Abstract
Cone snail venoms provide a largely untapped source of novel peptide drug leads. To enhance the discovery phase, a detailed comparative proteomic analysis was undertaken on milked venom from the mollusk-hunting cone snail, Conus textile, from three different geographic locations (Hawai'i, American Samoa and Australia's Great Barrier Reef). A novel milked venom conopeptide rich in post-translational modifications was discovered, characterized and named α-conotoxin TxIC. We assign this conopeptide to the 4/7 α-conotoxin family based on the peptide's sequence homology and cDNA pre-propeptide alignment. Pharmacologically, α-conotoxin TxIC demonstrates minimal activity on human acetylcholine receptor models (100 μM, <5% inhibition), compared to its high paralytic potency in invertebrates, PD50 = 34.2 nMol Kg-1. The non-post-translationally modified form, [Pro]2,8[Glu]16α-conotoxin TxIC, demonstrates differential selectivity for the α3β2 isoform of the nicotinic acetylcholine receptor with maximal inhibition of 96% and an observed IC50 of 5.4 ± 0.5 μM. Interestingly its comparative PD50 (3.6 μMol Kg-1) in invertebrates was ∼100 fold more than that of the native peptide. Differentiating α-conotoxin TxIC from other α-conotoxins is the high degree of post-translational modification (44% of residues). This includes the incorporation of γ-carboxyglutamic acid, two moieties of 4-trans hydroxyproline, two disulfide bond linkages, and C-terminal amidation. These findings expand upon the known chemical diversity of α-conotoxins and illustrate a potential driver of toxin phyla-selectivity within Conus.
Keywords: α-conotoxin, conopeptides, Conus textile, mass spectrometry, milked venom, radula tooth, Post-translational modifications, nAChR
1. Introduction
The genus Conus (Family: Conidae, Subfamily: Coninae) has maintained a position of predatory superiority within tropical marine ecosystems for some 50 million years. The genus attributes its evolutionary success to the development and delivery of a venomous cocktail. The cone snail injects this venom through a hypodermic needle-like radula harpoon that can penetrate deep into the dermis of its prey [19]. These venoms comprise a potent pharmacopeia of individual bioactive peptide constituents, commonly referred to as conotoxins or conopeptides. New estimates indicate that half-a-million distinct biologically active molecules are expressed within this genus alone [17, 38, 49].
Stemming from their intended predatory use as potent neurotoxic agents for prey immobilization [12, 21], the chemical complexity and functional side-chain variability of conopeptides has been studied, whereby providing tools for probing ion channel function and structure-activity relationships. The majority of research has focused on the clinical significance and therapeutic potential of conopeptides. This is evident by the number of conopeptide derived molecules in pre-clinical development or clinical phase trials for the treatment of a broad spectrum of conditions ranging from neuropathic pain and Alzheimer's, to Parkinson's and epilepsy [5, 20, 22, 30, 47, 55].
Early investigations using milked venom from piscivorous species has fuelled rapid identification of a number of novel bioactive peptides including αA-conotoxin OIVA from Conus obscurus [50] and ω-conotoxin SIIIA from Conus striatus [48]. Most notably, in 2004, the United States Food and Drug Administration (FDA) approved Prialt™ (ω-Conotoxin MVIIA), a peptide naturally expressed in the milked venom of Conus magus, for the treatment of chronic neuropathic pain [7, 26]. Unfortunately, due to this heightened medical interest, in conjunction with intensive exploitation by the ornamental shell-trade industry, the over-harvesting of Conus has the potential to result in the depletion of population densities [9, 16]. Subsequently, the need for a rapid and biosustainable approach to identify novel, clinically significant conopeptides while simultaneously reducing pressure on native snail populations has emerged.
In order to increase our understanding of molluscivorous milked venoms, the current study was initiated to perform a comprehensive investigation into the venomic multiplicity of Conus textile using Reverse-Phase High Performance Liquid Chromatographic (RP-HPLC2) profiling and mass spectral analysis of both duct (DV) and milked venoms (MV) collected from non-captive specimens representing diverse locations throughout the Pacific. This widely distributed tropical species currently represents one of the most well-studied molluscivore cone snail with countless molecular constituents, including 77 fully characterized conopeptides (see Table S1).
Our results from the combined ‘conovenomic’ approach revealed an unexpected level of diversity within the venom profiles of geographically unique populations of C. textile, and further demonstrated molecular consistency within individuals between the profiles of (i) MV, (ii) dissected whole DV, and (iii) Radula lumen Extract (RE). The comparison of geographically diverse venom profiles led to the identification of a previously uncharacterized peptide, α-conotoxin TxIC, which exhibits extensive post-translational modifications (PTMs). Here we demonstrate how PTM amino acids act as determining factors for selective targeting of the nicotinic acetylcholine receptor (nAChR) – a well-documented therapeutic target for the treatment of chronic neuropathic pain [8, 31, 44, 51, 52].
2. Materials and Methods
2.1 Snail milking
MV from non-captive C. textile was obtained within 24 hours of field collection. Envenomation was stimulated by the presence of live gastropods Morula marginalba, Strombus luhuanus or Cypraea caputserpentis. On extension of the cone snail's proboscis, a pipette fitted with a 5000 μL tip was depressed and placed near the upturned aperture, close to the foot of the prey. On subsequent firing of the radula, envenomation was observed by the release of excess venom, typically as a visible ‘cloud’, which was carefully aspirated to avoid dispersion. The collected MV was acidified (1% v/v TFA) and either frozen (-20°C) or lyophilized for later analysis. Aliquots of seawater (blanks) were collected and processed in the identical manner.
2.2 Dissected duct venom extract preparation
Whole venom ducts were dissected from live specimens, individually dried by Speed-Vac, weighed and homogenized to obtain a fine powder. A mixture of (1000 μL) 95% Solvent A (0.1% v/v TFA/aq.) and 5% Solvent B (90/10 v/v CH3CN/0.08% v/v TFA/aq.) was used as the extracting solvent at a standardized concentration (1 mg mL-1). Samples were vortexed (30 sec.), sonicated (10 min.) and then centrifuged (4,500g for 10 min.). The resulting supernatant was decanted, dried via Speed-Vac, weighed and then stored at −20°C until required. All materials were dissolved in 500 μL of the above solvent, sonicated (5 min.), filtered (0.2 μm membrane) and then centrifuged (12,000g for 10 min.) prior to chromatographic separation and analysis.
2.3 Chromatographic separation and analysis, RP-HPLC
Representative MV peptides and DV extracts were individually separated as follows: (i) Capillary Scale (Phenomenex; C18, 5 μm, 300 Å, 1.0 × 250 mm, flow 100 μL min.-1) – used for comparative RP-HPLC/UV profiling, quality control of peptide purity, peptide quantification and peptide co-elution experiments. (ii) Analytical Scale (Vydac; C18, 5 μm, 300 Å, 4.2 × 250 mm, flow 1 mL min.-1) – used for the isolation and purification of native peptides for MALDI-TOF-(TOF)-MS and Edman Degradation (Sections 2.4 & 2.5). (iii) Preparative Scale (Vydac; C18, 10 μm, 300 Å, 22 × 250 mm, flow 5 mL min.-1) – used for the desalting and preparative separation of native MV peptides for sequencing and pharmacological assay (Sections 2.9 & 2.12). Systems (i) and (ii) used a Waters 2695 Alliance RP-HPLC System interfaced with a 996 Waters Photo Diode Array Detector for automated sample analysis and detection. Data was acquired and analyzed using Waters Millennium32 (v3.2) software. Samples were eluted using a linear 1% min.-1 gradient of organic (90/10% v/v CH3CN/0.08% v/v aq. TFA) Solvent B against aqueous (0.1% v/v TFA aq.) Solvent A for 65 min., excluding a terminating high organic wash (80% Solvent B for 5 min.), and pre-equilibration step (5% Solvent B) for 10 min. prior to sample injection. Eluent was monitored from 200–300 nm and extracted at 214 nm. Preparative RP-HPLC/UV, system (iii), used a 625 Waters HPLC pump and controller interfaced with a 996 Waters Photo Diode Array Detector. Both gradient control and data acquisition were facilitated by the use of the Waters Millennium32 software. Filtered (Nylon 0.22 μm) crude MV and DV peptide extracts were manually loaded and eluted from the preparative scale column using the same 1% gradient at 5 mL min.-1 and monitored at 214 and 280 nm. Fractions were collected manually and stored at – 20°C or freeze-dried until required.
2.4 Direct ESI-MS infusion
AB/MDS-Sciex API 3000 triple quadrupole mass spectrometer (Thornhill, Ontario, Canada) was used in this investigation as previously described by Chun et al. [10]. The ESI-MS system was calibrated manually in positive mode with PPG 3000 (AB/MDS-Sciex) to achieve <5-ppm mass accuracy, as per manufacturer's protocol.
2.5 MALDI-TOF MS venom analysis
ZipTip™ or RP-HPLC/UV purified venom fractions in Solvent A (0.1% v/v aq. TFA) were mixed 1:1 with matrix solution (40 g L-1 2,5-dihydroxybenzoic acid (DHB) in 1:1 0.1% v/v aq. TFA: CH3CN) and 1 μL was spotted on a MTP 384 polished steel target plate (Bruker Daltonics). The spots were dried under a stream of N2 gas. Mass spectra were acquired on the Ultraflex III (Bruker Daltonics), controlled by the Compass 1.2 SR1 software package (Bruker Daltonics), in positive reflector mode from m/z 500 to 5000. Mass spectra were summed (400 to 1200 laser shots) until no further improvement to the signal to noise ratio of peaks was achieved. Peptide II Calibration Mix (Bruker Daltonics) was used for external calibration, with a mass accuracy of approximately 50-ppm. Analysis of the spectra was completed using FlexAnalysis v3.0 (Bruker Daltonics).
2.6 MALDI-TOF/TOF MS peptide sequencing and PTM characterization
The reduced venom peptide, α-conotoxin TxIC, was desalted via ZipTip™ (C18 reversed-phase media, Millipore) or RP-HPLC/UV isolated and spotted onto the target plate with DHB matrix, as described above (Section 2.5). Tandem mass spectra (MS/MS) were acquired in reflector positive LIFT mode on the UltraflexIII (Bruker Daltonics), externally calibrated with Peptide Calibration Mix II (Bruker Daltonics) with a MS/MS accuracy of 0.04 Da. FlexAnalysis v3.0 (Bruker Daltonics) was used for manual inspection and annotation of the LIFT-spectra. The RapiDeNovo module in BioTools (Bruker Daltonics) was used to make additional assignments to the amino acid sequence.
2.7 MALDI-TOF MS radula analysis
Conus textile radulae were collected from dissected radula sacs. Harpoons were prepared for MALDI-TOF MS analysis as described previously by Chun et al. [10].
2.8 Peptide reduction and thiol alkylation
For complete peptide reduction, Conus extracts and peptide(s) (typically 0.02–3 mg) were exposed to 100 μL of 200 mM Tris(2-carboxyethyl)phosphine (TCEP; Pierce Chemicals, USA) in 25 mM NH4OAc (pH 4.5) and heated for 5–30 min. at 50°C. Alkylation of the RP-HPLC purified, reduced peptide(s) was achieved by dissolving the peptide in 90-150 μL of 25 mM NH4OAc (pH 4.5) and adding 100 mM N-phenylmaleimide or Maleimide (Fluka, Switzerland) in isopropanol. Typically 20–40 fold excess (w/w) of the alkylating agent was used. Alkylation was allowed to proceed at 50°C for 15 min., prior to RP-HPLC/UV purification.
2.9 Sequencing – Edman degradation
Non-alkylated and Maleimide alkylated derivatives were applied to Polybrene-treated glass fiber support filters for automated Edman degradation on a gas-phase sequencer (Model 470A; Applied Biosystems, Foster City, CA, USA). Assignment of the amino acid sequence was essentially as described by Atherton et al. [2] and Matsudaira [34].
2.10 Peptide synthesis
α-Conotoxin TxIC [ROQC4CSHOAC10NVDHPγIC-NH2; γ = γ-carboxyglutamic acid (Gla), O = 4-trans hydroxyproline (Hyp)] was manually assembled using a 0.5 mmole scale and Fmoc SPPS via in-situ neutralization with 2 mmole Fmoc-amino acid per 10–30 min. single coupling, as adapted from Schnölzer et al. [46] and described in detail in Kapono et al. [26]. A Fmoc-Cys(Trt)-Rink-Amide MBHA Resin (0.55 meq. g-1; Peptides International, Louisville, KY, USA) was used for peptide assembly to give the desired C-terminal amide function. Side chain protecting groups were: Cys(Trt), Asp(tBu), Arg(Pbf) and Gln(Trt). Gla(otBu)2, His(Trt), Asn(otBu) and Hyp(tBu) (as supplied by Peptides International, Louisville, KY, USA). An additional 0.5 mmole scale synthesis of α-conotoxin TxIC was undertaken using orthogonally protected Cys(Acm) being placed in positions 4 and 10 in the above sequence. This synthesis used the same synthetic strategy as previously mentioned, but deviated using an alternative directive/sequential oxidation process (see below). Finally, a non-PTM amino acid variant of α-conotoxin TxIC, (i.e. [Pro]2,8[Glu]16α-conotoxin TxIC), was produced and folded using the same orthogonally Cys(Acm) protected scheme as above. Incorporated were the Fmoc amino acids Pro and Glu(OtBu) used as PTM substituents.
2.11 TFA cleavage
Fmoc peptidyl-resins were subjected to a cleavage with 82.5% v/v TFA in the presence of thioanisole (5% v/v), Phenol (5% v/v) H2O (5% v/v) and TIPS (2.5% v/v), acting as protecting group scavengers, for 2.5 h at 24°C. The resulting cleaved peptide material was recovered by filtration and cold t-butyl ether precipitation. Resulting crude peptide was stored at −20°C, as lyophilized powder, until required.
2.12 Random disulfide bond formation
Peptide oxidation was achieved with 15 mg of TCEP reduced C18 RP-HPLC purified peptide, using 0.1 M NH4HCO3 pH 8.7 (5 days stirring; room temp.). The oxidized peptide was preparative RP-HPLC/UV isolated and re-purified, and the oxidized molecular mass was verified by ESI-MS.
2.13 Directed disulfide bond formation
Cleaved α-conotoxin TxIC and [Pro]2,8[Glu]16α-conotoxin TxIC, both containing Cys(Acm) in positions 4 and 10, were RP-HPLC/UV purified, and then air oxidized, as above. Partially oxidized materials, as confirmed by ESI-MS, were then subjected to spontaneous thiol deprotection and disulfide bond formation. Deprotection was achieved by dissolving the partially folded peptide in 50% v/v acetic acid (1 mg mL-1) and by adding a solution of freshly saturated I2 in 50% v/v acetic acid to the stirring peptide (25% reaction vol.). Reaction was quenched after 5 min. with the addition of 10 μL aliquots of 1M Na2S2O3 until the stirring solution became clear, which was then followed by the addition of 200 μL TFA. Resulting acidified material was centrifuged (12,000g, 5 min.) and directly purified by preparative RP-HPLC/UV (as above) with mass confirmation provided by ESI-MS.
2.14 Genetic analysis of α-conotoxin TxIC
The venom duct and bulb of C. textile specimens (Great Barrier Reef, Australia) were removed by dissection and snap frozen in liquid N2. The secretory venom duct and bulb were ground and mRNA was extracted using a Dynabeads mRNA direct kit (Dynal, Norway). A cDNA library was created from this mRNA using a Marathon cDNA amplification kit (Clontech) as previously described [44]. All PCRs utilized the primers Uni α-1 (universal α-conotoxin primer) 5′ ATGGGCATGCGGATGATGTT 3′; and Uni α-2 (universal α-conotoxin primer) 5′ CGGAAAGTGAAGCAGGTCAG 3′, designed against conserved sequences found on the 5′ and 3′ conserved regions of known conotoxins. Reaction mixtures contained Taq polymerase (Roche) and deoxynucleotides in a buffer supplied by the manufacturer. Samples were incubated at 94°C for 2 min.; followed by 30 cycles of 94°C for 30 sec, an annealing step for 30 sec, 72°C for 45 sec; concluding with a final step of 72°C for 5 min. Amplification products were purified after separation on a 1.5% agarose gel using a Qiaquick gel extraction kit (Qiagen). The purified PCR products were transformed into competent INVαF′ Escherichia coli cells in accordance with the manufacturer's specifications (Invitrogen, Netherlands). Recombinant plasmids containing inserts of approximately 250 bp were sequenced by the di-deoxy chain termination method using the ABI PRISM Big-Dye Terminator Cycle Sequence Ready Reaction Kit (Perkin Elmer, USA). Sequences were analyzed on a Perkin Elmer 377 sequencer at the Australian Genome Research Facility (AGRF).
2.15 Pharmacology
2.15.1 Expression of voltage-gated ion channels in Xenopus laevis oocytes
For the expression of nAChR (α1, α3, α4, α5, β2, β4, γ; δ; ε) in Xenopus oocytes, the linearized plasmids were transcribed using the T7 or SP6 mMESSAGE-mMACHINE transcription kit (Ambion®, Carlsbad, CA, USA). The harvesting of stage V–VI oocytes from anaesthetized female Xenopus laevis frog was previously described by Liman et al. [29]. Oocytes were injected with 50 nL of cRNA at a concentration of 1 ng nL-1 using a micro-injector (Drummond Scientific®, Broomall, PA, USA). The oocytes were incubated in a solution containing (in mM): NaCl, 96; KCl, 2; CaCl2, 1.8; MgCl2, 2 and HEPES, 5 (pH 7.4), supplemented with 50 mg L-1 gentamycin sulfate.
2.15.2 Electrophysiological recordings
Two-electrode voltage-clamp recordings were performed at room temperature (18–22°C) using a Geneclamp 500 amplifier (Molecular Devices®, Downingtown, PA, USA) controlled by a pClamp data acquisition system (Axon Instruments®, Union City, CA, USA). Whole cell currents from oocytes were recorded 1–4 days after injection. Bath solution composition was (in mM): NaCl, 96; KCl, 2; CaCl2, 1.8; MgCl2, 2 and HEPES, 5 (pH 7.4). Voltage and current electrodes were filled with 3 M KCl. Resistances of both electrodes were kept between 0.5–1.5 MΩ. During recordings, the oocytes were voltage-clamped at a holding potential of −70 mV and super fused continuously with ND96 buffer via gravity-fed tubes at 0.1–0.2 mL min.-1, with 5 min. incubation times for the bath-applied peptides. Acetylcholine (ACh) was applied via gravity-fed tubes until peak current amplitude was obtained (1–3 sec.), with 1–2 min. washout periods between applications. Data was sampled at 500 Hz and filtered at 200 Hz. Peak current amplitude was measured prior to and following incubation of the peptide.
To assess the concentration-response relationships, data points were fitted with the Hill equation: y = 100/[1 + (EC50/[toxin])h], where y is the amplitude of the toxin-induced effect, EC50 is the toxin concentration at half maximal efficacy, [toxin] is the toxin concentration and h is the Hill coefficient. Comparison of two sample means was made using a paired Student's t test (p<0.05). All data is presented as mean ± standard error (S.E.M) of at least 4 independent experiments (n ≥ 4). All data was analyzed using pClamp Clampfit 10.0 (Molecular Devices®, Downingtown, PA, USA) and Origin 7.5 software (Originlab®, Northampton, MA, USA).
2.16 Whole animal bioassay
Standardized concentrations of native α-conotoxin TxIC (2.5, 5, 10, 20, 40, 80, 160, 320 and 640 pMol g-1) and synthetic [Pro]2,8[Glu]16α-conotoxin TxIC (0.16, 0.32, 0.64, 1.28, 2.56, 5.12 and 10.24 nMol g-1) in 10 μL volumes (PBS), were injected intramuscularly (IM - foot) into Hawaiian snakehead cowries (Cypraea caputserpentis) using a Hamilton 10 μL syringe at a depth of 2 mm. Following injection, animals were placed in glass Petri dishes filled with fresh aerated seawater, where paralysis was determined by the inability of the animal to cling to the substrate. Dosage and paralysis were recorded and plotted according to Reed and Muench [40], and the PD50 was extrapolated using GraphPad Prism Software (v5.02). All dose experiments were repeated in triplicate or greater (n ≥ 3).
3. Results
3.1 Conformational analysis of δ-conotoxin TxVIA
The MH+ 3035.4 Da was identified by ESI-MS from RP-HPLC/UV fractions of DV, confirming the identity of δ-conotoxin TxVIA (Calc. MH+ 3035.3 Da). TCEP reduction, RP-HPLC/UV separation and ESI-MS analyzed demonstrated a mass shift of 6 Da (Obs. Red. MH+ 3041.5 Da), which reflects disulfide content. This same material (∼300 nmole) was then subjected to 27 cycles of Edman degradation, and a single unambiguous sequence that corresponded to δ-conotoxin TxVIA W(-)KQSGEM(-)NLLDQN(-)(-)DGY(-)IVLV(-)T was obtained. ‘Blanks’ (-) were observed at degradative cycles 2, 9, 16, 17, 21 and 26. These corresponded to the 6 underivatized cysteine moieties as predicted within this specific sequence. This confirmed the expression of δ-conotoxin TxVIA – a species-specific duct venom ‘biomarker’ for all specimens examined in this study. The methionine sulfoxide derivative (+16 Da) of this peptide was also observed by MALDI-TOF MS (m/z 3051.3; Figs. 2A & B).
Fig. 2. MALDI-TOF MS mass spectra of of representative non-captive milked venoms from C. textile specimens obtained from: (A) Hawai'i; (B) American Samoa; and (C) the Great Barrier Reef, Australia.
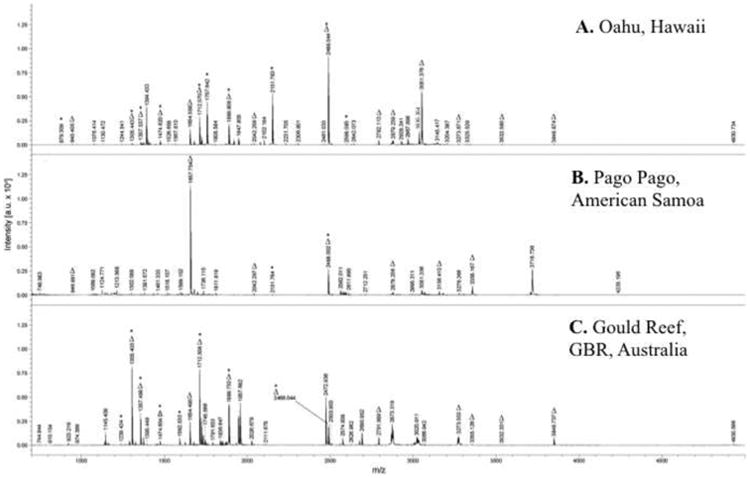
Peaks corresponding to known conopeptides (*) and peaks common to all 3 spectra (Δ) are indicated Table S2 provides an extensive list comparing all peaks observed in these venom profiles.
3.2 Geographic biodiversity - RP-HPLC/UV non-captive milked venom profiling
Typical representative MVs from non-captive specimens of C. textile demonstrate an abundance of chromatographic peaks ranging from 20–70 perMV profile (Fig. 1). Multiple MV samples from Hawai'i (n=30 specimens) and the Great Barrier Reef (n=50 specimens; GBR), Australia contained similar chromatographic content, with varying relative peptide concentrations as observed in the moderately hydrophilic region (25–40% CH3CN, 25 to 40 min.; see Figs. 1A & C, respectively). No observable RP-HPLC/UV correlation to cone snail sex or collection/milking season could be established from the MV profiles from any of the three locations examined (not shown).
Fig. 1.
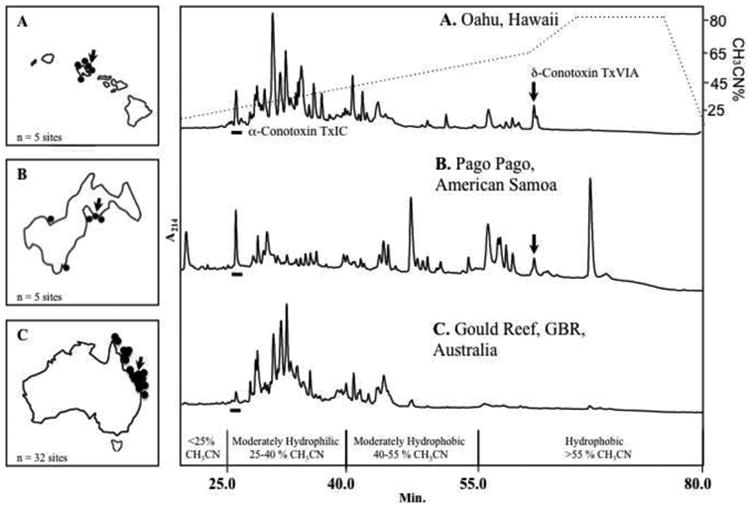
RP-HPLC/UV comparison of peptides from the milked venom of representative non-captive C. textile specimens sampled from different geographic locations in the South Pacific: (A) Hawai'i; (B) American Samoa; and (C) the Great Barrier Reef, Australia.
The expression of δ-conotoxin TxVIA (arrow), was detected in MV samples from Hawai'i (n=30, from 5 sites) and American Samoa (n=20, from 5 sites), but was absent from those from GBR, Australia (n=50, from 32 sites). α-Conotoxin TxIC (underscore) was detected in MV from all three locations, indicating its importance in the envenomation strategy of C. textile. MALDI-TOF MS analysis of these MV samples is shown in Fig. 2. In each map, indicated are cone snail collection sites (filled circle) and individual representative samples (arrow).
The moderate to hydrophobic RP-HPLC/UV profile region (40–80% CH3CN, 40 to 80 min.) accounted for >30% of milked venom variability, with the highest proportion of peak variability seen in the representative MV of non-captive specimens from American Samoa (n=20 specimens) (Fig. 1B). The hydrophobic δ-conotoxin TxVIA (Rt 65.3 min.) was observed in both non-captive Hawai'ian and American Samoan representative samples (see arrow in Fig. 1A & B respectively), but was absent in non-captive GBR Australian MV (Fig. 1C). One previously undocumented peak (underlined), with a retention time (Rt) = 26.6 min. (α-conotoxin TxIC), was observed in all three locations (n=100, total specimens examined) (Fig. 1A–C). This observed geographic commonality lends credence to the importance of that particular conopeptide.
3.3 Geographic biodiversity - Molecular mass profiling of non-captive milked venom
MALDI-TOF MS profiles of MV from 5 random specimens of non-captive C. textile from each geographic location cover the entirety of the examined mass range (Figs. 2A-C). In the examples illustrated, a combined total of 178 non-overlapping peaks were observed in the three MVs (Table 1 & Table S2). Of these, 42 peaks (∼24%) were common to two collection sites, with only 11 (∼6%) being common to all three. Importantly, 27 of the 178 non-captive MV constituents (∼15%) correlated to known C. textile peptides (Table 1; Tables S1 & S2), excluding potential PTM variants including C-terminal processing (Δ ± 1.0 Da for free-acid vs. amide). Expression of 2 of these known conopeptides, TxIA and TxIIIC, were previously confirmed by Edman degradation sequencing [4].
Table 1. MALDI-TOF MS analysis of C. textile MV data from different geographical locations, showing masses that correlate to know conopeptides.
| Obs. m/z | HI | AS | GBR | Mass correlation |
|---|---|---|---|---|
| 879.309 | ✓ | Leu-contryphan-Tx | ||
|
| ||||
| 1091.051 | ✓ | TeAr193 | ||
|
| ||||
| 1238.410 | ✓ | TeAr151 | ||
| 1239.434 | ✓ | |||
|
| ||||
| 1305.405 | ✓ | TxIIIC (Tx3.4) | ||
| 305.443 | ✓ | |||
|
| ||||
| 1357.499 | ✓ | Tx10b or Tx10c | ||
| 1357.537 | ✓ | |||
|
| ||||
| 1392.411 | ✓ | TxXIIIA | ||
|
| ||||
| 1447.593 | ✓ | Tx5b | ||
| 1448.557 | ✓ | |||
|
| ||||
| 1474.604 | ✓ | Tx5c | ||
| 474.620 | ✓ | |||
|
| ||||
| 1592.509 | ✓ | Tx3d | ||
|
| ||||
| 1656.531 | ✓ | Tx1c | ||
|
| ||||
| 1657.754 | ✓ | TxIA | ||
|
| ||||
| 1678.968 | ✓ | Tx-D0111 | ||
| 1679.718 | ✓ | |||
|
| ||||
| 1712.485 | ✓ | TxIIIA | ||
| 1712.570 | ✓ | |||
|
| ||||
| 1723.566 | ✓ | TxMLKM-021 | ||
| 1723.636 | ✓ | |||
| Obs. m/z | HI | AS | GBR | Mass correlation |
|---|---|---|---|---|
| 1757.642 | ✓ | TxMMSK-03 | ||
| 1756.444 | ✓ | |||
|
| ||||
| 1761.518 | ✓ | TxMRCL-D012 | ||
|
| ||||
| 1779.609 | ✓ | TxMMSK-04 | ||
|
| ||||
| 1844.578 | ✓ | Tx3e | ||
|
| ||||
| 1889.753 | ✓ | Tx3f (Tx3.5) | ||
| 1889.808 | ✓ | |||
|
| ||||
| 2151.783 | ✓ | Tx3h (TxMLKM-011) | ||
|
| ||||
| 2487.917 | ✓ | Convulsant Peptide | ||
| 2488.002 | ✓ | |||
| 2488.044 | ✓ | |||
|
| ||||
| 2642.073 | ✓ | Tx6.3 | ||
|
| ||||
| 2942.164 | ✓ | TxMKLT1-015 | ||
|
| ||||
| 2961.308 | ✓ | Tx05 | ||
|
| ||||
| 2995.234 | ✓ | Tx6.2 | ||
| 2995.311 | ✓ | |||
|
| ||||
| 3035.342 | ✓ | TxVIA (King-Kong 0) | ||
|
| ||||
| 3066.942 | ✓ | TxMLKLT1-0111 | ||
| 3067.313 | ✓ | |||
| 3067.339 | ✓ | |||
Obs. m/z are within ± 1.0 Da of the identified peptide mass to account for possible amide and free acid variants.
Abbreviations are: Hawai'i (HI); American Samoa (AS); and Great Barrier Reef, Australia (GBR).
3.4 Single specimen venom source consistency – Non-captive RP-HPLC/UV profiling
RP-HPLC/UV venom peptide profiles from a single non-captive specimen of C. textile (Oahu, Hawai'i) is shown in Figs. 3A–C. Results illustrate a high level of peptide Rt consistency between the whole duct venom (DV) extract, the milked venom (MV), and the Radula lumen Extract (RE). The relative abundance of peptides in the different venom extracts was highly variable as seen with the relative intensity of the hydrophobic δ-conotoxin TxVIA (Rt 65.3 min.; Figs. 3B & C).
Fig. 3. RP-HPLC/UV comparison of venom obtained from (A) Radula Lumen Extract; (B) Milked Venom; and (C) Duct Venom from a representative specimen of C. textile.
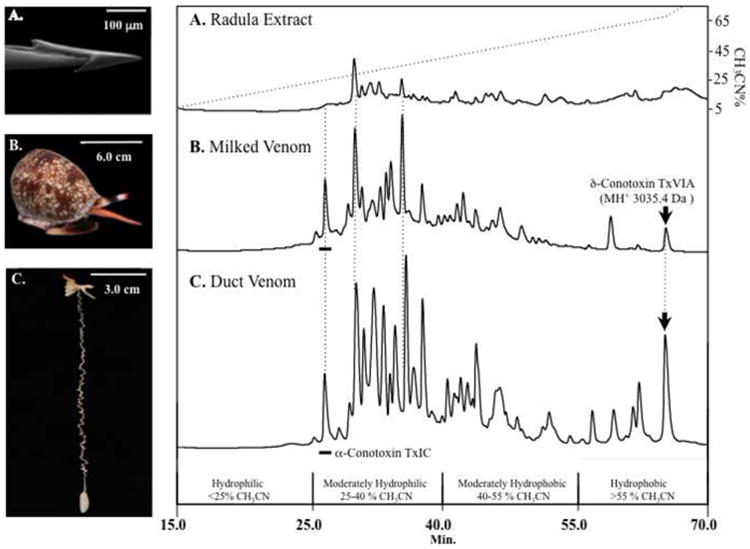
(A) The RE revealed limited similarity of peptides within the hydrophilic region to those in the hydrophilic regions of MV and DV. Note that α-conotoxin TxIC (underscore) and δ-conotoxin TxVIA (arrow) were not detected in the RE. Both (B) MV and (C) DV demonstrated a high level of peptide abundance and continuity. These samples were profiled via MALDI-TOF MS (Fig. 4).
3.4.1 Duct venom (DV) extracts
In the DV, δ-conotoxin TxVIA was one of the major components (Fig. 3C). The presence of α-conotoxin TxIC in DV (underscored, Fig. 3C) was observed at Rt = 26.6 min., corresponding to an identical elution profile from the MV of the same specimen (Fig. 3B) as well as in the representatives from geographic MV sampling (Fig. 1).
3.4.2 Milked venom (MV) extracts
In contrast to the DV, MV profiles naturally correspond to a more accurate representation of bioactive constituents used in prey immobilization. δ-Conotoxin TxVIA was one of only a few hydrophobic peptides observed in the MV (Fig. 3B), which exhibited direct correlation to the DV. These results are reiterated in the geographic specimens that exhibit few hydrophobic peptides (Fig. 1); most notably, Australian specimens – which may reflect venom processing. Highly abundant hydrophilic peptides within both DV and MV venom extract profiles were examined (Fig. 3B & C), which identified α-conotoxin TxIC (underscored, Rt = 26.6 min.).
3.4.3 Radula lumen (RE) extracts
Analysis of the RE demonstrated poorly resolved peaks with limited abundance (Fig. 3A), yet the two major peaks correlated with the two highly abundant peaks in the MV (Fig. 3B). Both peaks showed marked differences in the DV extract.
3.5 Single specimen venom source consistency – Non-captive molecular mass profiling
MALDI-TOF-MS was used to detect and compare identifiable peptide masses in extracts of the RE, MV and DV, from a single non-captive representative specimen of C. textile (Fig. 4). A total of 159 unique molecular masses were observed (Table S3). Common to all three extracts were 16 individual masses (∼10%). Showing the greatest similarity were the MV and DV profiles that shared 38 masses (∼24%). The RE mass spectrum, exhibiting masses within m/z 945 to 3850, contained fewer masses than the MV and DV. The MV mass spectrum was the most complex in peak number of the three.
Fig. 4. MALDI-TOF MS profiles of peptide extracts from a single representative Conus textile specimen obtained from: (A) Radula Lumen Extract; (B) Milked Venom and (C) Whole Duct Venom.
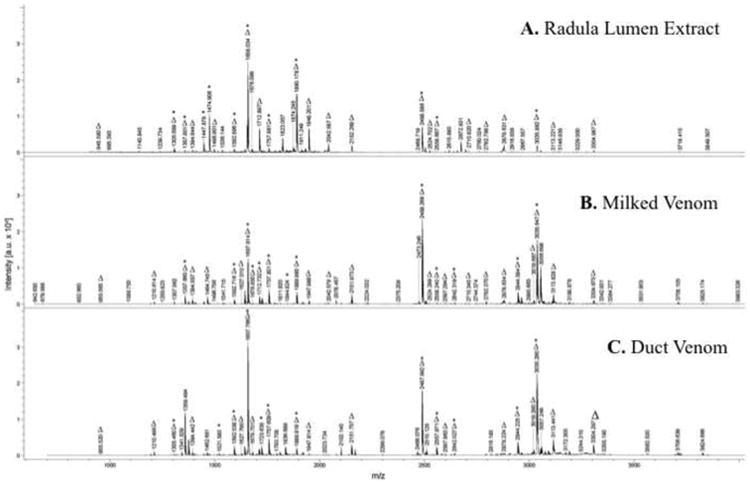
Peptides exhibiting common peaks are listed in Table 2 and indicated (Δ) in each profile. These data illustrate expressional consistency, although some unique peaks are observed from each venom source. Absence of α-conotoxin TxIC is apparent, but is suspected to be present as an in-source decay fragment. δ-Conotoxin TxVIA was observed at m/z 3035.6±0.2. Peaks corresponding to other known C. textile peptides are indicated (*). Table S1 provides an extensive list of observed peaks corresponding to known C. textile venom peptides.
Most notable are the high intensity masses observed in both the DV and MV extracts which correspond to important and previously characterized conopeptides (Table 2 & Fig. 4). The dominant peak in the MV and DV mass spectra (m/z 2488.2 ± 0.3) corresponds to the C. textile Convulsant peptide. This peak is also present in the RE mass spectrum. Other identified peptides (i.e., TxIA and TxIIIC) were also observed as major contributors in the geographic analysis (Table 1, Fig. 2).
Table 2. Comparative analysis of MALDI-TOF MS C. textile data from different venom sources (RE; MV; and DV).
| Obs. m/z | RE | MV | DV | Mass correlation |
|---|---|---|---|---|
| 1305.460 | ✓ | TxIIIC | ||
| 1305.528 | ✓ | |||
| 1305.689 | ✓ | |||
|
| ||||
| 1357.487 | ✓ | Tx10b/Tx10c | ||
| 1357.660 | ✓ | |||
| 1357.801 | ✓ | |||
|
| ||||
| 1392.422 | ✓ | TxXIIIA (TxMRCL-02) | ||
| 1392.533 | ✓ | |||
|
| ||||
| 1447.628 | ✓ | Tx5b | ||
| 1447.747 | ✓ | |||
| 1447.879 | ✓ | |||
|
| ||||
| 1474.753 | ✓ | Tx5c | ||
| 1474.906 | ✓ | |||
|
| ||||
| 1521.536 | ✓ | TxIIIB | ||
|
| ||||
| 1592.538 | ✓ | Tx3d | ||
| 1592.716 | ✓ | |||
| 1592.896 | ✓ | |||
|
| ||||
| 1642.756 | ✓ | Tx5d | ||
| 1642.927 | ✓ | |||
| 1643.013 | ✓ | |||
|
| ||||
| 1657.766 | ✓ | TxIA | ||
| 1657.914 | ✓ | |||
| 1658.034 | ✓ | |||
|
| ||||
| 1679.701 | ✓ | Tx-D0111 | ||
| 1679.850 | ✓ | |||
|
| ||||
| 1710.766 | ✓ | TxIIIA | ||
|
| ||||
| 1723.635 | ✓ | TxMLKM-021 | ||
| 1723.787 | ✓ | |||
| Obs. m/z | RE | MV | DV | Mass correlation |
|---|---|---|---|---|
| 1757.609 | ✓ | TxMMSK-03 | ||
| 1757.801 | ✓ | |||
| 1757.981 | ✓ | |||
|
| ||||
| 1844.824 | ✓ | Tx3e | ||
|
| ||||
| 1889.819 | ✓ | Tx3f | ||
| 1889.980 | ✓ | |||
| 1890.178 | ✓ | |||
|
| ||||
| 1931.063 | ✓ | TxVA | ||
|
| ||||
| 2151.757 | ✓ | Tx3h | ||
| 2151.973 | ✓ | |||
| 2152.196 | ✓ | |||
|
| ||||
| 2488.586 | ✓ | Convulsant Peptide | ||
| 2488.269 | ✓ | |||
| 2487.992 | ✓ | |||
|
| ||||
| 2557.971 | ✓ | Tx02 | ||
| 2558.240 | ✓ | |||
| 2558.667 | ✓ | |||
|
| ||||
| 2642.027 | ✓ | Tx6.3 | ||
| 2642.319 | ✓ | |||
|
| ||||
| 2944.225 | ✓ | TxMKLT1-015 | ||
| 2944.594 | ✓ | |||
|
| ||||
| 3011.221 | ✓ | King-Kong 2 | ||
|
| ||||
| 3035.290 | ✓ | TxVIA (KK-0) | ||
| 3035.647 | ✓ | |||
| 3035.892 | ✓ | |||
|
| ||||
| 3067.747 | ✓ | TxMKLT1-0111 | ||
|
| ||||
| 3107.684 | ✓ | Tx04 | ||
Obs. m/z are within ± 1.0 Da of the identified peptide mass to account for possible amide and free acid variants.
3.6 Mass spectrometric analysis of α-conotoxin TxIC disulfide bonds
A geographically consistent peptide (Rt = 26.6 min.; Fig. 3) was isolated from MV by RP-HPLC/UV. A single parent mass of MH+ 2080.8 Da was assigned by ESI-MS (Obs. [M+3H]+3 694.6 Da; data not shown). After TCEP reduction, the observed parent mass was MH+ 2084.7 Da, which indicated the presence of 2 disulfide bridges. This was confirmed by alkylation of the reduced material with N-phenylmaleimide, after which an increase in 692 Da was observed, corresponding to 4 alkylation groups (Δ 173 Da for each free thiol [6]).
3.7 CID analysis of α-conotoxin TxIC
The parent ion of α-conotoxin TxIC was not observed in MALDI-TOF MS analyses of geographic and source extracts (Figs. 2 & 4). This indicated decay of the parent ion, which is common in MALDI-TOF MS of peptides containing labile PTMs. Direct observation of this suspected decay was achieved via MALDI-TOF MS of the purified material in its reduced form (Fig. 5A). Two well resolved peaks, with a difference of 44 Da, indicated the presence of a carboxyl group. Similar decay of the parent ion could be reproduced with ESI-MS, a softer ionization method, by increasing the orifice potential voltage outside of the normal operating range (data not shown).
Fig. 5.
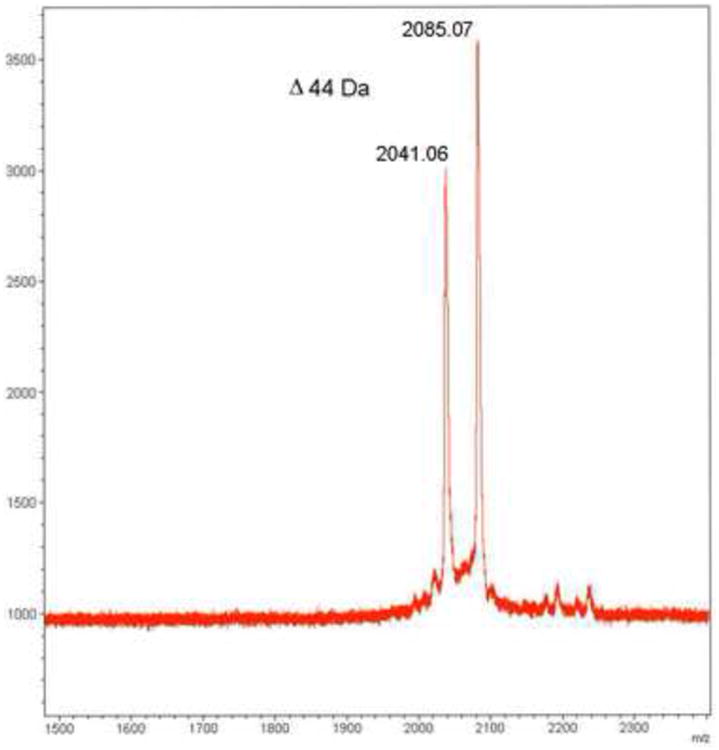
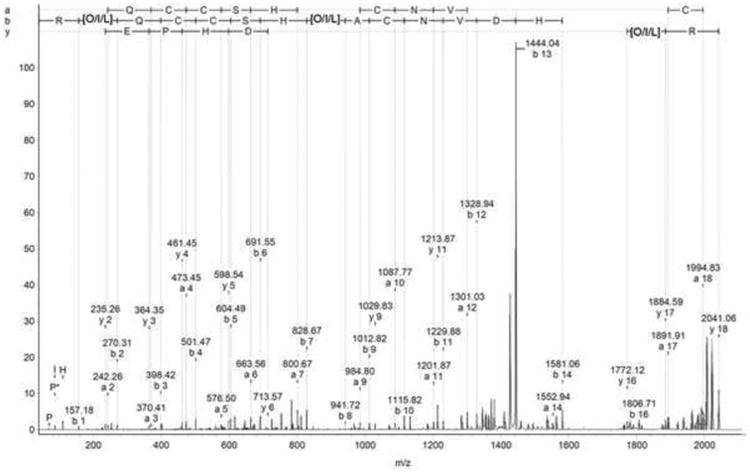
(A) The decarboxylation of TCEP Reduced α-conotoxin TxIC as seen by MALDI-TOF MS. Observed molecular masses differ by 44 Da. The ratio could be influenced by choice of MALDI-MS matrix (not shown). (B) CID analysis of α-conotoxin TxIC. Using overlapping a-, b-, and y-ion series, the sequence was interpreted to be R-[I/L/O]-Q-C-C-S-H-[I/L/O]-A-C-N-V-D-H-P-E-[I/L/O]-C. Fragment ions also indicated decarboxylation and heterogeneity of the C-terminal amino acid.
MALDI-TOF/TOF MS/MS fragmentation spectra of the suspected decarboxylated (m/z 2041.06; Fig. 5B) and ‘intact’ form (m/z 2805.07; data not shown) of TCEP reduced α-conotoxin TxIC were nearly indistinguishable. In both cases MS/MS fragmentation displayed a 44 Da deficient a-, b-, and y-ion series. Analysis of the overlapping ion series allowed for interpretation of the amino acid sequence as R-[I/L/O]2-Q-C-C-S-H-[I/L/O]8-A-C-N-V-D-H-P-E-[I/L/O]17-C. The isobaric nature of isoleucine and leucine (I and L) and their similarity in mass to 4-trans-hydroxylproline (O) (Δ 0.036 Da) resulted in ambiguities at positions 2, 8 and 17, which were resolved with Edman degradation (Section 3.8).
The CID fragment ions also indicated heterogeneity of the C-terminus, the predominant form being amidated. The presence of low abundant C-terminal free acid form was verified using a narrow precursor ion selection window of ± 0.5 Da. The MS/MS fragmentation spectrum was interpreted to have the same amino acid sequence as the amidated form (data not shown). Additional verification was obtained by comparing predicted and observed isotopic distribution patterns for the respective parent ions.
3.8 Edman degradation of α-conotoxin TxIC
RP-HPLC/UV purified N-phenylmaleimide thiol alkylated α-conotoxin TxIC, ∼300 nmole, was subjected to 20 cycles of automated Edman degradation. PTH-4-trans-hydroxylproline was added to the amino acid standard mixture to aid identification. Sequential degradation provided an unambiguous 18 amino acid sequence: RO2QCCSHO8ACNVDHP(-)16IC. Cycle 16 indicated a low recovery level PTH-Glu (<1 nmole) and was designated as a ‘blank cycle’ based on the expected sequential cycle yields observed within the normal degradative process.
Edman degradation assigned 17 of the 18 amino acids conclusively, including positions 2 and 8 as 4-trans-hydroxylproline and 17 as isoleucine. The presence of low levels of glutamic acid in cycle 16, combined with the observed mass difference of 44 Da (-CO2) under various mass spectrometric conditions, provided further corroborated the presence of a γ-carboxylated glutamic acid within α-conotoxin TxIC at position 16.
3.9 Genetic analysis of α-conotoxin TxIC
The peptide sequence was confirmed genetically by identifying the α-conotoxin transcript in C. textile venom duct cDNA (GBR, Australia) using oligonucleotides based on regions of conserved sequence in the pre-region and C-terminal flanking region of α-conotoxins [44]. The resulting 19 amino acid toxin- or mature-sequence region was deduced: RPQCCSHPACNVDHPEIC[R] (see Fig. 6). A C-terminal arginine was absent following Edman analysis; its inclusion would also be at variance with native molecular mass as observed by either ESI-MS, MALDI-TOF MS or by sequence analysis using MALDI-TOF/TOF MS LIFT.
Fig. 6. Genetic analysis of α-conotoxin TxIC and its comparison to α-conotoxins MII and Vc1.1.
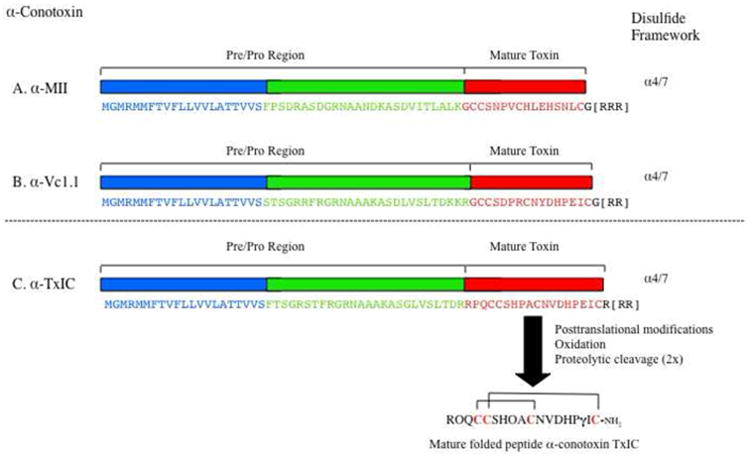
The Pre/Pro region of α-conotoxin TxIC demonstrates a high level of amino acid identity to established members of the 4/7 α-conotoxin family. N-terminal processing of α-conotoxin TxIC shares similarities with the processing of α-conotoxin Vc.1.1 required to produce the mature toxin. Similar mechanisms operate to process α-conotoxin MII. C-terminal brackets [ ] represent stop codons within the formal cDNA sequence.
The N-terminal pro-region of α-conotoxin TxIC also possesses an argininase site for mature conopeptide cleavage, an occurance common to conotoxin processing [24]. In α-conotoxin TxIC the pro-peptide cleavage site is located between two neighboring arginine moieties – see Fig. 6. This is confirmed by the recovery/identification of an N-terminal arginine in α-conotoxin TxIC both by MALDI-TOF/TOF MS LIFT and Edman Degradation. Independent identification of [Glu]16, via genetic analysis, conclusively confirmed the original parent PTM amino acid assignment.
3.10 Peptide synthesis of α-conotoxin TxIC
Based on the assigned C-terminal amide sequence above, α-conotoxin TxIC was synthetically constructed and folded by random air oxidation. Upon completion, the correct target molecular mass was observed by ESI-MS: Obs. MH+ 2080.9 Da (Calc. MH+ 2080.8 Da). RP-HPLC/UV demonstrated the presence of 3 peaks, with the two dominant peaks separated by <0.3 min. (Fig. 7A). All three peaks were determined to be disulfide isomers of α-conotoxin TxIC, as confirmed by ESI-MS. Separation of the two main isomers proved difficult. Changes to gradient, ion pair agents and column failed to achieve their full separation (data not shown).
Fig. 7.
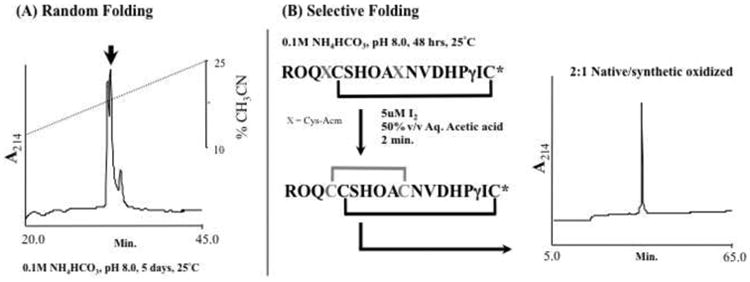
(A) Random folding of synthetic α-conotoxin TxIC. Production of isomeric materials was achieved upon random peptide folding, which when resolved by RP-HPLC/UV demonstrated difficulties in purification. (B) Selective folding of α-conotoxin TxIC and its co-elution with native material. Undertaking a separate synthetic, together with a step-wise directed folding strategy, allowed for the production of a single peak/isomer upon full oxidation. RP-HPLC/UV co-elution in oxidized and TCEP reduced from (not shown), demonstrated identical chromatographic behaviors.
A second synthesis was performed using the established disulfide connectivity pattern reported for the 4/7 α-conotoxin family [31]. This was achieved by incorporation of differential orthogonally protected cysteine moieties at positions 4 and 10 (Fig. 7B). This allowed for the step-wise production of a single oxidized peptide peak that retained target characteristics of mass (Obs. MH+ 2080.9 Da) and RP-HPLC/UV using a slower chromatographic gradient (0.5% min.-1), without production of the previously observed isomeric materials (Fig. 7A). Co-injection of the RP-HPLC/UV purified synthetic material with α-conotoxin TxIC isolated from the native milked venom, in a ratio of 1:2 respectively, demonstrated chromatographic homogeneity at 33.3 min., Fig. 7B. RP-HPLC/UV co-injection of the TCEP reduced material also demonstrated chromatographic homogeneity (not shown). This confirmed the original peptide sequence assignment, including C-terminal assignment, and provided an independent synthetic standard for RP-HPLC/UV.
3.11 Pharmacological assessment
Native α-conotoxin TxIC (≤100 μM) had minimal inhibitory action on human nicotinic acetylcholine receptor (nAChR) subunit combinations α4β2 (brain), α4β4 (neuronal), α7 (neuronal), αβγδ (muscle), αβγε (muscle) or the human peripheral nAChR subtypes α3β2, α3β2α5 and α3β4. Biological activity was observed with [Pro]2,8[Glu]16α-conotoxin TxIC, the synthetic non-PTM form (Fig. S1A & B). This analogue inhibited the human nAChR subtypes (α3β2, α3α5β2 and α3β4), with the α3β4 isoform demonstrating the highest sensitivity (IC50 2.1±0.2 μM) and exhibiting a 77% maximum inhibition. Isoform α3β2 demonstrated the highest level of inhibition (96%) with an IC50 of 5.4±0.5 μM (Table 4). No significant inhibition was observed at the remaining isoform targets (α4β2, α4β4, α7, αβγδ, αβγε) at concentrations up to 100 μM (Table 4).
Table 4. Pharmacological analysis of α-conotoxin TxIC and [Pro]2,8[Glu]16α-conotoxin TxIC on nAchR isoforms.
| nAChR subtypea | IC50b[μM] | Maximum inhibition % | PD50c [nMol Kg-1] | |
|---|---|---|---|---|
| α-Conotoxin TxIC | 34.2 | |||
| α3β2, α3α5β2, α3β4 | 100 | <5 | ||
| α4β2, α4β4, α7, αβγε, αβγε | 50 | 0 | ||
|
|
||||
| [Pro]2,8, [Glu]16α-Conotoxin TxIC | 3600 | |||
| α3β2 | 5.4 ± 0.5 | 96 | ||
| α3α5β2 | 4.9 ± 0.8 | 91 | ||
| α3β4 | 2.1 ± 0.2 | 77 | ||
| α4β2, α4β4, α7, αβγε, αβγε | ≥100 | <10 | ||
Human variants;
half-maximal inhibitory concentration (human);
paralytic dose (mollusk)
3.13 Whole animal bioassay
Whole animal bioassay using the native α-conotoxin TxIC demonstrated a dose-dependency in its ability to produce animal paralysis (see Fig. S2). α-Conotoxin TxIC produced total paralysis at 640 pMol g-1 (n=14), this dose did not cause lethality in test animals. A PD50 of 34.2 pMol g-1 (34.2 nMol Kg-1; whole snail weight) was determined for the native toxin (Table 4). The synthetic non-PTM analogue of α-conotoxin TxIC demonstrated an inability to cause paralysis at same PD50 concentration (n=15). Total paralysis was achieved with 10.24 nMol g-1 (n=7), this being ∼16× that of the native. A similar dose-dependent trend was observed (see Fig. S2), with the maximal dose used not causing animal lethality. A PD50 of 3.6 nMol g-1 (3.6 μMol Kg-1; whole snail weight) was calculated (Table 4), this PD50 concentration being ∼100× more than observed with the native PTM toxin.
4. Discussion
This study is the first reported analysis comparing the venom profile of a single specimen of Conus from milked venom (MV), to duct venom (DV), and finally to radula lumen extract (RE) by RP-HPLC/UV analysis and molecular mass profiling. This has provided greater insight into the complexity of molluscivorous venom from a single specimen through to the gross variation observed at a species level from wide-ranging geographical locations.
The importance of Conus textile to conopeptide research cannot be underestimated. Early studies on C. textile [21, 27, 36] made major contributions to the present understanding of conopeptide processing and expression [11, 23, 54], and led to the discovery of novel PTM amino acids [14, 41] and unique biological activities. The specie's relative abundance, population distribution and access has allowed for an intensive level of research, covering specimens collected from the Philippines [28], Red Sea [21], South China Sea [33], Japan [27, 36] and the GBR, Australia [15, 25]. In the present study, we have expanded this geographic range to include specimens collected from Hawai'i and American Samoa (Fig. 1 and 2), making this the largest geographic study of a single Conus species to date – this provides an indication to their regional biodiversity, an area poorly addressed in Conus research.
A ‘conovenomic’ comparison of MV from distant geographic locations, illustrates the true extent of venom peptide diversity present within C. textile, as alluded to by Lu et al. [33]. Our evidence further demonstrates the intraspecific venom variability identified in DV of other members of the genus [1, 15, 42, 43] – a characteristic that now extends to MV. This may increase earlier estimates of bioactive venom constituents [18, 35, 36, 49], and expand the therapeutic potential of Conus [5, 7, 20, 30, 47].
This is only further demonstrated with the identification of a novel, highly PTM-modified conopeptide, α-conotoxin TxIC, despite the intensive level of research that C. textile has received historically. The newly observed α-conotoxin demonstrates homology to Vc1.1 (∼66 %; Table 3), and an experimentally confirmed level of selectivity (in its non-PTM form) to human isoforms of neuronal type nAChR (Table 4), a well-documented target for the treatment of chronic neuropathic pain [8, 31, 44, 51].
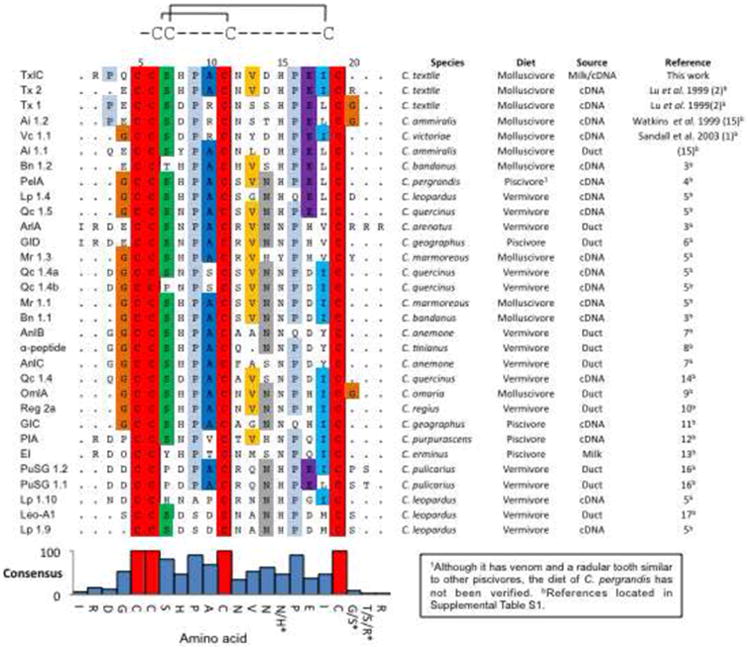
Table 3. Sequence comparison of α-conotoxin TxIC with members of the α-conopeptide 4/7 family.
All 31 peptides, including α-conotoxin TxIC, share a conserved cysteine pattern framework. Peptides most similar in sequence are those derived from C. textile and other molluscivores, while α-conopeptides from piscivores and vermivores share less sequence similarity. The overall sequence similarity to α-conotoxin TxIC is displayed graphically. This calculation is based on the total number of peptides similar to α-conotoxin TxIC and the number that exhibits conservation of the same specific amino acid and position in the sequence. For example, 3 peptides have a Pro in position 3; this is divided by 17 peptides that possess an amino acid in the equivalent position 3 of the α-conotoxin TxIC sequence; this gives a consensus of 18% for that position. Note that the α-4/7 cysteine pattern framework is solely used as the point of sequence alignment. bReferences listed are Supplemental Information – References II.
|
Within C. textile, considerable diversity is found primarily within the hydrophobic region of the RP-HPLC/UV venom profiles (Fig. 1), and prominently in specimens from American Samoa. Mass spectral analysis revealed a level of molecular mass consistency across all locations, however the relative proportion of venom constituents varies markedly (Fig. 2; Table 1; Table S2). Peptide expressional variability supports an important biological role in the predator envenomation strategy, a factor that may aid the discovery of a novel conopeptides, as illustrated with α-conotoxin TxIC.
To further refine this approach, the analysis of a single C. textile specimen facilitated the tracking of integral venom constituents from DV to MV and then RE. This strengthens the proteomic correlation, end-point detection and characterization of the essential secretory venom constituents used during envenomation [51]. It is these identified end-point MV peptides that have shown the strongest correlation to both potential biological activity and proven drug potential in piscivores [7, 26]. Using RP-HPLC/UV we established that α-conotoxin TxIC is represented in DV extract (Fig. 3C), and that expressional continuity continues through the MV (Fig. 3B). Interestingly α-conotoxin TxIC was absent from the RE (Fig. 3A). For this species at least, the radula lumen extract is not representative of the venom content as a whole – however some level of conopeptide continuity, by m/z comparison, is clearly observable (Table 2; Fig. 4).
α-Conotoxin TxIC represents a novel toxin that highlights some of the issues that have plagued conopeptide discovery and sequencing, necessitating a combined ‘conovenomic’ approach to obtain a thorough and accurate sequence analysis. The sole use of mass spectrometry for Conus venom analysis can lead to misidentification of sequence and molecular mass arising from inconsistencies associated with sample preparation, matrix selection and the ionization technique used. However we are unsure of the extent of PTM ‘miss assignment’ that may exist in the 1000's of conopeptides already documented in the publically available conopeptide database (http://www.conoserver.org). Yet, such errors are highly possible given the relative abundance and differential incorporation of PTM amino acids in conotoxins/conopeptides [10].
The ability to detect the presence of α-conotoxin TxIC only by its decarboxylated and fragmented products, and not its parent molecular mass, illustrates the susceptibility of the parent peptide to in-source decay. This process was exacerbated for α-conotoxin TxIC by the presence of the PTM amino acid γ-carboxyglutamic acid (Gla). Similar observations have been described involving the loss of the PTM Tyr-SO4 desulfation (-80 Da) in α-conotoxins PnIA (Conus pennaceus) and EpI (Conus episcopatus) [32, 53]. Although ESI-MS is less harsh, a similar degree of peptide degradation can occur if working outside normal operating parameters. Our observations highlight the necessary awareness that needs to be undertaken when dealing with complex and highly PTM processed DV extracts and native MVs.
Irrespective of MS technique, each method has advantages and disadvantages in the performance of mass detection and sequencing. If suspicion of PTMs arise, it is best to employ a combined approach for analysis and verification; this would include the use of molecular biology techniques along with more traditional biochemical methods such as Edman degradation. The latter technique becomes a necessity when considering the isobaric CID fragments generated from Hyp and Ile and Leu (± 0.04 Da). Three such unambiguous assignments were observed in the de-novo sequencing analysis of α-conotoxin TxIC (Fig. 5B). Residues were later confirmed and assigned by Edman degradation (Section 3.8) and via genetic analysis, as shown in Fig. 6 (Section 3.9).
Comprising of 18 amino acids with two disulfide bonds, 44% of residues display one or more forms of modification, α-conotoxin TxIC is one of the most highly PTM conopeptides documented to date. This novel 4/7 conopeptide (as represented by its cysteine framework) shares sequence homology with other α-conotoxins [18, 31] and demonstrates sequence commonality to other molluscivorous conopeptides, specifically Ai1.2 (C. ammiralis) and Vc1.1 (C. victoriae) (Table 3). This homology is seen explicitly in Vc1.1 in its native PTM form: [Hyp]6[Gla]14Vc1.1 [24], and more so within the last cysteine loop (89% homology; Table 3). Both C. ammiralis (Ai) and C. victoriae (Vc) are close relations of C. textile – with C. victoriae being endemic to Australia. Genetic analysis of α-conotoxin TxIC strengthens its relationship to the established 4/7 α-conotoxins, as seen by the homology of the pre/pro-peptide regions of α-conotoxins MII and Vc1.1 (Fig. 6) [17, 33, 39, 44].
As the majority of these illustrated α-conotoxin sequences are deduced from genomic sequences, the current inclusion of the observed PTMs at positions [Pro→Hyp]2, [Pro→Hyp]8 and [Glu→Gla]16 in Table 3 increases the known chemical diversity of α-conotoxin TxIC. Modifications may be affecting in vivo characteristics including kinetic properties and/or pharmacological selectivity of conotoxins. The absence of these PTMs in piscivorous 4/7 α-conotoxins, such as α-conotoxin MII (Conus magus), potentially indicates a phyla-selective targeting differentiation. By examining both the native α-conotoxin TxIC, as well as a synthetic unmodified isoform ([Pro]2,8[Glu]16α-conotoxin TxIC), we have here substantiated these claims, highlighting the role of PTMs in vivo. While remaining biologically active in whole animal bioassay, native α-conotoxin TxIC is inactive when tested human nAChR channel isoforms (Table 4). The synthetic unmodified α-conotoxin TxIC (i.e. [Pro]2,8[Glu]16α-conotoxin TxIC) however remains active at the nAChR, demonstrating isoform selectivity (Table 4). These features highlight the importance of PTMs in the selective targeting of α-conotoxins. This is further illustrated at a phylogenic level with the observed switching in potency in invertebrate models that represents the native prey target (Fig. S2).
The phyla-selectivity and reduced potency of the native α-conotoxin TxIC towards human nAChR isoforms demonstrates an underlying differentiation between receptor selectivity and pharmacodynamic properties in ion channels from different phyla. This is an area reiterated by the PTM of the nAchR in the Egyptian mongoose (Herpestes ichneumon) that provides a level of resistance to α-bungarotoxin [3]. Here, genomic and proteomic investigation into invertebrate nAchR isoforms, as well as other receptors (e.g. N-type (Cav2.2) calcium channels and GABAB [13]), may provide insight into the determinants for pharmacological potency and phyla selective characteristics observed commonly with Conus. Such information will provide insights into toxin target specificity enhancement though peptide bioengineering and the potential manipulation/incorporation of PTMs.
5. Conclusions
The complexity of MV profiles from specimens of C. textile revealed in this study serve to remind us of the extent we have underestimated the biodiversity and value of these unique venomous marine snails. The analysis of venoms necessitates a specifically tailored approach, requiring special attention to the analytical methods employed, and further accentuating the need to employ a combined ‘conovenomic’ approach. The lessons learned and approaches outlined will facilitate peptide prospecting, toxinological correlations and the future discovery of new, clinically relevant conopeptides.
Supplementary Material
Supplementary Fig. S1: Pharmacology of the non-post translationally modified α-conotoxin TxIC ([Pro]2,8[Glu]16α-conotoxin TxIC) (A) Resulting effect of 10 μM toxin application upon the action potential of different nAchR isoforms. (B) Resulting dose dependent inhibition of nAchR isoforms with increasing toxin concentration application.
Supplementary Fig. S2: Whole animal bioassay for α-conotoxin TxIC and its non-post translationally modified variant Pro]2,8[Glu]16α-conotoxin TxIC). Demonstration of dose dependency in causing animal paralysis.
Table S1. Characterized C. textile peptides.
Table S2. Observed C. textile milked venom constituents from Hawai'i, American Samoa and Great Barrier Reef, Australia.
Table S3. Comparative analysis of MALDI-TOF MS C. textile data from different peptide sources.
Highlights.
Single Cone snail analysis shows m/z continuity in duct, milk and radula extracts.
Geographic specimen analysis shows peptide diversity in the milked venoms.
α-Conotoxin TxIC represents a highly post-translationally modified conotoxin.
Post-translational modifications influence biological and phylogenetic selectivity.
Acknowledgments
We are indebted to Mr. David Bingham, the members of the Malacological Society of Australasia (Brisbane Branch), Mr. Harold “JJ” Jackson (Hawai'ian Malacological Society) and Mr. Don Barclay (American Samoa) for assistance in obtaining species described in this study. We wish to acknowledge the financial support from the American Heart Association (Scientist Development Award 0530204N to J-P.B.) and the University of Hawai'i Sea Grant College Program (J-P.B.) and USDA TSTAR (# 2009-34135-20067) (J-P. B.) & HATCH (HAW00595-R) (J-P.B.). This project in-part was supported by grants from the National Center for Research Resources (5 G12 RR003061-26) and the National Institute on Minority Health and Health Disparities (8 G12 MD007601-26) from the National Institutes of Health. JT was supported by the following grants: G.0433.12, G.A071.10N and G.0257.08 (F.W.O. Vlaanderen), EU –FP7 – MAREX, A portion of this work was submitted to The University of Queensland, Australia, (J-P.B.), The University of Melbourne, Australia (D.W.S.) and The University of Hawai'i, USA (Z.L.B.) as fulfillment for awarding a doctoral degrees.
Abbreviations
- αα
amino acid
- CH3CN
acetonitrile
- Acm
Acetamidomethyl
- API
Atmospheric pressure ionization
- DHB
Dihydroxybenzoic acid
- cDNA
complimentary DNA
- CHCA
α-Cyano-4-hydroxycinnamic acid
- CID
Collision induced dissociation
- DCM
Dichloromethane
- DIEA
N,N-diisopropylethylamine
- DMF
Dimethylformamide
- DV
Duct venom
- ESMS
Electro spray mass spectrometry
- FDA
Food and Drug Administration
- Fmoc
9-Fluorenylmethyloxycarbonyl
- Fmoc-Asn(Trt)-OH
N-alpha-9-Fluorenylmethoxycarbonyl-N-beta-Trityl-L-Asparagine
- Fmoc-Asp(OtBu)-OH
9-Fluorenylmethoxycarbonyl-L-Aspartic Acid-beta-t-Butyl Ester
- Fmoc-Arg(Pbf)-OH
N-alpha-9-Fluorenylmethoxycarbonyl-N-g-2,2,4,6,7-Pentamethyldihydrobenzofuran-5-Sulfonyl-L-Arginine
- Fmoc-Cys(Acm)-OH
9-Fluorenylmethoxycarbonyl-S-Acetamidomethyl-L-Cysteine
- Fmoc-Cys(Trt)-OH
9-Fluorenylmethoxycarbonyl-S-Trityl-L-Cysteine
- Fmoc-Gla(otBu)2-OH
N-Fmoc-L-γ-Carboxyglutamic Acid γ,γ-Di-t-Butyl Ester
- Fmoc-Gln(Trt)-OH
N-alpha-9-Fluorenylmethoxycarbonyl-N-gamma-Trityl-L-Glutamine
- Fmoc-His(Trt)-OH
N-alpha-9-Fluorenylmethoxycarbonyl-Nim-Trityl-L-Histidine
- Fmoc-Hyp(tBu)-OH
9-Fluorenylmethoxycarbonyl-O-t-Butyl-L-Hydroxyproline
- GBR
Great Barrier Reef
- (Gla)
γ-gammacarboxyglutamic acid
- HCTU
1H-Benzotriazolium-1-[bis(Dimethylamino)Methylene]-5-Chloro Hexafluorophosphate-(1-),3-Oxide
- IND
Investigational new drug
- MALDI-MS
Matrix assisted desorption/ionization Mass spectrometry
- MALDI-TOF-(TOF)-MS
Matrix assisted desorption/ionization-Time of light-(Time of flight)-Mass spectrometry
- Mr
average molecular mass
- mRNA
Messenger RNA
- MS
Mass spectrometry
- MV
Milked venom
- m/z
Mass to charge ratio
- nAChR
Nicotinic acetylcholine receptor
- PCR
Polymerase chain reaction
- PSD
Post-source decay
- PTM
Post-translational modification
- RE
Radula Extract
- RV
Radula Venom
- RP-HPLC
Reverse phase - High performance liquid chromatography
- Rt
Retention time
- SEM
Scanning electron micrograph
- SPPS
Solid phase peptide synthesis
- TCEP
Tris(2-carboxyethyl)phosphine
- TFA
Trifluoroacetic acid
- UV
Ultra-violet
- TIPS
Triisopropylsilane
Footnotes
Conflict of interest: Authors state that there is no conflict of interest.
Supplementary Information: References to Table 3.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abdel-Rahman MA, AbdelNabi IM, El-Naggar MS, Abbas OA, Strong PN. Intraspecific variation in the venom of the vermivorous cone snail Conus vexillum. Comp Biochem Physiol C Toxicol Pharmacol Comp Biochem Physiol C Toxicol Pharmacol. 2011;154:318–25. doi: 10.1016/j.cbpc.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 2.Atherton D, Fernandez J, DeMott M, Andrews L, Mische SM. Routine Protein Sequence Analysis Below Ten Picomoles: One Sequencing Facility's Approach. San Diego, California: Academic Press, Inc. (Harcourt Brace and Co.); 1993. [Google Scholar]
- 3.Barchan D, Kachalsky S, Neumann D, Vogel Z, Ovadia M, Kochva E, Fuchs S. How the mongoose can fight the snake: the binding site of the mongoose acetylcholine receptor. Proc Natl Acad Sci USA. 1992;89:7717–21. doi: 10.1073/pnas.89.16.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bingham JP. Novel Toxins From the Genus Conus - From Taxonomy to toxins. Brisbane: University of Queensland; 1998. [Google Scholar]
- 5.Bingham JP, Andrews EA, Kiyabu SM, Cabalteja CC. Drugs from Slugs. Part II-Conopeptide bioengineering. Chem Biol Interact. 2012;200:92–113. doi: 10.1016/j.cbi.2012.09.021. [DOI] [PubMed] [Google Scholar]
- 6.Bingham JP, Broxton NM, Livett BG, Down JG, Jones A, Moczydlowski EG. Optimizing the connectivity in disulfide-rich peptides: alpha-conotoxin SII as a case study. Anal Biochem. 2005;338:48–61. doi: 10.1016/j.ab.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Bingham JP, Mitsunaga E, Bergeron ZL. Drugs from slugs--past, present and future perspectives of omega-conotoxin research. Chem Biol Interact. 2010;183:1–18. doi: 10.1016/j.cbi.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 8.Carstens BB, Clark RJ, Daly NL, Harvey PJ, Kaas Q, Craik DJ. Engineering of conotoxins for the treatment of pain. Curr Pharm Des. 2011;17:4242–53. doi: 10.2174/138161211798999401. [DOI] [PubMed] [Google Scholar]
- 9.Chivian E, Roberts CM, Bernstein AS. The threat to cone snails. Science. 2003;302:391. doi: 10.1126/science.302.5644.391b. [DOI] [PubMed] [Google Scholar]
- 10.Chun JB, Baker MR, Kim DH, Leroy M, Toribo P, Bingham JP. Cone snail milked venom dynamics - a quantitative study of Conus purpurascens. Toxicon. 2012;60:83–94. doi: 10.1016/j.toxicon.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conticello SG, Gilad Y, Avidan N, Ben-Asher E, Levy Z, Fainzilber M. Mechanisms for evolving hypervariability: the case of conopeptides. Mol Biol Evol. 2001;18:120–31. doi: 10.1093/oxfordjournals.molbev.a003786. [DOI] [PubMed] [Google Scholar]
- 12.Cruz LJ, Ramilo CA, Corpuz GP, Olivera BM. Conus Peptides: Phylogenetic Range of Biological Activity. Biol Bull. 1992;183:159–64. doi: 10.2307/1542418. [DOI] [PubMed] [Google Scholar]
- 13.Cuny H, de Faoite A, Huynh TG, Yasuda T, Berecki G, Adams DJ. γ-Aminobutyric acid type B (GABAB) receptor expression is needed for inhibition of N-type (Cav2.2) calcium channels by analgesic α-conotoxins. J Biol Chem. 2012;287:23948–57. doi: 10.1074/jbc.M112.342998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czerwiec E, Kalume DE, Roepstorff P, Hambe B, Furie B, Furie BC, Stenflo J. Novel gamma-carboxyglutamic acid-containing peptides from the venom of Conus textile. FEBS J. 2006;273:2779–88. doi: 10.1111/j.1742-4658.2006.05294.x. [DOI] [PubMed] [Google Scholar]
- 15.Davis J, Jones A, Lewis RJ. Remarkable inter- and intra-species complexity of conotoxins revealed by LC/MS. Peptides. 2009;30:1222–7. doi: 10.1016/j.peptides.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 16.Duda TFJ, Bingham JP, Livett BG, Kohn AJ, Massilia GR, Schultz JR, Down J, Sandall D, Sweedler JV. How much at risk are cone snails? Science. 2004;303:955–7. doi: 10.1126/science.303.5660.955. [DOI] [PubMed] [Google Scholar]
- 17.Dutertre S, Jin AH, Kaas Q, Jones A, Alewood PF, Lewis RJ. Deep venomics reveals the mechanism for expanded peptide diversity in cone snail venom. Mol Cell Proteomics. 2013;12:312–29. doi: 10.1074/mcp.M112.021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dutertre S, Ulens C, Büttner R, Fish A, van Elk R, Kendel Y, Hopping G, Alewood PF, Schroeder C, Nicke A, Smit AB, Sixma TK, Lewis RJ. AChBP-targeted alpha-conotoxin correlates distinct binding orientations with nAChR subtype selectivity. EMBO J. 2007;26:3858–67. doi: 10.1038/sj.emboj.7601785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Endean R, Rudkin C. Further Studies of the Venoms of Conidae. Toxicon. 1965;69:225–49. doi: 10.1016/0041-0101(65)90021-8. [DOI] [PubMed] [Google Scholar]
- 20.Essack M, Bajic VB, Archer JA. Conotoxins that confer therapeutic possibilities. Mar Drugs. 2012;10:1244–65. doi: 10.3390/md10061244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fainzilber M, Gordon D, Hasson A, Spira ME, Zlotkin E. Mollusc-specific toxins from the venom of Conus textile neovicarius. Eur J Biochem. 1991;202:589–95. doi: 10.1111/j.1432-1033.1991.tb16412.x. [DOI] [PubMed] [Google Scholar]
- 22.Halai R, Craik DJ. Conotoxins: natural product drug leads. Nat Prod Rep. 2009;26:526–36. doi: 10.1039/b819311h. [DOI] [PubMed] [Google Scholar]
- 23.Hillyard DR, Olivera BM, Woodward S, Corpuz GP, Gray WR, Ramilo CA, Cruz LJ. A molluscivorous Conus toxin: conserved frameworks in conotoxins. Biochemistry. 1989;28:358–61. doi: 10.1021/bi00427a049. [DOI] [PubMed] [Google Scholar]
- 24.Jakubowski JA, Kelley WP, Sweedler JV. Screening for post-translational modifications in conotoxins using liquid chromatography/massspectrometry: an important component of conotoxin discovery. Toxicon. 2006;47:688–99. doi: 10.1016/j.toxicon.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 25.Jones J, Bingham JP, Gehrmann J, Bond T, Loughnan M, Atkins A, Lewis RJ, Alewood PF. Isolation and characterization of conopeptides by high-performance liquid chromatography combined with mass spectrometry and tandem mass spectrometry. Commun Mass Spectrom. 1996;10:138–43. doi: 10.1002/(SICI)1097-0231(19960115)10:1<138::AID-RCM442>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 26.Kapono CA, Thapa P, Cabalteja CC, Guendisch D, Collier AC, Bingham JP. Conotoxin truncation as a post-translational modification to increase the pharmacological diversity within the milked venom of Conus magus. Toxicon. 2013;70:170–78. doi: 10.1016/j.toxicon.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi J, Ohizumi Y, Nakamura H, Hirata Y. Pharmacological study on the venom of the marine snail Conus textile. Toxicon. 1981;19:757–62. doi: 10.1016/0041-0101(81)90072-6. [DOI] [PubMed] [Google Scholar]
- 28.Lev-Ram V, Olivera BM, Levitan IB. A toxin from the venom of the predator snail Conus textile modulates ionic currents in Aplysia bursting pacemaker neuron. Brain Res. 1994;640:48–55. doi: 10.1016/0006-8993(94)91856-2. [DOI] [PubMed] [Google Scholar]
- 29.Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–71. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- 30.Livett BG, Gayler KR, Khalil Z. Drugs from the sea: conopeptides as potential therapeutics. Curr Med Chem. 2004;11:1715–23. doi: 10.2174/0929867043364928. [DOI] [PubMed] [Google Scholar]
- 31.Livett BG, Sandall DW, Keays D, Down J, Gayler KR, Satkunanathan N, Khalil Z. Therapeutic applications of conotoxins that target the neuronal nicotinic acetylcholine receptor. Toxicon. 2006;48:810–29. doi: 10.1016/j.toxicon.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 32.Loughnan M, Bond T, Atkins A, Cuevas J, Adams DJ, Broxton NM, Livett BG, Down JG, Jones A, Alewood PF, Lewis RJ. Alpha-conotoxin EpI, a novel sulfated peptide from Conus episcopatus that selectively targets neuronal nicotinic acetylcholine receptors. J Biol Chem. 1998;273:15667–74. doi: 10.1074/jbc.273.25.15667. [DOI] [PubMed] [Google Scholar]
- 33.Lu BS, Yu F, Zhao D, Huang PT, Huang CF. Conopeptides from Conus striatus and Conus textile by cDNA cloning. Peptides. 1999;20:1139–44. doi: 10.1016/s0196-9781(99)00116-3. [DOI] [PubMed] [Google Scholar]
- 34.Matsudaira PT. Introduction A practical guide to protein and peptide purification for microsequencing. San Diego, California: Academic Press, Inc. (Harcourt Brace Jovanovich); 1989. [Google Scholar]
- 35.McIntosh JM, Santos AD, Olivera BM. Conus peptides targeted to specific nicotinic acetylcholine receptor subtypes. Annu Rev Biochem. 1999;68:59–88. doi: 10.1146/annurev.biochem.68.1.59. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura H, Kobayashi J, Ohizumi Y, Hirata Y. The occurrence of arachidonic acid in the venom duct of the marine snail conus textile. Experientia. 1982;38:897. doi: 10.1007/BF01953639. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura T, Yu Z, Fainzilber M, Burlingame AL. Mass spectrometric-based revision of the structure of a cysteine-rich peptide toxin with gamma-carboxyglutamic acid, TxVIIA, from the sea snail, Conus textile. Protein Sci. 1996;5:524–30. doi: 10.1002/pro.5560050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olivera BM, Rivier J, Clark C, Ramilo CA, Corpuz GP, Abogadie FC, Mena EE, Woodward SR, Hillyard DR, Cruz LJ. Diversity of Conus neuropeptides. Science. 1990;249:257–63. doi: 10.1126/science.2165278. [DOI] [PubMed] [Google Scholar]
- 39.Price-Carter M, Gray WR, Goldenberg DP. Folding of omega-conotoxins. 2. Influence of precursor sequences and protein disulfide isomerase. Biochemistry. 1996;35:15547–57. doi: 10.1021/bi9615755. [DOI] [PubMed] [Google Scholar]
- 40.Reed LJ, Muench S. A Simple Method of Estimating Fifty Percent Time Points. Am J Hyg. 1938;27:493–97. [Google Scholar]
- 41.Rigby AC, Lucas-Meunier E, Kalume DE, Czerwiec E, Hambe B, Dahlqvist I, Fossier P, Baux G, Roepstorff P, Baleja JD, Furie BC, Furie B, Stenflo JA. conotoxin from Conus textile with unusual posttranslational modifications reduces presynaptic Ca2+ influx. Proc Natl Acad Sci U S A. 1999;96:5758–63. doi: 10.1073/pnas.96.10.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rivera-Ortiz JA, Cano H, Marí F. Intraspecies variability and conopeptide profiling of the injected venom of Conus ermineus. Peptides. 2011;32:306–16. doi: 10.1016/j.peptides.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romeo C, Di Francesco L, Oliverio M, Palazzo P, Massilia GR, Ascenzi P, Polticelli F, Schininà ME. Conus ventricosus venom peptides profiling by HPLC-MS: a new insight in the intraspecific variation. J Sep Sci. 2008;31:488–98. doi: 10.1002/jssc.200700448. [DOI] [PubMed] [Google Scholar]
- 44.Sandall DW, Satkunanathan N, Keays DA, Polidano MA, Liping X, Pham V, Down JG, Khalil Z, Livett BG, Gayler KR. A novel alpha-conotoxin identified by gene sequencing is active in suppressing the vascular response to selective stimulation of sensory nerves in vivo. Biochemistry. 2003;42:6904–11. doi: 10.1021/bi034043e. [DOI] [PubMed] [Google Scholar]
- 45.Sasaki T, Feng ZP, Scott R, Grigoriev N, Syed NI, Fainzilber M, Sato K. Synthesis, bioactivity, and cloning of the L-type calcium channel blocker omega-conotoxin TxVII. Biochemistry. 1999;38:12876–84. doi: 10.1021/bi990731f. [DOI] [PubMed] [Google Scholar]
- 46.Schnölzer M, Alewood P, Jones A, Alewood D, Kent SBH. In situ neutralization in Boc-chemistry solid phase peptide synthesis. Int J Peptide Protein Res. 1992;40:180–93. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 47.Schroeder CI, Craik DJ. Therapeutic potential of conopeptides. Future Med Chem. 2012;4:1243–55. doi: 10.4155/fmc.12.70. [DOI] [PubMed] [Google Scholar]
- 48.Schroeder CI, Ekberg J, Nielsen KJ, Adams D, Loughnan ML, Thomas L, Adams DJ, Alewood PF, Lewis RJ. Neuronally selective μ-conotoxins from Conus striatus utilize an alpha-helical motif to target mammalian sodium channels. J Biol Chem. 2008;283:21621–8. doi: 10.1074/jbc.M802852200. [DOI] [PubMed] [Google Scholar]
- 49.Tayo LL, Lu B, Cruz LJ, Yates JR. Proteomic analysis provides insights on venom processing in Conus textile. J Proteome Res. 2010;9:2292–301. doi: 10.1021/pr901032r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teichert RW, Rivier J, Dykert J, Cervini L, Gulyas J, Bulaj G, Ellison M, Olivera BM. AlphaA-Conotoxin OIVA defines a new alphaA-conotoxin subfamily of nicotinic acetylcholine receptor inhibitors. Toxicon. 2004;44:207–14. doi: 10.1016/j.toxicon.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 51.Townsend A, Livett BG, Bingham JP, Truong HT, Karas JA, O'Donnell P, Williamson NA, Purcell AW, Scanlon D. Mass spectral identification of Vc1.1 and differential distribution of conopeptides in the venom duct of Conus victoriae. Effect of post-translational modifications and disulfide isomerisation on bioactivity. Int J Peptide Res and Therap. 2009;15:195–203. [Google Scholar]
- 52.Vincler M, Wittenauer S, Parker R, Ellison M, Olivera BM, McIntosh JM. Molecular mechanism for analgesia involving specific antagonism of alpha9alpha10 nicotinic acetylcholine receptors. Proc Natl Acad Sci U S A. 2006;103:17880–4. doi: 10.1073/pnas.0608715103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolfender JL, Chu F, Ball H, Wolfender F, Fainzilber M, Baldwin MA, Burlingame AL. Identification of tyrosine sulfation in Conus pennaceus conotoxins alpha-PnIA and alpha-PnIB: further investigation of labile sulfo- and phosphopeptides by electrospray, matrix-assisted laser desorption/ionization (MALDI) and atmospheric pressure MALDI mass spectrometry. J Mass Spectrom. 1999;34:447–54. doi: 10.1002/(SICI)1096-9888(199904)34:4<447::AID-JMS801>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 54.Woodward SR, Cruz LJ, Olivera BM, Hillyard DR. Constant and hypervariable regions in conotoxin propeptides. EMBO J. 1990;9:1015–20. doi: 10.1002/j.1460-2075.1990.tb08204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao WH, Bennett GJ. Synthetic omega-conopeptides applied to the site of nerve injury suppress neuropathic pains in rats. J Pharmacol Exp Ther. 1995;274:666–72. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1: Pharmacology of the non-post translationally modified α-conotoxin TxIC ([Pro]2,8[Glu]16α-conotoxin TxIC) (A) Resulting effect of 10 μM toxin application upon the action potential of different nAchR isoforms. (B) Resulting dose dependent inhibition of nAchR isoforms with increasing toxin concentration application.
Supplementary Fig. S2: Whole animal bioassay for α-conotoxin TxIC and its non-post translationally modified variant Pro]2,8[Glu]16α-conotoxin TxIC). Demonstration of dose dependency in causing animal paralysis.
Table S1. Characterized C. textile peptides.
Table S2. Observed C. textile milked venom constituents from Hawai'i, American Samoa and Great Barrier Reef, Australia.
Table S3. Comparative analysis of MALDI-TOF MS C. textile data from different peptide sources.


