Abstract
Dental caries is a multifactorial infectious disease and a major public health problem estimated to affect 60-90% of school children as well as a vast number of adults. The aim of this work was to define patterns of progression of the disease based on longitudinal data in contrast to using a cross-sectional assessment. dmft/DMFT scores were collected at ages 5, 12, 14, 16, 17, and 18 from 876 individuals. We tested our newly defined phenotypes for association with genetic variants in genes shown to be associated with caries. We generated genotyping data using Taqman chemistry in markers of genes involved in processes such as enamel formation and salivary contributions. Kallikrein 4 (KLK4) was found to show a significant association with the created phenotypes (p=0.0008 in a recessive model for low caries experience in the primary dentition vs. high caries experience in the primary dentition, and p=0.0004 in a recessive model for caries free primary dentition vs. high caries experience in the primary dentition).
Keywords: caries prediction, genetics, saliva, caries detection
BACKGROUND
Dental caries is a major public health problem and is estimated to affect 60 to 90 percent of school children as well as a vast number of adults [The World Health Organization, 2003]. Recent statistics from the CDC state that in the United States alone, 17.5% of children aged 5-19 years and 27.4% of adults aged 20-44 years are suffering from untreated dental caries [National Center for Health Statistics., 2016]. In Europe, a meta-analysis was conducted that showed caries rates in Europeans has been significantly increasing since the 18th century [Müller, and Hussein, 2017]. Additionally, tooth loss was associated with caries [Müller, and Hussein, 2017].
Caries is expected to increase in developing countries as diets change to include more processed and sugary foods [Petersen, 2004]. Another study showed that world oral health has not improved over the last 25 years, and as the world population continues to grow, so does the number of people affected by oral diseases [Kassebaum et al., 2015]. In 2015, it was reported that 3.5 billion persons have untreated oral diseases [Kassebaum et al., 2015]. Left untreated, caries can cause any number of complications, from pain and abscess to life-threatening orofacial swelling and cellulitis, requiring expensive surgery and potential for hospital stay. In rare cases (despite being as recent as 2008), individuals have gone blind [Moschos et al., 2005] and even died from complications surrounding dental decay [Owings, 2007]. For many patients, there are barriers preventing patients from accessing care such as lack of insurance, location, and cost, making prevention of dental disease imperative.
Dental caries is a chronic, multifactorial disease. Factors such as diet, host behavior, and oral microbiota contribute to the development of the disease. It is characterized by the gradual demineralization of enamel, dentin, and cementum. The localized area of destruction of these tissues is called the carious lesion. While caries lesions are diagnosed based on the mineral loss seen by the clinician, their formation requires oral bacteria from dental plaque, which use the sugars from one’s diet as energy [Fejerskov, and Kidd, 2008].
Currently, the best predictor of caries experience for a patient is whether or not the patient has had dental caries in the past. While this is a way to predict future experience and increase preventative care for at-risk patients, it is not the most practical tool for identifying individuals at risk for the first onset of the disease. We know that aside from diet and oral microbiota, the host itself plays a role in dental caries. For instance, the immune system of the host plays a part in the disease since caries triggers inflammation and is considered infectious. Arachidonate 15-lipoxygenase (ALOX15) and Beta-defensin 1 (DEFB1) are examples of immune response genes that may play a role in the inflammatory response triggered by carious lesions [Nandula et al., 2007]. Aquaporins have also been associated with dental caries. Aquaporins are water channel proteins that may be involved in formation of saliva or other salivary contributions to caries experience [Matsuki-Fukushima et al., 2008]. Host behavior may also contribute to the disease since the patient’s dietary preferences may contribute. For example, taste preferences may influence dietary behavior, which in turn may influence whether or not a patient develops caries. Studies have shown some taste genes to be associated with caries [Wendell et al., 2010]. There are also a range of enamel formation genes such as amelogenin, enamelin, tuftelin, and ameloblastin, which exhibit variation and association to dental caries [Patir et al., 2008; Ergoz et al., 2014].
To create new opportunities to identify risk factors for dental caries, we analyzed patterns of disease progression based on longitudinal caries experience data. We hypothesize that there are individuals that present particular disease progression patterns and that these patterns associate to specific caries risk factors. Our hope is that understanding the genetic etiology of dental caries can eventually translate to better prevention strategies and ultimately lower the rates of untreated disease.
SUBJECTS AND METHODS
The 876 human subjects came from Western Norway. Patients were unrelated, healthy, and had regular access to dental care. Caries experience (D3-5MFT/d3-5mft) data were obtained from patient’s dental records and collected as part of a routine dental examination. Concerning approximal lesions recorded during the radiographic examination, only dentin lesions were included (D/d3-5). DMFT/dmft scores were recorded at different ages and depending on the birth year of participants, they had four, five, or six DMFT scores overtime. Children born in 1994 had DMFT/dmft data recorded at ages 5, 12, 14, 16, 17, and 18. Children born in 1995 had DMFT/dmft data recorded at ages 5, 12, 14, 16, and 17. Finally, children born in 1996 had DMFT/dmft data recorded at ages 5, 12, 14, and 16. This study was approved by both the local Regional Committee for Medical Research Ethics (Norway) and the Institutional Review Board (IRB) of the University of Pittsburgh. Written informed consent was obtained from all participants. DMFT and dmft scores were not combined in this study. This study follows STROBE guidelines.
Dental caries was diagnosed using a modified World Health Organization protocol recommended for oral health surveys.[The World Health Organization, 2003] Teeth lost to trauma or primary teeth lost to exfoliation were not included in the final DMFT/dmft scores. When records indicated that teeth were extracted for orthodontic reasons or periodontal disease, or treatments were performed in sound teeth, these situations were not included in the final DMFT/dmft scores. Carious lesions were recorded based on both radiological- and clinical examinations, and included only decayed lesions into dentin (D/d3-5). Teeth missing or damaged due to other oral health conditions such as enamel hypoplasia were not included in the scoring.
The population included 451 female patients and 405 male patients. The longitudinal DMFT/dmft data were analyzed to determine if there were patterns in dental caries experience progression in the studied adolescents. Patients were clustered into groups based on their DMFT/dmft scores at each time point and sex. The mean DMFT/dmft score for the population was approximately 3.5. Differences observed in the caries phenotype groups between sexes were assessed by the chi-square test with statistical significance set at p < 0.05.
Our genotyping focused on 38 single nucleotide polymorphisms (SNPs) related to immune response, enamel formation, and dental development. These SNPs are listed in Table 1. We used the Genetic Power Calculator software to estimate statistical power [Purcell et al., 2003]. Using the total sample and individuals who are caries free as a comparison group, D’ of 1.0, and the frequency of the genetic marker 0.3, we would have 96% power to detect as association assuming alpha 0.05. Power may be lower in some subgroups because they are smaller, but many of the groups are still large enough to maintain it.
Table 1.
SNPs selected for genotyping.
| SNP | Chr | Gene | Comments | References |
|---|---|---|---|---|
| rs9701796 | 1 | TAS1R2 | Taste genes associated with caries experience | [Wendell et al., 2010] |
| rs946252 | 1 | TUFT1 | Contribute to enamel development and microhardness | [Deeley et al., 2008; Shaffer et al., 2011; Shimizu et al., 2012; Shaffer et al., 2015] |
| rs12640848 | 4 | ENAM | ||
| rs4694075 | 4 | AMBN | ||
| rs713598 | 7 | TAS2R38 | Taste genes associated with caries experience | [Wendell et al., 2010] |
| rs1726866 | 7 | TAS2R38 | ||
| rs10246939 | 7 | TAS2R38 | ||
| rs6862039 | 5 | BTF3 | Previous association with high caries experience | [Shimizu et al., 2013] |
| rs27565 | 5 | PART1 | ||
| rs17159702 | 7 | AQP1 | Salivary contributions | |
| rs11362 | 8 | DEFB1 | Potential role in inflammatory response | [Ozturk et al., 2010; Krasone et al., 2014] |
| rs1800972 | 8 | DEFB1 | ||
| rs3579129 | 12 | AQP5 | Potential involvement in immune response and salivary contributions | [Nandula et al., 2007; Matsuki-Fukushima et al., 2008; Anjomshoaa et al., 2015, 5] |
| rs1996315 | 12 | AQP6 | ||
| rs296763 | 12 | AQP6 | ||
| rs2878771 | 12 | AQP2 | ||
| rs3736309 | 12 | AQP5 | ||
| rs461872 | 12 | AQP2 | ||
| rs3741559 | 12 | AQP2 | ||
| rs467323 | 12 | AQP2 | ||
| rs1997532 | 14 | TRAV4 | Previous association with low caries experience | [Briseño-Ruiz et al., 2013] |
| rs7150049 | 14 | TRAV4 | ||
| rs8011979 | 14 | TRAV4 | ||
| rs10132091 | 14 | ESRRB | Individuals with dental caries have an over-representation of the T allele of rs55835922 (74% versus 54%; p=0.01). The SNP rs61742642 is a missense mutation (P386S), but its frequency was just slightly elevated in cases with dental caries (13% versus 9.5%). SNP rs35544003 is a synonymous change. | [Weber et al., 2014] |
| rs6574293 | 14 | ESRRB | ||
| rs1077430 | 14 | ESRRB | ||
| rs745011 | 14 | ESRRB | ||
| rs4903399 | 14 | Flanking ESRRB | ||
| rs2860216 | 14 | Flanking ESRRB | ||
| rs1676303 | 14 | Flanking ESRRB | ||
| rs55835922 | 14 | Flanking ESRRB | ||
| rs1997533 | 14 | TRAV4 | Previous association with low caries experience | [Briseño-Ruiz et al., 2013] |
| rs2619112 | 17 | ALOX15 | Inflammatory response | [Abbasoglu et al., 2015] |
| rs7217186 | 17 | ALOX15 | ||
| rs198968 | 19 | KLK4 | Previous protection against caries | [Wang et al., 2012a; Wang et al., 2012b; Abbasoglu et al., 2015] |
| rs2235091 | 19 | KLK4 | ||
| rs5997096 | 22 | TFIP11 | Contribute to enamel development and microhardness | [Lu et al., 2008; Shimizu et al., 2012] |
| rs946252 | X | AMELX |
Subjects were asked to spit and provide unstimulated saliva samples, which were collected and stored in Oragene DNA Self-Collection kits (DNA Genotek Inc.) at room temperature. DNA was extracted from this saliva according to standard protocol, without centrifugation. Genotyping was performed using Taqman chemistry on 875 Norwegian patients. We used the discreet dental caries groups defined in the initial analyses, and these comparisons are summarized in Table 2.
Table 2.
Examples of comparisons of created phenotypes based on dmft/DMFT scores.
| Comparison | Sample Size (n) |
|---|---|
| No Caries vs. Low Caries | 201 vs. 301 |
| No Caries vs. High Caries | 201 vs. 373 |
| Low Caries vs. High Caries | 301 vs. 373 |
| No primary caries (dmft) vs. Low primary caries (dmft) | 647 vs. 125 |
| No primary caries (dmft) vs. High primary caries (dmft) | 647 vs. 103 |
| Low primary caries (dmft) vs. High primary caries (dmft) | 125 vs. 103 |
| No Caries vs. Very High Caries | 201 vs. 128 |
| Very High Caries vs. Low Caries | 301 vs. 128 |
| Very High Caries vs. High Caries | 245 vs.128 |
| Acute Increase in DMFT vs. No Acute Increase in DMFT | 128 vs. 527 |
| Acute Increase in DMFT vs. No Caries | 128 vs. 201 |
With the phenotype groups created, PLINK software [Purcell et al., 2007] was used to compare patients genotypes and phenotypes and look for association between caries experience and the different SNPs tested. In PLINK, we chose the “genotypic (2df) test” to test five models: allelic (C vs. c), genotypic (CC vs. Cc vs. cc), Trend (C vs. c assuming no Hardy Weinberg), dominant (CC and Cc vs. cc), and recessive (CC vs. Cc and cc). A Bonferroni correction was implemented to correct for multiple comparisons.
RESULTS
The definitions of the caries phenotype we have been using in our studies [Shimizu et al., 2012; Briseño-Ruiz et al., 2013; Shimizu et al., 2013; Küchler et al., 2013; Küchler et al., 2014; Weber et al., 2014] considers the permanent dentition DMFT scores and includes individuals caries free (DMFT of zero), individuals with low caries experience, which would include subjects with a DMFT of one, two, or three, and individuals with a high caries experience, which would include subjects with a DMFT of four or more. We chose our high caries DMFT/dmft score as four or more due to the mean DMFT/dmft score for the population being 3.5. The characteristics of this clustering of patients are located in Appendix Table 1. A chi-square analysis showed no differences between sexes within each of these three groups. A graphical representation of what the longitudinal DMFT/dmft data for patients placed in each group is included in Figure 1.
Figure 1.
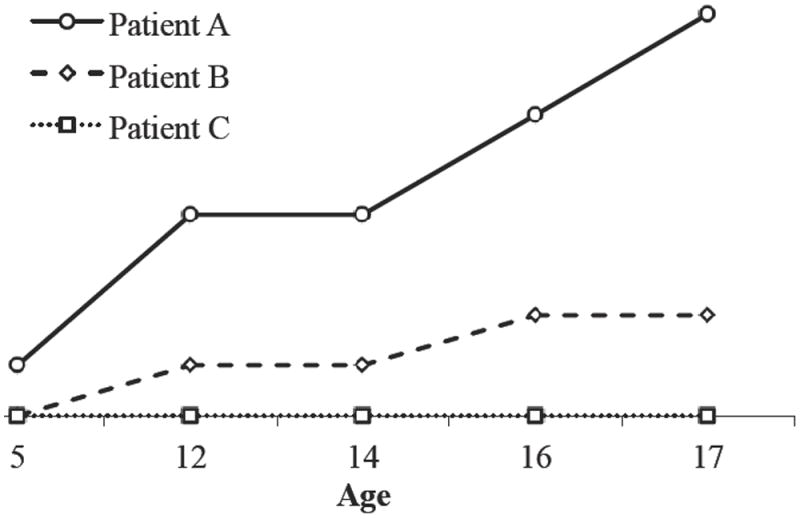
Graphical representation of selected Table 1 patients’ longitudinal DMFT data. Patient A exhibits high caries experience with a DMFT of four or more. Patient B has low caries experience and Patient C has no caries experience.
Next, the careful evaluation of the data showed that patients could be subdivided into more discrete groups. First patients from the high caries experience group were split into a high caries group with a DMFT of four or more and a very high caries group with a DMFT of eight or more. Eight was chosen as the DMFT cutoff since it is double the number used for high caries experience. These results are located in Appendix Table 1. A chi-square analysis showed no differences between sexes within each group. A graphical representation of what the longitudinal DMFT/dmft data for patients placed in each group is included in Appendix Figure 1.
We also separated patients based on whether or not they had an acute increase in caries experience between two time points. We called these acute increases “spikes” for short and looked to see if patients experienced more than one over time. 128 patients had a spike of four or more between two time points while the next most common spike was five and occurred in only 42 patients. Therefore, a spike was chosen as an increase in DMFT of four or more between two time points based on our previous determination of high caries experience and based on the distribution of increases between time points across the population. Patients were broken down into two groups, those with a spike and those with only a steady increase in DMFT. These results are located in Appendix Table 2. A chi-square analysis showed no differences between sexes within each group. A graphical representation of what the longitudinal DMFT/dmft data for patients placed in each group is included in Appendix Figure 1.
Our final stratifications of patients involved caries experience in the primary dentition at the age of five-time point. Again we looked at high, low, and no caries experience patients based on a dmft score of four or more, 1-3, or zero respectively. Our high caries experience was set at four or more due to the mean of the population dmft being 3.5. These results are located in Appendix Table 3. A chi-square analysis showed no differences between sexes within each group. A graphical representation of what the longitudinal DMFT/dmft data for patients placed in each group is included in Figure 2. We also found that some patients showed a decrease in DMFT/dmft scores between ages five and 12 and attributed this to normal loss of the caries affected primary dentition.
Figure 2.
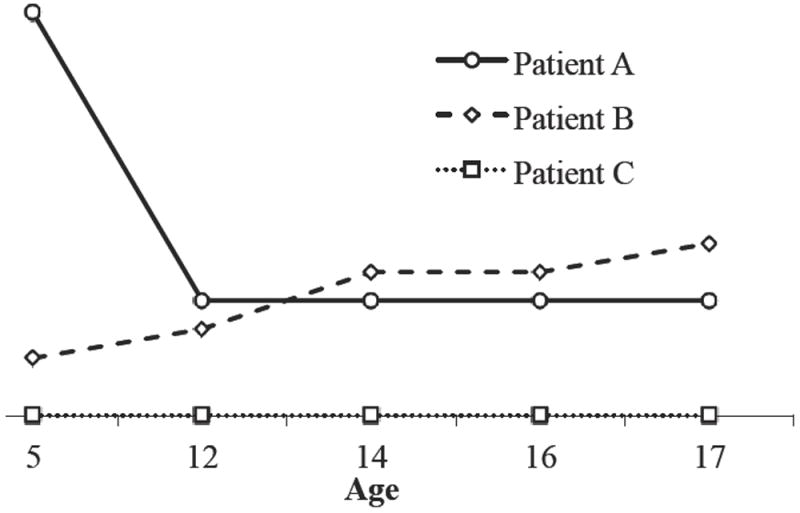
Graphical representation of selected Fehler! Verweisquelle konnte nicht gefunden werden. patients’ longitudinal dmft data. Patient A has a dmft score of 14 at age five and therefore has high caries experience in the primary dentition. Patient A also shows a decrease in caries experience between ages 5 and 12, which can be attributed to exfoliation of the primary dentition. Patient B has a dmft score of two at age five and so has low caries experience in the primary dentition. Patient C exhibits no caries experience.
Some SNPs were found to show an association with the created caries phenotypes. These are listed in Table 3 below. This table includes both nominal results (p≤.05) as well as the results that hold up after a Bonferroni correction where association was set at p=0.001 (.05/49). The results significant after Bonferroni are in bold. Kallikrein 4 (KLK4) was found to show a significant association with the created phenotypes (p=0.0008 in a recessive model for low caries experience in the primary dentition vs. high caries experience in the primary dentition, and p=0.0004 in a recessive model for caries free primary dentition vs. high caries experience in the primary dentition).
Table 3.
Nominal and significant results of association tests for created dental caries phenotypes.
| Comparison (Affected vs. Unaffected) | SNP | Test | Affected | Unaffected | ChiSq | p-value |
|---|---|---|---|---|---|---|
| No Caries vs. Low Caries | rs9701796 | Recessive | 22/271 | 6/186 | 4.097 | 0.04 |
| rs713598 | Genotype | 39/111/115 | 18/95/65 | 6.015 | 0.05 | |
| rs3736309 | Recessive | 15/229 | 3/154 | 4 | 0.04 | |
| rs7526319 | Allelic | 94/350 | 44/244 | 3.966 | 0.05 | |
| No Caries vs. High Caries | rs9701796 | Recessive | 28/326 | 6/186 | 4.88 | 0.03 |
| rs10246939 | Genotype | 87/133/92 | 29/99/49 | 10.64 | 0.005 | |
| Recessive | 87/225 | 29/148 | 8.255 | 0.004 | ||
| rs1996315 | Dominant | 222/141 | 138/56 | 5.505 | 0.03 | |
| rs4694075 | Dominant | 185/101 | 115/39 | 4.605 | 0.03 | |
| No Primary Caries vs. Low Primary Caries | rs296763 | Recessive | 11/110 | 28/574 | 3.891 | 0.05 |
| rs4903399 | Recessive | 1/114 | 30/545 | 4.222 | 0.04 | |
| rs2619112 | Genotype | 14/67/30 | 100/278/211 | 6.475 | 0.04 | |
| rs5997096 | Genotype | 15/54/30 | 133/235/125 | 6.182 | 0.04 | |
| Allelic | 84/114 | 501/485 | 4.64 | 0.03 | ||
| Trend | 84/114 | 501/485 | 4.534 | 0.03 | ||
| Recessive | 15/84 | 133/360 | 6.15 | 0.01 | ||
| No Primary Caries vs. High Primary Caries | rs11362 | Recessive | 19/59 | 74/410 | 4.001 | 0.04 |
| rs1800972 | Recessive | 4/45 | 6/288 | 5.562 | 0.02 | |
| rs1997533 | Recessive | 4/93 | 61/528 | 3.772 | 0.05 | |
| rs2235091 | Genotype | 20/32/40 | 55/253/281 | 12.68 | 0.002 | |
| Allelic | 72/112 | 363/815 | 5.062 | 0.02 | ||
| Trend | 72/112 | 363/815 | 4.88 | 0.03 | ||
| Recessive | 20/72 | 55/534 | 12.49 | 0.0004 | ||
| Low Primary Caries vs. High Primary Caries | rs296763 | Genotype | 2/42/52 | 11/40/70 | 6.137 | 0.05 |
| Recessive | 2/94 | 11/110 | 4.667 | 0.03 | ||
| rs4903399 | Recessive | 6/83 | 1/114 | 5.221 | 0.02 | |
| rs2235091 | Genotype | 20/32/40 | 7/52/57 | 11.38 | 0.003 | |
| Allelic | 72/112 | 66/166 | 4.85 | 0.03 | ||
| Trend | 72/112 | 66/166 | 5.282 | 0.02 | ||
| Recessive | 20/72 | 7/109 | 11.2 | 0.0008 | ||
| rs5997096 | Allelic | 72/62 | 84/114 | 4.102 | 0.04 | |
| Trend | 72/62 | 84/114 | 4.499 | 0.03 |
DISCUSSION
Despite previous studies using classifications such as high, low, or no caries experience to describe the disease in patients, to our knowledge this is the first attempt to subphenotype the disease based on patterns evident from longitudinal DMFT/dmft data. We created groups based on high, very high, low, spiking, decreasing, and lack of caries experience over time. Our goal is to be able to use these subphenotypes to better understand the progression of dental caries experience and as an additional tool in gene identification studies of the disease.
Because the DMFT/dmft data for patients was collected by different clinicians, there may have been some variation in how the DMFT/dmft index was used. Again, the index is not always an accurate measurement for the severity of the disease since it gives equal weight to each symptom and sign of the disease. This along with subjectivity of the diagnosing clinician could cause DMFT/dmft diagnosis to vary more among patients. However, because the DMFT/dmft index is usually the standard method of disease classification among clinicians, we feel that variation would not be high enough within the population to cause significant issues in subphenotyping.
Some patients in the first group born in 1994 had additional DMFT/dmft scores at ages 18. This could allow for patients in this group to have the opportunity to be classified as high caries or with a spike for instance, just because they had data collected at a later time. Patients in the other two groups may eventually have high caries at age 18, but unfortunately we did not have DMFT/dmft data for that time point for those groups. However, analyses of the data without these subjects did not substantially change our results (data not shown).
Our numbers for high caries experience and very high caries experience were a DMFT/dmft score of four or more and eight or more respectively. These numbers were based on the mean DMFT/dmft score for the population. While other numbers could have been chosen based on previous studies or predictions of greater populations, we feel that this was the best method for choosing the high caries cutoffs for our study. We chose a spike in caries experience to be an acute increase of four or more in the DMFT score between time points. Again, while other spike numbers could have been chosen, we felt that the best indication for an acute and significant spike should be that which would place them from the low to high caries group or high to very high caries groups.
We also included groups of patients that exhibited a decrease in the DMFT/dmft scores between ages five and twelve. Normally, DMFT/dmft scores should not decrease based on the nature of the index and how it is used. However, we know that exfoliation or loss of the primary dentition often happens between ages six and 12. It was not uncommon to see children with caries experience in the primary dentition that did not develop any or as many caries lesions in the permanent dentition. Therefore it was possible to have, a decrease in the score from age five, which was exclusively primary dentition (dmft), to age 12, which was exclusively permanent dentition (DMFT).
Our goal is to be able to use these dental caries subphenotypes to better understand the progression of the disease and as an additional tool in gene identification studies of the disease. Future studies will continue to investigate potential risk factors of the disease to better understand its etiology and perhaps aid in future prevention and treatment strategies.
We hypothesized that specific genes have a detectable influence in caries experience. Genes involved in saliva formation, immune response, behavior, and tooth development could contribute to risk of caries in patients, either independently or acting with other factors such as diet, oral microbiota, and fluoride exposure. Successfully identifying whether these genes are associated with caries experience could help provide insight into the etiology of the disease and perhaps reveal patterns of the disease that assist in the identification of individuals at risk for caries and aid in the development of new strategies of prevention.
PLINK was chosen as the software for data analysis. According to the PLINK site “The focus of PLINK is purely on analysis of genotype/phenotype data (Purcell et al., 2007).” Therefore, it is an applicable program for this study. Genotypic association tests were chosen, which provide an association test in a 2-by-3 table of disease-by-genotype. It provides a few different results including tests in a dominant and recessive model for the minor allele. A second option to these tests would have been running a non-parametric ANOVA with planned comparisons. This is because comparisons of the different phenotypic subsets were created previously. However, ANOVA is a better option when there are many comparisons, and here there were only fifteen comparisons, so the PLINK software was used.
There were a number of SNPs showing an association with the created phenotypes. After a Bonferroni correction, one SNP stood out as significant, rs2235091 (p=0.0008 in a recessive model for low primary caries vs. high primary caries experience, and p=0.0004 in a recessive model for no primary caries vs. high primary caries experience). This SNP is found in Kallikrein 4 (KLK4), an enamel development gene involved in enamel mineralization [Lu et al., 2008]. Previously, studies have found that KLK4 may play a protective role in dental caries experience [Wang et al., 2012a]. After genotyping the Norwegian population, rs2235091 was found to be significant in a recessive model as a protective factor for primary caries.
Additional genes were found to be nominally associated with the created caries phenotypes. Genes from the Aquaporin (AQP) family, which form water channel proteins, were found to be nominally associated with caries experience. aquaporin 5 (aqp5) has been shown in mice to play a role in salivary water contributions, as aqp5 deficient mice had reduced salivary flow [Culp et al., 2005]. In humans, the aquaporins function in producing normal salivary flow specifically from the parotid gland [Smith et al., 1999]. Studies of aquaporin and dental caries have led to findings that it may play a protective role, potentially because it helps the oral cavity with buffering against acids that may demineralize the teeth [Wang et al., 2012b; Anjomshoaa et al., 2015].
Taste gene SNPs rs10246939 in TAS1R2 and rs10246939 in TAS2R38 indicated that patients with the recessive allele may have a higher chance of having dental caries. Previous studies have shown associations between these genes and dental caries [Wendell et al., 2010]. TAS2R38 is a gene for a bitter taste receptor. While some studies have shown that this gene is associated with lower caries experience [Yildiz et al., 2016], it could be that patients in this study with the recessive allele may choose more acidic drinks and food that may be richer in sugars due to their bitter taste preferences, which may in turn lead to increased dental caries.
This study had some limitations. Although some significant associations were found between our selected SNPs and our created caries phenotypes, these do not necessarily mean that these genes directly cause dental caries. Further studies could be done to look at potential mechanisms and roles these genes play in dental caries experience, and evidence for association will help prioritize future effort aiming to identify the mechanisms underlying the expression of the disease. Additionally, associations may happen by chance, particularly with multiple comparisons. We attempted to correct for this using Bonferroni correction and other multiple testing correction approaches. Bonferroni is by many considered a strict test, and therefore may rule out associations that are otherwise significant. We did have a large number of nominally significant associations. However, we chose our SNPs based on previous studies linking them to dental caries experience and therefore expect there to be higher amounts of association than in a GWAS (genome-wide association study) using the same phenotypes. Also, by creating more discrete phenotypic groups, we aimed to find more specific associations.
To our knowledge, this is the first study to separate patients into more discrete groups based on dental caries experience defined from longitudinal data and the first to study acute increases in DMFT score between two time points. Our hope with doing so is to provide more power in our predictions of genetic influences of dental caries and perhaps lead to better ways of studying the disease based on more specific categorizations of phenotypes.
Acknowledgments
Funding for this study came from NIH/NIDCR R01-DE18914 and the University of Oslo and graduate support from the University of Pittsburgh School of Dental Medicine.
Appendix
Appendix Table 1.
Breakdown of patients based on DMFT scores in the permanent dentition.
| Group (Phenotype) | Females (n1) | Males (n2) | Total | p-value |
|---|---|---|---|---|
| Low Caries | 161 (18.8%) | 140 (16.4%) | 301 (35.2%) | 0.73 |
| High Caries (4-7) | 123 (34.7%) | 123 (34.7%) | 246 (69.5%) | 0.32 |
| Very High Caries (8+) | 59 (16.7%) | 49 (13.8%) | 108 (30.5%) | 0.66 |
| No Caries | 108 (12.6%) | 93 (10.8%) | 201 (23.4%) | 0.73 |
Appendix Table 2.
Breakdown of patients into those with a spike, those without a spike, and those with no caries experience.
| Group (Phenotype) | Females (n1) | Males (n2) | Total | p-value |
|---|---|---|---|---|
| Spike | 66 (7.7%) | 62 (7.2%) | 128 (14.9%) | 0.78 |
| No Spike | 277 (32.4%) | 250 (29.2%) | 527 (61.6%) | 0.93 |
| No Caries | 108 (12.6%) | 93 (10.9%) | 201 (23.5%) | 0.73 |
| Total | 451 (52.7%) | 405 (47.3%) | 856 | - |
Appendix Table 3.
Breakdown of patients based on dmft scores in the primary dentition.
| Group (Phenotype) | Females (n1) | Males (n2) | Total | p-value |
|---|---|---|---|---|
| Low Caries | 66 (7.6%) | 59 (6.8%) | 125 (14.5%) | 0.93 |
| High Caries | 41 (4.8%) | 43 (5.0%) | 84 (9.7%) | 0.48 |
| No Caries | 344 (40.2%) | 307 (36%) | 654 (75.8%) | 0.68 |
| Total | 454 (52.6%) | 409 (47.4%) | 863 | - |
Appendix Figure 1. Graphical representation of selected Table 1 and 2 patients’ longitudinal DMFT data.
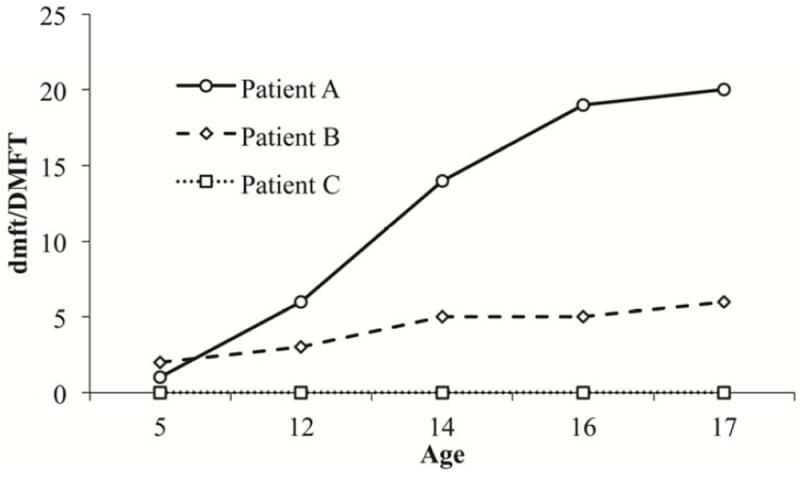
Patient A shows spikes of four or more between ages 5 and 12, 12 and 14, and between 14 and 16. Patient B fits into the high caries experience group and the very high caries experience group, but has no spike in disease experience between time points. Patient C has no caries experience.
Footnotes
There are no conflicts of interest to report.
Author Roles:
Jenny Søvik, Aida Mulic, and Anne Tveit participated in both sample and data collection and writing. Megan Weber participated in genotyping, data analysis, and writing. Alexandre Vieira participated in data analysis and writing. Kathleen Deeley, Jessalyn Forella, and Nicholas Shirey participated in genotyping and writing.
References
- The World Health Organization. The World Oral Health Report 2003 [Internet] [2013 Sep 15];World Oral Health Rep 2003. 2003 Available from: http://www.who.int/oral_health/publications/report03/en/)
- National Center for Health Statistics. Health, United States 2015: With Special Feature on Racial and Ethnic Health Disparities. [Internet] [2017 Feb 11];2016 Available from: https://www.cdc.gov/nchs/data/hus/hus15.pdf#060. [PubMed]
- Müller A, Hussein K. Meta-analysis of teeth from European populations before and after the 18th century reveals a shift towards increased prevalence of caries and tooth loss. Arch Oral Biol. 2017 Jan 1;73:7–15. doi: 10.1016/j.archoralbio.2016.08.018. [DOI] [PubMed] [Google Scholar]
- Petersen PE. Challenges to improvement of oral health in the 21st century — the approach of the WHO Global Oral Health Programme. Int Dent J. 2004 Dec 1;54:329–343. doi: 10.1111/j.1875-595x.2004.tb00009.x. [DOI] [PubMed] [Google Scholar]
- Kassebaum NJ, Bernabe E, Dahiya M, Bhandari B, Murray CJL, Marcenes W. Global Burden of Untreated Caries: A Systematic Review and Metaregression. J Dent Res. 2015 May;94:650–658. doi: 10.1177/0022034515573272. [DOI] [PubMed] [Google Scholar]
- Moschos MM, Brouzas D, Mezitis M, Zachariadis N. Visual loss due to a carious tooth. The Lancet. 2005;366:1504. doi: 10.1016/S0140-6736(05)67602-7. [DOI] [PubMed] [Google Scholar]
- Owings L. Toothache Leads to Boy’s Death [Internet] [2017 Feb 11];ABC News. 2007 Mar 7; Available from: http://abcnews.go.com/Health/Dental/story?id=2925584&page=1.
- Fejerskov O, Kidd E. Dental caries: the disease and its clinical management. John Wiley & Sons; 2008. [Google Scholar]
- Nandula SR, Amarnath S, Molinolo A, Bandyopadhyay BC, Hall B, Goldsmith CM, et al. Female mice are more susceptible to developing inflammatory disorders due to impaired transforming growth factor β signaling in salivary glands. Arthritis Rheum. 2007 Jun;56:1798–1805. doi: 10.1002/art.22715. [DOI] [PubMed] [Google Scholar]
- Matsuki-Fukushima M, Hashimoto S, Shimono M, Satoh K, Fujita-Yoshigaki J, Sugiya H. Presence and localization of aquaporin-6 in rat parotid acinar cells. Cell Tissue Res. 2008 Apr;332:73–80. doi: 10.1007/s00441-007-0558-4. [DOI] [PubMed] [Google Scholar]
- Wendell S, Wang X, Brown M, Cooper ME, DeSensi RS, Weyant RJ, et al. Taste Genes Associated with Dental Caries. J Dent Res. 2010 Nov 1;89:1198–1202. doi: 10.1177/0022034510381502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patir A, Seymen F, Yildirim M, Deeley K, Cooper ME, Marazita ML, et al. Enamel Formation Genes Are Associated with High Caries Experience in Turkish Children. Caries Res. 2008;42:394–400. doi: 10.1159/000154785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergoz N, Seymen F, Gencay K, Tamay Z, Deeley K, Vinski S, et al. Genetic variation in Ameloblastin is associated with caries in asthmatic children. Eur Arch Paediatr Dent Off J Eur Acad Paediatr Dent. 2014 Nov 8;15:211–6. doi: 10.1007/s40368-013-0096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Cherny S, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007 Sep;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Ho B, Deeley K, Briseno-Ruiz J, Faraco IM, Schupack BI, et al. Enamel Formation Genes Influence Enamel Microhardness Before and After Cariogenic Challenge. Plos One. 2012 Sep 24;7:e45022. doi: 10.1371/journal.pone.0045022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briseño-Ruiz J, Shimizu T, Deeley K, Dizak PM, Ruff TD, Faraco IM, et al. Role of TRAV locus in low caries experience. Hum Genet. 2013 Sep;132:1015–1025. doi: 10.1007/s00439-013-1313-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Deeley K, Briseño-Ruiz J, Faraco IM, Jr, Poletta FA, Brancher JA, et al. Fine-mapping of 5q12. 1–13.3 unveils new genetic contributors to caries. Caries Res. 2013;47:273–283. doi: 10.1159/000346278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küchler EC, Deeley K, Ho B, Linkowski S, Meyer C, Noel J, et al. Genetic mapping of high caries experience on human chromosome 13. BMC Med Genet. 2013;14:116. doi: 10.1186/1471-2350-14-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küchler EC, Feng P, Deeley K, Fitzgerald CA, Meyer C, Gorbunov A, et al. Fine mapping of locus Xq25. 1-27-2 for a low caries experience phenotype. Arch Oral Biol. 2014 May;59:479–486. doi: 10.1016/j.archoralbio.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber ML, Hsin H-Y, Kalay E, Brozkova DS, Shimizu T, Bayram M, et al. Role of estrogen related receptor beta (ESRRB) in DFN35B hearing impairment and dental decay. Bmc Med Genet. 2014 Jul 15;15:81. doi: 10.1186/1471-2350-15-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Papagerakis P, Yamakoshi Y, Hu JC-C, Bartlett JD, Simmer JP. Functions of KLK4 and MMP-20 in dental enamel formation. Biol Chem. 2008 Jun;389:695–700. doi: 10.1515/BC.2008.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Willing MC, Marazita ML, Wendell S, Warren JJ, Broffitt B, et al. Genetic and Environmental Factors Associated with Dental Caries in Children: The Iowa Fluoride Study. Caries Res. 2012a;46:177–184. doi: 10.1159/000337282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culp DJ, Quivey RG, Bowen WH, Fallon MA, Pearson SK, Faustoferri R. A mouse caries model and evaluation of AQP5-/- knockout mice. Caries Res. 2005;39:448–454. doi: 10.1159/000088179. [DOI] [PubMed] [Google Scholar]
- Smith JK, Siddiqui AA, Modica LA, Dykes R, Simmons C, Schmidt J, et al. Interferon-alpha upregulates gene expression of aquaporin-5 in human parotid glands. J Interferon Cytokine Res. 1999 Aug 19;:929–935. doi: 10.1089/107999099313479. [DOI] [PubMed] [Google Scholar]
- Wang X, Shaffer JR, Zeng Z, Begum F, Vieira AR, Noel J, et al. Genome-wide association Scan of dental caries in the permanent dentition. BMC Oral Health. 2012b;12:57. doi: 10.1186/1472-6831-12-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjomshoaa I, Briseno-Ruiz J, Deeley K, Poletta FA, Mereb JC, Leite AL, et al. Aquaporin 5 Interacts with Fluoride and Possibly Protects against Caries. Plos One. 2015 Dec 2;10:e0143068. doi: 10.1371/journal.pone.0143068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz G, Ermis RB, Calapoglu NS, Celik EU, Turel GY. Gene-environment Interactions in the Etiology of Dental Caries. J Dent Res. 2016 Jan;95:74–79. doi: 10.1177/0022034515605281. [DOI] [PubMed] [Google Scholar]
- Deeley K, Letra A, Rose EK, Brandon CA, Resick JM, Marazita ML, et al. Possible association of amelogenin to high caries experience in a guatemalan-mayan population. Caries Res. 2008;42:8–13. doi: 10.1159/000111744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer JR, Wang X, Feingold E, Lee M, Begum F, Weeks DE, et al. Genome-wide Association Scan for Childhood Caries Implicates Novel Genes. J Dent Res. 2011 Dec;90:1457–1462. doi: 10.1177/0022034511422910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer JR, Carlson JC, Stanley BOC, Feingold E, Cooper M, Vanyukov MM, et al. Effects of enamel matrix genes on dental caries are moderated by fluoride exposures. Hum Genet. 2015 Feb;134:159–167. doi: 10.1007/s00439-014-1504-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk A, Famili P, Vieira AR. The Antimicrobial Peptide DEFB1 Is Associated with Caries. J Dent Res. 2010 Jun;89:631–636. doi: 10.1177/0022034510364491. [DOI] [PubMed] [Google Scholar]
- Krasone K, Lace B, Akota I, Care R, Deeley K, Kuechler EC, et al. Genetic variation in the promoter region of beta-defensin 1 (DEFB 1) is associated with high caries experience in children born with cleft lip and palate. Acta Odontol Scand. 2014 Apr;72:235–240. doi: 10.3109/00016357.2013.822549. [DOI] [PubMed] [Google Scholar]
- Abbasoglu Z, Tanboga I, Kuechler EC, Deeley K, Weber M, Kaspar C, et al. Early Childhood Caries Is Associated with Genetic Variants in Enamel Formation and Immune Response Genes. Caries Res. 2015;49:70–77. doi: 10.1159/000362825. [DOI] [PMC free article] [PubMed] [Google Scholar]


