Abstract
Regulated proteolysis is a vital process that affects all living things. Bacteria use energy-dependent AAA+ proteases to power degradation of misfolded and native regulatory proteins. Given that proteolysis is an irreversible event, specificity and selectivity in degrading substrates is key. Specificity is often augmented through the use of adaptors that modify the inherent specificity of the proteolytic machinery. Regulated protein degradation is intricately linked to quality control, cell cycle progression, and physiological transitions. In this review, we highlight recent work that has shed light on our understanding of regulated proteolysis in bacteria. We discuss the role AAA+ proteases play during balanced growth as well as how proteases are mobilized during changes in growth. We present examples of how protease selectivity can be controlled in increasingly complex ways. We promote the concept that coupling a core recognition determinant to one or more modifying agents is a general theme for regulated protein degradation.
Introduction
Regulated protein degradation is a vital process that affects all biological pathways. Because proteolysis is an irreversible event, the cell must take great care to avoid degrading proteins indiscriminately. As a consequence, energy-dependent proteases are finely tuned cellular machines that recognize substrates with exquisite sensitivity and selectivity.
In eukaryotes, proteins targeted for degradation are modified by the covalent linkage of ubiquitin, a small protein that is appended to a lysine residue on the target protein, for review see (1). Following additional extension by the ubiquitin ligase families, the target protein is recognized and degraded by the proteasome. In bacteria, regulated proteolysis is carried out by energy-dependent AAA+ (ATPases associated with cellular activities) proteases that use the power of ATP hydrolysis to recognize, unfold, translocate, and degrade substrates. Several energy-dependent proteases exist in bacteria: Lon, ClpXP, ClpAP, ClpCP, ClpEP, HslUV, and FtsH (2–4). The importance of these AAA+ proteases is highlighted by the defects in viability and virulence of bacteria deficient in one or more proteases (5, 6). For instance, bacteria lacking Lon are known to be filamentous and more sensitive to ultraviolet radiation than their wild-type counterparts (7). In the alpha-proteobacterium Caulobacter crescentus, ClpXP is essential as mutants lacking this protease are arrested during the cell cycle (8). In the human pathogen Vibrio cholerae, Lon mutants were unable to compete with wild-type V. cholerae in colonizing the infant mouse intestine (9).
AAA+ proteases share a similar general architecture being composed of an ATPase and peptidase domain (10). In the case of Lon and FtsH, the two domains are encoded on a single polypeptide chain (11, 12). On the other hand, the Clp family of proteases encodes distinct hexameric ATPases, (either ClpA or ClpX in gram-negative bacteria and ClpC or ClpE in gram-positive bacteria) which associate with ClpP, a sequestered 14-subunit peptidase, to form ClpXP, ClpAP, ClpCP, or ClpEP (2, 13). ClpP alone has the ability to degrade small peptides, but in order to degrade larger, more stable substrates, ClpP must associate with an unfoldase that harvests the energy of ATP hydrolysis to power degradation (13, 14). This is separate from the properties of non-energy dependent proteases and peptidases that serve important recycling roles in the cell (15). The energy dependent AAA+ proteases will be the primary focus of this review.
Operational Rules of Proteolysis
Regardless of architecture or function, bacterial AAA+ proteases seem to follow similar operational rules. In the most general case, regulated proteolysis requires recognition of an initial degradation determinant (also known as degrons) followed by processive degradation of the polypeptide in an ATP dependent manner (Figure 1). The unfolding power and processivity of an AAA+ protease depends on both substrate and protease. For example, poorly folded substrates require less power to unfold (16). In addition, low complexity portions of a substrate protein can stall proteases, resulting in release of partially processed species (17, 18). By appending the same substrate with different degrons, the unfolding and processivity of the known bacterial AAA+ proteases classes were shown to vary over 2 orders of magnitude (19), with Lon being the “worse” unfoldase and ClpAP being the “best”. Because unfolding parameters can vary wildly depending on the specific fold of the substrate, we are cautious in generalizing these results to all substrates, but single molecule experiments have recently shown similar correlations between some of these machines (20–22).
Figure 1.
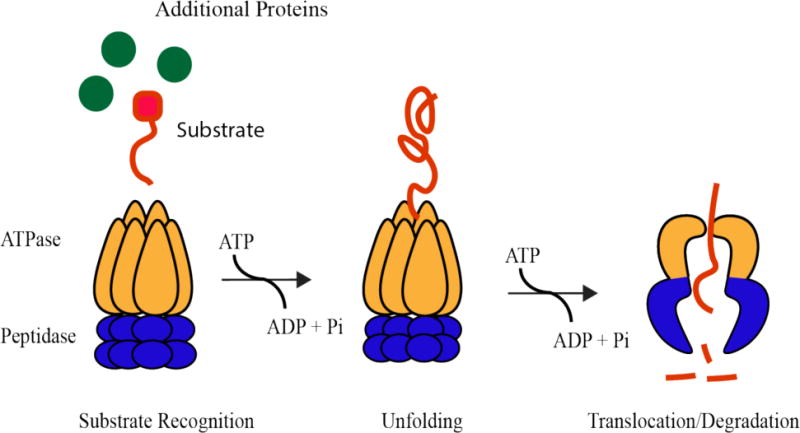
Energy-dependent proteases are composed of an ATP-hydrolysis active unfoldase domain and a chambered peptidase domain. Through successive rounds of ATP hydrolysis, a specific substrate protein is selected by the protease, unfolded by the ATPase domain, and translocated through a central pore to the peptidase chamber where it is degraded.
Due to the processive nature of these proteases, the most important governing feature in vivo is likely the pioneering round of substrate engagement, as once a substrate is committed for degradation it is unfolded and translocated relatively quickly (22). This initial commitment is a combination of the specificity of the protease for a given class of substrates and the ability of those substrates to be recruited to the protease. In order for any AAA+ protease to successfully degrade a substrate, there must be initial recognition of some determinant on the substrate for the protease to start pulling on. This intrinsic recognition can be modified through inhibition or activation by additional factors or the substrates themselves in complex ways dependent on the need of the cell. These needs can include quality control, as damaged or misfolded proteins must be cleared before they elicit toxic effects. On the other hand, energy-dependent proteases are also playing a major role in maintaining protein homeostasis during balanced growth and during physiological transitions, such as stationary phase or sporulation. Although the mechanisms proteases employ for these distinct arms of degradation may differ, the general theme of linking a core recognition determinant to a modifier seems to be common.
Proteases as Quality Control Responders
Bacteria live in a dynamic, constantly fluctuating environment where they are subject to proteotoxic stressors, such as heat or oxidative stress. Because stress conditions require a swift response, regulated proteolysis allows bacteria an effective way to get rid of damaged proteins rapidly, without having to wait for protein removal by dilution through cell division (14, 23). In addition, many stresses lead to the accumulation of misfolded proteins, a problem that needs to be addressed by the cell before lethal consequences ensue. The response to stress can be thought of as a competition between rescuing factors, such as chaperones or repair enzymes, which seek to restore proteins, and proteases, which seek to degrade them (Figure 2a). A central challenge for the cell therefore lies in determining when a protein is terminally damaged or misfolded and when rescue attempts should be abandoned for proteolysis (24).
Figure 2.
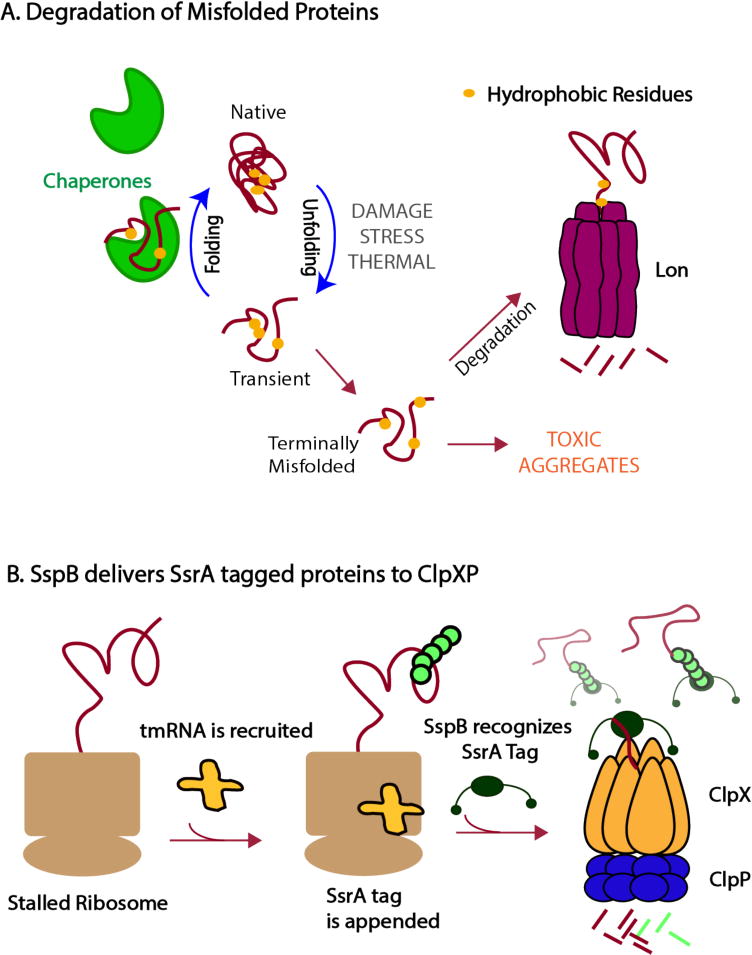
Proteases can survey protein quality in the cell (a) Competition between chaperones and proteases dictate the fate of proteins. Proteases, such as Lon, must be able to distinguish between normal protein dynamics with transient excursions into non-native states and terminally misfolded proteins that must be degraded before forming toxic aggregates. Lon recognizes hydrophobic motifs (shown as yellow circles) that are usually buried in the core of a native protein. These motifs are exposed more persistently for misfolded proteins than during the transient fluctuations of properly folded proteins, allowing Lon to recognize and degrade the terminally misfolded proteins. Chaperones contribute to this flux by binding misfolded proteins in an effort to refold them. (b) Following the process of trans-translation, in which a ssrA-tag is appended to incomplete polypeptides, the adaptor SspB binds tagged substrates and tethers them to ClpXP, enhancing the protease’s ability to degrade these substrates.
Recognizing failed quality products through specific tags: the ClpXP protease
Misfolded proteins are cleared by Lon in bacteria as well as in the mitochondrial matrix in eukaryotes (25, 26). In Escherichia coli, Lon is thought to be responsible for the degradation of approximately 50% of misfolded proteins (27), which suggests that the protease recognizes general motifs in misfolded proteins with little sequence specificity. By contrast, the bacterial ClpXP protease is far more selective and requires a specific degron sequence, such as the ssrA tag, in order to recognize a substrate (28, 29). These two enzymes exemplify different mechanisms used to ensure degradation of poor quality proteins.
One of the best-studied examples in bacteria of regulated proteolysis is the recognition of the ssrA tag by ClpXP following trans-translation, a mechanism by which stalled ribosomes are rescued upon recruitment of tmRNA and nascent polypeptides are tagged with the ssrA peptide (30). Because these stalled ribosomes often arise from damaged messenger RNAs that lack a stop codon, the resulting polypeptide products cannot be complete. Therefore, the presence of the ssrA tag is itself the signal for a poor-quality protein. Recognition of the ssrA tag by ClpXP is highly specific, with even a single amino acid substitution abolishing substrate recognition (28). The amount of ssrA-tagged proteins is staggering, with some estimates that an ssrA tag is appended in approximately 1 in every 20 translation events (31). During starvation ribosome stalling and mRNA cleavage is enhanced, resulting in an even further taxing of the trans-translation system (32). Activation of certain endogenous toxins, such as MazF and RelE, can induce rampant mRNA cleavage as well that is counteracted by tmRNA (33). In these cases, the need for clearance of ssrA-tagged proteins becomes even more urgent to eliminate the surge in truncated polypeptides.
The ClpXP protease is fully capable of degrading ssrA-tagged proteins. However, the adaptor protein SspB enhances the ability of ClpXP to recognize and degrade ssrA-tagged substrates (34). SspB forms a homodimer, with each subunit containing a substrate binding domain that binds ssrA-tagged proteins, and a disordered C-terminal tail that interacts with ClpX. Efficient delivery of ssrA-tagged substrates requires both tails on each subunit of SspB to interact with ClpX, which tethers substrates to the protease to increase effective substrate concentration (35). Thus, the ssrA tag is the fundamental protease recognition determinant with the SspB adaptor acting as a modifier of this recognition by serving as a passive tether (Figure 2b).
In addition to enhancing the ability of ClpXP to degrade ssrA-tagged substrates, SspB also promotes degradation of N-RseA, the N-terminal fragment of the stress response protein RseA (13, 36). During normal conditions, RseA binds σE, preventing it from activating transcription. However, during the envelope-stress response, RseA is cleaved by proteases, freeing the N-terminal segment of RseA in complex with σE. SspB then delivers N-RseA to the ClpXP protease, leaving σE free to upregulate the envelope-stress response. Remarkably, there is no clear sequence similarity between the region of N-RseA that interacts with SspB and the region of the SsrA tag that binds SspB, indeed, binding of N-RseA is in the opposite orientation as that of ssrA (36). Having a single adaptor bind multiple substrates would enable a coordinated response across several pathways and, perhaps not surprisingly, other examples of adaptors enabling degradation of several substrates are now emerging (37–39). However, this multiplexing would eventually reach an upper limit as the need for selectivity begins to outweigh the advantages of coordinated degradation.
Recognition of poor quality proteins without specific tagging: the Lon protease
The cellular response to an acute stress must often occur at timescales faster than transcription and translation. Importantly, if the stress affects some of this central machinery, such as the fidelity of the ribosome, then the consequences of this stress must be repaired prior to restarting normal growth. It can be argued that the Lon protease is uniquely suited to serve as a quality control protease due to its ability to broadly recognize misfolded proteins and its ability to be allosterically activated.
Quality control through regulated proteolysis requires bacteria to discriminate between fatally misfolded or damaged proteins and proteins that simply share features associated with compromised proteins, such as folding intermediates or normal transient fluctuations in protein structures. Compared to other energy dependent proteases, the Lon protease has the weakest unfolding capacity (19), but a surprisingly promiscuous substrate repertoire. Indeed, Lon seems to recognize hydrophobic residues on misfolded substrates that are typically buried in the native structure as its primary recognition determinant rather than any unique sequence motif (40) (Figure 2a) for review, see (41).
To properly survey protein quality, Lon must be able to distinguish between fatally misfolded proteins and those that are in intermediate folding states. Given that terminally misfolded proteins are kinetically trapped, it seems likely that the lifetime of the exposed hydrophobic regions may be a key determinant for this type of quality control surveillance. The shorter lifetime of exposed hydrophobic regions for a protein en route to the native state would set the lower boundary of timescales where Lon should recognize a ‘poor quality’ protein. By extension, this means that the Lon-substrate complex would need to have sufficiently transient kinetics to be compatible with this discrimination, otherwise, Lon would erroneously destroy proteins in the process of being folded or fluctuating with normal dynamics. Thus, by combining a low efficiency unfolding capacity with a broad recognition spectrum, the Lon protease gains selectivity in recognizing truly misfolded proteins. A similar model is thought to hold for eukaryotic quality control, where only persistent Hsp70 chaperone to a folding client recruits the CHIP ubiquitin ligase to target the client for ubiquitylation and degradation (42).
The ability of cells to use proteases and chaperones to ensure protein quality can also apply to the quaternary structure of complex assemblies. For example, individual subunits of protein complexes must be assembled in the correct stoichiometry to ensure function. Overflow of these subunits could be toxic, but AAA+ proteases are well-suited to destroy these unincorporated subunits. For example, degradation of the CcdA antitoxin by the Lon protease is inhibited when it is incorporated into the CcdAB complex (43) and degradation of subunits are often suppressed when complexes are fully assembled. In this respect, Lon is ensuring that active protein complexes maintain the proper stoichiometry.
Similarly, many protease substrates are DNA binding factors, including transcription factors, replication regulators, and components of polymerases (44–47). For some substrates, degradation is only evident when the substrate is not binding DNA, suggesting again that a surveillance of the proper active complex (in this case the degree of DNA-bound species) affords the cell the ability to ensure destruction of proteins when they are not performing their function. We caution that interpretation of this phenomenon is more complex as in some cases DNA can stimulate substrate recognition by the protease (46), and in the case of Lon, DNA can affect the protease directly (48, 49) and for review see (47).
The allosteric activation of Lon is an intriguing aspect to consider in light of its role in quality control in the cell and its fundamental broad specificity. Many enzymes exhibit substrate-activity relationships in line with the classic Michaelis-Menten equation. The ClpXP and ClpAP proteases fall into this class, where increasing substrate initially results in linear increases of degradation rates until the enzyme becomes saturated and maximum reaction velocity (vmax) is achieved (Figure 3a). In such cases, an underlying assumption is that the enzyme specific activity is unchanged as substrate is added.
Figure 3.
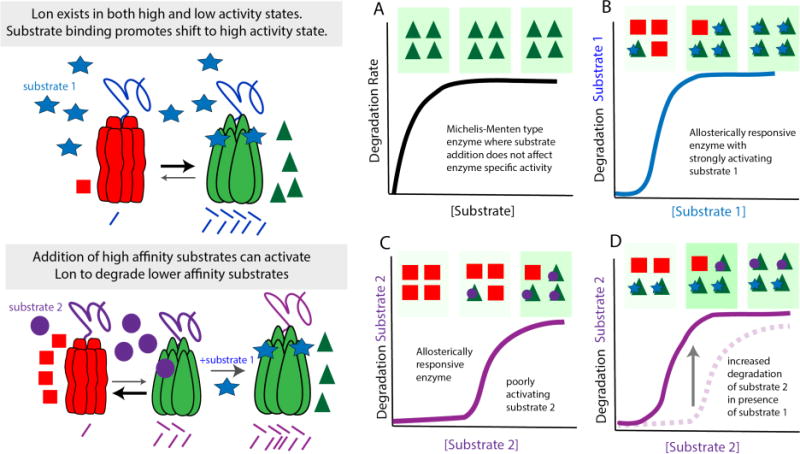
Lon is subject to allosteric regulation (a) Many proteases, such as ClpXP and ClpAP, adhere to typical Michaelis-Menten kinetics where adding increasing amounts of substrate leads to an increase in the rates of degradation until the protease becomes saturated and Vmax is reached resulting in the classic hyperbolic curve. (b) This contrasts with Lon which exhibits positive cooperativity upon increasing substrate concentration. The working model is that Lon exists as low (red triangles) and high activity (green squares) states with substrate binding promoting the highly active state. (b) In the case of substrate 1, which has a strong affinity for Lon, relatively low amounts of substrate are needed to shift Lon to the active state. (c) Substrate 2 binds Lon poorly and activation requires much higher concentrations of substrate 2. (d) If the concentration of substrate 2 is kept the same and the high affinity substrate 1 is added, Lon will be shifted to the active form, leading to robust degradation of the normally poorly degraded substrate 2.
By contrast, the Lon protease has long been known to respond to substrate concentration in a cooperative manner (50), with the working model that substrates allosterically activate the Lon protease and increase its proteolytic activity. Intriguingly it has been shown that substrates not only activate Lon for their own degradation, but can also serve to activate Lon for degradation of other substrates (51). Thus, the behavior of Lon in the presence of two substrates could lead to regulation conducive to quality control. For example, suppose substrate 1 has a higher affinity for Lon such that low concentrations of this substrate are needed to allosterically activate Lon, while substrate 2 can only activate Lon at a higher substrate concentration (Figure 3b–3c). Titration of these substrates would show that at the same concentrations, substrate 2 is degraded more slowly than 1. However, addition of substrate 1, even at concentrations lower than substrate 2, would cause shifting of the Lon population to the more active species resulting in more rapid degradation of substrate 2 (Figure 3d). Therefore, higher affinity substrates can effectively act as activators of lower affinity substrate degradation. In these cases, the basic recognition of the protease is modified by the presence of other substrates.
An intriguing extension of this biochemical framework is that it would result in the greatest activation of the Lon protease when demand is greatest in vivo. For example, during an acute proteotoxic stress, the rapid increase in misfolded proteins would result in allosteric activation of Lon to eliminate these misfolded proteins, but also to degrade fully active regulatory proteins as part of the stress response. Such a case was recently reported in Caulobacter, with the Lon dependent degradation of DnaA serving to halt cell cycle progression during proteotoxic stress (52). This type of regulation would also make sense for a protease such as Lon given its ability to recognize features found in all proteins (such as hydrophobic residues), as persistently high activation of Lon would inevitably result in the destruction of proteins not needed for quality control. Indeed overproduction of Lon in E. coli results in cell death, in part because of rampant degradation of antitoxin proteins (53). Finally, because Lon can be allosterically regulated by non-protein molecules such as DNA, it is tempting to speculate that a surge in Lon activity upon allosteric activation could also be deployed under additional stress responses, such as during genotoxic stress where extended exposure of single-stranded DNA is a unique flag for DNA damage.
Regulated proteolysis during balanced growth
Coordinating proteolysis with cell division and replication
The cell cycle involves a highly regulated sequence of events in which DNA is faithfully replicated and divided into daughter cells. Progression through the cell-cycle requires regulated proteolysis to ensure the timely degradation of regulatory proteins necessary to drive this irreversible process. In eukaryotes, the concerted activity of APC/C and SCF ubiquitin ligases enforce the selective tagging and ultimate degradation of many regulatory factors (54).
In Caulobacter crescentus, asymmetric cell division yields a motile, flagellated swarmer cell and a sessile stalked cell (23, 55). The stalked cell is replication-competent and can immediately commence DNA replication and enter the cell cycle. However, the swarmer cell must first shed its flagellum and grow a stalk before it can initiate chromosome replication. Thus, the swarmer to stalked transition, also known G1 to S transition, is coupled to DNA replication and is intricately linked to regulated protein degradation (56). At the center of this transition is the essential master regulator, CtrA which is responsible for regulating expression of approximately 100 genes (57–59). CtrA also binds to and inhibits the origin of replication in swarmer cells, preventing swarmer cells from initiating replication. Therefore, when it becomes time to resume DNA replication during the swarmer-to-stalked transition, CtrA must be eliminated. This occurs through dephosphorylation by the CckA pathway (60–62) and proteolysis by the ClpXP protease (8, 63).
Since the levels of ClpXP remain constant during the cell-cycle, additional mechanisms must exist to support the cyclic oscillations seen in CtrA levels during the cell cycle (8). Indeed, a tightly regulated series of events, involving the adaptor proteins CpdR, RcdA, PopA, and the second messenger cyclic di-GMP ensures the degradation of CtrA (and other substrates) and timely entrance into S phase (64).
Intriguingly, although CtrA degradation in vivo requires ClpXP as well as additional accessory factors, ClpXP can degrade CtrA without these adaptors in vitro (65, 66). This paradox was reconciled by data that showed that CpdR, RcdA, PopA can increase the rate of CtrA degradation in vitro as the addition of these factors lowered the Km 10-fold, keeping Vmax constant (66). Intriguingly, the assembly of the adaptors was shown to be essential during times when recognition of CtrA by ClpXP was less robust, such as when CtrA is bound to DNA and thereby inaccessible (67). This ensures that there is complete removal of CtrA to allow resumption of DNA synthesis as even a small amount of CtrA is enough to inhibit the origins of replication (66, 68).
Regulated protein degradation is linked to cell-cycle progression in other bacteria as well. In E. coli, FtsZ is an essential component of the cell division machinery. FtsZ polymerization is necessary to form the z-ring, the site where septation and cell division occurs. Studies in Bacillus subtilis determined that ClpX inhibits FtsZ polymerization in a Clp and ATP-independent manner (69). However, in E. coli ClpXP has been shown to degrade FtsZ directly, potentially functioning to promote the disassembly of FtsZ polymers (70, 71). Similarly, both ClpAP and ClpXP can degrade Caulobacter FtsZ in vitro as well as in vivo (72).
Diversifying proteolysis through hierarchies
Recently, CpdR, PopA, and RcdA have been shown to function as adaptors capable of degrading various classes of substrates in a hierarchical manner in Caulobacter (38) (Figure 4). The lowest level of the hierarchy consists of ClpXP alone, which can theoretically degrade many substrates limited only by the recognition determinants of those substrates. For example, trapping studies in E. coli and Caulobacter have identified hundreds of potential substrates, several of which are degraded by ClpXP alone in vitro (29, 73) and proteomic studies in Staphylococcus aureus illustrate the range of substrate degraded by the Clp family (74). The next tier consists of ClpXP and the adaptor CpdR. While traditional adaptors can bind their substrates directly, two-hybrid studies found that CpdR alone does not interact with its substrates strongly (37). Instead, CpdR primes ClpXP by binding to the N-terminal domain of the unfoldase domain, preparing ClpXP to engage its first class of substrates, which include McpA, a chemoreceptor (62), and the cyclic di-GMP phosphodiesterase PdeA (37, 75). This priming event opens ClpXP to an array of adaptors and substrates that would have been inaccessible before such as RcdA, which binds a CpdR-primed ClpX directly to establish the third tier of the hierarchy. In this way, RcdA acts like a canonical adaptor in tethering cargo to ClpXP to enhance delivery of a second class of substrates, including the developmental regulator TacA and various proteins of unknown function (38).
Figure 4.
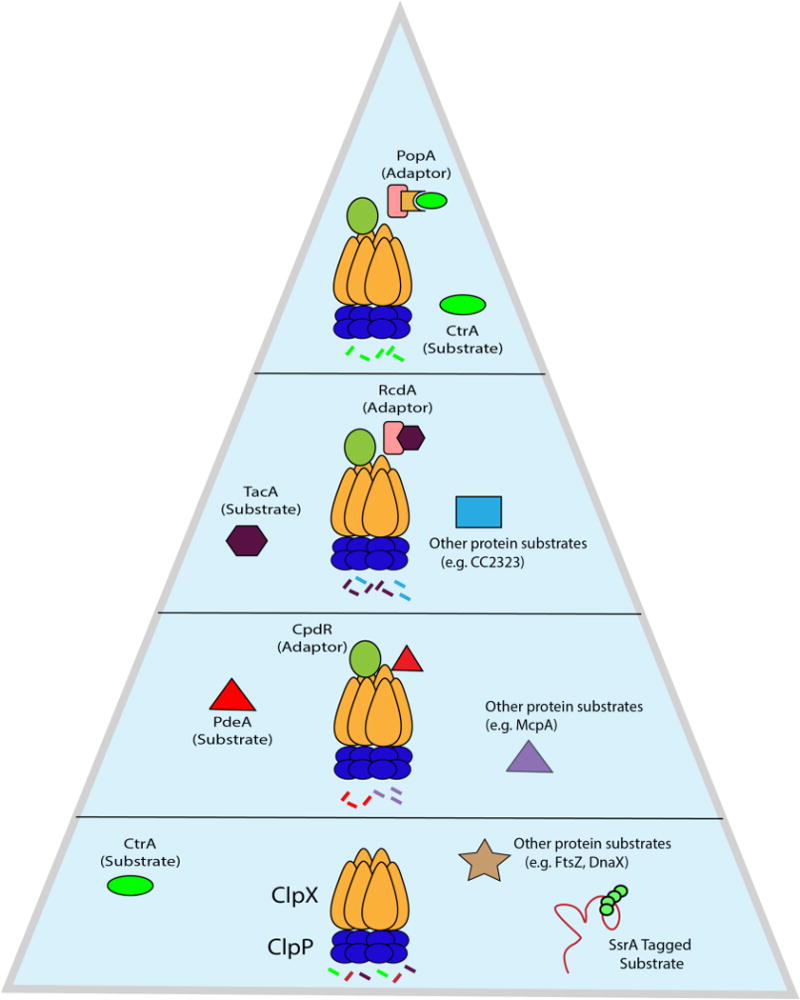
Adaptors assemble in a hierarchical manner to degrade various classes of substrates. ClpXP can degrade numerous substrates on its own, included ssrA-tagged proteins. During the G1 to S transition in Caulobacter, the adaptor CpdR first primes ClpXP, allowing it to recruit the first class of substrates (PdeA, McpA, etc) for degradation. The primed protease can now recruit another adaptor, RcdA, to degrade a second class of substrates, such as the transcription factor TacA and others. Finally, the adaptor PopA binds RcdA and in the presence of the second messenger cyclic di-GMP completes the hierarchy to deliver a third class of substrates, including the master regulator CtrA, to ClpXP. As the hierarchy is assembled and adaptors are added on to the protease, specificity increases. When ClpXP is limiting, this increase in specificity also comes at the cost of preventing degradation of other members of the substrate pool.
The pinnacle of the hierarchy requires the addition of the adaptor PopA bound to cyclic di-GMP, which culminates in the degradation of CtrA (38, 66, 76) and likely other substrates such as GdhZ and KidO (77, 78). Strikingly, bacterial two-hybrid experiments demonstrated that PopA alleles which cannot engage cyclic di-GMP still bind RcdA (38, 76). This suggests that even in the absence of delivery, PopA can compete with the RcdA-dependent degradation of substrates like TacA, suggesting that members of the hierarchy can act as both adaptors and anti-adaptors (38, 56). Cross species comparisons find that CpdR and RcdA are highly conserved in all alpha proteobacteria, but the presence of PopA is more restricted (79), leading to the speculation that CpdR/RcdA represent a more ancient aspect of this adaptor hierarchy. Interestingly, RcdA is essential in Agrobacterium tumefaciens (80), suggesting that this adaptor hierarchy might be more deeply woven into the essential physiology of other bacteria.
The costs and benefits of adaptors
Balanced growth requires regular predictable changes in protein levels to drive replication and division. Although AAA+ proteases can have different ranges of selectivity, there is a clear need to augment their specificity through other regulators. Adaptor proteins in their most basic form can be thought of as simple tethers that locally increase the concentration of potential protease substrates. However, there is a conflict in that adaptors which bind cargo weakly may not efficiently deliver substrates to the protease while adaptors that grip cargo too tightly could also hinder the substrate degradation by the protease. Therefore, there must be an appropriate tuning of the lifetimes of the different subcomplexes involved in adaptor-dependent handoff to ensure robust degradation. Below, we illustrate how the well-characterized SspB/ssrA system demonstrates this principle.
As described previously, the SspB adaptor binds ssrA-tagged substrates and delivers them to the ClpXP protease. Bulk measurements find that GFP tagged with ssrA binds SspB with a kon of ~ 5 μM−1s−1 at 30°C and a KD of ~ 50 nM yielding a ~4 second lifetime for this complex (81) (Figure 5). Single-turnover experiments suggest that the limiting step for degradation by ClpXP is substrate commitment, rather than translocation or proteolysis, with an estimate of ~30 seconds for tagged GFP at saturating concentrations (82). Therefore, cargo can be released from the adaptor’s grip 7-8 times during the time that ClpXP is establishing commitment. However, if a cargo binds SspB 10-fold more tightly, with the same on rate, then this new 40 second lifetime exceeds the ClpXP commitment time and may result in paradoxically slower proteolysis. Although such substrates have not yet been identified, this simple example illustrates the point that tighter binding of an adaptor to cargo may not result in more robust delivery to the protease and that an optimum balance likely exists between adaptor-cargo lifetimes and protease commitment timescales.
Figure 5.
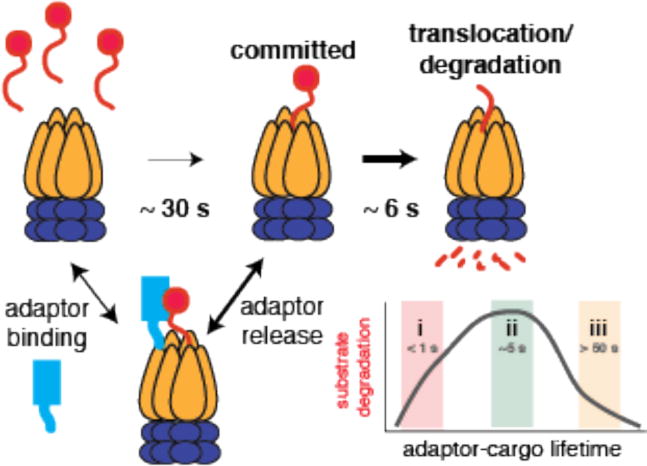
Substrate degradation by ClpXP is rate-limited by the commitment step, where the protease initially engages a target, rather than the unfolding or translocation steps, which are relatively fast. Commitment is estimated to be ~ 30 seconds for degradation of tagged GFP by ClpXP (Cordova, et al. Cell 2014). Tethering adaptors (like SspB and RcdA) enhance degradation of substrate but the strength of the interaction between the adaptor and substrate must be tuned to the commitment time for the protease (regime ii). Poor adaptor-cargo binding results in failure to deliver (regime i) but binding too tightly (regime iii) hinders substrate release during the commitment step for protease engagement of the substrate.
Adaptors can also serve as more than tethers. In addition to anchoring cargo to the protease, adaptor binding may also cause degrons in the cargo protein to be exposed, as described for YjbH/Spx (83) and RssB/RpoS (84, 85). Adaptors could also affect the protease itself. As described above, CpdR binds to ClpX and activates the ClpXP protease for degradation of the substrate PdeA, but CpdR on its own fails to bind PdeA (37). Binding of CpdR, or similar adaptors, may contribute to substrate engagement directly, e.g., providing additional low affinity contacts, or may affect ClpXP substrate engagement through allosteric changes of ClpX itself. Given our discussion above regarding the balance of protease commitment and adaptor-cargo lifetimes, it is tempting to consider that some adaptors may influence protease specificity through altering the commitment time of the protease rather than changing its ability to directly recognize a target.
Regulated proteolysis during changes in growth phase
Bacteria are constantly being challenged in their environments with changing conditions, such as nutrient deprivation, that necessitate a swift response. Regulated proteolysis is required for bacteria to undergo the necessary physiological transitions and adaptation needed for survival and persistence. An example of this is the transition from logarithmic growth to stationary phase, when growth slows and cells alter their metabolism to accommodate this change in phase (86). This transition requires σS, also known as RpoS, an alternative sigma factor that can compete with σ70 during stationary phase to significantly alter gene expression profiles. During logarithmic growth, RpoS is rapidly degraded to undetectable levels by ClpXP with the required assistance of the adaptor RssB (85, 87) (Figure 6a).
Figure 6.
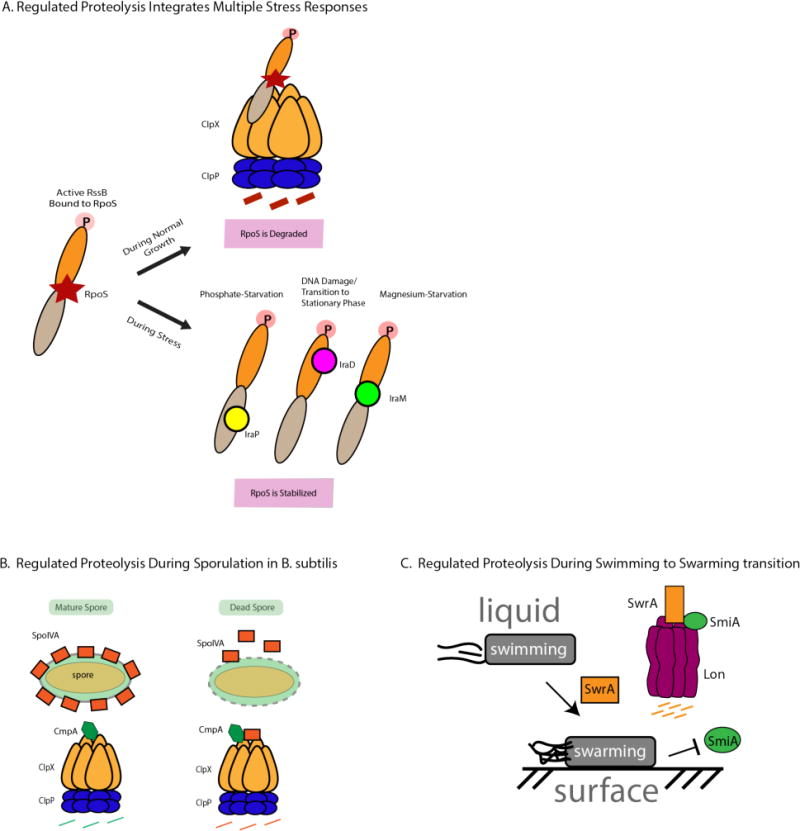
Regulated proteolysis is required during physiological transitions and changes in growth (a) When bacteria are actively growing in logarithmic phase, the alternative sigma factor RpoS is rapidly degraded by ClpXP in an RssB-mediated manner. When RssB is phosphorylated, it has high affinity for RpoS and can deliver it to ClpXP for degradation. Anti-adaptors bind to RssB in different binding modes dependent on the kind of stress the bacteria is encountering, preventing it from delivering RpoS for degradation. (b) Bacillus subtilis requires proteolysis by ClpXP to ensure proper spore envelope assembly. In cells with improperly assembled envelopes, the adaptor CmpA delivers SpoIVA to ClpXP for degradation, leading to lysis of the defective cell. If the spore envelope is properly assembled, the adaptor is targeted for degradation by ClpXP instead. (c) Lon-mediated degradation is required for proper motility during the transition from liquid to solid media. In liquid media, Lon degrades SwrA with the help of SmiA, an adaptor protein. Upon shift to solid media, degradation of SwrA is inhibited, leading to an increase in SwrA levels necessary for swarming on solid media.
During entry to stationary phase, RpoS becomes stabilized and produces a surge in transcription of RpoS regulated genes (88, 89). Inhibition of RssB activity is accomplished by a group of anti-adaptors, each specific for a particular stress response. In E. coli, three anti-adaptors have been identified, IraP (Inhibitor of RssB activity during phosphate starvation), IraD (Inhibitor of RssB activity during DNA damage), and IraM (Inhibitor of RssB activity during magnesium starvation). Interestingly, each anti-adaptor binds to RssB in a different binding mode (2, 90, 91). IraP binds at the C-terminal domain of RssB while IraD interacts with the N-terminal domain, and IraM has been shown to make interactions with both domains. This example shows how a family of distinct anti-adaptors, each binding at a characteristic location, can each prevent the degradation of RpoS, allowing the cell to mount a rapid universal program in response to a variety of stresses.
In nutrient-poor conditions, Bacillus subtilis initiates a sporulation program resulting in mature spores that can withstand harsh environmental conditions. The structural protein SpoIVA is required for proper assembly of the spore envelope. To ensure the fidelity of the sporulation program, Bacillus relies on regulated proteolysis as a quality control mechanism to remove spore envelopes that have been improperly assembled (92) (Figure 6b). In sporulation defective cells, CmpA, an adaptor, delivers SpoIVA to ClpXP for degradation, ultimately leading to the lysis and removal of the defective cell from the population. However, in cells that properly constructed the spore envelope, CmpA is degraded, preventing the degradation of SpoIVA and allowing cells to continue the process of sporulation.
Bacteria growing in a liquid environment use flagella to swim in three-dimensional space. When shifted to solid media, bacteria transition their mode of motility from swimming to swarming. In Bacillus subtilis, this transition requires Lon-dependent degradation of SwrA, a master regulator of flagellar biosynthesis (93) (Figure 6c). When Bacillus is in a liquid environment, Lon robustly degrades SwrA in the presence of SmiA, a Lon-specific adaptor protein. Lon was unable to degrade SwrA both in vivo as well as in vitro in the absence of SmiA. When Bacillus transitions to a solid surface, Lon-mediated degradation is inhibited via an unknown mechanism and SwrA levels increase, resulting in increased flagellar density which is necessary for swarming motility. Intriguingly, SmiA was the first Lon-specific adaptor to be discovered, but more recent work using a Lon trapping approach has led to the discovery of HspQ, a conserved, small heat shock protein, that can also enhance Lon-dependent degradation of known substrates in the gram-negative bacterium Yersinia pestis (39). Understanding how Lon activity can be controlled in so many different ways and the consequences of this regulation is an outstanding question clearly worth exploring.
Proteolytic responses in response to starvation
Amino acids are the building blocks of proteins and all organisms must be able to accurately assess amino acid levels to avoid costly interruptions in protein synthesis (94). In times of amino acid starvation, protein degradation could replenish amino acid pools. In eukaryotes, inhibition of the ubiquitin-proteasome system results in a lethal depletion of amino acid pools which could be rescued by externally supplementing with additional amino acids or reducing translation (95). Earlier studies in E. coli found that starvation causes an increase in protein degradation which mirrored the rate of synthesis of new proteins (96). However, very little is currently known about how starvation mechanistically leads to amino acid recycling in bacteria.
In bacteria amino acid starvation leads to increased cellular levels of the alarmone (p)ppGpp, eliciting what is known as the stringent response (97). Increased levels of (p)ppGpp lead to many downstream effects, effectively allowing the cell to divert resources away from translation and towards amino acid biosynthesis (98). One of the effects of (p)ppGpp accumulation is the inhibition of the enzymes that break down polyphosphate, leading to increased polyphosphate levels (99). Work from Kornberg and colleagues have shown that polyphosphate can stimulate the proteolysis of ribosome subunits and other proteins by the Lon protease (100–102). This has led to the provocative hypothesis that during amino acid stress, activation of the Lon protease via ppGpp/polyphosphate induction will reduce ribosome pools to slow down translation and replenish pools of amino acids. Although recent work suggests that polyphosphate activation of Lon proteolysis may not be a universal feature for all substrates (103), the ability of regulated proteases such as Lon to contribute to nutrient stress responses to improve survival under starvation conditions is a very appealing notion. Indeed, prior work has shown that loss of energy dependent proteases in bacteria yields compromised responses to starvation (104, 105).
Perspective
Energy dependent proteases can differ dramatically in architecture and substrate preference. However, a recurring theme is that regulated proteolysis requires two elements for robust controlled substrate degradation. All substrates must contain some type of recognition determinant that can be engaged by the protease in order to begin unfolding and processing. This determinant can be highly sequence dependent, as in the case of ssrA and ClpXP, or it can be more general, as seen with recognition of hydrophobic residues by Lon. To truly maintain regulation, these determinants are further elaborated by modifying factors such as adaptors that deliver substrates directly or prime the protease for substrate recognition. These modifiers can also be a property of the protein itself, e.g., protein dynamics that cause transient or extended display of residues normally in the hydrophobic core. Finally, substrates themselves can act as modifiers to alter the specific activity of a protease in an effort to mount an effective stress response.
From a therapeutic perspective, physiological consequences of protease loss are especially apparent during stressful situations, such as when pathogens invade their hosts. Therefore, these proteases would be opportune targets to explore for the development of future antibiotics. Indeed, recent studies have shown that small molecule inhibitors of both Clp and Lon family proteases can be highly efficacious for various bacteria (106–108). Perhaps most intriguingly is the fact that unconstrained activation of these proteases could be as toxic to the bacteria (or more so) than inhibition. As a recent illustration of this possibility, the ClpP activating acyldepsipeptide ADEP was able to eradicate Staphylococcus aureus even in conditions where the cells were tolerant of other antibiotics (109).
Acknowledgments
The authors wish to thank E. Strieter, all members of the Chien lab, and members that make up the Protein Homeostasis group in the Institute for Applied Life Sciences for invaluable feedback. Work in the Chien lab was provided in part by a grant from the National Institutes of Health (R01GM111706) to PC and from a Chemistry Biology Interface Program Training Grant (NIH T32GM08515) to SM.
Literature Cited
- 1.Strieter ER, Korasick DA. Unraveling the complexity of ubiquitin signaling. ACS Chem Biol. 2012;7(1):52–63. doi: 10.1021/cb2004059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gur E, Biran D, Ron EZ. Regulated proteolysis in Gram-negative bacteria — how and when? Nat Rev Microbiol. 2011;9(12):839–48. doi: 10.1038/nrmicro2669. [DOI] [PubMed] [Google Scholar]
- 3.Olivares AO, Baker TA, Sauer RT. Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat Rev Microbiol. 2016;14(1):33–44. doi: 10.1038/nrmicro.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 5.Breidenstein EBM, Janot L, Strehmel J, Fernandez L, Taylor PK, et al. The Lon Protease Is Essential for Full Virulence in Pseudomonas aeruginosa. PLoS One. 2012;7(11):e49123. doi: 10.1371/journal.pone.0049123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanemori M, Nishihara K, Yanagi H, Yura T. Synergistic roles of Hs1VU and other ATP-dependent proteases in controlling in vivo turnover of σ32 and abnormal proteins in Escherichia coli. J Bacteriol. 1997;179(23):7219–25. doi: 10.1128/jb.179.23.7219-7225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howard-Flanders P, Simson E, Theriot L. A locus that controls filament formation and sensitiviry to radiation in Escherichia coli K-12. Genetics. 1964 Feb;49:237–46. doi: 10.1093/genetics/49.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenal U, Fuchs T. An essential protease involved in bacterial cell-cycle control. EMBO J. 1998;17(19):5658–69. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogers A, Townsley L, Gallego-hernandez AL, Beyhan S, Kwuan L, Yildiz FH. The LonA Protease Regulates Biofilm Formation, Motility, Virulence, and the Type VI Secretion System in Vibrio cholerae. J Bacteriol. 2016;198(6):973–85. doi: 10.1128/JB.00741-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker TA, Sauer RT. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem Sci. 2006;31(12):647–53. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee I, Suzuki CK. Functional mechanics of the ATP-dependent Lon protease- lessons from endogenous protein and synthetic peptide substrates. Biochim Biophys Acta - Proteins Proteomics. 2008;1784(5):727–35. doi: 10.1016/j.bbapap.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langklotz S, Baumann U, Narberhaus F. Structure and function of the bacterial AAA protease FtsH. Biochim Biophys Acta - Mol Cell Res. 2012;1823(1):40–48. doi: 10.1016/j.bbamcr.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 13.Baker TA, Sauer RT. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta - Mol Cell Res. 2012;1823(1):15–28. doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottesman S. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol. 2003;19(1):565–87. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- 15.Rawlings ND, Barrett AJ, Finn R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016;44(D1):D343–50. doi: 10.1093/nar/gkv1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114(4):511–20. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- 17.Kraut DA. Slippery substrates impair ATP-dependent protease function by slowing unfolding. J Biol Chem. 2013;288(48):34729–35. doi: 10.1074/jbc.M113.512533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vass RH, Chien P. Critical clamp loader processing by an essential AAA+ protease in Caulobacter crescentus. Proc Natl Acad Sci U S A. 2013;110(45):18138–43. doi: 10.1073/pnas.1311302110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koodithangal P, Jaffe NE, Kraut DA, Fishbain S, Herman C, Matouschek A. ATP-dependent proteases differ substantially in their ability to unfold globular proteins. J Biol Chem. 2009;284(28):18674–84. doi: 10.1074/jbc.M900783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olivares AO, Nager AR, Iosefson O, Sauer RT, Baker TA. Mechanochemical basis of protein degradation by a double-ring AAA+ machine. Nat Struct Mol Biol. 2014;21(10):871–75. doi: 10.1038/nsmb.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maillard RA, Chistol G, Sen M, Righini M, Tan J, et al. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell. 2011;145(3):459–69. doi: 10.1016/j.cell.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aubin-Tam ME, Olivares AO, Sauer RT, Baker TA, Lang MJ. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell. 2011;145(2):257–67. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konovalova A, Søgaard-Andersen L, Kroos L. Regulated proteolysis in bacterial development. FEMS Microbiol Rev. 2014;38(3):493–522. doi: 10.1111/1574-6976.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mogk A, Huber D, Bukau B. Integrating protein homeostasis strategies in prokaryotes. Cold Spring Harb Perspect Biol. 2011;3(4):1–19. doi: 10.1101/cshperspect.a004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bezawork-Geleta A, Brodie EJ, Dougan DA, Truscott KN. LON is the master protease that protects against protein aggregation in human mitochondria through direct degradation of misfolded proteins. Sci Rep. 2015 Oct;5:17397. doi: 10.1038/srep17397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Melderen L, Aertsen A. Regulation and quality control by Lon-dependent proteolysis. Res Microbiol. 2009;160(9):645–51. doi: 10.1016/j.resmic.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 27.Chung CH, Goldberg AL. The product of the lon (capR) gene in Escherichia coli is the ATP-dependent protease, protease La. Proc Natl Acad Sci U S A. 1981;78(8):4931–35. doi: 10.1073/pnas.78.8.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flynn JM, Levchenko I, Seidel M, Wickner SH, Sauer RT, Baker TA. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc Natl Acad Sci U S A. 2001;98(19):10584–89. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell. 2003;11(3):671–83. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 30.Keiler KC, Waller PRH, Sauer RT. Role of a Peptide Tagging System in Degradation of Proteins Synthesized from Damaged Messenger RNA. Science (80-) 1996;271(5251):990–93. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 31.Lies M, Maurizi MR. Turnover of endogenous SsrA-tagged proteins mediated by ATP-dependent proteases in Escherichia coli. J Biol Chem. 2008;283(34):22918–29. doi: 10.1074/jbc.M801692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Yagi M, Morita T, Aiba H. Cleavage of mRNAs and role of tmRNA system under amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68(2):462–73. doi: 10.1111/j.1365-2958.2008.06167.x. [DOI] [PubMed] [Google Scholar]
- 33.Christensen SK, Pedersen K, Hansen FG, Gerdes K. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J Mol Biol. 2003;332(4):809–19. doi: 10.1016/s0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- 34.Levchenko I, Seidel M, Sauer RT, Baker TA. A Specificity-Enhancing Factor for the ClpXP Degradation Machine. Science (80-) 2000;289(5488):2354–56. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- 35.Bolon DN, Wah DA, Hersch GL, Baker TA, Sauer RT. Bivalent Tethering of SspB to ClpXP Is Required for Efficient Substrate Delivery: A Protein-Design Study. Mol Cell. 2004;13(3):443–49. doi: 10.1016/s1097-2765(04)00027-9. [DOI] [PubMed] [Google Scholar]
- 36.Levchenko I, Grant RA, Flynn JM, Sauer RT, Baker TA. Versatile modes of peptide recognition by the AAA+ adaptor protein SspB. Nat Struct Mol Biol. 2005;12(6):520–25. doi: 10.1038/nsmb934. [DOI] [PubMed] [Google Scholar]
- 37.Lau J, Hernandez-Alicea L, Vass RH, Chien P. A Phosphosignaling Adaptor Primes the AAA+ Protease ClpXP to Drive Cell Cycle-Regulated Proteolysis. Mol Cell. 2015;59(1):104–16. doi: 10.1016/j.molcel.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joshi KK, Bergé M, Radhakrishnan SK, Viollier PH, Chien P. An Adaptor Hierarchy Regulates Proteolysis during a Bacterial Cell Cycle. Cell. 2015;163(2):419–31. doi: 10.1016/j.cell.2015.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puri N, Karzai AW. HspQ Functions as a Unique Specificity-Enhancing Factor for the AAA+ Lon Protease. Mol Cell. 2017;66(5):672–683.e4. doi: 10.1016/j.molcel.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gur E, Sauer RT. Recognition of misfolded proteins by Lon, a AAA+ protease. Genes Dev. 2008;22(16):2267–77. doi: 10.1101/gad.1670908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gur E. The Lon AAA+ Protease. Regulated Proteolysis in Microorganisms. 2013;66:35–51. doi: 10.1007/978-94-007-5940-4_2. [DOI] [PubMed] [Google Scholar]
- 42.Arndt V, Rogon C, Höhfeld J. To be, or not to be - Molecular chaperones in protein degradation. Cell Mol Life Sci. 2007;64(19–20):2525–41. doi: 10.1007/s00018-007-7188-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Melderen L, Hoa M, Thi D, Lecchi P, Gottesman S, et al. ATP-dependent Degradation of CcdA by Lon Protease. J Biol Chem. 1996;271(44):27730–38. doi: 10.1074/jbc.271.44.27730. [DOI] [PubMed] [Google Scholar]
- 44.Kunová N, Ondrovičová G, Bauer JA, Bellová J, Ambro Ľ, et al. The role of Lon-mediated proteolysis in the dynamics of mitochondrial nucleic acid-protein complexes. Sci Rep. 2017;7(1):631. doi: 10.1038/s41598-017-00632-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu B, Lee J, Nie X, Li M, Morozov YI, et al. Phosphorylation of Human TFAM in Mitochondria Impairs DNA Binding and Promotes Degradation by the AAA+ Lon Protease. Mol Cell. 2013;49(1):121–32. doi: 10.1016/j.molcel.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kubik S, Wegrzyn K, Pierechod M, Konieczny I. Opposing effects of DNA on proteolysis of a replication initiator. Nucleic Acids Res. 2012;40(3):1148–59. doi: 10.1093/nar/gkr813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ambro L, Pevala V, Bauer J, Kutejová E. The influence of ATP-dependent proteases on a variety of nucleoid-associated processes. J Struct Biol. 2012;179(2):181–92. doi: 10.1016/j.jsb.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 48.Karlowicz A, Wegrzyn K, Gross M, Kaczynska D, Ropelewska M, et al. Defining the crucial domain and amino acid residues in bacterial Lon protease for DNA binding and processing of DNA-interacting substrates. J Biol Chem. 2017;292(18):7507–18. doi: 10.1074/jbc.M116.766709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chung CH, Goldberg AL. DNA stimulates ATP-dependent proteolysis and protein-dependent ATPase activity of protease La from Escherichia coli. Proc Natl Acad Sci U S A. 1982;79(3):795–99. doi: 10.1073/pnas.79.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waxman L, Goldberg AL. Selectivity of intracellular proteolysis: protein substrates activate the ATP-dependent protease (La) Science. 1986;232(4749):500–503. doi: 10.1126/science.2938257. [DOI] [PubMed] [Google Scholar]
- 51.Gur E, Sauer RT. Degrons in protein substrates program the speed and operating efficiency of the AAA+ Lon proteolytic machine. Proc Natl Acad Sci U S A. 2009;106(44):18503–8. doi: 10.1073/pnas.0910392106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jonas K, Liu J, Chien P, Laub MT. Proteotoxic stress induces a cell-cycle arrest by stimulating Lon to degrade the replication initiator DnaA. Cell. 2013;154(3):623–36. doi: 10.1016/j.cell.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Christensen SK, Maenhaut-Michel G, Mine N, Gottesman S, Gerdes K, Van Melderen L. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: Involvement of the yefM-yoeB toxin-antitoxin system. Mol Microbiol. 2004;51(6):1705–17. doi: 10.1046/j.1365-2958.2003.03941.x. [DOI] [PubMed] [Google Scholar]
- 54.Vodermaier HC. APC/C and SCF: Controlling each other and the cell cycle. Curr Biol. 2004;14(18):787–96. doi: 10.1016/j.cub.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 55.Skerker JM, Laub MT. Cell-cycle progression and the generation of asymmetry in Caulobacter crescentus. Nat Rev Microbiol. 2004;2(4):325–37. doi: 10.1038/nrmicro864. [DOI] [PubMed] [Google Scholar]
- 56.Joshi KK, Chien P. Regulated Proteolysis in Bacteria: Caulobacter. Annu Rev Genet. 2016;50:423–45. doi: 10.1146/annurev-genet-120215-035235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quon KC, Marczynski GT, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84(1):83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 58.Quon KC, Yang B, Domian IJ, Shapiro L, Marczynski GT. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci U S A. 1998;95(1):120–25. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laub MT, Chen SL, Shapiro L, McAdams HH. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci U S A. 2002;99(7):4632–37. doi: 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lori C, Ozaki S, Steiner S, Böhm R, Abel S, et al. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature. 2015;523(7559):236–39. doi: 10.1038/nature14473. [DOI] [PubMed] [Google Scholar]
- 61.Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, et al. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006;444(7121):899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- 62.Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci U S A. 2006;103(29):10935–40. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Domian IJ, Quon KC, Shapiro L. Cell-type specific phosphorylation and proteolysis of a transcriptional regulator controls the G1 to S transition in a bacterial cell cycle. Cell. 1997;90:415–24. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 64.McGrath PT, Iniesta AA, Ryan KR, Shapiro L, McAdams HH. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell. 2006;124(3):535–47. doi: 10.1016/j.cell.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 65.Chien P, Perchuk BS, Laub MT, Sauer RT, Baker TA. Direct and adaptor-mediated substrate recognition by an essential AAA+ protease. Proc Natl Acad Sci U S A. 2007;104(16):6590–95. doi: 10.1073/pnas.0701776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith SC, Joshi KK, Zik JJ, Trinh K, Kamajaya A, et al. Cell cycle-dependent adaptor complex for ClpXP-mediated proteolysis directly integrates phosphorylation and second messenger signals. Proc Natl Acad Sci. 2014;111(39):14229–34. doi: 10.1073/pnas.1407862111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gora KG, Cantin A, Wohlever M, Joshi KK, Perchuk BS, et al. Regulated proteolysis of a transcription factor complex is critical to cell cycle progression in Caulobacter crescentus. Mol Microbiol. 2013;87(6):1277–89. doi: 10.1111/mmi.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siam R, Marczynski GT. Cell cycle regulator phosphorylation stimulates two distinct modes of binding at a chromosome replication origin. EMBO J. 2000;19(5):1138–47. doi: 10.1093/emboj/19.5.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haeusser DP, Lee AH, Weart RB, Levin PA. ClpX Inhibits FtsZ Assembly in a manner that does not require its ATP hydrolysis-dependent chaperone activity. J Bacteriol. 2009;191(6):1986–91. doi: 10.1128/JB.01606-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Camberg JL, Hoskins JR, Wickner S. ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc Natl Acad Sci U S A. 2009;106(26):10614–19. doi: 10.1073/pnas.0904886106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Camberg JL, Hoskins JR, Wickner S. The Interplay of ClpXP with the cell division machinery in Escherichia coli. J Bacteriol. 2011;193(8):1911–18. doi: 10.1128/JB.01317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Williams B, Bhat N, Chien P, Shapiro L. ClpXP and ClpAP proteolytic activity on divisome substrates is differentially regulated following the Caulobacter asymmetric cell division. Mol Microbiol. 2014;93(5):853–66. doi: 10.1111/mmi.12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bhat NH, Vass RH, Stoddard PR, Shin DK, Chien P. Identification of ClpP substrates in Caulobacter crescentus reveals a role for regulated proteolysis in bacterial development. Mol Microbiol. 2013;88(6):1083–92. doi: 10.1111/mmi.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Michalik S, Bernhardt J, Otto A, Moche M, Becher D, et al. Life and Death of Proteins: A Case Study of Glucose-starved Staphylococcus aureus. Mol Cell Proteomics. 2012;11(9):558–70. doi: 10.1074/mcp.M112.017004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, et al. Regulatory Cohesion of Cell Cycle and Cell Differentiation through Interlinked Phosphorylation and Second Messenger Networks. Mol Cell. 2011;43(4):550–60. doi: 10.1016/j.molcel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Folcher M, Nicollier M, Schwede T, Amiot N, Duerig A. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beaufay F, Coppine J, Mayard A, Laloux G, De Bolle X, Hallez R. A NAD-dependent glutamate dehydrogenase coordinates metabolism with cell division in Caulobacter crescentus. EMBO J. 2015;34(13):1–15. doi: 10.15252/embj.201490730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Radhakrishnan SK, Pritchard S, Viollier PH. Coupling Prokaryotic Cell Fate and Division Control with a Bifunctional and Oscillating Oxidoreductase Homolog. Dev Cell. 2010;18(1):90–101. doi: 10.1016/j.devcel.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 79.Ozaki S, Schalch-Moser A, Zumthor L, Manfredi P, Ebbensgaard A, et al. Activation and polar sequestration of PopA, a c-di-GMP effector protein involved in Caulobacter crescentus cell cycle control. Mol Microbiol. 2014;94(3):580–94. doi: 10.1111/mmi.12777. [DOI] [PubMed] [Google Scholar]
- 80.Curtis PD, Brun YV. Identification of essential alphaproteobacterial genes reveals operational variability in conserved developmental and cell cycle systems. Mol Microbiol. 2014;93(4):713–35. doi: 10.1111/mmi.12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hersch GL, Baker TA, Sauer RT. SspB delivery of substrates for ClpXP proteolysis probed by the design of improved degradation tags. Proc Natl Acad Sci. 2004;101(33):12136–41. doi: 10.1073/pnas.0404733101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cordova JC, Olivares AO, Shin Y, Stinson BM, Calmat S, et al. Stochastic but highly coordinated protein unfolding and translocation by the ClpXP proteolytic machine. Cell. 2014;158(3):647–58. doi: 10.1016/j.cell.2014.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chan CM, Hahn E, Zuber P. Adaptor bypass mutations of Bacillus subtilisspx suggest a mechanism for YjbH-enhanced proteolysis of the regulator Spx by ClpXP. Mol Microbiol. 2014;93(3):426–38. doi: 10.1111/mmi.12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stüdemann A, Noirclerc-Savoye M, Klauck E, Becker G, Schneider D, Hengge R. Sequential recognition of two distinct sites in σS by the proteolytic targeting factor RssB and ClpX. EMBO J. 2003;22(16):4111–20. doi: 10.1093/emboj/cdg411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou Y, Gottesman S, Hoskins JR, Maurizi MR, Wickner S. The RssB response regulator directly targets σS for degradation by ClpXP. Genes Dev. 2001;15(5):627–37. doi: 10.1101/gad.864401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hengge R. Proteolysis of σS (RpoS) and the general stress response in Escherichia coli. Res Microbiol. 2009;160(9):667–76. doi: 10.1016/j.resmic.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 87.Jishage M, Ishihama A. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of sigma 70 and sigma 38. J Bacteriol. 1995;177(23):6832–35. doi: 10.1128/jb.177.23.6832-6835.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pratt LA, Silhavy TJ. The response regulator SprE controls the stability of RpoS. Proc Natl Acad Sci U S A. 1996;93(6):2488–92. doi: 10.1073/pnas.93.6.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Muffler A, Fischer D, Altuvia S, Storz G, Hengge-Aronis R. The response regulator RssB controls stability of the sigma(S) subunit of RNA polymerase in Escherichia coli. EMBO J. 1996;15(6):1333–39. [PMC free article] [PubMed] [Google Scholar]
- 90.Micevski D, Zammit JE, Truscott KN, Dougan DA. Anti-adaptors use distinct modes of binding to inhibit the RssB-dependent turnover of RpoS (σ(S)) by ClpXP. Front Mol Biosci. 2015 Apr;2:15. doi: 10.3389/fmolb.2015.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Battesti A, Hoskins JR, Tong S, Milanesio P, Mann JM, et al. Anti-adaptors provide multiple modes for regulation of the RssB adaptor protein. Genes Dev. 2013;27(24):2722–35. doi: 10.1101/gad.229617.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tan IS, Weiss CA, Popham DL, Ramamurthi KS. A Quality-Control Mechanism Removes Unfit Cells from a Population of Sporulating Bacteria. Dev Cell. 2015;34(6):682–93. doi: 10.1016/j.devcel.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mukherjee S, Bree AC, Liu J, Patrick JE, Chien P, Kearns DB. Adaptor-mediated Lon proteolysis restricts Bacillus subtilis hyperflagellation. Proc Natl Acad Sci U S A. 2014;112(1):1–6. doi: 10.1073/pnas.1417419112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517(7534):302–10. doi: 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Suraweera A, Münch C, Hanssum A, Bertolotti A. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell. 2012;48(2):242–53. doi: 10.1016/j.molcel.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mandelstam J. Turnover of protein in starved bacteria and its relationship to the induced synthesis of enzyme. Nature. 1957;179:1179–81. doi: 10.1038/1791179a0. [DOI] [PubMed] [Google Scholar]
- 97.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol. 2015;13(5):298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Srivatsan A, Wang JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol. 2008;11(2):100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 99.Kuroda A, Murphy H, Cashel M, Kornberg A. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1997;272(34):21240–43. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- 100.Kuroda A. Role of Inorganic Polyphosphate in Promoting Ribosomal Protein Degradation by the Lon Protease in E. coli. Science (80-) 2001;293(5530):705–8. doi: 10.1126/science.1061315. [DOI] [PubMed] [Google Scholar]
- 101.Nomura K, Kato J, Takiguchi N, Ohtake H, Kuroda A. Effects of inorganic polyphosphate on the proteolytic and DNA-binding activities of Lon in Escherichia coli. J Biol Chem. 2004;279(33):34406–10. doi: 10.1074/jbc.M404725200. [DOI] [PubMed] [Google Scholar]
- 102.Nomura K, Kato J, Takiguchi N, Ohtake H, Kuroda A. Inorganic polyphosphate stimulates lon-mediated proteolysis of nucleoid proteins in Escherichia coli. Cell Mol Biol. 2006;52(4):23–29. [PubMed] [Google Scholar]
- 103.Osbourne DO, Soo VW, Konieczny I, Wood TK. Polyphosphate, cyclic AMP, guanosine tetraphosphate, and c-di-GMP reduce in vitro Lon activity. Bioengineered. 2014;5(4):264–68. doi: 10.4161/bioe.29261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Reeve CA, Bockman AT, Matin A. Role of Protein Degradation in the Survival of Carbon-Starved Escherichia coli and Salmonella typhimurium. J Bacteriol. 1984;157(3):758–63. doi: 10.1128/jb.157.3.758-763.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Damerau K, John ACS. Role of Clp protease subunits in degradation of carbon starvation proteins in Escherichia coli. J Bacteriol. 1993;175(1):53–63. doi: 10.1128/jb.175.1.53-63.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fetzer C, Korotkov VS, Thänert R, Lee KM, Neuenschwander M, et al. A Chemical Disruptor of the ClpX Chaperone Complex Attenuates Multiresistant Staphylococcus aureus Virulence. Angew Chemie. 2017 doi: 10.1002/anie.201708454. [DOI] [PubMed] [Google Scholar]
- 107.Compton CL, Schmitz KR, Sauer RT, Sello JK. Antibacterial activity of and resistance to small molecule inhibitors of the clpp peptidase. ACS Chem Biol. 2013;8(12):2669–77. doi: 10.1021/cb400577b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Famulla K, Sass P, Malik I, Akopian T, Kandror O, et al. Acyldepsipeptide antibiotics kill mycobacteria by preventing the physiological functions of the ClpP1P2 protease. Mol Microbiol. 2016;101(2):194–209. doi: 10.1111/mmi.13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, et al. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature. 2013;503(7476):365–70. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]


