Abstract
Genomic, transcriptomic and proteomic databases indicate that the N-terminal 322 residues encoded by the presumptive LOC100996504 gene, which is adjacent to the ARHGEF18 guanine nucleotide exchange factor gene on chromosome 19, constitute the N-terminal portion of a 1361-residue isoform of ARHGEF18, dubbed LOCGEF-X3. LOCGEF-X3 arises from the use of a leukocyte-specific alternative transcriptional start site and splicing that bypasses the initial non-coding exon of the canonical 1015-residue ARHGEF18 isoform, p114. Eosinophil LOCGEF-X3 was amplified and cloned, recombinant LOCGEF-X3 was expressed, and anti-ARHGEF18 antibody was found to recognize a band in immunoblots of eosinophil lysates that co-migrates with recombinant LOCGEF-X3. PCR of eosinophils revealed minor amounts of transcripts for X4 and X5 isoforms of LOCGEF that arise from differential splicing and differ from the X3 isoform at their extreme N-termini. No p114 transcript or protein band was detected in eosinophils. Immunostaining with anti-ARHGEF18 antibody revealed relocalization of LOCGEF and RHOA from the periphery of round unstimulated eosinophils to the two poles of eosinophils polarized by treatment with IL5, CCL11, or IL33 in suspension. Canonical p114 ARHGEF18 has been implicated in maintenance of epithelial cell polarity. We suggest that the “LOC” portion of LOCGEF, which is unlike any other protein domain, has unique functions in control of polarity in activated eosinophils and other leukocytes.
Keywords: LOC100996504, guanine nucleotide exchange factor (GEF), granulocyte, p114-GEF, ARHGEF18, alternative transcriptional start
Introduction
ARHGEF18 is a widely-expressed Ras homology guanine nucleotide exchange factor (RhoGEF) known to enhance the activity of RHOA GTPase [1]. ARHGEF18 may also modulate RAC1 GTPase [2–4], although this activity is controversial [1, 5]. Two variants of human ARHGEF18 are annotated in UniProtKB, isoforms 1 and 2. The 114 kDa isoform 2 (hereafter called the p114 isoform) has been characterized and comprises a 1015-residue protein with guanine nucleotide exchange activity and the Dbl and pleckstrin homology domains characteristic of GEFs [1]. In medaka fish, mutation of ARHGEF18 results in dysregulation of RHOA-ROCK2 signaling that maintains neuro-epithelial apico-basal polarity and proliferation [4]. The effects of ARHGEF18 depletion by RNA interference have been characterized in breast cancer cells [6], endothelial cells [7] and various types of epithelial cells [6, 8]. ARHGEF18 is necessary for cell-cell junction maturation in endothelial and epithelial cells [7–9], intercellular communication of bronchial epithelial cells [10], and amoeboid movement of breast cancer cells and epithelial sheet closure after corneal wounding [6, 11]. Transfection of ARHGEF18 (p114) in fibroblasts led to increased production of reactive oxygen species [2] and increased RHOA activation [5]. Exogenous expression of the p114 isoform also demonstrated its role in organizing actomyosin cytoskeletal components, thereby regulating stress fiber formation [1, 5] and maintaining neuro-epithelial apico-basal polarity [4]. Arno et al. [12] recently reported five mutations in ARHGEF18 that are associated with retinal dystrophy in humans. The changes phenocopy effects of ARHGEF18 mutations in medaka fish [4], and thus the association has been attributed to the contribution of ARHGEF18 protein to the maintenance of apico-basal polarity of retinal cells.
Recently, global proteomics revealed that ARHGEF18 is present in human eosinophils [13]. The eosinophil is a blood granulocyte that is specified during hematopoietic differentiation [14]. The eosinophil has a unique repertoire of granular proteins and cell surface receptors and is prominent in inflammation driven by mediators of T helper cell type 2 immunity [15]. Eosinophils undergo rapid and profound morphological change following cytokine stimulation [16], going from ovoid to acorn-shaped and polarized in the space of 5 minutes. The shape change is accompanied by enhanced integrin-mediated adherence to extracellular matrix proteins [17] and is dependent on actomyosin cytoskeleton rearrangement [16]. The ARHGEF18-RHOA-ROCK2-MLC signaling axis regulates actomyosin contractility in other cell types [9, 11]. Roles of ARHGEF18 have not been studied in hematopoietic cells, and if and how ARHGEF18 contributes to mechanisms underlying the dramatic polarization of integrins and receptors to the nucleopod tip in eosinophils is not known.
We now describe previously uncharacterized isoforms of ARHGEF18 in eosinophils. The investigations arose out of recent isobaric labeling mass spectrometric study indicating, unexpectedly and inexplicably, that the abundance of a peptide from LOC100996504, annotated in UniProtKB [18] as a transmembrane protein, decreases acutely upon activation of eosinophils with interleukin-5 (IL5) [13]. A BLAST search [19] revealed multiple predicted ARHGEF18 isoforms containing part of LOC100996504 based on expressed sequences tags (ESTs) [20]. We dub these isoforms the “LOCGEFs.” By PCR and immunoblotting, LOCGEFs were the only isoforms of ARHGEF18 that we could detect in human eosinophils. The 1361-residue X3 isoform (NCBI XP_006722769.1) is the most abundant, and transcripts of two closely related isoforms (X4, XP_011526140.1 and X5, XP_011526141.1) are also present. We suggest that expression of LOCGEFs is controlled by a leukocyte-specific set of transcription factors acting on a site far upstream of the start site for the p114 isoform, present evidence that LOC100996504 as annotated in UniProtKB does not exist as a separate protein, and cast doubt on the existence of the ARHGEF18 Isoform 1 annotated in UniProtKB. Immunofluorescence microscopy revealed striking relocalization of LOCGEFs from the plasma membrane to the two poles upon polarization of eosinophils by IL5, IL33 and CCL11. The potential to make LOCGEFs is retained by metazoans as distant as zebrafish, suggesting that the “LOC” portion of LOCGEFs, which does not have significant sequence homology to known protein domains, endows LOCGEFs with unique functions in leukocyte activation.
Materials and Methods
Ethics Statement
Peripheral human blood eosinophils were purified to >98% purity by Percoll centrifugation and negative selection for neutrophils and monocytes as described previously [21]. The cells were received without identifying information in accord with a protocol approved (#2013-1570) by the University of Wisconsin-Madison Center for Health Sciences Institutional Review Board. Signed informed consent from the donor was obtained for the use of each sample in research.
Bioinformatics
BLAST
We used the blastp 2.6.1+ algorithm [19] (default parameters) to interrogate non-redundant human protein sequences for sequence similarity and the tblastn 2.6.1+ algorithm to interrogate translated nucleotide collection data (including GenBank+EMBL+DDBJ+ PDB+RefSeq sequences) to see whether there were orthologues in other species with significant homology to human LOCGEF-X3.
RNA-Seq mapping
We visualized RNA-Seq reads from the Illumina BodyMap 2.0 project [22] (NCBI GEO accession: GSE30611) mapped to human genome build hg19 using Integrative Genome Viewer (IGV) [23]. From these data, percentage spliced in (PSI) values were calculated; enabling us to determine predicted proportions of X3, X4 and X5 transcripts. Raw RNA-Seq datasets generated for neutrophils [24] (GSE40548) and Jurkat T lymphocytes [25] (GSE45428) were processed in the computational biology workflow platform Galaxy [26], and mapped to human genome build hg19 using the TopHat read alignment algorithm [27].
Pairwise sequence alignment
Clustal Omega (1.2.4) [28] was used to align NCBI sequences from H sapiens (XP_006722768.1) and M musculus (NP_001028550.2 and NP_598723.3). The mouse LOCGEF sequence was determined by mapping data from DBA/2J x C57BL/6J mouse RNA-Seq [29] to the genome using the online computational biology platform Galaxy [26]. The mapped reads were visualized using Integrated Genome Viewer [23], enabling determination of mouse LOCGEF isoforms. These isoforms were then verified through a process of specific amplification and sequencing (see below).
PCR and sequencing of eosinophils’ ARHGEF18 isoform transcripts
Preparation of human and mouse eosinophil cDNAs
Human RNA was extracted from peripheral blood eosinophils from donors using the RNeasy kit (Qiagen #74104). Mouse RNA was extracted from cultured bone marrow cells [30]. As previously described [31], cDNA was generated using the SuperScript® III First-Strand Synthesis System for RT-PCR kit (ThermoFisher #18080051) with random primers.
PCR to investigate transcript composition
We created primer pairs (Item Ia of Supp Data) specific for some of the ARHGEF18 isoforms (Table I) and used conventional PCR cycling to amplify target regions from eosinophil cDNA, that is, the 5′ end of the transcripts that harbor the major differences in human isoform sequences. Transcripts with premature truncations due to alternative splicing (found in X4 and X11) were not evaluated. Although RNA-Seq data suggested that only t1, X3, X4 and X5 are present in white blood cells (see below), the primer pairs created would also recognize t2, X2 and X6. Amplicons were sequenced. We created primer pairs specific for the “LOC” region of the predicted mouse orthologue of LOCGEF (Item Ib of Supp Data), and sequenced amplicons to verify our model.
Table I.
Predicted transcripts and isoforms of the gene ARHGEF18 accessed from the National Center for Biotechnology Information [20]. Transcripts and isoforms that we believe exist in various cell types are in bold. Asterisks indicate splicing models that result in premature truncations of the protein. For the Kozak sequence, initiating Met codons are underlined.
| Transcript | Isoform | Also known as | Number of residues | Kozak sequence |
|---|---|---|---|---|
|
t1: NM_015318.3 nX9: XM_011527840.1 nX10: XM_006722708.2 |
Isoform A: NP_056133.2/X6: XP_006722771.1 | p114Isoform 2 | 1015 | gtcggtatga |
| t2: NM_001130955.1 | Isoform B: NP_001124427.1 | Isoform 1 | 1173 | cagtaaatgg |
| nX1: XM_011527835.2 nX2: XM_005272464.4 nX3: XM_011527836.2 |
X1: XP_011526137.1 | 1426 | gcacgcatgg | |
| nX4: XM_011527837.2 | X2: XP_011526139.1 | 1370* | gcacgcatgg | |
|
nX5: XM_006722705.3 nX6: XM_006722706.3 |
X3: XP_006722769.1 | LOCGEF-X3 | 1361 | tctgccatgg |
| nX7: XM_011527838.2 | X4: XP_011526140.1 | LOCGEF-X4 | 1361 | gagctgatga |
| nX8: XM_011527839.2 | X5: XP_011526141.1 | LOCGEF-X5 | 1345 | gagagtatgg |
| nX11: XM_011527841.2 | X7: XP_011526143.1 | 959* | gcacgcatgg |
Expression of recombinant p114 and LOCGEF
Amplification of the LOCGEF transcript for cloning
The LOCGEF transcript is 4kb, and the 3′ half is GC-rich (68%). After failing with several strategies, we succeeded in assembling full-length cDNA by amplifying and joining four overlapping segments making use of internal restriction sites DraI, Bsu36I and BamHI (Item Ic of Supp Data, Supp Figure 1). The LOCGEF transcript was cloned with a His-tag sequence at the N-terminus into pET-Elmer (a variant of pET-28c) [32] and amplified in E. coli strain DH5α. The cloned product contained alleles of two single-nucleotide non-synonymous polymorphisms, designated as rs536249774 and rs2287918 in dbSNP [33]. Our cDNA encodes Y211 (exon L8, frequency <0.01; major allele would encode C) and R889 (exon 22 of full-length LOCGEF/exon 13 of p114, frequency of 0.83; minor allele with a frequency of 0.17 would encode Q) [33].
Amplification of p114, and protein expression and purification of LOCGEF and p114
After the LOCGEF construct was complete, the plasmid was used as template for amplifying the p114 sequence for cloning into pET-Elmer. Expression of both proteins was induced by IPTG in E. coli BL21 (DE3). After lysing the bacteria in urea buffer (8 M urea, 100 mM NaH2PO4, 10 mM Tris, 1 mM β-mercaptoethanol pH 8.0), the protein was purified on a Nickel-NTA column. Eluted protein was dialyzed into acetate buffer (100 mM, pH 2.88). Yields were low, but sufficient for use of the proteins as standards in immunoblotting.
Detection of LOCGEF protein in eosinophils
Preparation of cells
Human peripheral blood eosinophils were allowed to rest for 1 h in RPMI with 50 mM HEPES and 0.1 % HSA at 37 °C, before being stimulated for 5 minutes with 50 ng/ml IL5, 100 ng/ml IL33 or 30 ng/ml CCL11. Lysates of eosinophils were prepared using SDS buffer (62.5 mM Tris, 4 % SDS, 5 % glycerol, 4 M urea pH 6.8) with 700 mM β-mercaptoethanol. Samples were boiled in 25 μl buffer per million cells for 5 minutes.
Immunoblotting
Antibodies are listed in Item II of Supp Data. Samples loaded on gels were prepared from roughly 1 million cells. We used 8 % acrylamide gels to resolve lysates, before transferring protein overnight to PVDF membrane. Membranes were blocked in 1 % BSA, before being incubated in polyclonal rabbit or goat anti-ARHGEF18 antibody at 1 μg/ml in 0.1 % BSA in TBS-T, followed by incubation in 1:10,000 donkey anti-rabbit or bovine anti-goat IgG conjugated to HRP (Jackson ImmunoResearch). Bands were detected by enhanced chemiluminescence and compared to Precision Plus Protein Kaleidoscope Standard (BioRad). The PVDF membrane was Ponceau stained to evaluate equal loading of lysates. Densitometric analysis was performed using ImageJ [34]. Three representative donors are shown in Results. The experiment was performed several times, with seven biological replicates.
Immunofluorescent staining
Cells were stimulated with PBS, IL5, IL33 or CCL11 (eotaxin-1), final concentration of 50 ng/ml at 37 °C. Following the 10 minute incubation, 1 mM sodium vanadate was added and cells were fixed in 3.7 % paraformaldehyde for 10 minutes, and quenched with 0.1 M glycine for 10 minutes. Cells were washed and re-suspended in PBS and cytospun onto poly-L-lysine-coated (12 mm diameter) glass coverslips, with roughly 2.5×105 cells per coverslip. Cells were permeabilized using 0.5 % SDS in PBS for 15 minutes. After washing off the SDS, coverslips were blocked in 10 % BSA for one hour and then incubated overnight at 4 °C in primary antibodies (Item II of Supp Data) diluted in 2 % BSA, 0.1 % SDS in PBS. The following day, coverslips were washed in PBS, and incubated for 1 hour at room temperature with fluorophore-conjugated secondary antibodies specific for the species of the primary antibodies. After DAPI staining, coverslips were mounted on slides and sealed. Images were acquired using a Confocal Microscope (Nikon A1R-Si+ Confocal). Equal laser power settings and conversion gain were used for activated and unactivated cells; and also for control IgG staining.
Results
A leukocyte-specific form of ARHGEF18 exists in eosinophils
Discovery and evaluation of LOC100996504 peptides in eosinophils
Our investigation of LOC100996504 was motivated by comparative proteomic studies of resting and IL5-activated human eosinophils [13]. LOC100996504, which is annotated as an uncharacterized, putative transmembrane protein of 382 residues in UniProtKB [18], decreased nearly 3-fold in abundance over a 5-minute interval of IL5 stimulation. A comparison of this sequence to database sequences using BLAST [19], however, revealed that the N-terminal 322 residues of LOC100996504 corresponded to the initial 322 residues encoded by nX5 or nX6 transcripts of ARHGEF18 predicted by NCBI’s Eukaryotic Annotation Pipeline’s Gnomon-prediction models [20] (Table I). The eosinophil-derived peptides discovered by mass spectrometry [13] were common to both LOC100996504 as annotated in UniProtKB and putative 1361- or 1345-residue X3, X4, and X5 isoforms of ARHGEF18 encoded by the nX5, nX7, and nX8 transcripts (Table I). These findings, therefore, focused our attention on ARHGEF18.
Genomic analysis
The DNA sequence encoding the exons of LOC100996504 mapped to a region upstream of the gene annotated as ARHGEF18 on human chromosome 19 (Figure 1A–B). The putative LOC100996504 gene is divided into 10 exons, which herein we number L1-L10 to distinguish them from the 20 exons that comprise the canonical ARHGEF18 t1 for which the initiating methionine codon is in exon 2. The region between the splice site of exon L10 of LOC100996504, and exon 2 of ARHGEF18 is roughly 50 kB and contains non-coding exon 1 of ARHGEF18. Examination of the ENCODE database [35] for RNA polymerase ChIP-Seq hits in this region supported binding site motifs for three transcription factors, including PU.1, localizing near exon L1 and a different set of five transcription factors localizing near exon 1 (Figure 1A–B). No high-ranking transcription factor-binding sites were predicted close to the presumptive transcriptional start site of Isoform 1 (Figure 1 arrowhead), a predicted isoform that is discussed further below.
Figure 1. Schematic of determinants of ARHGEF18 isoforms.
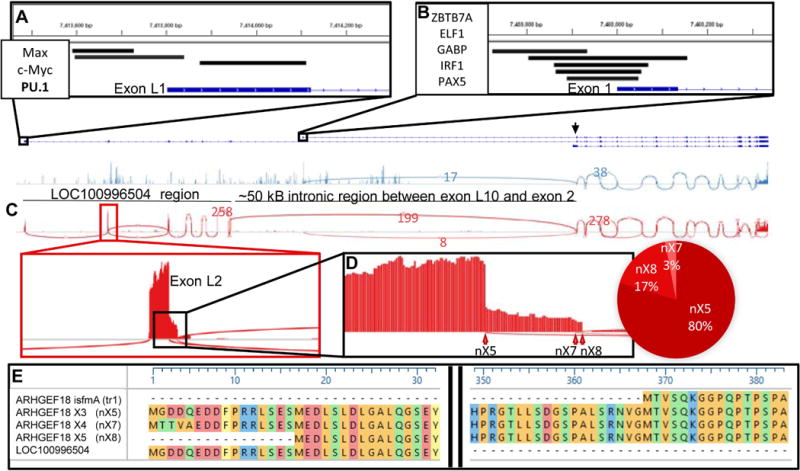
(A and B) Transcription factors predicted by ChIP-Seq to bind at initiating exons of LOCGEF (A) and p114 (B) are mapped onto genomic segments comprising the exons for p114, LOCGEF and theoretical Isoform 1 (predicted transcriptional start site indicated by a black arrowhead). (C) Sashimi plots mapping sequence-spanning reads from two Illumina BodyMap 2.0 datasets: white blood cells (red) and breast tissue (blue). Number of reads for selected splice events are shown. Plots have been sized to correspond to the genomic segments shown above. (D) Blow-up of exon L2 and percentage spliced in values for exon L2’s splice sites. (E) Initiating methionines and following sequences corresponding to p114, LOCGEF-X3, LOCGEF-X4 and LOCGEF-X5 are given along with sequence for LOC100996504 annotated in UniProtKB. The numbering above is in reference to residues of LOCGEF-X3. “MTVS…” of p114 is encoded in exon 2. “MGGD…” and “MTTV…” of LOCGEF-X3 and LOCGEF-X4 are encoded in exon L2. Use of an alternative initiating codon in exon L3 leads to the LOCGEF-X5 isoform beginning “MEDL….”
ARHGEF18 transcripts and predicted proteins
We assessed transcript evidence for NCBI-predicted ARHGEF18 isoforms by examination of publicly-available RNA-Seq data from Illumina’s BodyMap 2.0 dataset [22] (Figure 1C). Mapped reads showed that the transcript for the canonical 114 kDa form of ARHGEF18 (p114) was present in all 16 BodyMap tissue RNA-Seq datasets. In contrast, we found no transcript evidence supporting UniProtKB’s 382-residue form of LOC100996504 in any of them, nor was there evidence supporting transcripts t2 (ARHGEF18β in ProteomicsDB, Isoform 1 in UniProtKB), nX6, nX9, nX10, or nX11 (Table I). Instead, the mapped read data from white blood cells (WBC) supported three of NCBI’s predicted transcript models for ARHGEF18 in which exons L1-L10 of LOC100996504 are spliced to exon 2 of ARHGEF18. These vary in the 5' splice juncture in exon L2 (Figure 1D) and are designated nX5, nX7, and nX8; encoding predicted protein isoforms X3, X4 and X5 with differing N-terminal sequences (Table I and Figure 1E). We call these proteins “LOCGEFs.” The first 322 residues at the N-terminus of LOCGEF-X3 correspond to the N-terminal residues of UniProtKB’s LOC100996504 (Figure 1E). Analysis of the BodyMap data for PSI values [36] indicated that LOCGEF transcripts in white blood cells comprise mostly nX5 with lesser amounts of nX7 and nX8 (Figure 1D). Analyses of published RNA-Seq reads from neutrophils [24] and Jurkat T cells [25] revealed that >90 % of reads spanned the “exL10-ex2” junction compared to the “ex1-ex2” junction of the p114 transcript (data not shown). All of the neutrophil spanning reads supported the nX5 model, whereas of the Jurkat RNA-Seq reads, 91% fit the nX5 model and 9% the nX8 model.
Of the ARHGEF18 transcript models lacking premature stop codons, t1/nX9/nX10, nX5, nX6, and nX8 conform to the most-favored Kozak consensus sequence for translation initiation – gcc(A/G)ccATGg [37] (Table I). None of the RNA-Seq data, however, conformed to the nX6 model, which would encode the same protein as X3 but contains nucleotides from an extra exon between exons L1 and L2. Thus, the most abundant protein isoforms are likely to be generated from t1, nX5 and nX8 transcripts to yield, respectively, p114, X3 and X5. LOCGEF-nX7 encoding X4 has a less favorable Kozak sequence. Because of the confusing nomenclature for the transcript and isoform models, we suggest that the possible ARHGEF18 isoforms be called canonical p114 and LOCGEF-X3, LOCGEF-X4 and LOCGEF-X5 (Table I).
Proteomic support for LOCGEF isoforms in eosinophils and other leukocytes
Our global proteomic analysis of eosinophils [13] identified 11 unique peptide sequence matches (some overlapping each other) to the first 322 residues of the 382-residue LOC100996504 and 66 (also with overlaps) to the 1173-residue form of ARHGEF18 annotated as ARHGEF18β in Proteomics DB, Isoform B in NCBI, and Isoform 1 in UniProtKB (Supp Figure 2, Item III in Supp Data). Of the discovered unique peptide sequences, 65 are common to both Isoform 1 and 2. As in our analysis, publicly-available proteomic data curated by ProteomicsDB [38] describe LOC100996504 peptides in B- and T-lymphocytes, monocytes, and NK cells, whereas peptides from the p114 form of ARHGEF18 are found in 27 other samples as well as in these (Table II). Neither our nor ProteomicsDB’s datasets include peptides corresponding to the final 60 residues of UniProtKB’s LOC100996504 sequence, compared to the 83% coverage of the first 322 residues recorded by ProteomicsDB.
Table II.
Average normalized log10 values of ARHGEF18 and “LOC100996504” protein for 27 human tissues, based on iBAQ intensities. Data obtained from ProteomicsDB [37]. NR: none reported, N/A: not applicable. The ratio of LOC1009966504 to ARHGEF18 in tissues where both are present is calculated based on absolute values. The iBAQ intensities ratio for ARHGEF18 and LOC100996504 in eosinophils was 1.37.
| Tissue | ARHGEF18 (log10) | LOC100996504 (log10) | ARHGEF18 (Absolute x10-4) | LOC100996504 (Absolute x10-4) | Ratio (18/LOC) |
|---|---|---|---|---|---|
| cytotoxic T-cell | 5.47 | 4.34 | 29.6 | 2.2 | 13.7 |
| B-cell | 5.33 | 4.29 | 21.3 | 1.9 | 11.0 |
| monocyte | 5.17 | 4.21 | 14.7 | 1.6 | 9.1 |
| helper T-cell | 5.11 | 4.64 | 12.8 | 4.3 | 3.0 |
| NK cell | 4.59 | 3.43 | 3.9 | 0.26 | 14.5 |
| lymph node, osteosarcoma, breast, gut, pancreatic islet, spleen, uterus, testis, stomach, ovary, rectum, retina, brain, colon, pancreas, placenta, adrenal gland, colonic epithelium, liver, salivary gland, heart, lung, thyroid gland, skin, urinary bladder, blood platelet, proximal fluid | 2.3-5.06 | NR | 0.019-11.5 | N/A | N/A |
In the ProteomicsDB datasets, the ratios of abundances of LOC100996504 relative to ARHGEF18 in sources containing peptides for both ranged from 3 to 14.5 (Table II). Intensity-based estimates for these two entries in eosinophils varied by <30%, comparable to the minimal variation in intensities of the subunits of fibrinogen or hemoglobin [13], contaminating proteins for which subunit stoichiometry is 1:1:1 or 1:1. These data suggest that p114 and LOCGEF isoforms are both present in lymphocytes and monocytes, but exclusively LOCGEF isoforms are found in eosinophils.
Of the peptide sequence matches (PSMs) found in our proteomic analyses, two are common to Isoform 1 and the LOCGEFs but not to p114 (Supp Figure 3). The ProteomicsDB datasets also contain one PSM that is specific for Isoform 1 but not LOCGEF (Supp Figure 3); this PSM was identified in studies of A375 [39] and U2OS [40] cells. We note that while absence of proof is not proof of absence, the RNA-Seq data lack transcripts matching the full sequence of Isoform 1 as well as that of LOC100996504, and there are no proteomic data to support these models in eosinophils. We propose that PSMs attributed to LOC100996504 derive from LOCGEF and two of the three peptides considered specific for ARHGEF18 Isoform 1 derive from LOCGEF. We doubt that Isoform 1 exists. However, assessing the PSM mapping only to Isoform 1 would require a targeted proteomics approach.
In all three plausible models of LOCGEF, the ARHGEF18 isoform transcript comprises 29 exons in which exon L10 of LOC100996504 is spliced to exon 2 of ARHGEF18, i.e., in LOCGEF-X3 966 coding nucleotides extending from exon L1 to a truncated exon L10 (a sequence encoding the initial 322 residues of LOC100996504) are spliced to the 3117 nucleotides of the ARHGEF18 transcript (a sequence encoding 1039 residues) resulting in a protein of 1361 residues (Table I). The three models predict different initiating methionines due to differential splicing of exon L2 (Figure 1E). PSMs matching the peptide starting with the initiating methionine predicted for LOC100996504 (and by extension LOCGEF-X3), although not present in our dataset [13], were found in global proteomic analyses of leukemic [41] and Jurkat T cells [25]. However, because LOCGEF-X4 and LOCGEF-X5 were not annotated in the databases used for these analyses, one cannot conclude that the other N-terminal sequences were not present.
Interrogation of LOCGEF transcripts and proteins in eosinophils
We designed sets of primers spanning predicted exon-exon junctions (Item Ia in Supp Data) and sequenced PCR products amplified from peripheral blood eosinophil cDNA. Importantly, we found that the predicted exonL10-exon2 junction of LOCGEFs was present (Figure 2A). Primers specific for transcripts encoding p114 or Isoform 1 did not yield product when used to amplify eosinophil cDNA. Primers able to amplify transcripts nX5, nX6, nX7 or nX8 resulted in a dominant nX5 product encoding LOCGEF-X3 (Figure 2B, lane 3). In addition, PCR products encoding X4 or X5 were produced using primers that would amplify nX7 or nX8 but not nX5 (Figure 2B, lane 2). Sequencing indicated that both sequences were present with the nX8 sequence dominating (Figure 2C). We saw no evidence of nX6. Thus, these data show that ARHGEF18 transcripts nX5, nX7 and nX8 (encoding protein isoforms X3, X4, and X5 respectively) are present in eosinophils with an order of abundance of nX5>>nX8>nX7 (Figure 2B–C), which is in agreement with the proportion of reads mapping across splice junctions in the WBC dataset (Figure 1D).
Figure 2. Eosinophils have transcripts encoding LOCGEF-X3, -X4, and -X5.
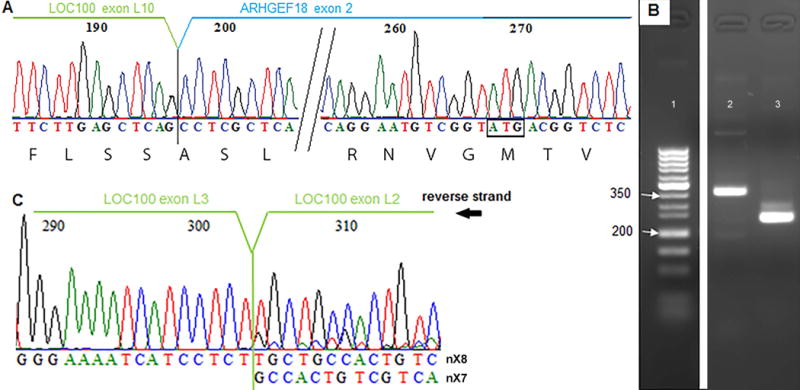
(A) Sequencing of LOCGEF amplicons demonstrating splicing of exon L10 exon 2 in eosinophil cDNA. (B) PCR of eosinophil cDNA demonstrating presence of nX5 and nX7/nX8 transcripts in eosinophils. Lane 1, exACTGene DNA ladder; lane 2, nX7/nX8 amplicons; lane 3, nX5/6/7/8 amplicons. The band in lane 2 is of the expected size for primers that amplify nX7, 358 bp; or nX8, 362 bp. The major band in lane 3 is of the expected amplicon size for nX5, 248 bp; and the minor band is of the expected amplicon size for nX7, 297 bp; or nX8, 301 bp. No band of the size expected for nX6, 411bp, was found. (C) Sequencing of the nX7/nX8 amplicons demonstrating presence of both nX7 and nX8, which cannot be distinguished by gel electrophoresis.
To produce standards for immunoblotting of ARHGEF18 isoforms in eosinophils using commercial anti-human p114 antibodies, we cloned full-length LOCGEF-X3 from eosinophil cDNA (Supp Figure 1), used the LOCGEF amplicon to generate an amplicon encoding p114, and expressed both proteins in E. coli. Polyclonal antibodies targeting epitopes within the regions of residues 612-802 (rabbit) or the C-terminus 1002-1015 (goat) of p114, which correspond to residues 958-1148 or 1348-1361 of LOCGEF-X3, recognized a protein of roughly 185 kDa in eosinophil lysate (Figure 3A–B). This band is larger than the molecular weight of 152 kDa calculated for LOCGEF-X3 by the ExPASy Compute Mw/pI tool [42], but migrates at the same position as the recombinant protein. A band co-migrating with recombinant p114, which runs at a size of 136 kDa relative to molecular weight markers, was not found in eosinophil lysate. Immunoblotting with rabbit anti-p114 of pairs of unactivated and IL5-treated eosinophils generated from three different donors revealed 185-kDa bands of similar intensities in both members of the pair (Figure 3C). Protein content of the PVDF membrane was visualized using Ponceau stain to demonstrate parity of total protein loaded (Figure 3D). Densitometric assessment attested to the near equivalence of paired lysates. The ratios shown indicate the IL5/PBS LOCGEF level, adjusted according to total protein loaded. These data are representative of seven biological replicates. One of the sample sets contained a minor immuno-reactive band of 165 kDa. This band was not found in any of the other samples. Since -X4 and -X5 have predicted molecular weights within 2 kDa of that of LOCGEF-X3, it is unlikely that the 165-kDa band represents an intact LOCGEF isoform.
Figure 3. LOCGEF protein is present in eosinophils.
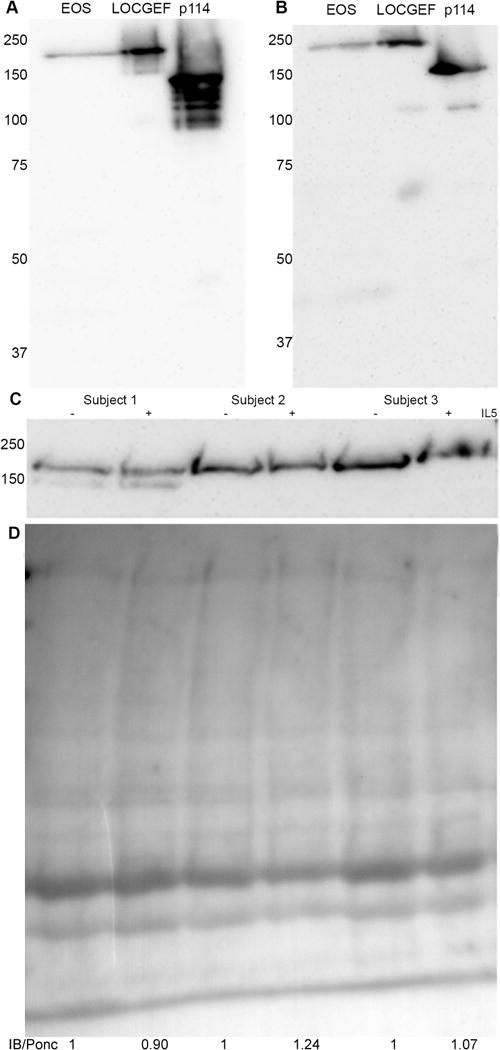
Immunoblotting with rabbit (A) or goat (B) anti-ARHGEF18 antibodies demonstrated a ~185 kDa band in eosinophils lysate (EOS) that co-migrates with recombinant LOCGEF-X3 and not recombinant p114. (C) Immunoblotting by rabbit anti-ARHGEF18 of sets of lysates from unstimulated (-) and IL5-stimulated (+) eosinophils from three separate donors. (D) Ponceau stain of the same transferred proteins, with ratios of levels of LOCGEF in IL5/untreated, determined by densitometric analysis adjusted according to total protein loading.
Microscopic localization of LOCGEF in unactivated and activated eosinophils
As described in the Introduction, p114 ARHGEF18 is important for establishing neuro-epithelial apico-basal cell polarity. Eosinophils are known to undergo rapid shape change and become polarized upon activation with a number of agonists, including IL5, CCL11 (eotaxin-1), and IL-33 [16, 43, 44]. To explore a possible role of LOCGEF in polarization, we localized goat anti-ARHGEF18 by confocal microscopy in unactivated blood eosinophils or eosinophils activated by one of these three agonists. Activation was carried out on suspended cells for 10 minutes, after which cells were fixed and collected by cytospinning prior to staining. LOCGEFs had a discontinuous distribution in the peri-membrane region of unactivated cells (Figure 4A). Polarization with IL5 or CCL11 entails movement of the nucleus to one end of the cell to form a structure called the nucleopod because the overlying plasma membrane is enriched in proteins that are markers of uropods in other leukocytes [16]. Upon activation with IL5, IL5 receptors become clustered at the tip of the nucleus, and activated JAK2, STAT1, and STAT5 are found in proximity to this cluster [16]. STAT3 is also part of the JAK-STAT signaling pathway and has heretofore not been localized in human eosinophils [45]. After activation with IL5, LOCGEF was concentrated in the nucleopod tip, partially overlapping with STAT3 that was similarly concentrated in the nucleopod (Figure 4B). LOCGEF, but not STAT3, also was enriched in apparent membrane protrusions at the opposite pole of the cell. Treatment with IL33 or CCL11 resulted in less dramatic polarization, i.e., movement of the nucleus into to one side of the cell without formation of a distinct nucleopod tip. LOCGEF was enriched in the half of the cells containing the nucleus and in the membrane protrusions at the opposite end (Figure 4C–D). To assess the significance of relocalization during activation, LOCGEF was co-stained with RHOA or RAC2, the major Rho and Rac family members in eosinophils [13]. RHOA had a peri-membranous distribution in unactivated eosinophils and became enriched along with LOCGEF (arrow) in the nucleopod and in protrusions at the opposite pole (arrowhead). In contrast, RAC2, as described previously in guinea-pig eosinophils [46], localized to granules and moved with the granules to one side on the cell upon activation. RAC2 did not co-localize with LOCGEF in either unactivated or activated eosinophils.
Figure 4. Immunofluorescence of LOCGEF in relation to RHOA, RAC2 or STAT3 in unstimulated (A), IL5- (B), IL33- (C) or CCL11-stimulated (D) eosinophils.
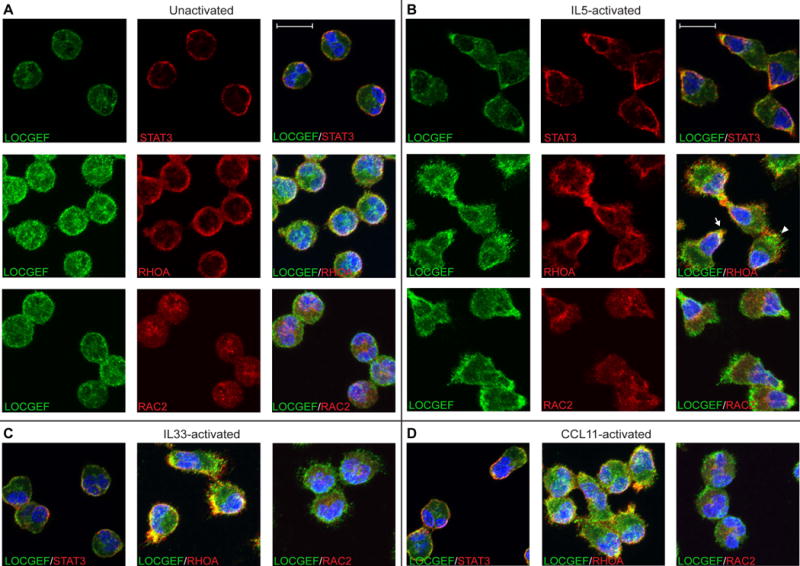
Cells were fixed in suspension, cytospun, stained, and examined by confocal microscopy. DAPI was used to stain the eosinophil nuclei. For STAT3 staining, single 0.3-μm slices taken 0.9-μm above the surface of the coverslip are shown. All other images are maximum intensity projections of 11-15 z-series slices. The arrow points to the nucleopod tip of an IL5-activated eosinophil, the arrowhead to the opposite pole. The scale bar is 10 μm.
LOCGEF orthologues
To identify model organisms amenable for genetic study of LOCGEF, we interrogated translated nucleotide collection data to identify sequences with homology to the 322 N-terminal residues of human LOC100996504 in other species. Pairwise sequence alignment revealed conservation of the “LOC” sequence in vertebrates, and examination of mouse and zebrafish genomes revealed conserved synteny of L1-L10 and 1-20 exons encoding full-length LOCGEF (NCBI’s “LOC100996504” and ARHGEF18 orthologue sequences in Item IV of Supp Data). The mouse equivalent of LOC100996504, A430078G23Rik, is upstream of the gene encoding ARHGEF18 on chromosome 8. Furthermore, A430078G23Rik is highly-expressed in T-lymphocytes, Natural Killer cells and various other leukocytes and their progenitors, including eosinophils [47]. Mapping of RNA-Seq data from DBA/2J x C57BL/6J mouse thymus [29] predicts 1405-residue and 1344-residue LOCGEF isoforms. We sequenced amplicons from mouse eosinophil cDNA to verify the sequences predicted by modeling (putative protein sequence given in Item V in Supp Data).
Discussion
Herein, we demonstrate that the major isoform of ARHGEF18 in human eosinophils is a 1361-residue protein, LOCGEF-X3, comprising the 1015-residues of p114 isoform and an additional 346 N-terminal residues. We further show that there are transcripts for two other potential extended ARHGEF18 isoforms, LOCGEF-X5 and LOCGEF-X4, differing at the extreme N-terminus. RNA-Seq and proteomics data indicate that LOCGEF isoforms are unique to leukocytes whereas the p114 isoform is ubiquitous in non-hematopoietic tissues. The p114 isoform was not detected in eosinophils either as transcript or protein.
Expression of the LOCGEF and p114 isoforms is presumably dictated by factors targeting two possible transcriptional start sites in exon L1 (for LOCGEF isoforms) and exon 1 (p114). The transcriptional start sites match ChIP-Seq data showing transcription factor clustering. PU.1 (SPI1), found by ChIP-Seq to bind close to exon L1 along with c-Myc and Max (Figure 1A), is an important regulator of hematopoiesis [48]. Eosinophils are specified early in hematopoiesis [14], and PU.1 is upregulated in eosinophil progenitor cells [49]. PU.1 is also abundant in differentiated peripheral blood eosinophils [13]. In contrast, the transcription factors predicted to bind in the vicinity of exon 1 of the shorter ARHGEF18 isoform (p114) are more widely-expressed [50]. Inasmuch as LOCGEFs may participate in various Rho GTPase-regulated events that are important during hematopoiesis (reviewed in [51]), it will be of interest to study the contribution of LOCGEF function during hematopoietic differentiation as well as in differentiated hematopoietic cells.
The discrepancy between immunoblotting, which demonstrated no differences in abundance of LOCGEF in unstimulated and IL5-stimulated eosinophils, and mass spectrometry of isobaric labeled peptides, which reported such a difference for LOC10096504, prompted us to review the spectra of the two LOC100996504 peptides and 24 other ARHGEF18 peptides that were found in our analysis [13]. Of the 26 peptides, the only one that had greater than 2-fold absolute fold change was SCESWR in the “LOC” region, encompassing residues 85-91 of LOCGEF-X3. This peptide was 10.7-fold higher in resting eosinophils compared to IL5-activated eosinophils. Isobaric labeling entails a workflow in which eosinophil samples are subjected individually to freeze-thaw cycles and sonication followed by trypsinization and labeling of tryptic peptides by one of a set of 10 isobaric tags. No detergent was introduced to disrupt protein-membrane interactions during trypsin digestion. In a Kyte-Doolittle plot [52] of LOCGEF-X3, SCESWR is in a hydrophobic region of LOCGEF. We speculate that this region associates with remodeling membrane in activated eosinophils and thus becomes resistant to trypsinization, thus explaining the decrement of SCESWR in digests of activated cells and discrepancy between immunoblotting and mass spectrometry. It should be stressed that the Kyte-Doolittle plot of LOCGEF-X3 did not reveal a stretch of hydrophobic residues long enough to transverse a membrane. This potentiality is attributed in UniProt to residues 342-362 of 382-residue LOC100996504, i.e., residues that are not present in the 322-residue segment of LOC100996504 found in LOCGEF-X3.
SMART domain prediction software [53] picked out the Dbl homology (DH) and pleckstrin homology (PH) domains that comprise a previously-established RhoGEF domain in the ARHGEF18 segment of the LOCGEFs. The “LOC” sequence was not identified as having significant homology with a known domain. There are over 70 recognized RhoGEFs with a diversity of N-terminal domains, many of which influence function (reviewed in [54]). The presence of a unique “LOC” domain at the N-terminus of LOCGEFs is consistent with the design of other RhoGEFs.
The functions of the p114 isoform of ARHGEF18 in regulation of actin cytoskeletal organization and maintenance of apico-basal polarity have been studied in a variety of non-hematopoietic cells, as described in the Introduction. Therefore, we localized LOCGEF in eosinophils before and after polarization induced by agonists that signal via three different types of receptors—JAK/STAT-associated receptors in the case of IL5 [16], G-protein-coupled receptors in the case of CCL11 [55], and TNF-type receptors in the case of IL33 [56]. All three agonists caused relocalization of LOCGEF and RHOA from peri-membranous distributions around the circumference of unstimulated eosinophils to the two poles of polarized cells. Shape change and formation of the nucleopod in response to IL5 are blocked by cytochalasin, indicating a requirement for remodeling of the actin cytoskeleton [16]. The relocalization of LOCGEF to the two poles of eosinophil upon activation suggest a role for LOCGEF in modulation of such cytoskeletally-driven shape change [16]. Eosinophil polarization is important for adherence of circulating eosinophils to inflamed vessels in intravital experiments [57], and RHOA and RHOA-associated coiled-coil kinase 2 (ROCK2) have previously be shown to have roles in uropod detachment of eosinophils and neutrophils during migration [58, 59]. We propose, therefore, that just as ARHGEF18-RHOA-ROCK2-MLC signaling axis regulates actomyosin contractility in other cell types [9, 11], LOCGEF acts upon RHOA in eosinophils to drive shape change prior to eosinophil adhesion and movement of eosinophils upon adhesion. We note that LOCGEF is not the only RHOA GTPase-regulating guanine nucleotide exchange factor in eosinophils—ARHGEF1 and 2 are both present in equal abundance [13], and it is likely the LOCGEF cooperates with the others in spatiotemporal regulation of RHOA activity. In contrast, there was no overlap of LOCGEF with RAC2, which is a leukocyte-specific guanine nucleotide exchange factor that regulates granular NADPH oxidase and superoxide formation [46], in the three stimulatory conditions. Rather, the distribution of RAC2 is suggestive of a role regulating actin scaffolding in the granulomeric region.
Further studies of LOCGEF protein expression in hematopoietic cells of mice and zebrafish will be required for determining if these are suitable model organisms in which to assess functions of LOCGEFs. Sites reporting knock-outs of canonical ARHGEF18 in mice report pre-weaning lethality (European Mouse Mutant Archive, MGI:2142567). Lining up the 1405-residue mouse and 1361-residue human LOCGEFs, 230 of the 322 N-terminal residues were identical, and only three gaps needed to be introduced (data not shown). Murine LOCGEF has 38 extra N-terminal residues compared to the human LOCGEF-X3, with the initiating methionine in exon L1 rather than in L2. It should be feasible to do selective knock-down of LOCGEF in leukocytes and leave canonical p114 ARHGEF18 intact.
Supplementary Material
Summary sentence.
A set of ARHGEF18 isoforms is specific for eosinophils and other leukocytes.
Acknowledgments
The authors have been supported by National Institute of Health grants P01 HL088594 (Nizar Jarjour, PI) and R01 AI125390 (Deane Mosher and Josh Coon, co-PIs). Emily Wilkerson received training grant support from T32 HL007899. We used the Biochemistry Optical Core at UW-Madison, and are grateful to Elle Grevstad for her assistance. We thank Doug Annis for help with molecular biology and protein chemistry. Mats Johansson, Doug Annis and Valeriu Bortnov provided useful comments on the manuscript. Finally, we thank Zhong-Jian Shen for providing cDNA from mouse eosinophils.
Abbreviations
- ARHGEF18
Rho guanine nucleotide exchange factor 18
- BLAST
Basic local alignment search tool
- BSA
bovine serum albumin
- βME
β-mercaptoethanol
- cDNA
complementary DNA
- ChIP-Seq
chromatin immunoprecipitation-sequencing
- DAPI
4′,6-diamidino-2-phenylindole
- dbSNP
database of single nucleotide polymorphisms
- GEF
guanine nucleotide exchange factor
- HRP
horse radish peroxidase
- HSA
human serum albumin
- IBAQ
intensity based absolute quantification
- IL5
interleukin-5
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- JAK
Janus kinase
- MAPK
mitogen-activated protein kinase
- MGI
Mouse Genome Informatics
- MLC
myosin light chain
- NCBI
National Center for Biotechnology Information
- PBS
phosphate buffered saline
- PSI
percentage spliced in
- PSM
peptide-sequence match
- PVDF
polyvinylidene fluoride
- RT-PCR
reverse transcription polymerase chain reaction
- SDS
sodium dodecyl sulfate
- STAT
signal transducer and activator of transcription
- TMT
Tandem Mass Tag
Footnotes
Authorship
KBT: conception, design, experiments, data analysis, writing manuscript
EMW: design, experiments, data analysis, writing manuscript
ASH: data analysis
FJF: design, experiments, data analysis
HMS: design, experiments, data analysis
FEB: design, experiments, data analysis
JJC: conception, design, data analysis
DFM: conception, design, data analysis, writing manuscript
Conflicts of interest disclosure
The authors declare no conflict of interest.
References
- 1.Blomquist A, Schworer G, Schablowski H, Psoma A, Lehnen M, Jakobs KH, Rumenapp U. Identification and characterization of a novel Rho-specific guanine nucleotide exchange factor. Biochem J. 2000;352(Pt 2):319–25. [PMC free article] [PubMed] [Google Scholar]
- 2.Niu J, Profirovic J, Pan H, Vaiskunaite R, Voyno-Yasenetskaya T. G Protein betagamma subunits stimulate p114RhoGEF, a guanine nucleotide exchange factor for RhoA and Rac1: regulation of cell shape and reactive oxygen species production. Circ Res. 2003;93:848–56. doi: 10.1161/01.RES.0000097607.14733.0C. [DOI] [PubMed] [Google Scholar]
- 3.Artym VV, Swatkoski S, Matsumoto K, Campbell CB, Petrie RJ, Dimitriadis EK, Li X, Mueller SC, Bugge TH, Gucek M, Yamada KM. Dense fibrillar collagen is a potent inducer of invadopodia via a specific signaling network. J Cell Biol. 2015;208:331–50. doi: 10.1083/jcb.201405099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herder C, Swiercz JM, Muller C, Peravali R, Quiring R, Offermanns S, Wittbrodt J, Loosli F. ArhGEF18 regulates RhoA-Rock2 signaling to maintain neuro-epithelial apico-basal polarity and proliferation. Development. 2013;140:2787–97. doi: 10.1242/dev.096487. [DOI] [PubMed] [Google Scholar]
- 5.Nagata K, Inagaki M. Cytoskeletal modification of Rho guanine nucleotide exchange factor activity: identification of a Rho guanine nucleotide exchange factor as a binding partner for Sept9b, a mammalian septin. Oncogene. 2005;24:65–76. doi: 10.1038/sj.onc.1208101. [DOI] [PubMed] [Google Scholar]
- 6.Terry SJ, Elbediwy A, Zihni C, Harris AR, Bailly M, Charras GT, Balda MS, Matter K. Stimulation of cortical myosin phosphorylation by p114RhoGEF drives cell migration and tumor cell invasion. PloS One. 2012;7:e50188. doi: 10.1371/journal.pone.0050188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tornavaca O, Chia M, Dufton N, Almagro LO, Conway DE, Randi AM, Schwartz MA, Matter K, Balda MS. ZO-1 controls endothelial adherens junctions, cell-cell tension, angiogenesis, and barrier formation. J Cell Biol. 2015;208:821–38. doi: 10.1083/jcb.201404140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X, Jin D, Durgan J, Hall A. LKB1 controls human bronchial epithelial morphogenesis through p114RhoGEF-dependent RhoA activation. Mol Cell Biol. 2013;33:2671–82. doi: 10.1128/MCB.00154-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terry SJ, Zihni C, Elbediwy A, Vitiello E, Leefa Chong San IV, Balda MS, Matter K. Spatially restricted activation of RhoA signalling at epithelial junctions by p114RhoGEF drives junction formation and morphogenesis. Nat Cell Biol. 2011;13:159–66. doi: 10.1038/ncb2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaritsky A, Tseng YY, Rabadan MA, Krishna S, Overholtzer M, Danuser G, Hall A. Diverse roles of guanine nucleotide exchange factors in regulating collective cell migration. J Cell Biol. 2017;216:1543–1556. doi: 10.1083/jcb.201609095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim M, A MS, Ewald AJ, Werb Z, Mostov KE. p114RhoGEF governs cell motility and lumen formation during tubulogenesis through a ROCK-myosin-II pathway. J Cell Sci. 2015;128:4317–27. doi: 10.1242/jcs.172361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arno G, Carss KJ, Hull S, Zihni C, Robson AG, Fiorentino A, Consortium, U. K. I. R. D. Hardcastle AJ, Holder GE, Cheetham ME, Plagnol V, Consortium, N. B.-R. D. Moore AT, Raymond FL, Matter K, Balda MS, Webster AR. Biallelic Mutation of ARHGEF18, Involved in the Determination of Epithelial Apicobasal Polarity, Causes Adult-Onset Retinal Degeneration. Am J Hum Gen. 2017;100:334–342. doi: 10.1016/j.ajhg.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkerson EM, Johansson MW, Hebert AS, Westphall MS, Mathur SK, Jarjour NN, Schwantes EA, Mosher DF, Coon JJ. The Peripheral Blood Eosinophil Proteome. J Proteome Res. 2016;15:1524–33. doi: 10.1021/acs.jproteome.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori Y, Iwasaki H, Kohno K, Yoshimoto G, Kikushige Y, Okeda A, Uike N, Niiro H, Takenaka K, Nagafuji K, Miyamoto T, Harada M, Takatsu K, Akashi K. Identification of the human eosinophil lineage-committed progenitor: revision of phenotypic definition of the human common myeloid progenitor. J Exp Med. 2009;206:183–93. doi: 10.1084/jem.20081756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spencer LA, Weller PF. Eosinophils and Th2 immunity: contemporary insights. Immunol Cell Biol. 2010;88:250–6. doi: 10.1038/icb.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han ST, Mosher DF. IL-5 induces suspended eosinophils to undergo unique global reorganization associated with priming. Am J Resp Cell Mol. 2014;50:654–64. doi: 10.1165/rcmb.2013-0181OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansson MW, Annis DS, Mosher DF. alpha(M)beta(2) integrin-mediated adhesion and motility of IL-5-stimulated eosinophils on periostin. Am J Resp Cell Mol. 2013;48:503–10. doi: 10.1165/rcmb.2012-0150OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breuza L, Poux S, Estreicher A, Famiglietti ML, Magrane M, Tognolli M, Bridge A, Baratin D, Redaschi N, UniProt C. The UniProtKB guide to the human proteome. Database. 2016 doi: 10.1093/database/bav120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.Thibaud-Nissen FSA, Murphy T, DiCuccio M, Kitts P. The NCBI Handbook [Internet] Bethesda (MD): National Center for Biotechnology Information (US); 2013. Eukaryotic Genome Annotation Pipeline. [Google Scholar]
- 21.Liu LY, Bates ME, Jarjour NN, Busse WW, Bertics PJ, Kelly EA. Generation of Th1 and Th2 chemokines by human eosinophils: evidence for a critical role of TNF-alpha. J Immunol. 2007;179:4840–8. doi: 10.4049/jimmunol.179.7.4840. [DOI] [PubMed] [Google Scholar]
- 22.Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, Kim T, Misquitta-Ali CM, Wilson MD, Kim PM, Odom DT, Frey BJ, Blencowe BJ. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–93. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- 23.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright HL, Thomas HB, Moots RJ, Edwards SW. RNA-seq reveals activation of both common and cytokine-specific pathways following neutrophil priming. PloS One. 2013;8:e58598. doi: 10.1371/journal.pone.0058598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheynkman GM, Shortreed MR, Frey BL, Smith LM. Discovery and mass spectrometric analysis of novel splice-junction peptides using RNA-Seq. Mol Cell Proteomics. 2013;12:2341–53. doi: 10.1074/mcp.O113.028142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Eberhard C, Gruning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellaker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assuncao JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–94. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dyer KD, Moser JM, Czapiga M, Siegel SJ, Percopo CM, Rosenberg HF. Functionally competent eosinophils differentiated ex vivo in high purity from normal mouse bone marrow. J Immunol. 2008;181:4004–9. doi: 10.4049/jimmunol.181.6.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turton KB, Annis DS, Rui L, Esnault S, Mosher DF. Ratios of Four STAT3 Splice Variants in Human Eosinophils and Diffuse Large B Cell Lymphoma Cells. PloS One. 2015;10:e0127243. doi: 10.1371/journal.pone.0127243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maurer LM, Tomasini-Johansson BR, Ma W, Annis DS, Eickstaedt NL, Ensenberger MG, Satyshur KA, Mosher DF. Extended binding site on fibronectin for the functional upstream domain of protein F1 of Streptococcus pyogenes. J Biol Chem. 2010;285:41087–99. doi: 10.1074/jbc.M110.153692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017;18(1):529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Consortium, E. P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–15. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–48. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, Mathieson T, Lemeer S, Schnatbaum K, Reimer U, Wenschuh H, Mollenhauer M, Slotta-Huspenina J, Boese JH, Bantscheff M, Gerstmair A, Faerber F, Kuster B. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–7. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 39.Pirmoradian M, Budamgunta H, Chingin K, Zhang B, Astorga-Wells J, Zubarev RA. Rapid and deep human proteome analysis by single-dimension shotgun proteomics. Mol Cell Proteomics. 2013;12:3330–8. doi: 10.1074/mcp.O113.028787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larance M, Ahmad Y, Kirkwood KJ, Ly T, Lamond AI. Global subcellular characterization of protein degradation using quantitative proteomics. Mol Cell Proteomics. 2013;12:638–50. doi: 10.1074/mcp.M112.024547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mertins P, Qiao JW, Patel J, Udeshi ND, Clauser KR, Mani DR, Burgess MW, Gillette MA, Jaffe JD, Carr SA. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods. 2013;10:634–7. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Method Mol Biol. 1999;112:531–52. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 43.Choi EN, Choi MK, Park CS, Chung IY. A parallel signal-transduction pathway for eotaxin- and interleukin-5-induced eosinophil shape change. Immunology. 2003;108(2):245–56. doi: 10.1046/j.1365-2567.2003.01565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cherry WB, Yoon J, Bartemes KR, Iijima K, Kita H. A novel IL-1 family cytokine, IL-33, potently activates human eosinophils. J Allergy Clin Immunol. 2008;121(6):1484–90. doi: 10.1016/j.jaci.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burnham ME, Koziol-White CJ, Esnault S, Bates ME, Evans MD, Bertics PJ, Denlinger LC. Human airway eosinophils exhibit preferential reduction in STAT signaling capacity and increased CISH expression. J Immunol. 2013;191(6):2900–6. doi: 10.4049/jimmunol.1300297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lacy P, Mahmudi-Azer S, Bablitz B, Gilchrist M, Fitzharris P, Cheng D, Man SF, Bokoch GM, Moqbel R. Expression and translocation of Rac2 in eosinophils during superoxide generation. Immunology. 1999;98(2):244–52. doi: 10.1046/j.1365-2567.1999.00873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Graaf CA, Choi J, Baldwin TM, Bolden JE, Fairfax KA, Robinson AJ, Biben C, Morgan C, Ramsay K, Ng AP, Kauppi M, Kruse EA, Sargeant TJ, Seidenman N, D’Amico A, D’Ombrain MC, Lucas EC, Koernig S, Baz Morelli A, Wilson MJ, Dower SK, Williams B, Heazlewood SY, Hu Y, Nilsson SK, Wu L, Smyth GK, Alexander WS, Hilton DJ. Haemopedia: An Expression Atlas of Murine Hematopoietic Cells. Stem Cell Rep. 2016;7:571–82. doi: 10.1016/j.stemcr.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voso MT, Burn TC, Wulf G, Lim B, Leone G, Tenen DG. Inhibition of hematopoiesis by competitive binding of transcription factor PU.1. PNAS USA. 1994;91:7932–6. doi: 10.1073/pnas.91.17.7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fulkerson PC. Transcription Factors in Eosinophil Development and As Therapeutic Targets. Front Med. 2017;4:115. doi: 10.3389/fmed.2017.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hurlin PJ, Ayer DE, Grandori C, Eisenman RN. The Max transcription factor network: involvement of Mad in differentiation and an approach to identification of target genes. Cold Spring Harbor Symposia on Quantitative Biology. 1994;59:109–16. doi: 10.1101/sqb.1994.059.01.014. [DOI] [PubMed] [Google Scholar]
- 51.Cancelas JA, Williams DA. Rho GTPases in hematopoietic stem cell functions. Curr Opin Hematol. 2009;16:249–54. doi: 10.1097/MOH.0b013e32832c4b80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–32. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 53.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43:D257–60. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–80. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 55.Zimmermann N, Hogan SP, Mishra A, Brandt EB, Bodette TR, Pope SM, Finkelman FD, Rothenberg ME. Murine eotaxin-2: a constitutive eosinophil chemokine induced by allergen challenge and IL-4 overexpression. J Immunol. 2000;165(10):5839–46. doi: 10.4049/jimmunol.165.10.5839. [DOI] [PubMed] [Google Scholar]
- 56.Pinto SM, Nirujogi RS, Rojas PL, Patil AH, Manda SS, Subbannayya Y, Roa JC, Chatterjee A, Prasad TS, Pandey A. Quantitative phosphoproteomic analysis of IL-33-mediated signaling. Proteomics. 2015;15(2–3):532–44. doi: 10.1002/pmic.201400303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alblas J, Ulfman L, Hordijk P, Koenderman L. Activation of Rhoa and ROCK are essential for detachment of migrating leukocytes. Mol Biol Cell. 2001;12:2137–45. doi: 10.1091/mbc.12.7.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adachi T, Vita R, Sannohe S, Stafford S, Alam R, Kayaba H, Chihara J. The functional role of rho and rho-associated coiled-coil forming protein kinase in eotaxin signaling of eosinophils. J Immunol. 2001;167:4609–15. doi: 10.4049/jimmunol.167.8.4609. [DOI] [PubMed] [Google Scholar]
- 59.Bahaie NS, Hosseinkhani MR, Ge XN, Kang BN, Ha SG, Blumenthal MS, Jessberger R, Rao SP, Sriramarao P. Regulation of eosinophil trafficking by SWAP-70 and its role in allergic airway inflammation. J Immunol. 2012;188(3):1479–90. doi: 10.4049/jimmunol.1102253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


