Abstract
High-mannose-type N-glycans are an important component of neutralizing epitopes on HIV-1 envelope glycoprotein gp120. They also serve as signals for protein folding, trafficking, and degradation in protein quality control. A number of lectins and antibodies recognize high-mannose-type N-glycans, and glycan array technology has provided an avenue to probe these oligomannose-specific proteins. We describe in this paper a top-down chemoenzymatic approach to synthesize a library of high-mannose N-glycans and related neoglycoproteins for glycan microarray analysis. The method involves the sequential enzymatic trimming of two readily available natural N-glycans, the Man9GlcNAc2Asn prepared from soybean flour and the sialoglycopeptide (SGP) isolated from chicken egg yolks, coupled with chromatographic separation to obtain a collection of a full range of natural high-mannose N-glycans. The Asn-linked N-glycans were conjugated to bovine serum albumin (BSA) to provide neoglycoproteins containing the oligomannose moieties. The glycoepitopes displayed were characterized using an array of glycan-binding proteins, including the broadly virus-neutralizing agents, glycan-specific antibody 2G12, Galanthus nivalis lectin (GNA), and Narcissus pseudonarcissus lectin (NPA).
Graphical Abstract
INTRODUCTION
Among the three major classes of the asparagine-linked (N-linked) glycans, including complex type, high-mannose type, and hybrid type, the high-mannose-type N-glycans play unique roles in a number of important biological recognition processes. For example, glycosylation of proteins with special high-mannose N-glycans serves as a signal for protein folding, trafficking, and degradation in protein quality control.1,2 The HIV envelope glycoprotein gp120 typically carries 20–25 N-glycans and the majority of them are of high-mannose type.3–7 The heavy glycosylation constitutes a strong defensive mechanism for the virus to evade host immune recognition,8,9 and the high-mannose glycans also serve as ligands for the DCSIGN mediated viral transmission from dendritic cells at the mucosal infection sites to T-lymphocytes in secondary lymphoid organs.10,11 In addition, the high-mannose glycans on HIV are also important antigenic structures and ligands for HIV-neutralizing antibodies and lectins. Indeed, the identification of a number of glycan-dependent broadly neutralizing antibodies (bnAbs) from HIV-infected individuals, such as 2G12, PG9/PG16, PGT128, and 10–1074, strongly suggests that the “glycan shield” of HIV virions display important targets for immunological intervention against viral infections.12–19
Like HIV-1, virtually all other human viruses also decorate their virions with carbohydrate moieties of cellular origin. For example, the spike glycoprotein of the severe acute respiratory syndrome coronavirus (SARS-CoV) has 23 potential N-linked glycosylation sites with high-mannose-type glycans (Man5–9GlcNAc2) as an essential component.20 The human cytomegalovirus (HCMV) envelope glycoproteins are extensively glycosylated and some of the ectodomains carry predominantly pauci- and high-mannose glycans.21 In order to explore whether viruses of distinct phylogenetic origins, such as HIV-1, SARS-CoV, and HCMV, express conserved glyco-determinants that are suitable for broad-spectrum virus neutralization, we have recently started to apply glycan microarray technology to probe the specific interactions between glycans and glycan-dependent virus-neutralizing antibodies and lectins.22 Our preliminary study has shown that Galanthus nivalis lectin (GNA), a high-mannose specific lectin, has very broad virus-neutralization activity against various human viruses, suggesting that there are shared high-mannose determinants on these viruses.22 Nevertheless, a detailed characterization of the molecular mechanism underlying the specific lectin–glycan and antibody–carbohydrate antigen interactions requires easy access to a comprehensive panel of structurally well-defined high-mannose type N-glycans and related glycoconjugates. Different oligomannoses and high-mannose-type N-glycans have been synthesized by multistep chemical synthesis.23,24 More recently, Ito and co-workers have reported a top-down chemoenzymatic approach to construct a library of high-mannose N-glycans. In this approach, a large common precursor of high-mannose N-glycan was synthesized first, in which the terminal mannose moieties on the three arms were selectively protected by a monosaccharide moiety (glucose, GlcNAc, and galactose) or an isopropylidene group, and then it was converted to a library of high-mannose N-glycans with varied size through deprotection and selective glycosidase trimming.25,26 While this method was sophisticated to make the precise oligomannose structures, it would be difficult for nonspecialists to follow the multistep protocols and the approach would not be easy to scale up. On the other hand, natural glycans can be an alternative and readily accessible sources for carbohydrate arrays as cleverly demonstrated by Cummings and co-workers.27–30
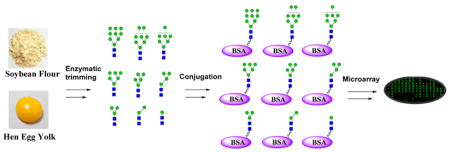
We describe in this paper a top-down chemoenzymatic approach that allows facile synthesis of a library of high-mannose-type N-glycans and related neoglycoproteins by sequential enzymatic trimming of two readily available natural N-glycans, the Man9GlcNAc2Asn prepared from soybean flour and the sialoglycopeptide (SGP) isolated from chicken egg yolks. The separated Asn-linked N-glycans were conjugated to bovine serum albumin (BSA) to provide neoglycoproteins containing the corresponding oligomannose moieties. Carbohydrate microarray analyses of these glyco-conjugates revealed critical virus-neutralizing epitopes; notably a Man5GlcNAc2 glycoform was identified as a highly reactive ligand for the broad-spectrum virus-neutralizing agent Galanthus nivalis lectin (GNA).
RESULTS AND DISCUSSION
Top-Down Chemoenzymatic Synthesis of Fmoc Protected Man4–9GlcNAc2Asn Glycans Starting from Man9GlcNAc2Asn
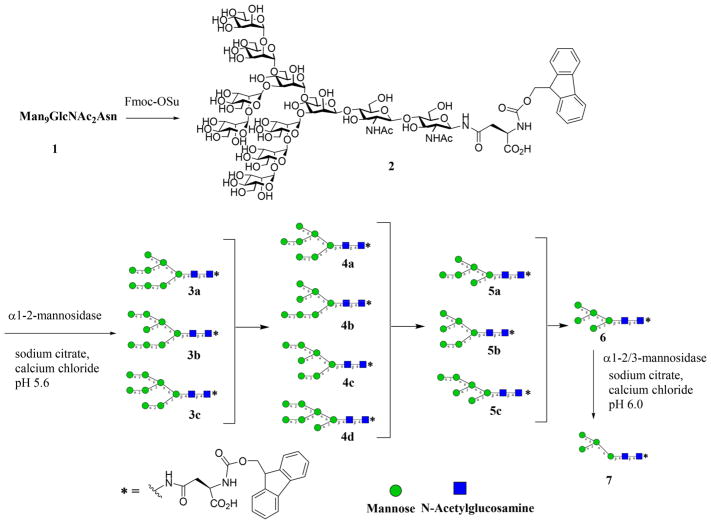
To have easy access to a panel of high-mannose N-glycans, we sought to use a top-down chemoenzymatic approach to generate a mixture of oligomannose intermediates by processing the readily available high-mannose glycan Man9GlcNAc2Asn with specific α-mannosidases and then to separate the intermediate oligomannoses by chromatographic methods. The Man9GlcNAc2Asn was readily prepared from soybean flour following the previously reported procedures.31,32 Briefly, crude soybean agglutinin, a glycoprotein carrying a full-size high-mannose N-glycan (Man9GlcNAc2Asn), was obtained by fractional precipitation of soybean flour. Then it was thoroughly digested with Pronase to afford Man9GlcNAc2Asn after size-exclusion chromatography. Man9GlcNAc2Asn was tagged with a 9-fluorenylmethyl (Fmoc) as a chemical handle to facilitate further purification as well as downstream isomer separation. Fmoc was selected due to its ease of attachment/removal, its excellent UV-activity, and high degree of hydrophobicity, which affords better differentiation of individual glycans in the context of high-performance liquid chromatography (HPLC) separation.33
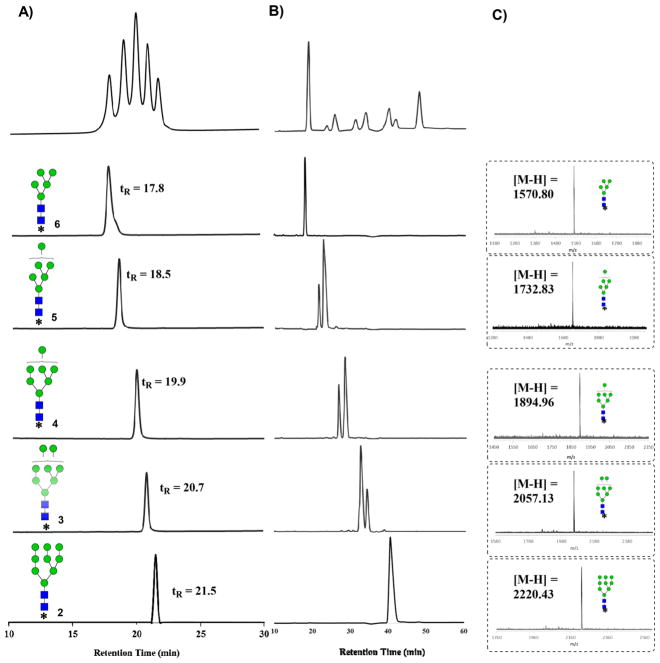
Since Man9GlcNAc2Asn contains four Manα-1,2-Man linkages, we chose an α1,2-mannosidase from Bacteroides thetaiotaomicron (a human gut bacterial symbiont)34 to digest the glycan precursor, which would be expected to generate Man5-Man8GlcNAc2-Asn-Fmoc intermediates under a controlled digestion (Scheme 1). We sought to capture the intermediate oligomannoses by controlling the enzyme concentration and carefully monitoring the progress of reaction. We discovered that at very low enzyme concentrations (i.e., 2 ng/mL), the intermediate Man6–8GlcNAc2 glycans could be captured yielding a mixture of oligomannoses under the controlled digestion. As expected, prolonged digestion of the precursor with the human α1,2-mannosidase yielded Man5GlcNAc2 as the sole product due to the complete hydrolysis of the four α1,2-Man linkages in the precursor. Careful normal-phase (NP)-HPLC on an amide-bonded column enabled an efficient separation of the Fmoc tagged oligomannose glycans (Man5–9GlcNAc2-Asn-Fmoc, 2–6) on the basis of their mannose components (Figure 1A). The individual oligomannoses separated appeared as single peaks on an analytical NP-HPLC, and their compositional homogeneity was verified by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) analysis (Figure 1). Interestingly, while analysis by NP-HPLC and MALDI-TOF MS established the purity and compositional homogeneity of each individual oligomannose, high-performance anion-exchange chromatography coupled with pulsed amperometric detection (HPAEC-PAD) analysis revealed that the oligomannoses Man6–8 were actually present as two or more isomers that could not be resolved on conventional NP-HPLC (Figure 1B). On the other hand, an isomeric Man4GlcNAc2Asn-Fmoc (7) was obtained by selective trimming of Man5GlcNAc2Asn-Fmoc (6) with an α1,2/3-mannosidase that can efficiently and selectively remove one of the α1,3-mannose moiety linked to the core β-mannose residue35 (Scheme 1).
Scheme 1.
Top-Down Chemoenzymatic Synthesis of Fmoc Tagged Oligomannose Glycans Man4-9GlcNAc2Asn-Fmoc Starting from the Man9GlcNAc2Asn Precursor
Figure 1.
HPLC, MALDI-TOF MS, and Dionex HPAEC-PAD analysis of the N-glycan intermediates. (A) Separation of oligomannose glycans 2–6 by NP-HPLC; (B) HPAEC-PAD analysis of the isolated oligomannoses; and (C) MALDI-TOF MS analysis of the isolated oligomannoses.
Top-down Chemoenzymatic Synthesis of the Pauci-Mannose (Man1–3GlcNAc2Asn) Glycan Library from the Sialoglycopeptide (SGP)
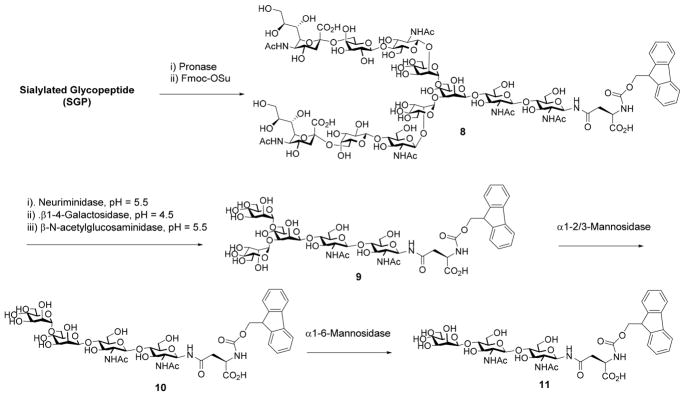
The construction of the pauci-mannose (Man1–3GlcNAc2Asn) glycan library was achieved by enzymatic digestion of a biantennary complex type sialoglycopeptide (SGP) with several exoglycosidases. The sialoglycopeptide (SGP) was prepared from chicken egg yolks following the previously reported procedures.36–38 Digestion of SGP with Pronase gave the Asn-linked complex type N-glycan, which was then tagged with Fmoc to provide the Fmoc tagged biantennary complex type N-glycan, SCT-Asn-Fmoc (8) (Scheme 2). Enzymatic trimming of the SCT-Asn-Fmoc with α2,6/8-neuraminidase, β1,4-galactosidase, and β-N-acetyl-glucosaminidase in a one-pot fashion by adjusting the pH after each enzyme addition afforded the Man3GlcNAc2–Asn-Fmoc (9), following the previously reported procedure37 (Scheme 2). The pauci-mannose glycans Man2GlcNAc2Asn-Fmoc (10) and Man1GlcNAc2Asn-Fmoc (11) were synthesized by enzymatic digestion of Man3GlcNAc2Asn-Fmoc (9) using theα1,2/3-mannosidase and the α1,6-mannosidase, respectively (Scheme 2). It should be noted that treatment of Man3GlcNAc2Asn-Fmoc (9) with the α1,6-mannosidase would not remove the α1,6-linked mannose moiety in the presence of the α1,3-mannose residue, due to the substrate specificity of this α1,6-mannosidase.35
Scheme 2.
Digestion of Sialylated Complex-Type Glycan to Provide Pauci-Mannose Glycans Man1-3GlcNAc2Asn-Fmoc
Functionalization of the Respective Oligomannose Glycans with a Maleimide Moiety
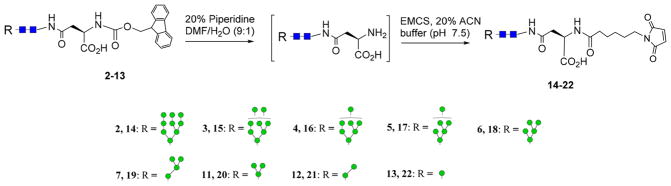
Subsequent to the separation of the oligomannose glycans, the Fmoc tag was removed by treatment of the Fmoc tagged oligomannoses (2–13) with 20% piperidine in DMF/water to give the unmasked oligomannoses. Then the free primary amine was functionalized with a maleimide moiety by reaction of the respective intermediate with N-ε-malemidocaproyl-oxysuccinimide ester (EMCS) to give the maleimide-tagged oligomannoses (14–22), respectively (Scheme 3), which were ready for coupling with a thiol-functionalized carrier protein to make neoglycoproteins.
Scheme 3.
Synthesis of Maleimide-Tagged High-Mannose Type N-Glycans
Synthesis of Oligomannose–BSA Conjugates
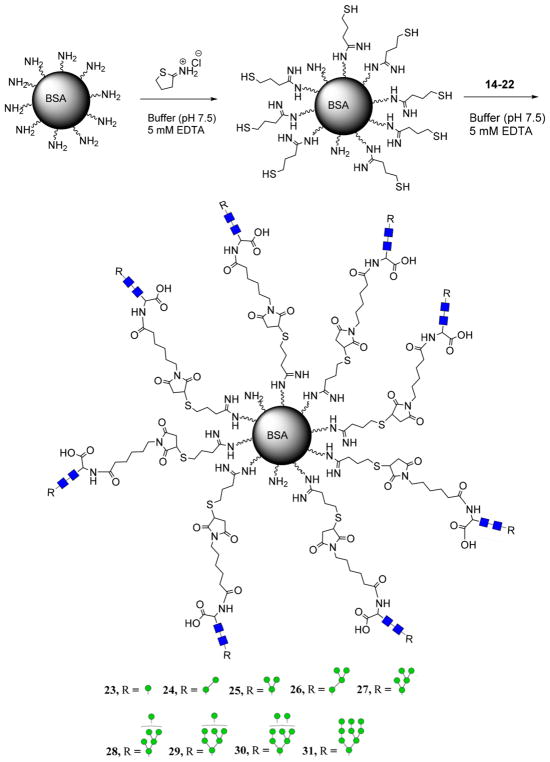
Bovine serum albumin (BSA) was chosen as the carrier protein given its well-established surface chemistry and resolved antigenicity. BSA was first functionalized with sulfhydryl groups by reaction with the Traut’s reagent that reacted with the solvent-exposed lysine groups, following our previously reported procedure.39 Incubation of thiol-functionalized BSA with the maleimide-tagged oligomannoses (14–22) yielded the corresponding oligomannose glycan–BSA conjugates (23–31) (Scheme 4). The oligomannose–BSA conjugates were purified by size-exclusion chromatography.
Scheme 4.
Sulfhydryl Functionalization of Native BSA and Subsequent Formation of the Oligomannose–BSA Conjugates
To determine the oligomannose loading, we first performed an MALDI-TOF MS analysis of the synthetic glycan/BSA conjugates. However, the preliminary analysis did not give useful information and the signals appeared as a broad peak, from which we could not deduce the loading of the oligomanoses on the protein. Both the introduction of the thiol groups by reaction with Traut’s reagent and the conjugation of the resulting thiolated BSA with the oligomannose through the thiol-maleimide reaction would give heterogeneous mixtures, which in combination contributes the high heterogeneity of the final conjugates, resulting in poor resolution in the MALDI-TOF MS analysis. We then measured the mannose contents in the glycoconjugates by a phenol-sulfuric acid carbohydrate assay, using mannose as a standard. Based on the carbohydrate content measurement, the loading of oligomannoses was estimated to be 8–12 copies of oligomannose per BSA molecule on average, respectively, depending on the size of the oligomannose moiety.
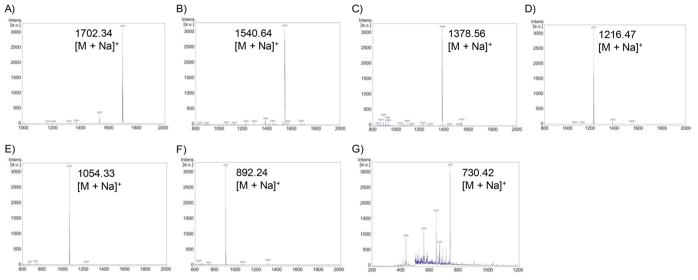
In addition, the identity of the oligomannose loaded on the glycoconjugates was also confirmed by MALDI-TOF MS analysis of the glycans released by enzymatic reactions. Briefly, the BSA-glycoconjugates were treated with enzyme Endo-A, which hydrolyzes the β1,4-glycosidic bond between the two GlcNAc moieties in the N-glycan. The released N-glycans were analyzed by MALDI-TOF-MS (Figure 2). The results clearly indicated that the N-glycans released from the BSA-glycoconjugates (25–31) are the expected oligomannoses (from Man3GlcNAc to Man9GlcNAc), respectively. However, MALDI-TOF MS of the relatively small oligosaccharides released from the ManGlcNAc 2 –BSA (23) and Man2GlcNAc2–BSA (24) conjugates fell in the low molecular mass range with high background, making it difficult to identify the expected ManGlcNAc and Man2GlcNAc signals.
Figure 2.
MALDI-TOF MS analysis of the oligomannoses released from the glycan-BSA conjugates (25–31). (A) Man9GlcNAc from glycoconjugate 25; (B) Man8GlcNAc from glycoconjugate 26; (C) Man7GlcNAc from glycoconjugate 27; (D) Man6GlcNAc from glycoconjugate 28; (E) Man5GlcNAc from glycoconjugate 29; (F) Man4GlcNAc from glycoconjugate 30; and (G) Man3GlcNAc from glycoconjugate 31.
Carbohydrate Microarray Epitope Mapping Studies
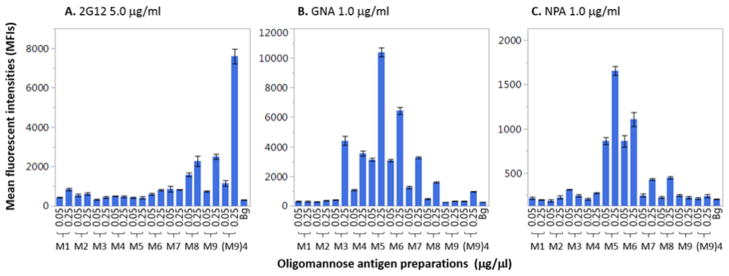
The oligomannose–BSA conjugates were tested against an array of oligomannose binding proteins in a microarray format. The oligomannose–BSA conjugates were directly printed onto the epoxy-functionalized microarray slides as we previously described39,40 and the oligomannose-binding proteins were incubated with each array. The oligomannose-binding proteins chosen included broadly virus-neutralizing agents, antibody 2G12, Galanthus nivalis lectin (GNA), and Narcissus pseudonarcissus lectin (NPA). The two lectins, NPA and GNA, are specific for terminal Manα1,6Man moieties and Manα1,3Man/Manα1,6Man linkages respectively,22 while 2G12 targets the high-mannose patch on the envelope glycoprotein gp120 of HIV by recognizing the terminal Manα1,2Man linkages in the oligomannose cluster.12,13,23 the synthesis of a tetravalent galactose scaffold bearing four Man9 glycans conjugated to a T-helper peptide, [(M9)4-TH], was described in our previous report.41
We found that the intensity of the 2G12 recognition of the oligomannose–BSA conjugates roughly corresponded to the number of Manα1,2 linkages present. The affinity of 2G12 for the oligomannose–BSA conjugates decreased for conjugates bearing fewer Manα1, 2 linkages. As expected, the gp120-mimicking (M9)4-TH controls exhibited very strong binding, while among the BSA conjugates, compounds 31 and 30 (M9 and M8-BSA) showed the best recognition (Figure 3). The far lower binding of the BSA conjugates as compared to the controls can be attributed to the mode of oligomannose glycan presentation. The high-mannose patch of gp120 and the tetravalent (M9)4-TH conjugate present the oligomannose glycans in a dense, clustered arrangement optimal for 2G12 binding. In the case of lectin binding, the oligomannose–BSA conjugates with higher access to the α1, 6-linked mannose generally bound more strongly to NPA, with compounds 27 and 28 (Man5-BSA and Man6-BSA) displaying the highest affinity (Figure 3). Recognition of the BSA conjugates by GNA appeared to be dependent upon the accessibility of the target Manα1, 3-Man linkages. GNA had little affinity for compound 31 (M9-BSA); however, recognition improved as access to the core Manα1,3Man linkages became more available. It follows that the two smallest pauci-mannose constituents (M1 and M2) had no affinity for GNA as they both lack Manα1,3Man linkages (Figure 3). The availability of the oligomannose-based neoglycoporteins enabled the assessment of the glycan specificity of the two lectins.
Figure 3.
Binding profiles of mannose-reactive proteins 2G12, GNA, and NPA to oligomannose–BSA conjugates. The glycoconjugates were spotted at 0.05 μg/μL and 0.25 μg/μL. Glycan-binding activities of antibody/lectin were shown as means of fluorescent intensities (MFIs) of triplicate microspots. Each error bar was constructed using one standard deviation from the mean of triplicate detections. The background (Bg) signal served as the negative control. The symbols M1 to M9 represent the neoglycoproteins ManGlcNAc2Asn-BSA to Man9GlcNAc2-BSA (Compounds 23–31).
CONCLUSION
A facile construction of a library of high-mannose-type N-glycans was achieved by a top-down chemoenzymatic approach by enzymatic trimming of two readily available natural N-glycans coupled with chromatographic separation. The method provides an easy access to a comprehensive panel of oligomannose structures ranging from Man1GlcNAc2 to Man9GlcNAc2 oligosaccharides and the related oligomannose–BSA conjugates. The usefulness of the synthetic oligomannose–BSA conjugates was exemplified by the identification of the novel glycoepitopes of several mannose-specific lectins and the HIV-neutralizing antibody 2G12 in a microarray setting. Identifying new mannose-binding proteins and antibodies, especially those with the GNA-like broad-spectrum virus-neutralizing activities, may lead to new insight in the design and development of vaccines and therapeutic agents targeting HIV-1 and other oligomannose-expressing pathogens.
EXPERIMENTAL PROCEDURES
Materials and General Methods
Untoasted soy flour was obtained from Archer-Daniel-Midlands (ADM) (Chicago, IL). α1-2-mannosidase from Bacteroides thetaiotaomicron was expressed following the previously described procedure.34 Neuraminidase, α1-2/3-mannosidase, α1-6-mannosidase, and β-N-acetyl-glucosaminidase were purchased from New England Biolabs. β1-4-galactosidase was purchased from Sigma. Reverse-phase analytical high-performance liquid chromatography was performed using a Waters Alliance e2956 HPLC system using a Waters XBridge C18 column (4.6 × 250 mm, 3.5 μm). Solvent A was water containing 0.1% trifluoroacetic acid (TFA). Solvent B was acetonitrile containing 0.1% TFA. Normal phase HPLC was performed with a YMC Amide-bonding NH2 column (4.6 × 250 mm, 3.5 μm). Preparative HPLC was performed using a Waters (600e) HPLC system. The RP preparative column used was a Waters XBridge Shield C18 column. HILIC-HPLC was performed using a Waters BEH Glycan column (4.6 × 250 mm, 3 μm), with 100 mM ammonium formate (pH 4.6) [solvent A] and acetonitrile (solvent B). A linear gradient was used for HILIC-HPLC from 22 → 44%A in 80 min. Electrospray ionization mass spectrometry was performed using a Thermo Q Exactive LC/MS. Matrix-assisted laser desorption ionization with time-of-flight detection was performed using a Bruker UltrafleXtreme MALDI TOF/TOF Mass Spectrometer with a dihydroxybenzoic acid/dimethylamide (DHB/DMA) matrix. High-performance anion-exchange chromatography with pulsed-amperometric detection (HPAEC-PAD) was performed using a Dionex 9000 system equipped with a Carbopac PA200 anion-exchange column (Thermo). Fast Protein Liquid Chromatography (FPLC) was performed using a General Electric (GE) Akta Explorer equipped with a GE Sephadex G-25 column.
Preparation of Man9GlcNAc2Asn and Its Fmoc-Tagged Derivative
Man9GlcNAc2Asn was prepared by enzymatic digestion of soybean agglutinin (SBA) and subsequent size-exclusion chromatographic purification. Crude SBA was obtained from untoasted soybean flour following a modified method from that reported by our group and others.31,32 Briefly, 0.8 kg untoasted soybean flour was suspended in 4 L 0.9% saline solution containing 0.02% sodium azide. The pH of the suspension was adjusted to 4.6 using 2 M hydrogen chloride, and vigorously stirred at 4 °C for 2 h. The suspension was centrifuged at 8000 rpm for 20 min, and to the supernatant was added 30% (NH4)2SO3 and the solution was stirred for 2 h at 4 °C. Again the solution was centrifuged (8000 rpm for 20 min), and ammonium sulfate was added to the supernatant to a final concentration of 60%. The solution was stirred vigorously overnight and centrifuged at 8500 rpm for 30 min. The pellet was dissolved in 400 mL water, and the crude SBA was dialyzed against 0.2% saline with 3 changes of dialysis solution (2 h for each). The SBA was dialyzed against running water overnight. The dialyzed protein was centrifuged (8000 rpm/20 min) and the pH of the supernatant was adjusted to 2. Pepsin from porcine gut mucosa was added (1:20 protein to enzyme as assessed by Nanodrop estimation) and the solution was shaken at 37 °C overnight to digest the proteins to peptides/glycopeptides. The pH of the solution was adjusted to pH 8 with 2 M NaOH. The solvent was reduced in vacuo to 60 mL, and CaCl2 and NaN3 was added to final concentrations of 5 mM and 0.02%, respectively. Pronase was added (1:50 enzyme to protein ratio) and the solution was incubated overnight at 55 °C. The release of Man9GlcNAc2Asn was monitored by high-performance anion exchange chromatography coupled with pulsed amperometric detection (HPAEC-PAD). The enzyme digestion was lyophilized and purified by size-exclusion chromatography (Sephadex G50) with monitoring by the phenol-sulfuric acid test (for carbohydrates). The carbohydrate positive fractions were pooled and lyophilized to yield crude Man9GlcNAc2Asn, which was further purified by C18 solid phase extraction (SPE) eluted by water. To a solution of Man9GlcNAc2Asn (20 mg, 10 μmol) in PBS (1 mL, pH 7.4) was added Fmoc-OSu (20 mg, 59 μmol) dissolved in dimethylformamide (200 μL). The solution was shaken at RT for 4 h, and the product was precipitated by the addition of a 10-fold excess of cold (−20 °C) acetone. The crude product 2 was dissolved in water and purified by RP-HPLC (0–90% B in 30 min, tR = 16 min), yielding Man9GlcNAc2Asn-Fmoc (2) (19.6 mg, 88% yield). MALDI-TOF: Calcd for C89H135N4O60 [M + H]+, 2221.03; found: m/z 2221.17.
Preparation of the Asn-Linked Complex Type N-Glycan from the Sialoglycopeptide (SGP) and its Fmoc Derivative
SGP was isolated from hen egg yolk following the previously reported procedure.36–38 The purified SGP was dissolved in 80 mM Tris buffer (pH 8.0) containing 5 mM CaCl2 and Pronase was added (final concentration 0.1 mg/mL), and the digestion was incubated at 55 °C overnight. The released SCT-Asn glycan was purified by G50 SEC, monitored by the phenol-sulfuric acid test. The carbohydrate positive fractions were pooled and lyophilized. Semipure SCT-Asn was further purified by C18 solid phase extraction (SPE) eluted by water. To a solution of SCT-Asn (30 mg, 12.8 μmol) in PBS (300 μL, pH 7.4) was added Fmoc-OSu (30 mg, 89 μmol) dissolved in dimethylformamide (300 μL). The solution was shaken at RT for 4 h, and the target was precipitated by the addition of a 10-fold excess of cold (−20 °C) acetone. The crude product 8 was dissolved in water and purified by RP-HPLC (0–90% B in 30 min, tR = 15 min), yielding the target compound 8 (29 mg, 89% yield). ESI-MS: Calcd for C103H155N8O66 [M + H]+, 2559.90; found: m/z 2559.34.
Chemoenzymatic Preparation of High-Mannose N-Glycans (3–6) through α-1,2-Mannosidase Digestion of Man9GlcNAc2Ans-Fmoc (2)
To a 2 mM solution of 2 (27 mg, 12.2 μmol) in sodium citrate buffer (50 mM, pH 5.6) containing CaCl2 (10 mM) was added α1,2-mannosidase (final concentration = 2 ng/μL). The solution was incubated at RT for 30 min, while monitoring by HPAEC-PAD. Before HPAEC-PAD analysis, the Fmoc was removed by incubating 1 μL of the enzymatic digestion with 1 μL 0.5 NaOH at 60 °C for 5 min. After achieving optimal digestion to yield the oligomannose intermediates, the reaction was quenched by addition of excess 0.1% TFA. The individual oligomannose glycans were separated by NP-HPLC, using a gradient of 22–41%B in 40 min. The separated oligomannose glycans were desalted using RP-HPLC, yielding respective oligosaccharides in milligram quantities (2, 3 mg, 1.40 μmol, 16%; 3, 4 mg, 1.95 μmol, 21%; 4, 5 mg, 2.64 μmol, 26%; 5, 4 mg, 2.31 μmol, 21%; 6, 3 mg, 1.91 μmol, 16%; 19 mg total recovered, 82% estimated yield [based on Man7]).
Compound 3 (Man8GlcNAc2Asn-Fmoc)
NP-HPLC: tR = 20.7 min. MALDI-TOF MS: Calcd for C83H125N4O55 [M + H]+, 2057.71; found: m/z 2057.13. HPAEC-PAD: tR = 35.67 min.
Compound 4 (Man7GlcNAc2Asn-Fmoc)
NP-HPLC: tR = 19.9 min. MALDI-TOF MS: Calcd for C77H115N4O50 [M + H]+, 1895.66; found: m/z 1894.96. HPAEC-PAD: tR = 29.4 min.
Compound 5 (Man6GlcNAc2Asn-Fmoc)
NP-HPLC: tR = 18.5 min. MALDI-TOF MS (Negative mode): Calcd for C71H103N4O45 [M - H]−, 1732.59; found: m/z 1732.83. HPAEC-PAD: tR = 22.7 min.
Compound 6 (Man5GlcNAc2Asn-Fmoc)
NP-HPLC: tR = 17.8 min. MALDI-TOF MS (Negative mode): Calcd for C65H93N4O40 [M - H]−, 1570.45; found: m/z 1570.80. HPAEC-PAD: tR = 15.9 min.
Synthesis of Man4GlcNAc2Asn-Fmoc (7) from glycan Man5Asn-Fmoc (6)
To a solution of 6 (10 mg, 6.37 μmol) in sodium citrate buffer (50 mM, pH 6.0) containing CaCl2 (10 mM) was added α-1,2/3-mannosidase (final concentration, 1 μg/μL). The enzymatic reaction mixture was incubated at 37 °C overnight. The complete conversion of Man5 to Man4 was monitored by HPAEC-PAD and ESI-MS. The product was purified by RP-HPLC (0–90% B, tR = 16 min), yielding the target compound 7 (8.1 mg, 90% yield). MALDI-TOF MS (positive mode): Calcd for C59H83N4O35 [M + H]+, 1410.32; found: m/z 1409.67.
Synthesis of the Man3GlcNAc2Asn-Fmoc Derivative (11)
The starting material 8 (20 mg, 7.8 μmol) was digested to the core Man3GlcNAc2-Asn-Fmoc core 11, following a modified version of the previously reported one-pot enzymatic procedure.37 Briefly, 8 (20 mg, 7.8 μmol) was dissolved in 50 mM sodium acetate buffer (50 mM, pH 5.5, pH adjusted with 0.5 M NaOH). To the solution was added neuraminidase (60 U) and the mixture was incubated at 37 °C. After complete removal of sialic acid, as determined by HPAEC-PAD/ESI-MS analysis, the pH was adjusted to 4.5 with 1 M HCl and β-galactosidase (32 U) was added. The solution was incubated at 37 °C overnight. After removal of terminal galactose, the pH was adjusted to 5.5, β-N-acetyl-glucosaminidase (0.2 U) was added, and the solution was again incubated overnight at 37 °C. The product was purified by RP-HPLC to provide the Man3GlcNAc2Asn-Fmoc derivative (11) (9.1 mg, 94%). MALDI-TOF MS (Positive mode): Calcd for C53H75N4O30 [M + H]+, 1247.45; found: m/z 1247.92.
Synthesis of Man2GlcNAc2Asn-Fmoc (12) from Compound 11
To a solution of 11 (7 mg, 5.6 μmol) in sodium citrate buffer (50 mM, pH 6.0) was added α-1,2/3-mannosidase (960 U) and the solution was incubated overnight at 37 °C. The product was purified by RP-HPLC to afford the Man2GlcNAc2Asn-Fmoc (12) (6 mg, 98% yield). MALDI-TOF MS (Positive mode): Calcd for C47H65N4O25 [M + H]+, 1085.39; found: m/z 1085.79.
Synthesis of Man1GlcNAc2Asn-Fmoc (13) from Compound 12
To a solution of 12 (3 mg, 2.7 μmol) in sodium citrate buffer (50 mM, pH 6.0) was added α-1,6-mannosidase (1200 U) and the solution was incubated overnight at 37 °C. The product was purified by RP-HPLC to give the Man1GlcNAc2Asn-Fmoc (13) (2.3 mg, 92% yield). ESI-MS: Calcd for C41H55N4O20 [M + H]+, 923.34; found: m/z 923.10.
General Procedure for Removal of the Asn-Linked Fmoc Group in the Fmoc-Tagged N-Glycans
Fmoc-tagged compounds 2–10 were dissolved in a small amount of water. Piperidine in DMF was added to give a final concentration of 20% in 1:9 water/DMF. The reaction mixture was shaken at room temperature for 30 min, when HPLC indicated the completion of deprotection. The reaction mixture was neutralized with acetic acid and lyophilized. The product was precipitated using cold 80% acetone. The white precipitate was collected, dried, and used in the next step without further purification.
Preparation of Maleimide-Functionalized Oligomannose Glycans (14–22)
General Procedures
The respective Asn-linked oligomannose glycan was dissolved in 1× PBS (final concentration = 25 mg/mL). Then a solution of 8 equiv of N-ε-malemidocaproyl-oxysuccinimide ester (EMCS) in acetonitrile was added. The solution was shaken at room temperature. When RP-HPLC (0–50%B in 15 min) indicated the completion of the reaction (1–3 h), the solution was diluted with water containing 0.1% TFA and the product was purified by RP-HPLC to give the respective maleimide-functionalized oligomannose.
Compound 14 (Man1GlcNAc2Asn-Maleimide)
From 13 (3 mg, 4.2 μmol), recovered yield (14): 3.2 mg, 84%. RP-HPLC: tR = 14.8 min. MALDI-TOF MS (positive mode): Calcd for C36H56N5O21 [M + H]+, 894.35; found: m/z 894.14.
Compound 15 (Man2GlcNAc2Asn-Maleimide)
From 12 (3 mg, 3.4 μmol), recovered yield (15): 3.0 mg, 83%. RP-HPLC: tR = 14.9 min. MALDI-TOF MS (positive mode): Calcd for C42H66N5O26 [M + H]+, 1057.00; found: m/z 1057.64.
Compound 16 (Man3GlcNAc2Asn-Maleimide)
From 11 (3 mg, 2.9 μmol), recovered yield (16): 3.0 mg, 86%. RP-HPLC: tR = 14.9 min. MALDI-TOF MS (positive mode): Calcd for C48H75N5O31 [M + H]+, 1219.14; found: m/z 1219.47.
Compound 17 (Man4GlcNAc2Asn-Maleimide)
From 7 (3 mg, 2.5 μmol), recovered yield (17): 3.1 mg, 89%. RP-HPLC: tR = 15.0 min. MALDI-TOF MS (positive mode): Calcd for C54H85N5NaO36 [M + Na]+, 1402.49; found: m/z 1402.12.
Compound 18 (Man5GlcNAc2Asn-Maleimide)
From 6 (3 mg, 2.2 μmol), recovered yield (18): 3.0 mg, 88%. RP-HPLC: tR = 15.0 min. MALDI-TOF MS (positive mode): Calcd for C60H95N5NaO41 [M + Na]+, 1564.54; found: m/z 1564.23.
Compound 19 (Man6GlcNAc2Asn-Maleimide)
From 5 (3 mg, 1.9 μmol), recovered yield (19): 2.7 mg, 84%. RP-HPLC: tR = 15.1 min. MALDI-TOF MS (positive mode): Calcd for C66H105N5NaO46 [M + Na]+, 1726.59; found: m/z 1726.88.
Compound 20 (Man7GlcNAc2Asn-Maleimide)
From 4 (3 mg, 1.7 μmol), recovered yield (20): 2.9 mg, 90%. RP-HPLC: tR = 15.1 min. MALDI-TOF MS (positive mode): Calcd for C72H115N5NaO51 [M + Na]+, 1888.65; found: m/z 1888.79.
Compound 21 (Man8GlcNAc2Asn-Maleimide)
From 3 (3 mg, 1.6 μmol), recovered yield (21): 2.8 mg, 84%. RP-HPLC: tR = 15.2 min. MALDI-TOF MS (positive mode): Calcd for C78H125N5NaO56 [M + Na]+, 2050.70; found: m/z 2050.12.
Compound 22 (Man9GlcNAc2Asn-Maleimide)
From 2 (3 mg, 1.5 μmol), recovered yield (22): 2.9 mg, 88%. RP-HPLC: tR = 15.2 min. MALDI-TOF MS (positive mode): Calcd for C84H135N5NaO61 [M + Na]+, 2212.75; found: m/z 2212.78.
Sulfhydryl Derivitization of Bovine Serum Albumin (BSA)
Sulfhydryl groups were installed on the surface of BSA following a procedure previously reported by our groups.39 In brief, 20 mg of BSA was dissolved in 1× PBS containing 5 mM EDTA. Twenty equivalents of Traut’s reagent was added, and the mixture was shaken at room temperature for 2 h. The thiol-derivatized BSA was purified by FPLC (G25). The protein positive fractions (Bradford assay) were pooled and concentrated. The quantity of recovered protein (18 mg) was estimated by Nanodrop analysis. Free sulfhydryl groups loaded on the surface of BSA was quantified by Ellman’s reagent against an L-cysteine standard curve. The sulfhydryl-containing BSA was used immediately in the next step to avoid disulfide formation and protein aggregation.
Conjugation of Maleimide-Tagged Oligomannose Glycans with Sulfhydryl-Derivatized BSA
Sulfhydryl-derivitized BSA (21) (4 mg, 2 μmol sulfhydryl groups) was dissolved in a phosphate buffer (20 mM, pH 7.2) containing EDTA (5 mM). Maleimide-tagged glycan (2 μmol) was added and the solution was shaken at room temperature for 3 h and purified by FPLC (G-25) using a 10 mM phosphate buffer (pH 6.6). Protein positive fractions were pooled, and carbohydrate-loading was estimated by phenol-sulfuric acid assay, quantified against a mannose monosaccharide standard curve. The loading of carbohydrates ranged from 12% to 20% for oligomannoses (Man5GlcNAc2 to Man9GlcNAc2) by weight, which corresponds to 6–10 copies of oligomannose per BSA molecule on average, respectively.
Release of the Oligomannoses from the BSA-Glycoconjugates by Endo-A Treatment and MALDI-TOF MS Analysis
A mixture of glycoconjugate (100 μg) and Endo-A enzyme (5 μg) in a buffer (PBS, 100 mM, pH 6.0, 100 μL) was incubated at 37 °C for 5 h. The reaction mixture was quenched by adding 0.1% TFA solution (1 mL) and then the released glycans were subject to purification on a HyperSep Hypercarb SPE Cartridge (Thermo), following the previously reported method.42 Briefly, the column was washed with water (2 × 1 mL) to remove salts and then with 30% (v/v) acetonitrile (2 × 1 mL) containing 0.1% TFA to elute the oligomannose N-glycan. The purified oligomannose N-glycans were analyzed by MALDI-TOF MS to confirm the identity of the released N-glycans. MALDI-TOF MS of the released N-glycan from glycoconjugate 25: Calcd for Man3GlcNAc, M = 707.25 Da; found (m/z), 730.42 [M + Na]+. MALDI-TOF MS of the released N-glycan from glycoconjugate 26: Calcd for Man4GlcNAc, M = 869.30 Da; found (m/z), 892.24 [M + Na]+.
MALDI-TOF MS of the released N-glycan from glycoconjugate 27: Calcd for Man5GlcNAc, M = 1031.35 Da; found (m/z), 1054.33 [M + Na]+.
MALDI-TOF MS of the released N-glycan from glycoconjugate 28: Calcd for Man6GlcNAc, M = 1193.41 Da; found (m/z), 1216.47 [M + Na]+.
MALDI-TOF MS of the released N-glycan from glycoconjugate 29: Calcd for Man7GlcNAc, M = 1355.46 Da; found (m/z), 1378.56 [M + Na]+.
MALDI-TOF MS of the released N-glycan from glycoconjugate 30: Calcd for Man8GlcNAc, M = 1517.51 Da; found (m/z), 1540.64 [M + Na]+.
MALDI-TOF MS of the released N-glycan from glycoconjugate 31: Calcd for Man9GlcNAc, M = 1679.57 Da; found (m/z), 1702.34 [M + Na]+.
Carbohydrate Microarray Analysis Using the Oligomannose-Conjugates
Carbohydrate microarray application was performed as previously described.39,40 In brief, carbohydrate antigens of various complexities were dissolved in phosphate-buffered saline (PBS) and spotted onto SuperEpoxy 2 Protein slides (ArrayIt Corporation, Sunnyvale, CA). Immediately before use, the printed microarray slides were washed in 1× PBS at RT for 5 min, and blocked with 1%BSA–PBS at RT for 30 min. They were then incubated with 50 μL of antibodies or lectins (as described in Figure 2 caption) at RT for 1 h followed by washing and then incubated with titrated secondary antibodies or streptavidin conjugated with Alexa647 at RT for 30 min. The stained slides were rinsed five times and spun dry at room temperature before scanning for fluorescent signals. The ScanArray5000A Microarray Scanner (PerkinElmer Life Science, Boston, MA) was used to scan the stained microarrays. Fluorescent intensity values for each array spot and its background were calculated using ScanArray Express software (PerkinElmer Life Science, Boston, MA).
Acknowledgments
This work was supported in part by NIH grants R01AI113896 (to L.X.W.), R56AI118464 (to D.W.), and R21AI124068 (to D.W.).
ABBREVIATIONS
- SGP
Sialoglycopeptide
- BSA
Bovine serum albumin
- GNA
Galanthus nivalis lectin
- HIV
Human immunodeficiency virus
- bnAbs
Broadly neutralizing antibodies
- SARS-CoV
Severe acute respiratory syndrome coronavirus
- HCMV
Human cytomegalovirus
- RP-HPL
CReverse-Phase High-performance liquid chromatography
- NP-HPLC
Normal-phase High-performance liquid chromatography
- MALDI-TOF MS
Matrix-assisted laser desorption ionization-time-of-flight mass spectrometry
- NPA
Narcissus pseudonarcissus lectin
Footnotes
Notes
The authors declare no competing financial interest.
References
- 1.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 2.Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Go EP, Liao HX, Alam SM, Hua D, Haynes BF, Desaire H. Characterization of Host-Cell Line Specific Glycosylation Profiles of Early Transmitted/Founder HIV-1 gp120 Envelope Proteins. J Proteome Res. 2013;12:1223–1234. doi: 10.1021/pr300870t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Go EP, Chang Q, Liao HX, Sutherland LL, Alam SM, Haynes BF, Desaire H. Glycosylation site-specific analysis of clade C HIV-1 envelope proteins. J Proteome Res. 2009;8:4231–4242. doi: 10.1021/pr9002728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Go EP, Irungu J, Zhang Y, Dalpathado DS, Liao HX, Sutherland LL, Alam SM, Haynes BF, Desaire H. Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes’ accessibility. J Proteome Res. 2008;7:1660–1674. doi: 10.1021/pr7006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonomelli C, Doores KJ, Dunlop DC, Thaney V, Dwek RA, Burton DR, Crispin M, Scanlan CN. The Glycan Shield of HIV Is Predominantly Oligomannose Independently of Production System or Viral Clade. PLoS One. 2011;6:e23521. doi: 10.1371/journal.pone.0023521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doores KJ, Bonomelli C, Harvey DJ, Vasiljevic S, Dwek RA, Burton DR, Crispin M, Scanlan CN. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc Natl Acad Sci U S A. 2010;107:13800–13805. doi: 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reitter JN, Means RE, Desrosiers RC. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 9.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 10.Geijtenbeek TB, Kwon DS, Torensma R, van Vliet SJ, van Duijnhoven GC, Middel J, Cornelissen IL, Nottet HS, KewalRamani VN, Littman DR, et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 11.Pohlmann S, Baribaud F, Doms RW. DC-SIGN and DC-SIGNR: helping hands for HIV. Trends Immunol. 2001;22:643–646. doi: 10.1016/s1471-4906(01)02081-6. [DOI] [PubMed] [Google Scholar]
- 12.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 14.Walker LM, Sok D, Nishimura Y, Donau O, Sadjadpour R, Gautam R, Shingai M, Pejchal R, Ramos A, Simek MD, et al. Rapid development of glycan-specific, broad, and potent anti-HIV-1 gp120 neutralizing antibodies in an R5 SIV/HIV chimeric virus infected macaque. Proc Natl Acad Sci U S A. 2011;108:20125–20129. doi: 10.1073/pnas.1117531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PN, Spencer DI, Seaman MS, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A. 2012;109:E3268–3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritchie G, Harvey DJ, Feldmann F, Stroeher U, Feldmann H, Royle L, Dwek RA, Rudd PM. Identification of N-linked carbohydrates from severe acute respiratory syndrome (SARS) spike glycoprotein. Virology. 2010;399:257–269. doi: 10.1016/j.virol.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma S, Wisner TW, Johnson DC, Heldwein EE. HCMV gB shares structural and functional properties with gB proteins from other herpesviruses. Virology. 2013;435:239–249. doi: 10.1016/j.virol.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D, Tang J, Tang J, Wang LX. Targeting N-glycan cryptic sugar moieties for broad-spectrum virus neutralization: progress in identifying conserved molecular targets in viruses of distinct phylogenetic origins. Molecules. 2015;20:4610–4622. doi: 10.3390/molecules20034610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang SK, Liang PH, Astronomo RD, Hsu TL, Hsieh SL, Burton DR, Wong CH. Targeting the carbohydrates on HIV-1: Interaction of oligomannose dendrons with human monoclonal antibody 2G12 and DC-SIGN. Proc Natl Acad Sci U S A. 2008;105:3690–3695. doi: 10.1073/pnas.0712326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuo I, Wada M, Manabe S, Yamaguchi Y, Otake K, Kato K, Ito Y. Synthesis of monoglucosylated high-mannose-type dodecasaccharide, a putative ligand for molecular chaperone, calnexin, and calreticulin. J Am Chem Soc. 2003;125:3402–3403. doi: 10.1021/ja021288q. [DOI] [PubMed] [Google Scholar]
- 25.Koizumi A, Matsuo I, Takatani M, Seko A, Hachisu M, Takeda Y, Ito Y. Top-down chemoenzymatic approach to a high-mannose-type glycan library: synthesis of a common precursor and its enzymatic trimming. Angew Chem, Int Ed. 2013;52:7426–7431. doi: 10.1002/anie.201301613. [DOI] [PubMed] [Google Scholar]
- 26.Fujikawa K, Koizumi A, Hachisu M, Seko A, Takeda Y, Ito Y. Construction of a high-mannose-type glycan library by a renewed top-down chemo-enzymatic approach. Chem - Eur J. 2015;21:3224–3233. doi: 10.1002/chem.201405781. [DOI] [PubMed] [Google Scholar]
- 27.Song X, Lasanajak Y, Rivera-Marrero C, Luyai A, Willard M, Smith DF, Cummings RD. Generation of a natural glycan microarray using 9-fluorenylmethyl chloroformate (FmocCl) as a cleavable fluorescent tag. Anal Biochem. 2009;395:151–160. doi: 10.1016/j.ab.2009.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song X, Xia B, Stowell SR, Lasanajak Y, Smith DF, Cummings RD. Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem Biol. 2009;16:36–47. doi: 10.1016/j.chembiol.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song X, Lasanajak Y, Xia B, Heimburg-Molinaro J, Rhea JM, Ju H, Zhao C, Molinaro RJ, Cummings RD, Smith DF. Shotgun glycomics: a microarray strategy for functional glycomics. Nat Methods. 2011;8:85–90. doi: 10.1038/nmeth.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song X, Ju H, Lasanajak Y, Kudelka MR, Smith DF, Cummings RD. Oxidative release of natural glycans for functional glycomics. Nat Methods. 2016;13:528–534. doi: 10.1038/nmeth.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lis H, Sharon N. Soybean agglutinin–a plant glycoprotein. Structure of the carbohydrate unit. J Biol Chem. 1978;253:3468–3476. [PubMed] [Google Scholar]
- 32.Wang LX, Ni J, Singh S, Li H. Binding of high-mannose-type oligosaccharides and synthetic oligomannose clusters to human antibody 2G12: implications for HIV-1 vaccine design. Chem Biol. 2004;11:127–134. doi: 10.1016/j.chembiol.2003.12.020. [DOI] [PubMed] [Google Scholar]
- 33.Makimura Y, Kiuchi T, Izumi M, Dedola S, Ito Y, Kajihara Y. Efficient synthesis of glycopeptide-alpha-thioesters with a high-mannose type oligosaccharide by means of tert-Boc-solid phase peptide synthesis. Carbohydr Res. 2012;364:41–48. doi: 10.1016/j.carres.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 34.Zhu Y, Suits MD, Thompson AJ, Chavan S, Dinev Z, Dumon C, Smith N, Moremen KW, Xiang Y, Siriwardena A, et al. Mechanistic insights into a Ca2+-dependent family of alpha-mannosidases in a human gut symbiont. Nat Chem Biol. 2010;6:125–132. doi: 10.1038/nchembio.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong-Madden ST, Landry D. Purification and characterization of novel glycosidases from the bacterial genus Xanthomonas. Glycobiology. 1995;5:19–28. doi: 10.1093/glycob/5.1.19. [DOI] [PubMed] [Google Scholar]
- 36.Seko A, Koketsu M, Nishizono M, Enoki Y, Ibrahim HR, Juneja LR, Kim M, Yamamoto T. Occurence of a sialylglycopeptide and free sialylglycans in hen’s egg yolk. Biochim Biophys Acta, Gen Subj. 1997;1335:23–32. doi: 10.1016/s0304-4165(96)00118-3. [DOI] [PubMed] [Google Scholar]
- 37.Sun B, Bao W, Tian X, Li M, Liu H, Dong J, Huang W. A simplified procedure for gram-scale production of sialylglycopeptide (SGP) from egg yolks and subsequent semi-synthesis of Man3GlcNAc oxazoline. Carbohydr Res. 2014;396:62–69. doi: 10.1016/j.carres.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 38.Liu L, Prudden AR, Bosman GP, Boons GJ. Improved isolation and characterization procedure of sialylglycopeptide from egg yolk powder. Carbohydr Res. 2017;452:122–128. doi: 10.1016/j.carres.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D, Dafik L, Nolley R, Huang W, Wolfinger RD, Wang LX, Peehl DM. Anti-Oligomannose Antibodies as Potential Serum Biomarkers of Aggressive Prostate Cancer. Drug Dev Res. 2013;74:65–80. doi: 10.1002/ddr.21063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L, Cummings RD, Smith DF, Huflejt M, Campbell CT, Gildersleeve JC, Gerlach JQ, Kilcoyne M, Joshi L, Serna S, et al. Cross-platform comparison of glycan microarray formats. Glycobiology. 2014;24:507–517. doi: 10.1093/glycob/cwu019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ni J, Song H, Wang Y, Stamatos NM, Wang LX. Toward a carbohydrate-based HIV-1 vaccine: synthesis and immunological studies of oligomannose-containing glycoconjugates. Bioconjugate Chem. 2006;17:493–500. doi: 10.1021/bc0502816. [DOI] [PubMed] [Google Scholar]
- 42.Packer NH, Lawson MA, Jardine DR, Redmond JW. A general approach to desalting oligosaccharides released from glycoproteins. Glycoconjugate J. 1998;15:737–747. doi: 10.1023/a:1006983125913. [DOI] [PubMed] [Google Scholar]