Abstract
Purpose
To analyze how automatic segmentation translates in accuracy and precision to morphology and relaxometry compared with manual segmentation and increases the speed and accuracy of the work flow that uses quantitative magnetic resonance (MR) imaging to study knee degenerative diseases such as osteoarthritis (OA).
Materials and Methods
This retrospective study involved the analysis of 638 MR imaging volumes from two data cohorts acquired at 3.0 T: (a) spoiled gradient-recalled acquisition in the steady state T1ρ-weighted images and (b) three-dimensional (3D) double-echo steady-state (DESS) images. A deep learning model based on the U-Net convolutional network architecture was developed to perform automatic segmentation. Cartilage and meniscus compartments were manually segmented by skilled technicians and radiologists for comparison. Performance of the automatic segmentation was evaluated on Dice coefficient overlap with the manual segmentation, as well as by the automatic segmentations’ ability to quantify, in a longitudinally repeatable way, relaxometry and morphology.
Results
The models produced strong Dice coefficients, particularly for 3D-DESS images, ranging between 0.770 and 0.878 in the cartilage compartments to 0.809 and 0.753 for the lateral meniscus and medial meniscus, respectively. The models averaged 5 seconds to generate the automatic segmentations. Average correlations between manual and automatic quantification of T1ρ and T2 values were 0.8233 and 0.8603, respectively, and 0.9349 and 0.9384 for volume and thickness, respectively. Longitudinal precision of the automatic method was comparable with that of the manual one.
Conclusion
U-Net demonstrates efficacy and precision in quickly generating accurate segmentations that can be used to extract relaxation times and morphologic characterization and values that can be used in the monitoring and diagnosis of OA.
© RSNA, 2018
Introduction
Osteoarthritis (OA) is a leading cause of chronic disabilities in the United States. OA of the knee is one of the most common forms of arthritis, which causes substantial social and economic impact. Conservative estimates of its prevalence in the U.S. population indicate that 26.9 million U.S. adults are affected (1). Its prevalence is on the rise, with incidences expected to increase to 59 million by 2020 (2).
Magnetic resonance (MR) imaging–based compositional quantitative data (relaxometry) and morphologic quantitative data have become central imaging metrics for studying long-range outcomes in OA (3). MR imaging–based quantification of articular cartilage volume and thickness has been widely investigated (4,5). Additionally, T1ρ and T2 mappings have shown the ability to reveal the risk of posttraumatic OA after anterior cruciate ligament (ACL) injury and reconstruction (6,7). Despite the evidence on the value of these quantitative MR imaging techniques for the assessment and tracking of OA (8–10), one of the obstacles in the clinical translation of these promising techniques is the time-consuming image postprocessing, particularly joint and musculoskeletal tissue segmentation, which is often performed manually or semiautomatically (8,10) and is affected by inter- and intrauser variability (11).
In the past few years, major efforts have been undertaken to develop automatic algorithms for the extraction of quantitative OA relaxometry and morphology data (12–14). However, there is still a lack of commonly accepted and widely distributed methods to solve this task (11). There is a crucial need for the development of a fully automatic knee segmentation method that is quick, accurate, precise, and able to reliably extract relaxation times and morphologic features from MR imaging data.
Deep neural networks, a subset of machine learning techniques, have become a popular method for solving a variety of computational problems, many of which are concerned with image analysis and have recently been applied to radiology and OA images (15–17). Deeper layers of a convolutional neural network can extract detailed lower-level information from the original image, which is very appealing for problems in radiology.
To our knowledge, our article presents one of the first examples of deep convolution neural networks to automatically segment and classify different subcompartments of the knee at MR imaging—femoral cartilage (FC), lateral tibial cartilage (LTC), medial tibial cartilage (MTC), patellar cartilage (PC), lateral meniscus (LM), and medial meniscus (MM). Putting our results in the perspective of OA research and clinical application, the aims of our study were to (a) analyze how our automatic segmentation performances translate in accuracy and precision to morphology and relaxometry in OA compared with manual segmentations and (b) increase the speed and accuracy of the work flow that uses quantitative MR imaging to study knee degenerative diseases such as OA.
Materials and Methods
Our retrospective study was performed in accordance with the regulations of the Committee for Human Research of the home institution prior to scanning. All subjects provided written informed consent. Part of this study was funded by GE Healthcare IT Business. Data were collected before collaboration with GE Healthcare, and the authors had control of the information and data at all points of the study.
Subjects
Two separate imaging data sets (both acquired at 3.0 T) were used for training, validating, and testing our model. The first data set consisted of 464 spoiled gradient-recalled acquisition in the steady state T1ρ-weighted image volumes (termed the “T1ρ-weighted data set”) that were acquired from three research studies and included patients with ACL injuries, patients with OA (Kellgren-Lawrence [KL] grade > 1), and control subjects. This T1ρ-weighted data set was used for the relaxometry analysis, as T1ρ and T2 maps were available for these subjects. We used the baseline and 12-month follow-up studies for 49 of these subjects whose condition was longitudinally stable (KL score = 0 across time points) to assess the precision of the proposed method in extracting T1ρ and T2 values. The second data set consisted of 174 three-dimensional (3D) double-echo steady-state (DESS) volumes (termed “DESS data set”) that were acquired from the Osteoarthritis Initiate Dataset (OAI) (18) and included data obtained in both patients with OA and control subjects at baseline and 12 months. This data set was used for morphometry analysis, as it was of higher spatial resolution than the T1ρ-weighted data set. Breakdown of subject demographics and MR imaging parameters for the two data sets are summarized in Table 1, and a flowchart of the data selection can be viewed in Figure 1.
Table 1.
Data Set Demographic Breakdown

Note.—Unless otherwise specified, data are means, with ranges in parentheses.
*A magnetization prepared angle-modulated partitioned k-space spoiled gradient-echo snapshots sequence with the following parameters: repetition time msec/echo time msec, 9/2.6; field of view, 14 cm; matrix, 256 × 128; section thickness, 4 mm; bandwidth, 62.5 kHz; and final image resolution, 0.56 × 0.56 × 4 mm.
†Data are numbers of patients, with percentages in parentheses.
‡Data are means, with 95% confidence intervals in parentheses.
§Performed with the following parameters: 16.2/4.7; field of view, 14 cm; matrix, 307 × 348; bandwidth, 62.5 kHz; and final image resolution, 0.346 × 0.346 × 0.7 mm.
Figure 1:
Data flow and exclusion process from the data sets used in this study. ACL = anterior cruciate ligament, DESS = double-echo steady state, OA = osteoarthritis, OAI = Osteoarthritis Initiate Dataset.
Image Annotation and Relaxometry/Morphology Data Extraction
The T1ρ-weighted data set was manually segmented in-house by skilled technicians, with the supervision of radiologists and by using an in-house MATLAB-based program (Mathworks, Natick, Mass), only after they had passed strict training where their segmentations met the metric standard proposed by Carballido-Gamio et al (10) in morphology analysis and the standard of Li et al (19) for relaxation time analysis. The DESS data set was manually segmented; images and segmentation masks are available through the OAI (20). Manual annotations were performed for the identification of the FC, LTC, MTC, PC, LM, and MM. Relaxation time analysis was performed in the T1ρ-weighted data set. T1ρ and T2 maps were computed on a pixel-by-pixel basis by using a two-parameter Levenberg-Marquardt monoexponential (21). The mean T1ρ and T2 values were calculated for each compartment by using the respective segmentation method’s mask. Cartilage morphologic analysis was performed in the DESS data set. Thicknesses were computed for each point in the bone-cartilage interfaces and were transferred to the bone surfaces by using a previously presented method (10). Corresponding anatomic points were computed on the basis of 3D shape descriptors to register bones with affine and elastic transformations and were then used to perform a point-to-point comparison of cartilage thickness values (V.P., with 8 years of experience).
Model Architecture
The neural network model chosen for this problem is based on the U-Net architecture, which has previously shown promising results in the tasks of segmentation, particularly for medical images (15,22–25), and has fewer trainable parameters than the other popular segmentation architecture, SegNet (26). The U-Net architecture can be viewed in Figure E1 (online). The network takes a full image section as input and then, through a series of trainable weights, creates the corresponding section segmentation mask (22).
Our U-Net model uses a weighted cross-entropy loss function between the true segmentation value and the output for our model. The weighted cross-entropy function was used to account for the class imbalance of the volume that cartilage and meniscus compartments make up compared with the entire MR imaging volume. Details on this equation can be viewed in Appendix E1 (online).
To build the U-Net models, data in subjects from both the T1ρ-weighted and the DESS sets were divided into training, validation, and time-point testing sets with a 70/20/10 split and were then broken down into their respective two-dimensional (2D) sections to be used as inputs for the two sequence models. The time-point testing set for both data sets consisted of only follow-up studies corresponding to baseline studies in the training and validation data sets. This time-point holdout data set was used as validation for the precision of the automatic segmentation longitudinally. A full breakdown of the T1ρ-weighted and DESS training, validation, and time-point testing data according to diagnostic group (ACL, OA, control) can be viewed in Table 2. The full 3D segmentation map was then generated by stacking the predicted 2D sections for a subject and then taking the largest 3D-connected component for each compartment class.
Table 2.
Demographics in Training, Validation, and Testing Data Sets
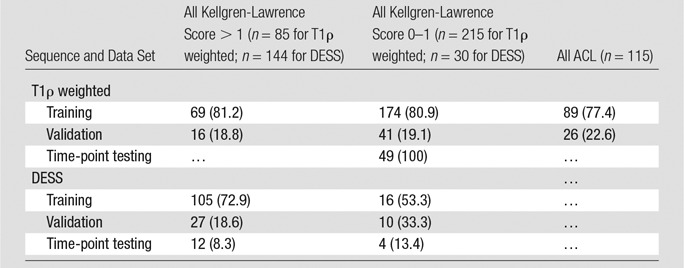
Note.—Data are numbers of patients, with percentages in parentheses.
All U-Net models were implemented in Native TensorFlow, version 1.0.1 (Google, Mountain View, Calif). Model selection was made by using the 1-standard-error rule on the validation data set (27) (B.N., with 3 years of experience). For full learning specifications and learning curves of the U-Net, see Table E1 and Figure E2 (both online).
Model Performances Evaluation and Statistical Analysis
A multicompartment model for each data set was created for predicting the four cartilage compartments, two meniscus compartments, overall cartilage, and overall meniscus. Segmentation performances of these models were gauged by using the Dice coefficient ( ), where T is the true manual segmentation map and P is the predicted segmentation map (28). For the T1ρ-weighted model, the automatic segmentations’ ability to evaluate T1ρ and T2 relaxation times was compared with that of the manual segmentation by using Pearson correlation and a two-sided t test to examine any associated differences. Relaxation times between time points for manual and automatic segmentation were evaluated by taking the absolute mean difference in relaxation times between time points for both manual and automatic segmentation and then comparing the difference between the two methods by using a two-sided t test. The same statistical method was used in the DESS data set for comparing the two methods’ ability to extract volume and thickness. All statistical tests were performed by using MATLAB (Mathworks, Natick, Mass) at the α < .05 level.
), where T is the true manual segmentation map and P is the predicted segmentation map (28). For the T1ρ-weighted model, the automatic segmentations’ ability to evaluate T1ρ and T2 relaxation times was compared with that of the manual segmentation by using Pearson correlation and a two-sided t test to examine any associated differences. Relaxation times between time points for manual and automatic segmentation were evaluated by taking the absolute mean difference in relaxation times between time points for both manual and automatic segmentation and then comparing the difference between the two methods by using a two-sided t test. The same statistical method was used in the DESS data set for comparing the two methods’ ability to extract volume and thickness. All statistical tests were performed by using MATLAB (Mathworks, Natick, Mass) at the α < .05 level.
Results
Automatic Segmentation Performances
Figure 2 shows predicted section examples from the model’s overall cartilage and meniscus predictions of the T1ρ-weighted and DESS test data sets (Fig 2, B, D) compared with the manual segmentations (Fig 2, A, C). Mean validation Dice coefficients calculated for predicting overall cartilage and meniscus in the T1ρ-weighted data set were 0.742 (95% confidence interval [CI]: 0.720, 0.764) for cartilage and 0.767 (95% CI: 0.743, 0.791) for meniscus. Mean validation Dice coefficients for cartilage and meniscus in the DESS data set were 0.867 (95% CI: 0.859, 0.875) for cartilage and 0.833 (95% CI: 0.821, 0.845) for meniscus. The multicompartment analysis for the T1ρ-weighted and DESS data sets also had strong Dice scores, which can be viewed in Table 3. For both the T1ρ-weighted and DESS data sets, Dice coefficient performances did not statistically differ between subjects with different KL grades, showing robustness of the method across the OA spectrum. However, it should be noted that the Dice scores between the different subject diagnostic cohorts for the T1ρ-weighted data did differ for validation data in two compartments. For the FC, ACL validation subjects performed 5.2% and 5.5% better than OA and control, respectively (P = .003). For the PC, ACL validation subjects performed 16.2% and 12.7% better than OA and control, respectively (P < .001).
Figure 2:
Example MR images show comparison between, A–C, manual segmentation and B, C, automatic segmentation predicted by using the U-net convolutional neural network. DESS = double-echo steady state, RES = resolution.
Table 3.
Dice Coefficient Results

Note.—Data are means, with 95% confidence intervals in parentheses. FC = femoral cartilage, LM = lateral meniscus, LTC = lateral tibial cartilage, MM = medial meniscus, MTC = medial tibial cartilage, PC = patellar cartilage, TP = time point.
In processing the automated segmentation maps for each training volume, a single subject volume from the T1ρ-weighted data set took 2.5 seconds to generate, while a volume from the DESS data set took 8 seconds.
Automatic Segmentation Extraction: T1ρ and T2 Relaxation Times
T1ρ and T2 relaxation times across all cartilage compartments showed no associated differences between the manual and automatic segmentations for training and validation in the T1ρ-weighted data set (validation T1ρ for FC: P = .825; for LTC: P = .464; for MTC: P = .858; and for PC: P = .669; and validation T2 for FC: P = .574; for LTC: P = .337; for MTC: P = .974; and for PC: P = .696). T1ρ and T2 relaxation times between manual and automatic segmentations had strong correlations for validation, with R values of 0.8767 and 0.894, respectively. The mean absolute difference between segmentation methods for validation T1ρ and T2 relaxation times were 2.1 and 1.54 msec, respectively. A full breakdown of these relaxometry metrics by cartilage compartments and training and validation can be viewed in Table 4, as well as in the scatterplots and Bland-Altman plots available in Figure 3. As for the longitudinal testing data analysis, there was no statistically associated difference between the manual and automatic segmentations’ relaxometry time point changes (T1ρ for FC: P = .355; for LTC: P = .235; for MTC: P = .695; and for PC: P = .584; and T2 for FC: P = .057; for LTC: P = .390; for MTC: P = .091; and for PC: P = .018 [not significant with Bonferroni correction]), showing comparable precision in manual and automatic procedures. A full breakdown of longitudinal testing comparison can be viewed in Table 5.
Table 4.
Results of Relaxometry and Morphology Analysis according to Data Set

Note.—Statistical significance is determined at the P < .05 level. FC = femoral cartilage, LTC = lateral tibial cartilage, MTC = medial tibial cartilage, PC = patellar cartilage.
Figure 3:
Scatterplots and Bland-Altman plots show comparison of T1p (top) and T2 (bottom) relaxation times produced from manual and automatic segmentation methods. (Note that the mean difference and standard errors of the mean of the Bland-Altman plot are calculated by using the entire data set, not between compartments.) FC = femoral cartilage, LTC = lateral tibial cartilage, MTC = medial tibial cartilage, PC = patellar cartilage, SD = standard deviation, Train = training.
Table 5.
Results of Relaxometry and Morphology Analysis according to Time Point

Note.—Statistical significance is determined at the P < .05 level. FC = femoral cartilage, LTC = lateral tibial cartilage, MTC = medial tibial cartilage, PC = patellar cartilage.
Automatic Segmentation Extraction: Thickness and Volume
Thickness and volume of training and validation data sets between manual and automatic segmentations showed strong linear relationships and correlation across all cartilage compartments. For validation data, the average thickness correlation across compartments was 0.9349, with an average absolute mean difference across compartments of 0.20195 mm, which was lower than image resolution. Again, for validation, the average volume correlation across compartments was 0.9384, with an average absolute mean differences across compartments of 510.134 mm3. A full breakdown of these morphologic metrics by cartilage compartment can be viewed in Table 4, as well as in the scatterplots and Bland-Altman plots available in Figure 4. Again, for the longitudinal testing data analysis, there was no associated difference in thickness or volume between the manual and automatic segmentations’ volume and thickness time-point changes (thickness for FC: P = .654; for LTC: P = .664; for MTC: P = .678; and for PC: P = .404; and volume for FC: P = .412; for LTC: P = .376; for MTC: P = .619; and for PC: P = .661). A full breakdown of longitudinal testing comparison can be viewed in Table 5. It should be noted that for some compartments, our U-Net tended to overestimate volume and thickness.
Figure 4:
Scatterplots and Bland-Altman plots show comparison of volumetric (top) and thickness (bottom) calculations produced from manual and automatic segmentation methods. (Note that the mean difference and standard errors of the mean of the Bland-Altman plot are calculated by using the entire data set, not between compartments.) FC = femoral cartilage, LTC = lateral tibial cartilage, MTC = medial tibial cartilage, PC = patellar cartilage, SD = standard deviation, Train = training.
Discussion
Our study’s results show insight into the application of deep neural networks within the field of musculoskeletal research. All metrics for model evaluation (Dice coefficients, relaxation times, morphology, speed) are competitive with or outperform current state-of-the-art automatic or semiautomatic segmentation methods.
Previous automatic or semiautomatic methods for knee tissue segmentation and relaxometry and morphology detection include a combination of model-based, atlas-based, and machine learning–based approaches (11). A popular state-of-the-art atlas-based approach is proposed by Dam et al (29), which uses a multiatlas registration accompanied by k-nearest neighbors (17,30). This approach requires multiple time-consuming steps to achieve the final segmentation, such as multiatlas registration and feature computation and selection. Dam et al used data in 88 subjects from the OAI. While it is unknown which patients Dam et al used, a loose comparison between their validation Dice coefficient results and our U-Net’s can be made by using a two-sample t test under the assumption that the two samples have equivalent distributions as a result of the same imaging parameters. The two methods show no associated difference between compartments, with the exception of the lateral tibia and femoral condyle (for MTC: P = .087; for LTC: P < .001; for FC: P < .001; for PC: P = .119; for MM: P = .339; and for LM: P = .052). Dam et al outperformed the U-Net by 3.7% for the lateral tibia (0.86 ± 0.034 [standard deviation] vs 0.822 ± 0.071), while our U-Net outperformed Dam et al by 5% for the femoral condyle (0.878 ± 0.033 vs 0.828 ± 0.044). Dam et al also compared volumes between manual and automatic segmentations for medial tibial and femoral compartments, overestimating the total segmentation volume by 14%. Our U-Net model overestimated volume by an average of about 12% across all compartments. For both methods, volume overestimation is not necessarily a flaw, as long as strong, linear correlations exist, allowing for bias adjustment.
Aside from the robust accuracy performances, our U-Net model has the distinct advantage over previous automatic/semiautomatic segmentation methods in that it is an “end-to-end” method. There is no pipeline that requires extensive image pre- and postprocessing and registration, resulting in noteworthy improvements in performance time.
A pattern that should be noted in our results is that the DESS data set outperformed the T1ρ-weighted data set across all compartments. We believe there are two main reasons for this. First, the T1ρ-weighted data set had a smaller sample size on a per-section basis than the DESS data set, which usually leads to less robust modeling. Second, because the T1ρ-weighted data were of lower spatial resolution, each misplaced voxel will decrease the Dice coefficient 2.25 times more than the Dice coefficient for a misplaced voxel in the DESS data set.
While our neural network model does show promising performance advantages in comparison to manual and other automatic segmentation methods, there is room for improvement in accuracy. Using the Dice coefficient as the loss function would be desirable for training (as well as a potential combination of volume and thickness measurements); however, the current configuration of TensorFlow does not allow for this gradient calculation. Another limitation with the current method is that it uses only four cartilage plates and two meniscus plates, whereas more information can be inferred about OA with more detailed subregions of the meniscus and cartilage. Finally, there was a lack of a real ground truth. Our accuracies are calculated assuming manual segmentation as the reference standard, which may change because of user variability. However, our presented results show longitudinal precision that proves the robustness of our algorithm, independent of the ground truth definition.
Using state-of-the-art convolutional neural networks, we were able to produce fast, accurate, and precise automatic segmentations of cartilage and meniscus compartments that are invariant across patients with OA. Our method also has substantial computational speed advantages. Additionally, our models have demonstrated efficacy in extracting relaxation times and morphologic features that can be used in the prediction and monitoring of joint degeneration in OA. This demonstrates our model’s interchangeability with manual segmentation, allowing clinicians to make quicker and more accurate OA inferences, representing an important step toward the clinical translation of quantitative MR imaging techniques.
Summary
We aim to analyze how automatic segmentation performances translate in accuracy and precision to morphology and relaxometry in osteoarthritis compared with manual segmentations and increase the speed and accuracy of the work flow that uses quantitative MR imaging to study knee degenerative diseases.
Implications for Patient Care
■ Morphologic and biochemical composition quantitative MR imaging have shown clinical relevance in the diagnosis and monitoring of osteoarthritis; accurate and precise automated segmentation will allow for rapid extraction of these values and their application to clinical management and research.
■ Automatic segmentation, morphology, and relaxometry allow for the timely incorporation of key parameters in the process that uses MR imaging to study degenerative diseases of the knee.
■ We demonstrate a data-driven approach’s interchangeability with manual segmentation, allowing clinicians to make quicker and more accurate diagnoses and representing an important step toward the clinical translation of quantitative MR imaging techniques.
APPENDIX
SUPPLEMENTAL FIGURES
Received September 29, 2017; revision requested December 4; revision received December 14; final version accepted January 9, 2018.
Study supported by GE Healthcare IT Business and the Arthritis Foundation (ACL Proof of Feasibility, Trial 6157). S.M. supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01 AR046905) and the National Institutes of Health (K99AR070902, P50-AR060752).
This content is solely the responsibility of the authors and does not necessarily reflect the views of the National Institutes of Health, the National Institute of Arthritis and Musculoskeletal and Skin Diseases, or the Arthritis Foundation.
Disclosures of Conflicts of Interest: B.N. Activities related to the present article: this trial was supported in part by funding from GE Healthcare IT Business. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. V.P. Activities related to the present article: this trial was supported in part by funding from GE Healthcare IT Business. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. S.M. Activities related to the present article: this trial was supported in part by funding from GE Healthcare IT Business. Activities not related to the present article: is a consultant for Smith Research, has grants or grants pending from GE Healthcare, receives royalties from the University of California Office of the President. Other relationships: disclosed no relevant relationships.
Abbreviations:
- ACL
- anterior cruciate ligament
- CI
- confidence interval
- DESS
- double-echo steady state
- FC
- femoral cartilage
- KL
- Kellgren-Lawrence
- LM
- lateral meniscus
- LTC
- lateral tibial cartilage
- MM
- medial meniscus
- MTC
- medial tibial cartilage
- OA
- osteoarthritis
- PC
- patellar cartilage
- 3D
- three-dimensional
- 2D
- two-dimensional
References
- 1.Lawrence RC, Felson DT, Helmick CG, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part II. Arthritis Rheum 2008;58(1):26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence RC, Helmick CG, Arnett FC, et al. Estimates of the prevalence of arthritis and selected musculoskeletal disorders in the United States. Arthritis Rheum 1998;41(5):778–799. [DOI] [PubMed] [Google Scholar]
- 3.Gong J, Pedoia V, Facchetti L, Link TM, Ma CB, Li X. Bone marrow edema-like lesions (BMELs) are associated with higher T1rho and T2 values of cartilage in anterior cruciate ligament (ACL)-reconstructed knees: a longitudinal study. Quant Imaging Med Surg 2016;6(6):661–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindsey CT, Narasimhan A, Adolfo JM, et al. Magnetic resonance evaluation of the interrelationship between articular cartilage and trabecular bone of the osteoarthritic knee. Osteoarthritis Cartilage 2004;12(2):86–96. [DOI] [PubMed] [Google Scholar]
- 5.Eckstein F, Kunz M, Schutzer M, et al. Two year longitudinal change and test-retest-precision of knee cartilage morphology in a pilot study for the osteoarthritis initiative. Osteoarthritis Cartilage 2007;15(11):1326–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su F, Hilton JF, Nardo L, et al. Cartilage morphology and T1ρ and T2 quantification in ACL-reconstructed knees: a 2-year follow-up. Osteoarthritis Cartilage 2013;21(8):1058–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osaki K, Okazaki K, Takayama Y, et al. Characterization of biochemical cartilage change after anterior cruciate ligament injury using T1ρ mapping magnetic resonance imaging. Orthop J Sports Med 2015;3(5):2325967115585092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carballido-Gamio J, Bauer J, Lee KY, Krause S, Majumdar S. Combined image processing techniques for characterization of MRI cartilage of the knee. Conf Proc IEEE Eng Med Biol Soc 2005;3:3043–3046. [DOI] [PubMed] [Google Scholar]
- 9.Joseph GB, Hou SW, Nardo L, et al. MRI findings associated with development of incident knee pain over 48 months: data from the osteoarthritis initiative. Skeletal Radiol 2016;45(5):653–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carballido-Gamio J, Bauer JS, Stahl R, et al. Inter-subject comparison of MRI knee cartilage thickness. Med Image Anal 2008;12(2):120–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pedoia V, Majumdar S, Link TM. Segmentation of joint and musculoskeletal tissue in the study of arthritis. MAGMA 2016;29(2):207–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue N, Doellinger M, Fripp J, Ho CP, Surowiec RK, Schwarz R. Automatic model-based semantic registration of multimodal MRI knee data. J Magn Reson Imaging 2015;41(3):633–644. [DOI] [PubMed] [Google Scholar]
- 13.Zhang K, Lu W, Marziliano P. Automatic knee cartilage segmentation from multi-contrast MR images using support vector machine classification with spatial dependencies. Magn Reson Imaging 2013;31(10):1731–1743. [DOI] [PubMed] [Google Scholar]
- 14.Shim H, Chang S, Tao C, Wang JH, Kwoh CK, Bae KT. Knee cartilage: efficient and reproducible segmentation on high-spatial-resolution MR images with the semiautomated graph-cut algorithm method. Radiology 2009;251(2):548–556. [DOI] [PubMed] [Google Scholar]
- 15.Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convoutional neural networks. Radiology 2017;284(2):574–582. [DOI] [PubMed] [Google Scholar]
- 16.Prasoon A, Petersen K, Igel C, Lauze F, Dam E, Nielsen M. Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. Med Image Comput Comput Assist Interv 2013;16(Pt 2):246–253. [DOI] [PubMed] [Google Scholar]
- 17.Antony J, McGuinness S, O’Connor NE, Moran K. Quantifying radiographic knee osteoarthritis severity using deep convolutional neural networks. Presented at the 23rd International Conference on Pattern Recognition (ICPR), Cancun, Mexico, December 4–8, 2016; 1195–1200.
- 18.Osteoarthritis Initiative . National Institute of Arthritis and Musculoskeltal and Skin Diseases. https://www.niams.nih.gov/Funding/Funded_Research/Osteoarthritis_Initiative/. Published 2016. Accessed March 12, 2018.
- 19.Li X, Pedoia V, Kumar D, et al. Cartilage T1ρ and T2 relaxation times: longitudinal reproducibility and variations using different coils, MR systems and sites. Osteoarthritis Cartilage 2015;23(12):2214–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osteoarthritis Initiative . UCSF Data Release. iMorphics. https://oai.epi-ucsf.org/datarelease/iMorphics.asp. Accessed March 12, 2018.
- 21.Pei M, Nguyen TD, Thimmappa ND, et al. Algorithm for fast monoexponential fitting based on Auto-Regression on Linear Operations (ARLO) of data. Magn Reson Med 2015;73(2):843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. arXiv:1505.04597. https://arxiv.org/abs/1505.04597. Published May 18, 2015. Accessed March 12, 2018.
- 23.Novikov AA, Lenis D, Major D, Hladůvka J, Wimmer M, Bühler K. Fully Convolutional Architectures for Multi-Class Segmentation in Chest Radiographs. arXiv:1701.08816. https://arxiv.org/abs/1701.08816. Published January 30, 2017. Accessed March 12, 2018. [DOI] [PubMed]
- 24.Erden B, Gamboa N, Wood S. 3D Convolutional Neural Network for Brain Tumor Segmentation. http://cs231n.stanford.edu/reports/2017/pdfs/526.pdf. Accessed March 12, 2018.
- 25.Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R. Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med 2017 Jul 21. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Badrinarayanan V, Kendall A, Cipolla R. SegNet: a deep convolutional encoder-decoder archtiecture for image segmentation. arXiv:1511.00561. https://arxiv.org/abs/1511.00561. Published November 2, 2015. Accessed March 12, 2018. [DOI] [PubMed]
- 27.Hastie T, Tibshirani R, Friedman J. The elements of statistical learning. New York, NY: Springer, 2009. [Google Scholar]
- 28.Zou KH, Warfield SK, Bharatha A, et al. Statistical validation of image segmentation quality based on a spatial overlap index. Acad Radiol 2004;11(2):178–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dam EB, Lillholm M, Marques J, Nielsen M. Automatic segmentation of high- and low-field knee MRIs using knee image quantification with data from the osteoarthritis initiative. J Med Imaging (Bellingham) 2015;2(2):024001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heimann T, Morrison BJ, Styner MA, Niethammer M, Warfield SK. Segmentation of knee images: a grand challenge. SKI10. http://www.ski10.org/ski10.pdf. Published 2010. Accessed March 12, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






