-digoxigenin-POD (Roche) antibodies were used. After antibody incubation, the hybridization signals were detected using TSA Plus Fluorescein Fluorescence System (Perkin Elmer) for green signals or TSA Plus Cy5 Fluorescence System (Perkin Elmer) for red signals. DAPI staining was performed by mounting the slices with 1µg/ml DAPI shortly before observing the in situ hybridization signals. Images were taken with a Zeiss LSM700 confocal microscope equipped with a 20 × 0.8NA dry objective. Laser excitation was 405 nm (DAPI), 488 nm (Fluorescein) and 633 nm (Cy5).
Double RNA FISH was used to check the mRNA expression of two genes in the same cells. Processed sections were hybridized with a mixture of two gene-specific probes, one labelled with digoxigenin and the other with fluorescein. The slices were incubated with anti-fluorescein-POD (Roche) and subsequently detected with TSA Plus Fluorescein Fluorescence System, giving green signals. After the first TSA reaction, 3% H2O2 (Sigma) was applied to quench peroxidase activity (1 hour incubation in 3% H2O2 was found to sufficiently quench all peroxidase activity of the first antibody). The slices were further incubated with anti-digoxigenin-POD antibody and detected by TSA Plus Cy5 Fluorescence System (Perkin Elmer), resulting in red signals.
RNA FISH and Immunohistochemistry
RNA FISH was carried out as described above. After TSA-Cy5 reaction to reveal the mRNA hybridization signals, the sections were washed in PBST (PBS containing 0.3% v/v Triton X-100), and then blocked in PBS-Blocking buffer (PBS containing 1.0% bovine serum albumin, 0.2% powdered skim milk, and 0.3% Triton X-100) for 30 min at room temperature. The sections were then incubated with Alexa Fluor® 488 conjugated GFP antibody (1:100 dilution) (A-21311, Molecular Probes) overnight at 4 °C. The slides were washed in PBST for 3 times, 5 min each and observed using a Zeiss LSM700 confocal microscope.
Plasmid Construction and Plant Transformation
GFP Fusion with Full Length CDC20 and CCS52B mRNA
The MultiSite Gateway® Three-Fragment Vector Construction system (Invitrogen) was used to generate plasmid constructs. For pCDC20.1::GFP-CDC20.1, a 2,417 bp promoter upstream of CDC20.1 ATG was amplified using genomic DNA as template with primers CDC20_promoter_F and CDC20_promoter_R. The PCR product was inserted into pDONR™ P4-P1R by BP reaction, resulting in 1R4-pCDC20. The enhanced GFP (EGFP) coding sequence was amplified using primers GFP_gateway_F and GFP_ gateway_R, and the product was inserted into pDONR™ 221 by BP reaction, resulting in 221-GFP. A 3,161bp genomic fragment containing the whole coding sequence of CDC20.1 as well as 1,115bp 3’ region was amplified with primers CDC20_DNA_F and CDC20_DNA_R, and the PCR product was inserted into pDONR™ P2R-P3, resulting in 2R3-gCDC20. The three entry constructs was incorporated into the binary vector pB7m34-GW by LR reaction. Similar strategy was applied to CCS52B. The primers used for CCS52B promoter were CCS52B_promoter_F and CCS52B_promoter_R; for coding region as well as 3’ region were CCS52B_DNA_F and CCS52B_DNA_R, and the constructs were named as 1R4-pCCS52B and 2R3-gCCS52B, respectively. pCDC20.1::GFP-CDC20.1 and pCCS52B::GFP-CCS52B were transformed into Arabidopsis wild-type Col-0 as well as nuclear reporter line H2B-RFP (Col-0 background) via Agrobacterium mediated transformation.
To construct CDC20 cDNA fused with GFP, the full length cDNA including 5’ and 3’ UTR was first amplified from meristem cDNA library using primers CDC20_cDNA_F and CDC20_cDNA_R. GFP was amplified with primers GFP_F and GFP_R. CDC20 cDNA and GFP fragments were ligated into pBluescript SK(−), resulting in SK-GFP-cCDC20, which was then incorporated into pB7m34-GW with CDC20 promoter and Nos terminator by LR reaction.
CDC20 5’UTR and 3’UTR Deletions
For 5’UTR deletion analysis, CDC20 promoter was amplified with primers CDC20_promoter_F and CDC20_promoter_NoUTR_R. The PCR products were inserted into pDONR™ P4-P1R by BP reaction, resulting in 1R4-pCDC20_No5’UTR. 1R4-pCDC20_ No5’UTR was further introduced into the binary vector pB7m34-GW with 221-GFP and 2R3-gCDC20 by LR reaction. For 3’UTR deletion analysis, CDC20 genomic sequence without 3’UTR was amplified with primers CDC20_KpnI and CDC20_SalI. CDC20 terminator was amplified with primers CDC20_SalI_1 and CDC20_BamHI. The two fragments were ligated into pBluescript SK(−), and the resulting plasmid was used as template for PCR with primers CDC20_DNA_F and CDC20_DNA_R. The PCR product was inserted into pDONR™ P2R-P3, resulting in 2R3-gCDC20_NoUTR. 1R4-pCDC20, 221-GFP and 2R3-gCDC20_NoUTR were ligated into pB7m34-GW by LR reaction.
CDC20 Coding Sequence Deletions
Fusion PCR was used to generate CDC20 ORF deletion constructs. Two PCR fragments with 25 bp overlapping were amplified with specific primers (Table S2). The PCR products were mixed and used as templates for a second round of PCR using primers GFP_GW1_F and GFP-CDC20_ GW1_R. The product was inserted into pDONR™ 221 by BP reaction, and further incorporated into pB7m34-GW with CDC20 promoter and Nos terminator by LR reaction.
Observation of Fluorescent Reporter Expression by Confocal Microscopy
Shortly after bolting (stem length ~ 1 cm), the shoot apex was dissected and the fully developed flowers were carefully removed in order to expose the SAM. The meristem was then transferred to a square box containing fresh MS medium (Duchefa Biochemie - MS basal salt mixture) supplemented with vitamins (myoinositol 100 µg/ml, nicotinic acid 1 µg/ml, pyridoxine hydrochloride 1 µg/ml, thiamine hydrochloride 1 µg/ml, glycine 2 µg/ml) and 1% sucrose in order to keep the meristem alive during observation. Viewed-stacks of SAMs were acquired with either a Zeiss LSM700 with 20 × NA 1.0 water dipping objective or a Leica SP8 with 25 × NA 1.0 water dipping objective. 3D rendering was carried out using either Zen (Zeiss) or LAS × (Leica) confocal microscope software. The cell boundaries of the SAM were revealed by 0.1% propidium iodide (PI) staining for 5 min. Laser excitations were 488 nm (PI, GFP) and 555nm or 561nm (RFP). GFP fluorescence intensity was measured in Fiji ImageJ. To display the fluorescence intensity as shown in Figures 5 and S7, the fluorescence pictures were edited with the LUT editor plugin in Fiji ImageJ.
For MG132 treatment, dissected meristems were emerged in liquid MS medium containing DMSO (Mock) or 50 µM MG132 (C2211 Sigma) for 2 hours. For time lapse experiment, dissected meristems were kept in MS medium (Duchefa) supplemented with vitamins and sucrose. The meristems were kept in growth chamber under long day conditions (16 h/8 h light/dark period) at 20 °C, and were taken out for confocal imaging at each time point.
QUANTIFICATION AND STATISTICAL ANALYSIS
For cell number quantification, ~10 serial sections with 8 µm thickness that could cover the whole meristem were used to count the number of mitotic cells expressing different cell cycle genes. Confocal pictures were used to count the number of cells expressing MBD-GFP, GFP-CDC20 and GFP-CCS52B. The fluorescence intensity was measured in Fiji.
Supplementary Material
Supplementary Dataset. The expression patterns of core cell cycle genes in Arabidopsis SAM by mRNA in situ hybridisation. Related to Figure 1.
Table S1. Expression level and patterns of cell cycle genes in Arabidopsis dissected shoot apical meristem (SAM) by RNA-seq and in situ hybridisation. Shown are TPM values adapted from Yang et al (2016). Related to Figure 1.
Table S2. Primers used in this study. Related to STAR Methods.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Digoxigenin-AP, Fab fragments | Sigma | 11093274910 ROCHE |
| Anti-Fluorescein-POD, Fab fragments | Sigma | 11426346910 ROCHE |
| Anti-Digoxigenin-POD, Fab fragments | Sigma | 11207733910 ROCHE |
| Alexa Fluor® 488 conjugated GFP antibody | Molecular Probes | A-21311 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Gateway® BP Clonase® II Enzyme mix | Thermofisher | 11789020 |
| Gateway® LR Clonase® II Enzyme mix | Thermofisher | 11791100 |
| MG132 | Sigma | C2211 |
| DAPI | Sigma | D9542 |
| Hydrogen peroxide solution | Sigma | H1009 |
| Critical Commercial Assays | ||
| DIG RNA Labeling Kit (SP6/T7) | Sigma | 11175025910 |
| TSA Fluorescein | Perkin Elmer | NEL701A001KT |
| TSA Cyanine 5 | Perkin Elmer | NEL705A001KT |
| Deposited Data | ||
| Unprocessed in situ hybridization images of cell cycle genes | Mendeley Data | http://dx.doi.org/10.17632/krzr6yvx25.1 |
| Unprocessed confocal images | Mendeley Data | http://dx.doi.org/10.17632/m2d3wr3zh5.1 |
| Experimental Models: Organisms/Strains | ||
| Arabidopsis: GFP-MBD | Hamant et al., 2008 | N/A |
| Arabidopsis: CYCB1;1-GFP | Reddy et al., 2005 | N/A |
| Arabidopsis: SUN2-GFP | Oda and Fukuda, 2011; Varas et al., 2015 | N/A |
| Oligonucleotides | ||
| Primers for making in situ probes and cloning, see Table S2 | This paper | N/A |
| Recombinant DNA | ||
| pCDC20::GFP-gCDC20 | This paper | N/A |
| pCCS52B::GFP-gCCS52B | This paper | N/A |
| pUBQ10::GFP-gCDC20 | This paper | N/A |
| pUBQ10::GFP-gCCS52B | This paper | N/A |
| pCDC20::GFP-cCDC20 | This paper | N/A |
| pCDC20::GFP-gCDC20Δ5’UTR | This paper | N/A |
| pCDC20::GFP-gCDC20Δ3’UTR | This paper | N/A |
| pCCS52B::GFP-gCCS52BΔ5’UTR | This paper | N/A |
| pCCS52B::GFP-gCCS52BΔ3’UTR | This paper | N/A |
| Software and Algorithms | ||
| Fiji | N/A | http://imagej.net/Fiji |
Acknowledgments
We would like to thank Jonathon Pines (The Institute of Cancer Research, London), David Ron (Cambridge Institute for Medical Research, University of Cambridge), Yrjö Helariutta and Henrik Jönsson (The Sainsbury Laboratory at Cambridge University), and Olivier Hamant (Plant Reproduction and Development Laboratory, INRA, ENS Lyon) for advice and insightful discussions. We also thank David E. Evans (Oxford Brookes University), Susan Armstrong (University of Birmingham), and Xinnian Dong (Duke University) for sharing seeds. We are grateful to Christoph Schuster for support with in situ hybridisation, Benoit Landrein for suggestions for confocal microscope analysis, Pawel Roszak for help with root sectioning, Alexis Peaucelle, Charles Melnyk, Paul Tarr, Pau Formosa Jordan and all members of the Meyerowitz Lab at the California Institute of Technology for helpful conversations. We appreciate Barbara Di Fiore and Anja Hagting (The Gurdon Institute, University of Cambridge), and Lisa Willis (The Sainsbury Laboratory at Cambridge University) for comments and careful reading of the manuscript. This work was funded by the Gatsby Charitable Trust (through fellowship GAT3395/DAA). E.M.M. is supported by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation (through grant GBMF3406). R.W. and W.Y. are supported by the Leverhulme Trust (grant RPG-2015-285).
Footnotes
Author Contributions
W.Y. and R.W. conceived the project; W.Y. and R.W. designed and performed the experiments; W.Y., R.W., and E.M.M. analysed and interpreted the data; W.Y., R.W., and E.M.M. wrote the manuscript.
Supplemental Information includes six figures, two tables, one dataset and can be found with this article online.
References
- Bahar Halpern K, Caspi I, Lemze D, Levy M, Landen S, Elinav E, Ulitsky I, Itzkovitz S. Nuclear Retention of mRNA in Mammalian Tissues. Cell Rep. 2015;13:2653–2662. doi: 10.1016/j.celrep.2015.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum AR, Haimovich G, Singer RH. In the right place at the right time: visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol. 2015;16:95–109. doi: 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebolla A, Vinardell JM, Kiss E, Oláh B, Roudier F, Kondorosi A, Kondorosi E. The mitotic inhibitor CCS52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J. 1999;18:4476–4484. doi: 10.1093/emboj/18.16.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudreuse D, Nurse P. Driving the cell cycle with a minimal CDK control network. Nature. 2010;468:1074–1079. doi: 10.1038/nature09543. [DOI] [PubMed] [Google Scholar]
- de Almeida Engler J, De Veylder L, De Groodt R, Rombauts S, Boudolf V, De Meyer B, Hemerly A, Ferreira P, Beeckman T, Karimi M, et al. Systematic analysis of cell-cycle gene expression during Arabidopsis development. Plant J. 2009;59:645–660. doi: 10.1111/j.1365-313X.2009.03893.x. [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Inzé D. The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 2007;8:655–665. doi: 10.1038/nrm2227. [DOI] [PubMed] [Google Scholar]
- Dewitte W, Murray JA. The plant cell cycle. Annu. Rev. Plant Biol. 2003;54:235–264. doi: 10.1146/annurev.arplant.54.031902.134836. [DOI] [PubMed] [Google Scholar]
- Di Fiore B, Pines J. Emi1 is needed to couple DNA replication with mitosis but does not regulate activation of the mitotic APC/C. J. Cell Biol. 2007;177:425–437. doi: 10.1083/jcb.200611166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 2016;17:756–770. doi: 10.1038/nrm.2016.126. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell. 1998;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- Fobert PR, Coen ES, Murphy GJ, Doonan JH. Patterns of cell division revealed by transcriptional regulation of genes during the cell cycle in plants. EMBO J. 1994;13:616–624. doi: 10.1002/j.1460-2075.1994.tb06299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R, Beretta A, Sironi L, Musacchio A, Lucchini G, Piatti S. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 2001;20:6648–6659. doi: 10.1093/emboj/20.23.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimitsu K, Grimaldi M, Yamano H. Cyclin-dependent kinase 1-dependent activation of APC/C ubiquitin ligase. Science. 2016;352:1121–1124. doi: 10.1126/science.aad3925. [DOI] [PubMed] [Google Scholar]
- Gaillochet C, Lohmann JU. The never-ending story: from pluripotency to plant developmental plasticity. Development. 2015;142:2237–2249. doi: 10.1242/dev.117614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S, Skaar JR, Pagano M. APC/C- and Mad2-mediated degradation of Cdc20 during spindle checkpoint activation. Cell Cycle. 2009;8:167–171. doi: 10.4161/cc.8.1.7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O, Heisler MG, Jonsson H, Krupinski P, Uyttewaal M, Bokov P, Corson F, Sahlin P, Boudaoud A, Meyerowitz EM, et al. Developmental patterning by mechanical signals in Arabidopsis. Science. 2008;322:1650–1655. doi: 10.1126/science.1165594. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Johnston RC, Smith DL, Murray AW. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J. Cell Biol. 2000;148:871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra R, Sabatini S. Plant and animal stem cells: similar yet different. Nat. Rev. Mol. Cell Biol. 2014;15:301–312. doi: 10.1038/nrm3790. [DOI] [PubMed] [Google Scholar]
- Heyman J, Van den Daele H, De Wit K, Boudolf V, Berckmans B, Verkest A, Alvim Kamei CL, De Jaeger G, Koncz C, De Veylder L. Arabidopsis ULTRAVIOLET-B-INSENSITIVE4 maintains cell division activity by temporal inhibition of the anaphase-promoting complex/cyclosome. Plant Cell. 2011;23:4394–4410. doi: 10.1105/tpc.111.091793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5'-untranslated regions of eukaryotic mRNAs. Science. 2016;352:1413–1416. doi: 10.1126/science.aad9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iampietro C, Bergalet J, Wang X, Cody NA, Chin A, Lefebvre FA, Douziech M, Krause HM, Lécuyer E. Developmentally regulated elimination of damaged nuclei involves a Chk2-dependent mechanism of mRNA nuclear retention. Dev. Cell. 29:468–481. doi: 10.1016/j.devcel.2014.03.025. [DOI] [PubMed] [Google Scholar]
- Iwata E, Ikeda S, Matsunaga S, Kurata M, Yoshioka Y, Criqui MC, Genschik P, Ito M. GIGAS CELL1, a novel negative regulator of the anaphase-promoting complex/cyclosome, is required for proper mitotic progression and cell fate determination in Arabidopsis. Plant Cell. 2011;23:4382–4393. doi: 10.1105/tpc.111.092049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa D, Pines J. The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature. 2015;517:631–634. doi: 10.1038/nature13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, Peters JM. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 2003;22:6598–6609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer ER, Gieffers C, Hölzl G, Hengstschläger M, Peters JM. Activation of the human anaphase-promoting complex by proteins of the CDC20/Fizzy family. Curr. Biol. 1998;8:1207–1210. doi: 10.1016/s0960-9822(07)00510-6. [DOI] [PubMed] [Google Scholar]
- Lammens T, Boudolf V, Kheibarshekan L, Zalmas LP, Gaamouche T, Maes S, Vanstraelen M, Kondorosi E, La Thangue NB, Govaerts W, et al. Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc. Natl. Acad. Sci. USA. 2008;105:14721–14726. doi: 10.1073/pnas.0806510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander AD. Pattern, growth, and control. Cell. 2011;144:955–969. doi: 10.1016/j.cell.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lécuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lipshitz HD, Smibert CA. Mechanisms of RNA localization and translational regulation. Curr. Opin. Genet. Dev. 2000;10:476–488. doi: 10.1016/s0959-437x(00)00116-7. [DOI] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, de Jager SM, Gruissem W, Murray JAH. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 2005;41:546–566. doi: 10.1111/j.1365-313X.2004.02319.x. [DOI] [PubMed] [Google Scholar]
- Meyerowitz EM. Genetic control of cell division patterns in developing plants. Cell. 1997;88:299–308. doi: 10.1016/s0092-8674(00)81868-1. [DOI] [PubMed] [Google Scholar]
- Minajigi A, Froberg JE, Wei C, Sunwoo H, Kesner B, Colognori D, Lessing D, Payer B, Boukhali M, Haas W, Lee JT. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 2015;349(6245) doi: 10.1126/science.aab2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat. Cell Biol. 2008;10:1411–1420. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y, Fukuda H. Dynamics of Arabidopsis SUN proteins during mitosis and their involvement in nuclear shaping. Plant J. 2011;66:629–641. doi: 10.1111/j.1365-313X.2011.04523.x. [DOI] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat. Rev. Mol. Cell Biol. 2011;12:427–438. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Prinz S, Hwang ES, Visintin R, Amon A. The regulation of Cdc20 proteolysis reveals a role for APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr. Biol. 1998;8:750–760. doi: 10.1016/s0960-9822(98)70298-2. [DOI] [PubMed] [Google Scholar]
- Qiao R, Weissmann F, Yamaguchi M, Brown NG, VanderLinden R, Imre R, Jarvis MA, Brunner MR, Davidson IF, Litos G, et al. Mechanism of APC/CCDC20 activation by mitotic phosphorylation. Proc. Natl. Acad. Sci. USA. 2016;113:2570–2578. doi: 10.1073/pnas.1604929113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy GV, Heisler MG, Ehrhardt DW, Meyerowitz EM. Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development. 2004;131:4225–4237. doi: 10.1242/dev.01261. [DOI] [PubMed] [Google Scholar]
- Reimann JD, Freed E, Hsu JY, Kramer ER, Peters JM, Jackson PK. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell. 2001;105:645–655. doi: 10.1016/s0092-8674(01)00361-0. [DOI] [PubMed] [Google Scholar]
- Santos SD, Wollman R, Meyer T, Ferrell J. Spatial positive feedback at the onset of mitosis. Cell. 2012;149:1500–1513. doi: 10.1016/j.cell.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar S, Gorbsky GJ. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat. Rev. Mol. Cell Biol. 2015;16:82–94. doi: 10.1038/nrm3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM. Patterns in Plant Development. New York: Cambridge University Press; 1989. [Google Scholar]
- Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarayre S, Vinardell JM, Cebolla A, Kondorosi A, Kondorosi E. Two classes of the CDh1-type activators of the anaphase-promoting complex in plants: novel functional domains and distinct regulation. Plant Cell. 2004;16:422–434. doi: 10.1105/tpc.018952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouze P, Rombauts S, Inzé D. Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell. 2002;14:903–916. doi: 10.1105/tpc.010445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, Hollunder J, Eeckhout D, Persiau G, Van De Slijke E, Stals H, Van Isterdael G, Verkest A, Neirynck S, Buffel Y, et al. Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 2010;6:397. doi: 10.1038/msb.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varas J, Graumann K, Osman K, Pradillo M, Evans DE, Santos JL, Armstrong SJ. Absence of SUN1 and SUN2 proteins in Arabidopsis thaliana leads to a delay in meiotic progression and defects in synapsis and recombination. Plant J. 2015;81:329–346. doi: 10.1111/tpj.12730. [DOI] [PubMed] [Google Scholar]
- Vanstraelen M, Baloban M, Da Ines O, Cultrone A, Lammens T, Boudolf V, Brown SC, De Veylder L, Mergaert P, Kondorosi E. APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proc. Natl. Acad. Sci. USA. 2009;106:11806–11811. doi: 10.1073/pnas.0901193106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Autran D, Traas J. Developmental control of cell division patterns in the shoot apex. Plant Mol. Biol. 2000;43:569–581. doi: 10.1023/a:1006464430936. [DOI] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell. 2005;123:1337–1349. doi: 10.1016/j.cell.2005.09.042. [DOI] [PubMed] [Google Scholar]
- Yang W, Schuster C, Beahan CT, Charoensawan V, Peaucelle A, Bacic A, Doblin MS, Wightman R, Meyerowitz EM. Regulation of Meristem Morphogenesis by Cell Wall Synthases in Arabidopsis. Curr. Biol. 2016;26:1404–1415. doi: 10.1016/j.cub.2016.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol. Cell. 2007;27:3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Zhang S, Chang L, Alfieri C, Zhang Z, Yang J, Maslen S, Skehel M, Barford D. Molecular mechanism of APC/C activation by mitotic phosphorylation. Nature. 2016;533:260–264. doi: 10.1038/nature17973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Dataset. The expression patterns of core cell cycle genes in Arabidopsis SAM by mRNA in situ hybridisation. Related to Figure 1.
Table S1. Expression level and patterns of cell cycle genes in Arabidopsis dissected shoot apical meristem (SAM) by RNA-seq and in situ hybridisation. Shown are TPM values adapted from Yang et al (2016). Related to Figure 1.
Table S2. Primers used in this study. Related to STAR Methods.