Abstract
A refinement protocol based on physics-based techniques established for water soluble proteins is tested for membrane protein structures. Initial structures were generated by homology modeling and sampled via molecular dynamics simulations in explicit lipid bilayer and aqueous solvent systems. Snapshots from the simulations were selected based on scoring with either knowledge-based or implicit membrane-based scoring functions and averaged to obtained refined models. The protocol resulted in consistent and significant refinement of the membrane protein structures similar to the performance of refinement methods for soluble proteins. Refinement success was similar between sampling in the presence of lipid bilayers and aqueous solvent but the presence of lipid bilayers may benefit the improvement of lipid-facing residues. Scoring with knowledge-based functions (DFIRE and RWplus) was found to be as good as scoring using implicit membrane-based scoring functions suggesting that differences in internal packing is more important than orientations relative to the membrane during the refinement of membrane protein homology models.
Keywords: Membrane protein structure prediction, homology models, molecular dynamics simulation, implicit membrane model
INTRODUCTION
Proteins have evolved over billions of years to form highly complex structures, which give rise to a diverse range of functions.1,2 The atomistic details of their structure are of great importance to develop a deeper understanding of their function as well as to develop novel pharmaceutical strategies in the treatment of diseases. Experimental structure prediction methods including X-ray techniques,3 Nuclear Magnetic Resonance (NMR) spectroscopy4,5 and cryo-Electron Microscopy (cryo-EM) methods6 have continuously improved over the last decades and have resulted in extensive structural information about proteins. However, the experimental methods of today cannot keep up with the vast number of genes encoding for proteins in living organisms that are being discovered at a rapid rate.
Computational structure predictions is an alternative to overcome the experimental limitations.7 Initially, ab initio methods were developed to perform structure prediction purely by computational sampling from extended chains just based on the information of the amino acid sequence of a given protein along with physical models to capture the energetics of proteins.8–10 Although these methods have succeeded in some cases, they remain computationally expensive and generally do not provide accurate predictions for most proteins.11 Much more successful has been the use of structural templates from known structures.12–14 In homology modeling, template structures are used to predict the structures of a given target protein based on sequence similarity between the template and the target proteins.15 More sophisticated approaches assemble structures in a piecewise fashion using structural fragments from a variety of known structures.16,17
Template-based modeling often results in good models that are at least topologically correct and often approach native structures for part of a given model. Nevertheless, it remains challenging to reach experimental accuracy throughout a given model. To improve model accuracy, refinement methods are being developed that start from homology models and bring them closer to the true native structure. Generally, the idea is that refinement methods either rely on general knowledge about protein structures encoded in statistical potentials18–21 or employ physics-based methods to drive a given homology model towards the native state. Physics-based methods that apply molecular dynamics (MD) simulations with extensive sampling22–28 have been most successful to date in achieving consistent refinement of soluble proteins29, although the best-performing physics-based refinement methods also incorporate statistical potentials for scoring and structure selection.24,25
The structure prediction of membrane proteins follows similar ideas but the still limited number of available experimental structures of membrane proteins hinders accurate template-based modeling. Moreover, refinement methods have not been applied extensively to membrane protein structures even when it is possible to build initial models via homology to known structures. The refinement of membrane protein structures could in principle follow the same protocols used in the refinement of soluble proteins, but it may be expected that sampling methods targeting proteins in aqueous environments do not generate representative ensembles of membrane-interacting proteins. Some studies have combined statistics-based refinement methods based on water soluble proteins with physics-based approaches specific to the membrane proteins and/or experimental results during membrane protein structure refinement.30–33 The combination of homology modeling with the application of various reconstruction techniques for the loop regions can also increase the accuracy of the structures for membrane proteins.34 Moreover, scoring functions based on statistical potentials for membrane proteins have not been developed as extensively as scoring functions for water soluble proteins.35–38 This may impact the ability to identify the most native-like structures from an ensemble of models generated during sampling. However, in one study by Gao et al.39 knowledge-based scoring functions meant for soluble proteins performed well in discriminating membrane as well. One approach to account for the membrane environment during scoring is to use physically motivated implicit membrane models along with atomistic force fields40–42. In past comparisons, such implicit membrane models have performed equally good or better than knowledge based scoring functions,40 suggesting that such scoring functions may be useful for membrane structure prediction and refinement.
Here, we are exploring how an MD-based refinement protocol that has been successful for soluble proteins23,25 could be extended to the refinement of membrane protein structures that were built via homology. We applied a modified protocol where proteins were solvated in explicit lipid bilayers with different lipid types instead of aqueous solvent, but we also compared with simply using only aqueous solvent. As in our previous protocol, we carried out extensive sampling via MD.23,25 Structures from the trajectories were then selected using different scoring functions. We tested membrane-specific scoring functions based on the Heterogeneous Dielectric Generalized Born implicit membrane model, version 3 (HDGBv3)43,44, and the HDGB van der Waals model (HDGBvdW)45 developed by us as well as commonly used knowledge-based scoring functions the Distance-scaled, Finite Ideal-gas REference (DFIRE)38 and side-chain orientation dependent potential derived from Random-Walk reference state (RWplus)36. The HDGB model models the membrane as a variable dielectric continuum based on the generalized Born formalism in combination with a solvent accessible surface area (SASA) approximation for the non-polar solvation free energy. The HDGBvdW model is a recent extension of HDGB that adds a van der Waals term to more accurately account for non-polar attractive interactions within the membrane. The HDGB-based models were tested as scoring functions before and found to perform similarly or better than knowledge-based approaches.40 DFIRE is a widely used distance-dependent knowledge-based statistical potential to discriminate native-like states of proteins. RWplus is another commonly used knowledge based potential using a hybrid model of distance and orientation-dependent potentials derived from the structural databases. Previous studies have established that both RWplus and DFIRE are effective scoring functions in native-like model selection.36,46,47 We otherwise followed our established refinement protocol for soluble proteins, which included averaging of the selected structures and further refinement with respect to their local stereochemistry using the local Protein structure REFinement via Molecular Dynamics (locPREFMD)48 method.
Ideally, we would have liked to test our protocol blindly during the Critical Assessment of protein Structure Prediction (CASP) competition,7 but the number of membrane protein structure targets in CASP has not been sufficient to date. Instead, we applied the refinement protocol to eight integral membrane protein targets (six α-helical and two β-barrel), where native structures were available and where we could build homology models using related structures.
The overall finding is that we were able to achieve a similar level of refinement for the membrane proteins as for soluble proteins. In the following we will explain the protocol in more detail and discuss how different solvent environments and different scoring functions employed during the refinement affected the results.
METHODS
Test systems
We tested our structure refinement protocol with eight membrane protein structures (six α-helical and two β-barrel) with known structures in the Protein Data Bank (PDB):49 aquaporin (PDB ID: 1j4n), bacteriorhodopsin (1py6), outer membrane protein X - OMPX (1qj8), CXCR4 chemokine receptor (3odu), adenosine A2A receptor (3vg9), proteorhodopsin (4hyj), salmonella typhi outer membrane protein F - OMPF (4kr8), and delta opioid receptor (4n6h). Homology models for each protein were generated using structures from homologous proteins. Alignments were obtained from the Position-Specific Iterated Basic Local Alignment Search Tool (PSI-BLAST) web server50 and models were built using MODELLER version 9.1515 based on the sequence alignment provided by the PSI-BLAST server. The generated homology structures used as input here are available upon request. Table 1 provides an overview of the target and template proteins used in the homology modeling along with their reported resolution values in Å. The models from MODELLER were refined further using locPREFMD to improve the local stereochemistry. The resulting structures were the initial homology models used in this study. The homology models built in this manner deviated from the true native structures by Cα root mean square deviation (RMSD) values between 1.9 and 5 Å (see Table 2).
Table 1.
Summary of the template and target proteins.
| Target PDB code |
Secondary structure |
Number of residues |
Target resolution (Å) |
Template PDB code |
Template resolution (Å) |
Sequence indentity (%) |
HL for target (Å) |
HL for homology (Å) |
Lipid type |
|---|---|---|---|---|---|---|---|---|---|
| 1j4n | α-helical | 249 | 2.2 | 1z98 | 2.1 | 44 | 24 | 24 | DMPC |
| 1py6 | α-helical | 227 | 1.8 | 5ahy | 2.2 | 35 | 32 | 29 | DPPC |
| 1qj8 | β-barrel | 148 | 1.9 | 2n2l | - | 44 | 24 | 24 | DMPC |
| 3odu | α-helical | 302 | 2.5 | 4ea3 | 3.0 | 31 | 28 | 27 | DPPC |
| 3vg9 | α-helical | 297 | 2.7 | 4gbr | 4.0 | 32 | 29 | 20 | DLPC |
| 4hyj | α-helical | 258 | 2.3 | 3ddl | 1.9 | 33 | 25 | 23 | DMPC |
| 4kr8 | β-barrel | 340 | 3.1 | 4gcp | 2.0 | 57 | 11 | 10 | DLPC |
| 4n6h | α-helical | 303 | 1.8 | 4ea3 | 3.0 | 60 | 32 | 27 | DPPC |
Resolutions in Å are given for crystal structures as reported from experiment. A resolution value is not available for the solution NMR structure 2n2l. Sequence identities were obtained from the PSI-BLAST Web server. Hydrophobic lengths (HL) were calculated by MEMHLength program for the target protein and homology models after the equilibration in DPPC bilayer. The lipid type indicates the lipids used in the second set of explicit bilayer simulations (see Methods).
Table 2.
Initial values of GDT-HA, RMSD, MolProbity and SphereGrinder and change after refinement from sampling in the presence of DPPC lipid bilayers.
| 1j4n | 1py6 | 1qj8 | 3odu | 3vg9 | 4hyj | 4kr8 | 4n6h | Average | |
|---|---|---|---|---|---|---|---|---|---|
| GDT-HA | |||||||||
| Initial | 63.55 | 59.58 | 51.69 | 43.13 | 43.43 | 47.14 | 66.25 | 62.38 | 54.64±3.33 |
| Refined (Δ) RWplus | 1.61 | 3.86 | 11.65 | 1.32 | −2.18 | 1.59 | −1.25 | 3.63 | 2.53±1.50 |
| Refined (Δ) DFIRE | 1.31 | 3.97 | 10.98 | 1.08 | −2.35 | 1.69 | −1.18 | 3.71 | 2.40±1.44 |
| Refined (Δ) HDGBv3 | 1.21 | 3.42 | 10.98 | 1.82 | −2.44 | 1.91 | −1.18 | 3.21 | 2.37±1.42 |
| Refined (Δ) HDGBvdW | 1.31 | 3.53 | 10.81 | 1.74 | −2.10 | 1.59 | −1.99 | 3.63 | 2.32±1.43 |
| RMSD [Å] | |||||||||
| Initial | 3.52 | 1.87 | 3.53 | 4.97 | 4.69 | 4.13 | 2.07 | 2.08 | 3.36±0.43 |
| Refined (Δ) RWplus | −0.14 | −0.08 | −0.47 | −0.12 | −0.02 | −0.06 | −0.06 | −0.24 | −0.15±0.05 |
| Refined (Δ) DFIRE | −0.11 | −0.08 | −0.47 | −0.11 | −0.02 | −0.06 | −0.05 | −0.24 | −0.14±0.05 |
| Refined (Δ) HDGBv3 | −0.13 | −0.07 | −0.46 | −0.13 | −0.02 | −0.06 | −0.04 | −0.24 | −0.14±0.05 |
| Refined (Δ) HDGBvdW | −0.14 | −0.08 | −0.45 | −0.13 | −0.01 | −0.05 | −0.03 | −0.23 | −0.14±0.05 |
| MolProbity | |||||||||
| Refined RWplus | 0.97 | 0.64 | 0.65 | 1.60 | 1.01 | 0.69 | 0.86 | 0.50 | 0.87±0.12 |
| Refined DFIRE | 0.93 | 0.58 | 0.65 | 1.61 | 0.95 | 0.74 | 0.82 | 0.50 | 0.85±0.12 |
| Refined HDGBv3 | 0.93 | 0.64 | 0.65 | 0.84 | 0.96 | 0.81 | 0.84 | 0.50 | 0.77±0.06 |
| Refined HDGBvdW | 0.91 | 0.64 | 0.65 | 0.71 | 0.93 | 0.74 | 0.84 | 0.53 | 0.74±0.05 |
| SphereGrinder | |||||||||
| Initial | 83.62 | 89.29 | 71.53 | 72.64 | 69.50 | 75.68 | 82.38 | 89.96 | 79.33±2.85 |
| Refined (Δ) RWplus | 0.0 | 0.95 | 12.15 | −1.63 | −0.17 | 1.57 | 3.92 | 2.47 | 2.41±1.52 |
| Refined (Δ) DFIRE | −0.43 | 0.95 | 12.5 | −1.08 | 0.0 | −1.13 | 1.05 | 0.89 | 1.59±1.59 |
| Refined (Δ) HDGBv3 | −0.43 | 0.47 | 11.11 | −0.72 | 0.89 | −0.68 | 1.66 | 1.06 | 1.67±1.38 |
| Refined (Δ) HDGBvdW | 0.0 | 0.47 | 12.15 | −0.9 | −0.17 | −0.91 | 1.81 | 0.71 | 1.65±1.53 |
Values for GDT-HA, RMSD, and SphereGrinder are for models with respect to the native structure. For refined models using different scoring functions for structure selection (RWplus, DFIRE, HDGBv3, HDGBvdW) the changes ΔGDT-HA, ΔRMSD and ΔSphereGrinder relative to the initial values are given. MolProbity scores are only given for the refined models. In addition to individual values, the averages over all eight proteins (1j4n, 1py6, 1qj8, 3odu, 3vg9, 4hyj, 4kr8 and 4n6h) for each row are given in the last column.
System setup for refinement
The initial models were oriented along the membrane with a Monte Carlo (MC) optimization protocol described in previous work40 using the Implicit Membrane Model 1 (IMM1) energy function.51 Once optimally positioned and oriented, each protein was solvated either in cubic boxes of explicit water or in rectangular boxes with an explicit lipid bilayer of either dipalmitoylphosphatidylcholine (DPPC), dimyristoylphosphatidylcholine (DMPC), or dilauroylphosphatidylcholine (DLPC) and surrounded by explicit water on either side using the CHARMM-GUI Membrane Builder module.52–54 The sizes of the aqueous solvent systems varied between 68 and 77 Å allowing at least 9 Å between the proteins and the edge of the box. The lipid bilayer systems varied between 60 and 75 Å for the x- and y-dimensions and between 72 and 110 Å for the z-dimension depending of the protein size. A water layer of 10 Å was presented on either side of the membrane surface. We tested different variations of the refinement protocol with an explicit lipid bilayer, where we used either DPPC for all targets or varied the lipid type based on predicted hydrophobic lengths of the membrane proteins calculated using the MEMHLength program.55 The lipid choice followed the experimental hydrocarbon region thicknesses reported by Kucerka et al.;56 21.7 Å for DLPC, 25.7 Å for DMPC (measured at a temperature of 30˚C), and 28.5 Å for DPPC (measured at a temperature of 50˚C). Hydrophobic lengths were predicted for homology models after equilibration in DPPC lipid bilayer (see Table 1). We also reported the values for the target crystal structure for comparison in Table 1. Hydrophobic lengths for target proteins were predicted to be in a reasonable range between 24 and 32 Å, except for 4kr8. MEMHLength underestimates the thickness of 4kr8 as 11 Å in comparison to the experimental hydrophobic length of OmpF, which was reported to be around 20 Å.57 We note that the resulting values for the homology models significantly underestimate the hydrophobic lengths obtained from the native structures in some cases (see Table 1) as a result of inaccuracies in the homology models. However, we used the hydrophobic lengths predicted from the homology models here to reflect the conditions of blind structure prediction. K+ or Cl− ions were added to the system for charge neutralization. The proteins, lipid, water and any ions were represented in atomistic detail using the CHARMM c36 force field for lipids58 and proteins59 and the TIP3P60 model for water.
The bilayer systems were first subjected to 3,000 steps of energy minimization consisting of 1,500 steps of steepest descent (SD) and 1,500 steps of adopted basis Newton-Raphson (ABNR) algorithms. Minimization was followed by 400 ps equilibration as prescribed by the CHARMM-GUI server.52,53 A six-step equilibration procedure was applied with restraints for proteins, lipids, water molecules and ions. The systems were relaxed gradually by decreasing force constants from 10 to 0.1 kcal/mol/Å2 for protein backbones, from 5.0 to 0.0 kcal/mol/Å2 for protein side chains, from 2.5 to 0.1 kcal/mol/Å2 for water and lipid molecules during the equilibration. For ions, restraints with a force constant of 10 kcal/mol/Å2 were applied for the first 25 ps of the equilibration. Water restraints were applied to prevent water molecules from moving to the hydrophobic core of the bilayer. Lipid restraints were applied to keep lipid tails and head groups in the hydrophobic and polar regions, respectively. The first 50 ps of the equilibration were performed using Langevin dynamics at constant volume and temperature and the rest of the equilibration was run at constant pressure and temperature at a pressure of 1 bar and temperature of 323.15 K for DPPC and 303.15 K for DLPC and DMPC.
The systems in aqueous solvent were neutralized with Na+ or Cl− ions. The difference in cations between the bilayer and aqueous solvent systems reflects default counterions in CHARMM-GUI but is not expected to affect the refinement results. All systems were initially minimized with 50 steps of SD and 500 steps of ABNR algorithms while applying restraints on water and ions with a 5 kcal/mol/Å2 force constant. The systems were further equilibrated for around 30,000 steps by gradually heating to 298 K using restraints on Cα and Cβ atoms with a force constant of 0.5 kcal/mol/Å2. A time step of 1 fs was used for equilibration.
Sampling via molecular dynamics simulations
Molecular dynamics (MD) simulations were carried out to generate conformational ensembles for each target. For each system, ten replicas with a total of 2 μs simulation time were carried out, similar to the amount of sampling that we applied in the refinement of soluble proteins during CASP23. Cα atoms were restrained during the simulations with a force constant of 0.025 kcal/mol/Å2 to avoid large deviations from the initial structures. Lennard-Jones interactions were switched between 10 and 12 Å. The Particle-Mesh Ewald algorithm was used for the calculation of the long range electrostatic potentials. For lipid bilayer simulations, Langevin dynamics was applied with a friction term of 0.01 ps−1 under a semi isotropic NPT ensemble at a temperature of 323 K for DPPC and 303 K for DLPC and DMPC and a pressure of 1 bar using an MC Barostat. Simulations in water were performed using Langevin dynamics with a friction term of 0.01 ps−1 and a temperature of 298 K. Lennard-Jones interactions were applied with a 9 Å cutoff switching between 8 and 9 Å for the simulations in water. The SHAKE algorithm was used to constrain bonds involving hydrogen atoms. All the simulations were performed using Chemistry at HARvard Molecular Mechanics (CHARMM)61 version c42a1 with OpenMM62 on GPU machines. The Multiscale Modeling Tools for Structural Biology (MMTSB) Tool Set63 was used to control the CHARMM simulations and also to carry out analysis of the generated models.
Structure selection via scoring and averaging
The same structure selection and averaging procedure that we used in CASP1025 and CASP1123 was also applied here. For each protein, 20,000 snapshots were extracted and scored. The scores were calculated using HDGBv3 and HDGBvdW-based scoring functions as well as RWplus and DFIRE for comparison. In all cases, energies were calculated for the oriented proteins as extracted from the simulations. For water simulations, the orientations of the models were optimized using the MC-based optimization protocol40 before HDGB scores were calculated. We note that RWplus and DFIRE are orientation-independent since they do not consider a membrane environment. HDGB and HDGBvdW scores were also calculated at the membrane widths matching with the hydrophobic lengths of the proteins calculated using MEMHLength program. Root mean squared deviations from the initial model (iRMSD) were calculated for each snapshot and used as a second scoring criterion. Structures were then filtered to extract those with the smallest normalized energy score and iRMSD values as described in our previous papers.23,25
The selected subsets of structures were then averaged to obtain a single structure. Each average structure was minimized with 1,000 steps of ABNR minimization with 2 kcal/mol/Å2 restraints on Cα and Cβ atoms. In a final refinement step, the locPREFMD procedure was applied to improve the local structural quality without changing the position of the Cα atoms.
RESULTS AND DISCUSSIONS
In this study, we expanded our established protein structure refinement protocol for aqueous proteins to membrane proteins. We compared sampling via MD in aqueous solvent, the standard protocol for soluble proteins, with sampling in the presence of explicit lipid bilayers. When selecting structures from the MD simulations we tested implicit membrane-based scoring functions as well as knowledge-based scoring functions that are commonly used for scoring soluble proteins. The refinement protocol was tested on eight targets where experimental structures are available and homology models could be built using related structures. We tested the protocol with using either DPPC lipids for all targets or adjusting the lipid type based on predicted hydrophobic lengths of the proteins. The main metrics for analyzing the results were Global Distance Test-High Accuracy (GDT-HA) scores and RMSD values after refinement with respect to the native structures. GDT-HA scores calculate the number of residues that are accurately superimposed within a set of short cutoff distances. In contrast to RMSD, GDT-HA is insensitive to large deviations of unstructured regions and it focuses on the similarity in the structured regions. In addition, we also report on MolProbity64 and SphereGrinder65 scores after refinement as additional metrics. The Molprobity score provides a quality assessment for the protein structures using various validation criteria based on known structures. SphereGrinder focuses on correct local packing by evaluating the percentage of residues that are within a certain RMSD value from a reference structure in spheres around each of the residues.
Overall refinement success
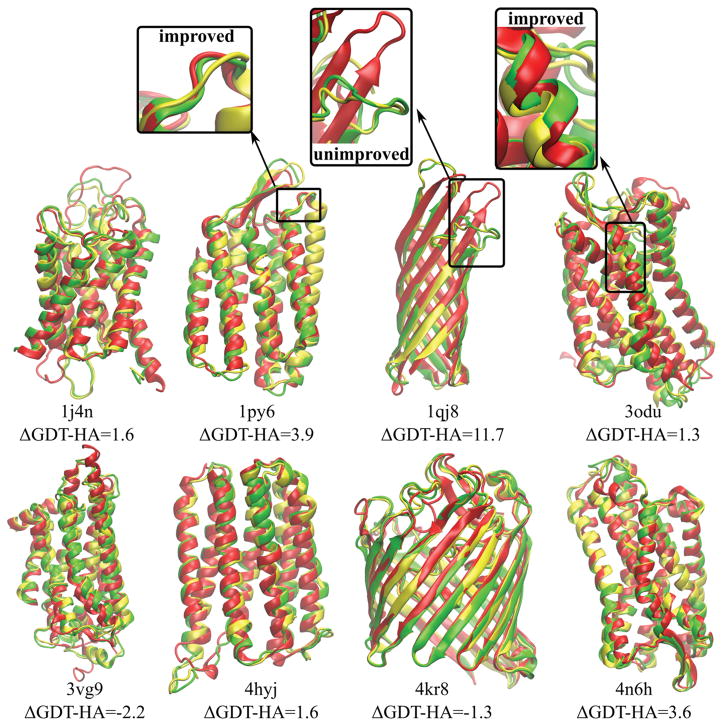
Tables 2–4 summarize the overall results of the refinement protocol for the eight proteins studied in this work. Most structures could be refined with either sampling in aqueous solvent or lipid bilayer environments. On average, the models were refined by 2–3 GDT-HA units and 0.13–0.15 Å Cα RMSD. Refined models have low MolProbity scores (0.74–0.95 on average) and exhibited modest improvements in SphereGrinder scores by 1–2 units. This is similar to what has been achieved in the refinement of soluble proteins23,24 and shows that structure refinement of membrane protein homology models via MD is also possible. The initial homology models and the refined structures, both superimposed onto the native structures, are shown in Figure 1. One structure (1qj8) was improved significantly by 9–12 GDT-HA units and 0.3–0.5 Å Cα RMSD depending on the protocol. For 3vg9, the GDT-HA scores decreased after refinement in all protocol variants. For 4kr8, the GDT-HA scores decreased when sampling involved lipid bilayers. However, the crystal structures of these two proteins have the lowest resolutions (2.7 Å for 3vg9 and 3.1 Å for 4kr8, see Table 1) and therefore, the native reference structure is less reliable than for the other targets.
Table 4.
GDT-HA, RMSD, MolProbity and SphereGrinder after refinement from sampling in the presence of aqueous solvent.
| 1j4n | 1py6 | 1qj8 | 3odu | 3vg9 | 4hyj | 4kr8 | 4n6h | Average | |
|---|---|---|---|---|---|---|---|---|---|
| ΔGDT-HA | |||||||||
| RWplus | 3.62 | 3.97 | 8.95 | 1.74 | −0.42 | 3.28 | 1.62 | 0.08 | 2.86±1.04 |
| DFIRE | 3.82 | 4.52 | 8.61 | 1.99 | −0.42 | 3.92 | 1.03 | 0.33 | 2.98±1.03 |
| HDGBv3 | 3.52 | 5.07 | 9.46 | 1.57 | −0.08 | 3.71 | 0.88 | 0.24 | 3.04±1.21 |
| HDGBvdW | 2.61 | 4.41 | 8.61 | 1.24 | −0.42 | 3.28 | 0.74 | −0.50 | 2.50±1.07 |
| ΔRMSD [Å] | |||||||||
| RWplus | −0.10 | −0.07 | −0.33 | −0.09 | −0.05 | −0.12 | −0.14 | −0.15 | −0.13±0.03 |
| DFIRE | −0.12 | −0.08 | −0.33 | −0.07 | −0.06 | −0.12 | −0.15 | −0.15 | −0.14±0.03 |
| HDGBv3 | −0.14 | −0.10 | −0.32 | −0.06 | −0.05 | −0.10 | −0.12 | −0.14 | −0.13±0.03 |
| HDGBvdW | −0.13 | −0.10 | −0.33 | −0.06 | −0.07 | −0.11 | −0.12 | −0.13 | −0.13±0.03 |
| MolProbity | |||||||||
| RWplus | 1.01 | 0.71 | 0.83 | 1.59 | 0.84 | 0.81 | 0.71 | 0.60 | 0.89±0.11 |
| DFIRE | 0.98 | 0.66 | 0.83 | 1.52 | 0.84 | 0.90 | 0.86 | 0.50 | 0.89±0.10 |
| HDGBv3 | 0.89 | 1.02 | 1.02 | 1.57 | 0.90 | 0.84 | 0.84 | 0.50 | 0.95±0.11 |
| HDGBvdW | 0.84 | 0.89 | 1.05 | 1.59 | 0.92 | 0.84 | 0.91 | 0.50 | 0.95±0.11 |
| ΔSphereGrinder | |||||||||
| RWplus | 0.88 | −1.19 | 7.98 | −1.44 | −0.17 | 2.02 | 4.82 | 2.47 | 1.92±1.13 |
| DFIRE | 0.66 | −0.24 | 7.98 | −1.63 | −0.88 | −1.13 | 1.35 | 1.24 | 0.92±1.08 |
| HDGBv3 | 1.53 | −0.72 | 6.59 | −1.63 | −0.17 | −1.13 | 0.9 | 1.24 | 0.83±0.92 |
| HDGBvdW | 0.66 | −0.72 | 6.59 | −1.44 | −0.35 | −1.36 | 1.51 | 1.77 | 0.83±0.93 |
Changes ΔGDT-HA, ΔRMSD and ΔSphereGrinder after refinement relative to the initial values as in Table 2. Averages for aqueous solvent over all eight proteins (1j4n, 1py6, 1qj8, 3odu, 3vg9, 4hyj, 4kr8 and 4n6h) with statistical uncertainties are given in the last column.
Figure 1.
Native structures of target proteins (red) vs. homology models (yellow) and refined models (green) using the RWplus scoring function for filtering. ΔGDT-HA values indicate the change after refinement from the initial homology models with respect to the native structures. Six proteins out of eight give positive ΔGDT-HA indicating an improvement in the secondary structures. Examples of improved and unimproved structural parts inside the boxes are shown in more detail.
There does not appear to be a strong overall trend with respect to α-helical and β-barrel structures, suggesting that refinement via our protocol is equally suitable for both types of membrane proteins. To understand in more detail, how different secondary structural elements were refined, we calculated per-residue improvements in RMSD values as shown in Table 5. We find that the largest improvements are seen in the unstructured (coil) regions whereas β-sheets and α-helical structures were refined to a lesser degree. This finding is in contrast to our previous work, where we concluded that in the refinement of soluble proteins23 the improvement of unstructured regions was more difficult. It is not entirely clear why we come to different conclusions here, but one explanation could be that the initial homology models for membrane proteins have better-preserved α-helical and β-strand regions than typical homology models for soluble proteins. In support of this argument, initial RMSD values for the unstructured regions are around twice as large as those of the α-helical and β-strand regions (see Table 5). Therefore, there is more room to improve the loops than the α-helical and β-strand regions. It may also be that at least in the simulations with the lipid bilayers, the presence of the lipids hinders rearrangements of the α-helices and β-strands whereas the sampling of unstructured regions that typically face the water are more easily accomplished.
Table 5.
Change in RMSD [Å] after refinement for different protein parts.
| PDB code | Helical | Extended | Coil | Membrane | Water | Ligand binding pocket |
|---|---|---|---|---|---|---|
| Initial models – RMSD [Å] | ||||||
| 1j4n | 1.33 | - | 4.26 | 1.32 | 4.64 | - |
| 1py6 | 1.07 | 3.52 | 2.20 | 0.92 | 2.18 | 0.78 |
| 1qj8 | - | 2.16 | 4.82 | 1.59 | 4.04 | - |
| 3odu | 2.57 | 2.65 | 5.97 | 1.97 | 4.76 | 3.30 |
| 3vg9 | 2.41 | - | 5.43 | 1.91 | 4.60 | 3.31 |
| 4hyj | 2.20 | - | 5.47 | 1.64 | 4.67 | 1.58 |
| 4kr8 | 0.80 | 0.72 | 2.22 | 0.83 | 2.29 | - |
| 4n6h | 1.21 | 1.27 | 2.59 | 0.79 | 2.08 | 1.05 |
| Ave. | 1.66±0.72 | 2.06±1.11 | 4.12±1.56 | 1.37±0.17 | 3.66±0.44 | 2.00+1.22 |
| DPPC bilayer simulations – ΔRMSD [Å] | ||||||
| 1j4n | −0.08 | - | −0.13 | −0.08 | −0.14 | - |
| 1py6 | −0.10 | −0.03 | −0.17 | −0.09 | −0.14 | −0.05 |
| 1qj8 | - | −0.30 | −0.56 | −0.17 | −0.57 | - |
| 3odu | −0.08 | −0.26 | −0.20 | −0.13 | −0.09 | −0.12 |
| 3vg9 | 0.06 | - | −0.02 | −0.06 | 0.13 | −0.02 |
| 4hyj | 0.08 | - | −0.23 | 0.09 | −0.09 | 0.23 |
| 4kr8 | −0.22 | 0.05 | −0.03 | 0.06 | −0.08 | - |
| 4n6h | −0.18 | −0.14 | −0.18 | −0.11 | −0.24 | −0.08 |
| Ave. | −0.07±0.04 | −0.14±0.07 | −0.19±0.06 | −0.06±0.03 | −0.15±0.07 | −0.01±0.06 |
| Water simulations – ΔRMSD [Å] | ||||||
| 1j4n | −0.10 | - | −0.10 | −0.09 | −0.14 | - |
| 1py6 | −0.10 | −0.01 | −0.17 | −0.11 | −0.10 | 0.02 |
| 1qj8 | - | −0.23 | −0.40 | −0.11 | −0.46 | - |
| 3odu | 0.00 | −0.07 | −0.26 | −0.05 | −0.06 | 0.27 |
| 3vg9 | −0.01 | - | −0.03 | −0.06 | 0.03 | −0.05 |
| 4hyj | 0.00 | - | −0.28 | 0.02 | −0.17 | 0.20 |
| 4kr8 | −0.23 | 0.00 | −0.17 | −0.01 | −0.20 | - |
| 4n6h | −0.07 | −0.20 | −0.16 | −0.04 | −0.14 | −0.04 |
| Ave. | −0.07±0.03 | −0.10±0.05 | −0.20±0.04 | −0.05±0.02 | −0.15±0.05 | 0.08±0.07 |
Residue-based RMSD and ΔRMSD values are shown for the initial homology models and final models after refinement. The rows labeled “Ave.” report the average values over proteins for each part of the structures for the initial models and for the refined models in DPPC and in water separately. Homology and final models were superimposed onto the experimental structure using Cα atoms for the calculation of residue based RMSD values. ΔRMSD values for each residue were then calculated by subtracting residue-based RMSD of homology models from that of the final models. Secondary structures were determined using DSSP for the experimental structures and residue-based RMSD values for helical, extended and coil regions were averaged over residues for each region. Residues for the membrane and water regions were determined with respect to the z-positions of the center of mass of each residue. Taking 28.5 Å as the membrane width of DPPC bilayer, the z-positions locate between +14.25 and −14.25 Å were assigned to the membrane region and residues outside of that range were assigned to the water region. The last column shows the change in RMSD for residues with the center of mass within 10 Å distance from the ligand center of mass. The coordinates of ligands were taken from the corresponding target crystal structures.
Effect of simulation environment
Our original refinement protocol was established for water soluble proteins and therefore simulations were performed in a water environment. For membrane proteins, the natural environment consists of lipid membranes. Therefore, we ran MD simulations in the presence of lipid bilayers, but we also compared with simulations that were run in aqueous solvent without a lipid bilayer. The simulations with lipid bilayers were carried out initially with default DPPC bilayers for all proteins, but we also tested whether choosing different lipid types that match the predicted lengths of the hydrophobic regions in the proteins would lead to different results. Based on the MEMHLength method developed by us, we predicted lengths of the hydrophobic regions that varied from 11 to 29 Å (see Table 1) so that there may be a significant mismatch with DPPC bilayers that have a hydrophobic width of 28.5 Å.
The results for simulations with DPPC lipid bilayers are given in Table 2. Refinement with lipid bilayers with different lipid types are compared with the DPPC results in Table 3, and results with only aqueous solvent are given in Table 4. Overall, the extent of refinement is very similar. There are slightly higher improvements in GDT-HA scores when sampling the protein structures in water only than in lipid bilayers, and the improvements in RMSD and SphereGrinder metrics are slightly lower in water while MolProbity scores are also slightly better in the refined models from the bilayer simulations. However, the differences are small compared to the statistical uncertainties. Replacing DPPC with shorter lipids for proteins with short hydrophobic lengths did also not significantly change the overall results, although the results for individual proteins did vary (see Table 3). Therefore, the main conclusion is that, at least within the context of our refinement protocol, the choice of the environment during the MD simulations is not critical.
Table 3.
ΔGDT-HA, ΔRMSD, MolProbity and ΔSphereGrinder values after refinement from sampling in the presence of lipid bilayers with different membrane widths.
| 1j4n | 1qj8 | 3vg9 | 4hyj | 4kr8 | Ave. | Ave. DPPC | |
|---|---|---|---|---|---|---|---|
| ΔGDT-HA | |||||||
| RWplus | −1.90 | 12.67 | −2.35 | 3.28 | −2.94 | 1.75±2.95 | 2.28±2.46 |
| DFIRE | −1.80 | 13.01 | −2.44 | 3.60 | −2.35 | 2.00±2.97 | 2.09±2.35 |
| HDGBv3 | −2.10 | 12.33 | −1.34 | 3.50 | −2.35 | 2.01±2.79 | 2.10±2.36 |
| HDGBvdW | −2.51 | 12.33 | −1.85 | 3.60 | −1.69 | 1.98±2.81 | 1.92±2.36 |
| ΔRMSD [Å] | |||||||
| RWplus | −0.08 | −0.47 | −0.07 | −0.15 | −0.04 | −0.16±0.08 | −0.15±0.08 |
| DFIRE | −0.08 | −0.49 | −0.07 | −0.14 | −0.05 | −0.17±0.08 | −0.14±0.08 |
| HDGBv3 | −0.09 | −0.47 | −0.08 | −0.14 | −0.04 | −0.16±0.08 | −0.14±0.08 |
| HDGBvdW | −0.08 | −0.47 | −0.07 | −0.14 | −0.04 | −0.16±0.08 | −0.14±0.08 |
| MolProbity | |||||||
| RWplus | 0.95 | 0.53 | 0.99 | 0.87 | 0.68 | 0.80±0.09 | 0.84±0.07 |
| DFIRE | 0.91 | 0.65 | 1.02 | 0.87 | 0.71 | 0.83±0.07 | 0.82±0.06 |
| HDGBv3 | 0.95 | 0.53 | 0.90 | 0.84 | 0.71 | 0.79±0.08 | 0.84±0.05 |
| HDGBvdW | 0.95 | 0.53 | 1.02 | 0.87 | 0.71 | 0.82±0.09 | 0.81±0.05 |
| ΔSphereGrinder | |||||||
| RWplus | −0.21 | 9.03 | 0.18 | 1.8 | 3.92 | 2.94±1.67 | 3.49±2.29 |
| DFIRE | −0.21 | 9.72 | 0.18 | −0.23 | 0.9 | 2.07±1.92 | 2.40±2.55 |
| HDGBv3 | 0.0 | 8.33 | 0.89 | −0.23 | 0.45 | 1.89±1.62 | 2.51±2.19 |
| HDGBvdW | −0.43 | 9.03 | 1.07 | −0.23 | 0.6 | 2.01±1.78 | 2.58±2.43 |
Lipid types were selected according to the hydrophobic length of the homology model (see ‘Lipid Type’ in Table 1). Average values over the five proteins with varying lipid types are compared (column labeled by Ave.) with averages for DPPC simulations (Ave. DPPC) for the same set of proteins (1j4n, 1qj8, 3vg9, 4hyj and 4kr8).
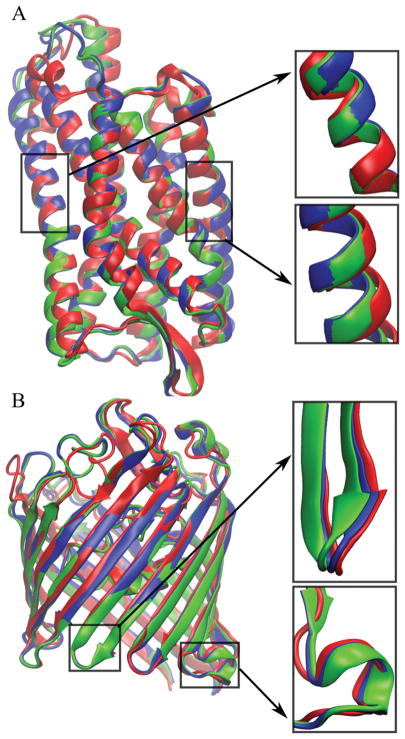
We would have expected an advantage of running the MD simulations with lipid bilayers as the natural environment of the membrane environments. To understand better why the overall results did not bear this out, further analysis was carried out. Table 5 shows the improvements in per-residue RMSD values for residues within the membrane bilayer and in the water phase. Generally, the structural improvements were greater in the water-exposed parts of the structures Average improvements were −0.15 Å for residues in water vs. −0.06 Å in the membrane with little overall difference between the bilayer and water simulations. However, the targets for which there were significant improvements in the membrane region (1j4n, 1py6, 1qj8, 3odu, 4n6h), based on average per-residues RMSD improvements of about −0.1 or better, the improvement was greater in the bilayer simulations. Especially 1qj8, 3odu, and 4n6h were refined more in the membrane region in the bilayer simulations. On the other hand, for targets 4hyj and 4kr8, there were larger improvements for water-exposed residues in the simulations without a bilayer. Figure 2 shows examples of improved parts for 4n6h and 4kr8 with both DPPC and water simulations. For 4n6h, the DPPC simulations provide more improvements in the membrane region, while for 4kr8, the water region was improved more with simulations using aqueous solvent only. It appears, therefore, that lipid bilayer environments may offer an advantage for the refinement of lipid-facing regions. However, for some targets (4hyj and 4kr8), where neither water nor lipid bilayer environments led to improvements in lipid-facing residues, the structures actually became worse in the presence of the lipid bilayer. It is possible that the slow kinetics of rearrangements of structural elements within the membrane bilayer plays a role here and that much longer simulations are needed to realize improvements in the lipid-facing residues for these residues when simulating in the presence of a lipid bilayer. We also note, that, at least for the α-helical bundle 4hyj, the use of DMPC instead of DPPC to match a shorter hydrophobic length leads to more significant improvements in the structure (see Table 3). Based on this analysis, one additional conclusion may be that more significant refinement of lipid-facing residues in membrane proteins is more likely when the MD simulations are carried out with lipid bilayers.
Figure 2.
Structures of target proteins (red), final models from the DPPC bilayer simulations (green), final models from the water simulations (blue) after the refinement using the RWplus scoring function for 4n6h (A) and 4kr8 (B). Details of the differences selected structure parts in the membrane and water regions are shown in the boxes.
We also performed a residue-based RMSD analysis for ligand binding sites of proteins that have ligands in the target crystal structures to determine whether those residues could be refined as much as the rest of the structure in the absence of such ligands. Table 5 shows that, on average, the ligand binding site environment did not change when sampling in the presence of a lipid bilayer and actually became worse with the aqueous solvent simulations. This is in contrast to modest improvements in the rest of the residues in both, bilayer and water environments as discussed above. Similar results were obtained in an earlier study, where even in the presence of ligands, refinement of the ligand binding site environment in G-protein coupled receptors (GPCRs) was less successful than other parts of the structures.66 This suggests that it is challenging to refine more flexible parts of membrane proteins, such as ligand binding sites of GPCRs.
Effect of scoring function
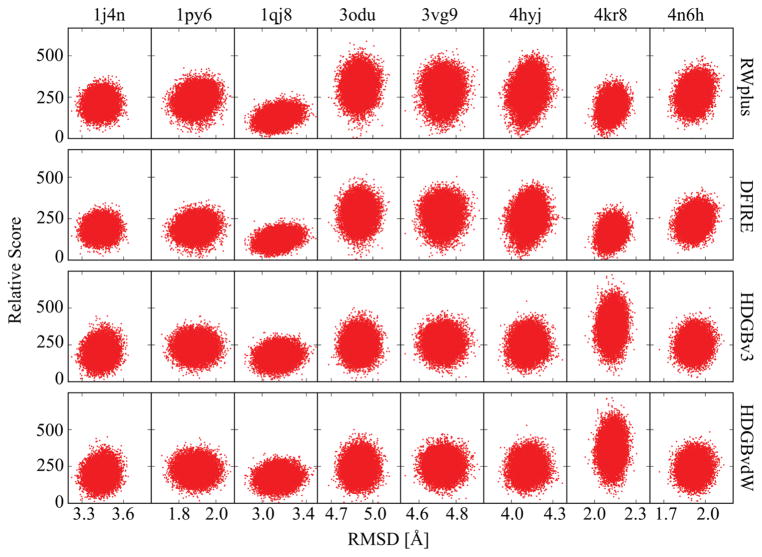
The refinement protocol involves the selection of snapshots from the MD sampling for subsequent structure averaging. Unexpectedly, there is not a large difference with different scoring functions (see Tables 2–4). In particular, the knowledge-based scoring functions RWplus and DFIRE performed as good as or better than the HDGB-based functions but the differences are again not significant when considering the statistical uncertainties. The HDGBv3 and HDGBvdW variants performed similarly well and we did not find a significant effect of using either a membrane width corresponding to DPPC for all targets or using membrane widths corresponding to the predicted hydrophobic lengths of each protein (see Tables S1 and S2). The apparent lack of sensitivity of the results to the scoring function is an interesting finding. This suggests that at least in the context of our refinement protocol, the consideration of the membrane environment is not the most essential factor for selecting structures for ensemble averaging. One explanation may be that the use of restraints keeps all of the generated structures in sufficiently similar orientations and conformations relative to the membrane and the key distinguishing factor between different snapshots may be subtleties of internal packing arrangements, which is captured relatively well with knowledge-based potentials such as DFIRE and RWplus.67 To further analyze this point, we compared the scoring of all decoys extracted from the simulations vs. RMSD from the native structure. The resulting scatter plots are shown in Figure 3 and correlation coefficients for the relation between scores and RMSD are given in Table 6. Overall, there is not a strong correlation, which may be expected because of the limited sampling in the presence of weak positional restraints. However, to the degree that there is any correlation, RWplus and DFIRE actually gave significantly higher positive correlations (0.15 vs. 0.05 on average). This contrasts an earlier study, where we scored Rosetta-generated membrane-structure decoys that spanned a larger variety of conformations generated without consideration of the membrane40. In that case, the HDGB-based scoring functions were correlated significantly better with the distance from the native structure than DFIRE.
Figure 3.
Score values vs. Cα RMSD from the native structure for each target with different scoring functions for snapshots extracted from the MD simulations with DPPC lipid bilayers. scattering plots. Scores are given relative to the respective minimum energy values. Energy values for the HDGB-based scores are given in units of kcal/mol.
Table 6.
Correlations between scores and RMSD for the simulation with DPPC.
| 1j4n | 1py6 | 1qj8 | 3odu | 3vg9 | 4hyj | 4kr8 | 4n6h | Average | |
|---|---|---|---|---|---|---|---|---|---|
| RWplus | 0.07 | 0.18 | 0.29 | 0.04 | 0.03 | 0.18 | 0.22 | 0.20 | 0.15±0.03 |
| DFIRE | 0.04 | 0.17 | 0.26 | 0.05 | 0.05 | 0.17 | 0.22 | 0.17 | 0.14±0.04 |
| HDGBv3 | 0.10 | −0.00 | 0.15 | 0.03 | 0.01 | 0.06 | 0.07 | 0.08 | 0.06±0.02 |
| HDGBvdW | 0.09 | −0.02 | 0.13 | 0.02 | 0.01 | 0.05 | 0.07 | 0.06 | 0.05±0.02 |
Spearman’s rank correlations coefficients between scores and RMSD were calculated for each protein. The averages over all proteins are given in the last column with statistical uncertainties.
Amount of sampling vs. refinement
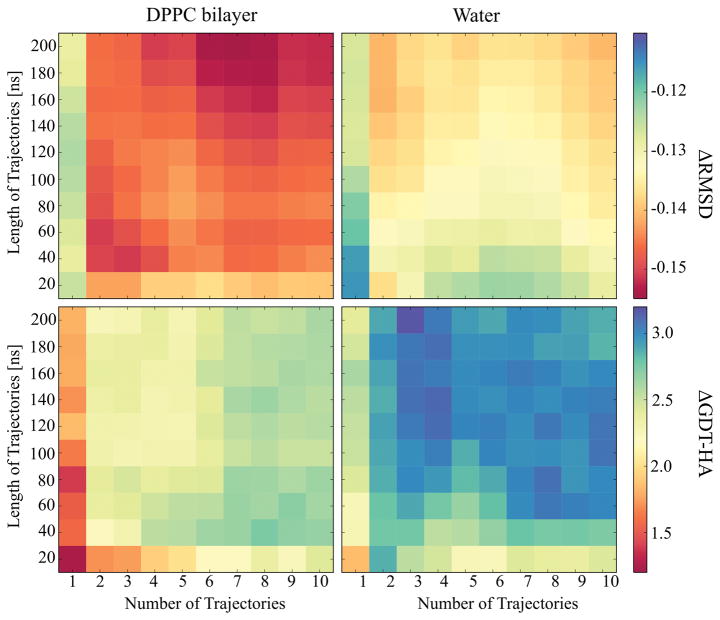
The analysis of our CASP10 and CASP11 results for soluble proteins suggested that combining sampling from multiple simulations provided benefits but that very long simulations did not necessarily increase the success of refinement. In CASP12, however, the analysis of 200 ns long simulations with a maximum of 20 replicas did result in increased GDT-HA scores with longer sampling.46 In the case of membrane proteins, we expected that more sampling may be needed because of the slow relaxation of lipid molecules. Figure 4 shows the changes in average ΔRMSD and ΔGDT-HA values as a function of simulation time and the number of simulations up to a maximum of 10 simulations over 200 ns each. Again, we find that multiple simulations are better than a single simulation and for DPPC simulations, there appears to be an additional benefit of running more than five replicates. In the DPPC simulations, sampling of 40 ns or more per replica gave similar improvements in GDT-HA scores, but RMSD values increased more significantly when simulations exceeded 100 ns. Therefore, it may be that much longer simulations could provide additional benefits for simulations with lipid bilayers. However, the simulations with water show little difference beyond a few replicates simulated for 60 ns or more suggesting that much longer simulations may not offer additional advantages for the membrane proteins studied here.
Figure 4.
Average ΔRMSD (top) and ΔGDT-HA (bottom) values plotted over the simulation time against the number of trajectories for the simulations in DPPC lipid bilayer (left) and in water (right). The RMSD and GDT-HA values were calculated for the final structures using RWplus score for the structure selection and averaged over the proteins.
CONCLUSION
In this study, we applied our refinement protocol for soluble proteins to eight membrane proteins covering both α-helical and β-barrel structures. We find that MD-based refinement of homology models for such membrane proteins is possible and results in similar improvements in terms of GDT-HA scores and RMSD values as seen in the refinement of soluble proteins in previous studies.23,25 Six out of eight proteins were refined, indicating furthermore consistent structural improvements. One structure was refined quite significantly, by around 11.5 units and no structure was made much worse than the initial model. This is also consistent with the refinement of soluble proteins seen previously.23,25
In order to reflect the different environment of membrane proteins, the sampling via MD included explicit lipid bilayers and the scoring of snapshots to select a subset for averaging involved implicit membrane-based scoring functions. We found, however, that within the context of our refinement protocol, sampling in simple aqueous solvent and scoring with knowledge-based functions RWplus and DFIRE resulted in similar degrees of refinement. However, based on a more detailed analysis it appears that the use of explicit bilayer may offer some benefit in the refinement of lipid-facing residues.
Overall, this study confirms the utility of physics-based refinement methods for protein structures and demonstrates that membrane protein structures can be subjected to such protocols with similar success. As in the refinement of soluble proteins, the degree of refinement still remains modest and a key challenge is how to expand sampling to achieve more significant refinement. The use of restraints has been necessary to prevent partial unfolding and larger deviations away from the native structure in longer simulations,22–24,26,46 but the restraints are also limiting how much structures can be refined. Overcoming this challenge is expected to benefit the refinement of soluble proteins as well as the membrane proteins.
Protein structure prediction and refinement of membrane proteins has not been explored as widely as for soluble proteins. The lack of template structures has hindered comparative modeling efforts and CASP has not provided a large number of targets where prediction methods could be tested blindly. We hope that this situation will change as structural biology efforts continue to focus on membrane structures and that there will be expanded opportunities to test and validate structure prediction and refinement methods for membranes such as the methods described here.
Supplementary Material
Acknowledgments
This work was funded by the National Institutes of Health Grant R01 GM084953.
References
- 1.Redfern OC, Dessailly B, Orengo CA. Exploring the structure and function paradigm. Curr Opin Struct Biol. 2008;18:394–402. doi: 10.1016/j.sbi.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whisstock JC, Lesk AM. Prediction of protein function from protein sequence and structure. Q Rev Biophys. 2003;36:307–340. doi: 10.1017/s0033583503003901. [DOI] [PubMed] [Google Scholar]
- 3.Shi YG. A glimpse of structural biology through X-Ray crystallography. Cell. 2014;159:995–1014. doi: 10.1016/j.cell.2014.10.051. [DOI] [PubMed] [Google Scholar]
- 4.Opella SJ, Marassi FM. Structure determination of membrane proteins by NMR spectroscopy. Chem Rev. 2004;104:3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wuthrich K. Protein-structure determination in solution by NMR-spectroscopy. J Biol Chem. 1990;265:22059–22062. [PubMed] [Google Scholar]
- 6.Kuhlbrandt W. Cryo-EM enters a new era. Elife. 2014;3:e03678. doi: 10.7554/eLife.03678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feig M. Computational protein structure refinement: almost there, yet still so far to go. Wires Comput Mol Sci. 2017;7:e1307. doi: 10.1002/wcms.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardin C, Pogorelov TV, Luthey-Schulten Z. Ab initio protein structure prediction. Curr Opin Struct Biol. 2002;12:176–181. doi: 10.1016/s0959-440x(02)00306-8. [DOI] [PubMed] [Google Scholar]
- 9.Lee J, Wu S, Zhang Y. Ab initio protein structure prediction. In: Daniel JR, editor. From protein structure to function with bioinformatics. Dordrecht: Springer; 2009. pp. 3–25. [Google Scholar]
- 10.Liwo A, Lee J, Ripoll DR, Pillardy J, Scheraga HA. Protein structure prediction by global optimization of a potential energy function. Proc Natl Acad Sci USA. 1999;96:5482–5485. doi: 10.1073/pnas.96.10.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonneau R, Baker D. Ab initio protein structure prediction: Progress and prospects. Annu Rev Biophys Biomol Struct. 2001;30:173–189. doi: 10.1146/annurev.biophys.30.1.173. [DOI] [PubMed] [Google Scholar]
- 12.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 13.Blundell TL, Sibanda BL, Sternberg MJE, Thornton JM. Knowledge-based prediction of protein structures and the design of novel molecules. Nature. 1987;326:347–352. doi: 10.1038/326347a0. [DOI] [PubMed] [Google Scholar]
- 14.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 15.Sali A, Blundell TL. Comparative protein modeling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 16.Rohl CA, Strauss CEM, Misura KMS, Baker D. Protein structure prediction using rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on a genomic scale. Proc Natl Acad Sci U S A. 2004;101:7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharya D, Cheng JL. i3Drefine software for protein 3D structure refinement and its assessment in CASP10. PLoS One. 2013;8:e69648. doi: 10.1371/journal.pone.0069648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chopra G, Summa CM, Levitt M. Solvent dramatically affects protein structure refinement. Proc Natl Acad Sci U S A. 2008;105:20239–20244. doi: 10.1073/pnas.0810818105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Barz B, Zhang JF, Xu D, Kosztin I. Selective refinement and selection of near-native models in protein structure prediction. Proteins: Struct Funct Bioinf. 2015;83:1823–1835. doi: 10.1002/prot.24866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu J, Xie L, Honig B. Structural refinement of protein segments containing secondary structure elements: Local sampling, knowledge-based potentials, and clustering. Proteins: Struct Funct Bioinf. 2006;65:463–479. doi: 10.1002/prot.21085. [DOI] [PubMed] [Google Scholar]
- 22.Chen JH, Brooks CL. Can molecular dynamics simulations provide high-resolution refinement of protein structure? Proteins: Struct Funct Bioinf. 2007;67:922–930. doi: 10.1002/prot.21345. [DOI] [PubMed] [Google Scholar]
- 23.Feig M, Mirjalili V. Protein structure refinement via molecular-dynamics simulations: What works and what does not? Proteins: Struct Funct Bioinf. 2016;84:282–292. doi: 10.1002/prot.24871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mirjalili V, Feig M. Protein structure refinement through structure selection and averaging from molecular dynamics ensembles. J Chem Theory Comput. 2013;9:1294–1303. doi: 10.1021/ct300962x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mirjalili V, Noyes K, Feig M. Physics-based protein structure refinement through multiple molecular dynamics trajectories and structure averaging. Proteins: Struct Funct Bioinf. 2014;82:196–207. doi: 10.1002/prot.24336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raval A, Piana S, Eastwood MP, Dror RO, Shaw DE. Refinement of protein structure homology models via long, all-atom molecular dynamics simulations. Proteins: Struct Funct Bioinf. 2012;80:2071–2079. doi: 10.1002/prot.24098. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Liang Y, Zhang Y. Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure. 2011;19:1784–1795. doi: 10.1016/j.str.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu J, Fan H, Periole X, Honig B, Mark AE. Refining homology models by combining replica-exchange molecular dynamics and statistical potentials. Proteins: Struct Funct Bioinf. 2008;72:1171–1188. doi: 10.1002/prot.22005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Modi V, Dunbrack RLJ. Assessment of refinement of template-based models in CASP11. Proteins: Struct Funct Bioinf. 2016;84(Suppl 1):260–281. doi: 10.1002/prot.25048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alford RF, Leman JK, Weitzner BD, Duran AM, Tilley DC, Elazar A, Gray JJ. An integrated framework advancing membrane protein modeling and design. PLoS Comput Biol. 2015;11:e1004398. doi: 10.1371/journal.pcbi.1004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barth P, Wallner B, Baker D. Prediction of membrane protein structures with complex topologies using limited constraints. Proc Natl Acad Sci U S A. 2009;106:1409–1414. doi: 10.1073/pnas.0808323106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leelananda SP, Lindert S. Iterative molecular dynamics-rosetta membrane protein structure refinement guided by Cryo-EM densities. J Chem Theory Comput. 2017;13:5131–5145. doi: 10.1021/acs.jctc.7b00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leman JK, Ulmschneider MB, Gray JJ. Computational modeling of membrane proteins. Proteins: Struct Funct Bioinf. 2015;83:1–24. doi: 10.1002/prot.24703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen KYM, Sun JM, Salvo JS, Baker D, Barth P. High-Resolution Modeling of Transmembrane Helical Protein Structures from Distant Homologues. PLoS Comput Biol. 2014;10:e1003636. doi: 10.1371/journal.pcbi.1003636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Zhang Y. A novel side-chain orientation dependent potential derived from random-walk reference state for protein fold selection and structure prediction. PLoS One. 2010;5:e15386. doi: 10.1371/journal.pone.0015386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou HY, Skolnick J. GOAP: A generalized orientation-dependent, all-atom statistical potential for protein structure prediction. Biophys J. 2011;101:2043–2052. doi: 10.1016/j.bpj.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou HY, Zhou YQ. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao C, Stern HA. Scoring function accuracy for membrane protein structure prediction. Proteins: Struct Funct Bioinf. 2007;68:67–75. doi: 10.1002/prot.21421. [DOI] [PubMed] [Google Scholar]
- 40.Dutagaci B, Wittayanarakul K, Mori T, Feig M. Discrimination of native-like states of membrane proteins with implicit membrane-based scoring functions. J Chem Theory Comput. 2017;13:3049–3059. doi: 10.1021/acs.jctc.7b00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forrest LR, Woolf TB. Discrimination of native loop conformations in membrane proteins: Decoy library design and evaluation of effective energy scoring functions. Proteins: Struct, Func, Genet. 2003;52:492–509. doi: 10.1002/prot.10404. [DOI] [PubMed] [Google Scholar]
- 42.Yuzlenko O, Lazaridis T. Membrane protein native state discrimination by implicit membrane models. J Comput Chem. 2013;34:731–738. doi: 10.1002/jcc.23189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mirjalili V, Feig M. Interactions of amino acid side-chain analogs within membrane environments. J Phys Chem B. 2015;119:2877–2885. doi: 10.1021/jp511712u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanizaki S, Feig M. A generalized Born formalism for heterogeneous dielectric environments: application to the implicit modeling of biological membranes. J Chem Phys. 2005;122:124706. doi: 10.1063/1.1865992. [DOI] [PubMed] [Google Scholar]
- 45.Dutagaci B, Sayadi M, Feig M. Heterogeneous dielectric generalized Born model with a van der Waals term provides improved association energetics of membrane-embedded transmembrane helices. J Comput Chem. 2017;38:1308–1320. doi: 10.1002/jcc.24691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heo L, Feig M. What maked is difficult to refine protein models further via molecular dynamics simulations? Proteins: Struct Funct Bioinf. 2018;86:177–188. doi: 10.1002/prot.25393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olson MA, Lee MS. Application of replica exchange umbrella sampling to protein structure refinement of nontemplate models. J Comput Chem. 2013;34:1785–1793. doi: 10.1002/jcc.23325. [DOI] [PubMed] [Google Scholar]
- 48.Feig M. Local protein structure refinement via molecular dynamics simulations with locPREFMD. J Chem Inf Model. 2016;56:1304–1312. doi: 10.1021/acs.jcim.6b00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lazaridis T. Effective energy function for proteins in lipid membranes. Proteins: Struct, Func, Genet. 2003;52:176–192. doi: 10.1002/prot.10410. [DOI] [PubMed] [Google Scholar]
- 52.Jo S, Kim T, Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One. 2007;2:e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jo S, Lim JB, Klauda JB, Im W. CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu EL, Cheng X, Jo S, Rui H, Song KC, Davila-Contreras EM, Qi YF, Lee JM, Monje-Galvan V, Venable RM, Klauda JB, Im W. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J Comput Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dutagaci B, Feig M. Determination of hydrophobic lengths of membrane proteins with the HDGB implicit membrane model. J Chem Inf Model. 2017;57:3032–3042. doi: 10.1021/acs.jcim.7b00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kucerka N, Nieh MP, Katsaras J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim Biophys Acta, Biomembr. 2011;1808:2761–2771. doi: 10.1016/j.bbamem.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 57.O’Keeffe AH, East JM, Lee AG. Selectivity in lipid binding to the bacterial outer membrane protein OmpF. Biophys J. 2000;79:2066–2074. doi: 10.1016/S0006-3495(00)76454-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Pastor RW. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Best RB, Zhu X, Shim J, Lopes P, Mittal J, Feig M, MacKerell AD., Jr Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. J Chem Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 61.Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eastman P, Friedrichs MS, Chodera JD, Radmer RJ, Bruns CM, Ku JP, Beauchamp KA, Lane TJ, Wang LP, Shukla D, Tye T, Houston M, Stich T, Klein C, Shirts MR, Pande VS. OpenMM 4: A reusable, extensible, hardware independent library for high performance molecular simulation. J Chem Theory Comput. 2013;9:461–469. doi: 10.1021/ct300857j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feig M, Karanicolas J, Brooks CL. MMTSB tool set: Enhanced sampling and multiscale modeling methods for applications in structural biology. J Mol Graphics Modell. 2004;22:377–395. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 64.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lukasiak P, Antczak M, Ratajczak T, Blazewicz J. SphereGrinder - reference structure-based tool for quality assessment of protein structural models. IEEE Int Conf Bioinf Biomed; 2015. pp. 665–668. [Google Scholar]
- 66.Lee GR, Seok C. Galaxy7TM: flexible GPCR-ligand docking by structure refinement. Nucleic Acids Res. 2016;44:W502–W506. doi: 10.1093/nar/gkw360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feig M, Brooks CL., III Evaluating CASP4 predictions with physical energy functions. Proteins. 2002;49:232–245. doi: 10.1002/prot.10217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.