Abstract
The extensive post-translational modifications of the envelope spikes of the human immunodeficiency virus (HIV) present considerable challenges and opportunities for HIV vaccine design. These oligomeric glycoproteins typically have over thirty disulphide bonds, around a hundred N-linked glycosylation sites and are functionally dependent on protease cleavage within the secretory system. The resulting mature structure adopts a compact fold with the vast majority of its surface obscured by a protective shield of glycans which can be targeted by broadly neutralizing antibodies. Despite the notorious heterogeneity of glycosylation, rare B-cell lineages can evolve to utilize and cope with viral glycan diversity and these structures therefore present promising targets for vaccine design. The latest generation of recombinant envelope spike mimetics contain reengineered post-translational modifications to present stable antigens to guide the development of broadly neutralizing antibodies by vaccination.
Keywords: virus, glycosylation, vaccine, immunogen, glycans
Introduction
The immense variation of the genome of HIV presents a considerable obstacle to the development of an effective, sterilizing vaccine. The failure of classical vaccination strategies has driven advances in our understanding of the molecular basis of viral infectivity and the host immune response. Lines of enquiry have included investigating correlates of protection arising from the RV144 trial such as the role of non-neutralizing antibodies [1–3]. Here, however, we focus on vaccination strategies aiming to develop immunogens capable of eliciting a protective broadly neutralizing antibody (bnAb) response.
Innovations towards the development of bnAb-eliciting immunogens have included the production of native-like trimers that mimic the envelope glycoprotein (Env) spikes expressed on the surface of the virion [4–7]. These soluble glycoproteins have revolutionized our understanding of the viral glycoprotein structure and have stimulated the design of new immunogens. Encouragingly, these mimetics bind to a growing number of bnAbs isolated from infected patients and they are being investigated as a platform for the next-generation of immunogens. Armed with the detailed structure of the envelope spike mimetics, detailed knowledge of the intricate network of post-translational modifications has been revealed [8–15]. Exploiting and targeting the post-translational modifications of the viral spike has enabled the design of improved immunogens that are able to elicit protective neutralizing antibodies against a narrow but growing range of viral isolates [16–18].
Engineering the envelope spike
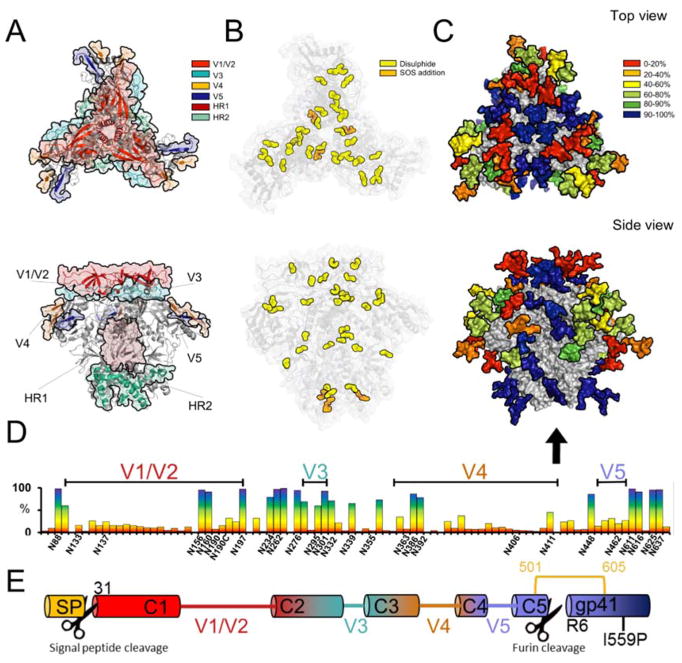
Env is the sole viral protein expressed on the surface of the HIV virion and antibodies capable of binding the functional form of the viral spike can effectively neutralize viral infectivity [19–25]. As a class I viral fusion glycoprotein, Env consists of a trimer of heterodimers comprising gp120, which consists of five constant domains interspersed with hypervariable loops (Figure 1A), responsible for receptor recognition, and the transmembrane fusion glycoprotein, gp41. Env enables viral particle recognition of CD4+ T-cells, binding to the chemokine co-receptor, and ultimately fusion of the viral membrane with that of the target cell. To generate infectious viral particles, the viral spike must be extensively post-translationally modified by the host cell. These modifications include the formation of disulphide bonds (Figure 1B), extensive N-linked glycosylation (Figure 1C and 1D) and two proteolytic cleavage events, one after the signal peptide and the other between gp120 and gp41 (Figure 1E)[26].
Figure 1. Representation of the post-translational modifications of BG505 clade A envelope glycoprotein.
Models were generated using the cryo-EM structure of BG505 SOSIP.664 PDB ID:5ACO [12]. A) 3D representation of the variable loops on gp120 and the heptad repeats of gp41. B) Canonical disulphide bonds found in BG505[36] with the additional stabilizing disulphide bond found in BG505 SOSIP.664 shown in orange. C) Conservation map of the glycans of BG505 SOSIP.664. The glycans were coloured according to their conservation across 4000 Env strains Huang et al.[44]. BG505 crystal structure with N-linked glycans modelled by Behrens et al.[52] D) The frequency of potential N-linked glycan sites across the Env sequence with the PNGs found in BG505 SOSIP.664 labelled on the X axis. E) Schematic showing the locations of proteolytic cleavage for the signal peptide and the furin cleavage site.
Successful attempts to recombinantly mimic the Env-based epitopes of broadly neutralizing antibodies have involved solubilizing and stabilizing glycoprotein trimers utilising post-translational modifications and amino acid substitutions. Crucially, native-like Env trimers have been generated by the addition of a disulphide bond between gp120 and gp41 subunits and an isoleucine to proline mutation in gp41 which stabilise the prefusion conformation of Env. These immunogens are termed “SOSIPs” [5,27,28]. These stable soluble mimetics are not in themselves sufficient to generate a broad and protective immune response. Strategies to improve the breadth and potency of the antibody response has included the design of immunogens to stimulate the precursor lineage of potent bnAbs, for example by the targeted elimination of key glycans and the hyperstabilization of trimers through disulphide bond engineering [27–31].
Disulphides in infection and hyperstabilization
The quaternary structure of natively folded viral spikes requires the correct formation of disulphide bonds in the mammalian secretory pathway. Slow folding events allow time for the extensive network of disulphide bonds to form and to “shuffle” using protein disulphide isomerase (PDI) to ensure the optimal disulphide bond configuration is achieved [32–34]. The correct pairing of disulphides is an essential requirement for infectious Env, as by knocking out PDIs in vitro the resultant protein is able to bind to CD4 but not undergo the conformational changes required for membrane fusion[35]. The incorrect formation of disulphides is also promoted when viral spike mimetics remain uncleaved by furin, this not only impacts the resultant quaternary structure of the viral spike it also perturbs the glycan shield and therefore bnAb binding[36].
With the structural characterisation by X-ray crystallography and mass spectrometry using the BG505 SOSIP.664 construct it has been possible to map the locations of the network of disulphide bonds[36,37]. As well as understanding the locations of the disulphide bonds in 3-dimensional space it is also possible to utilise the biophysical analysis to design next-generation hyperstabilized constructs that employ additional disulphide bonds to minimise the intrinsic flexibility and instability of Env. Torrents de la Peña et al. describe additional mutations to the original BG505 SOSIP.664 of which the most prominent is the addition of an interprotomer gp120 disulphide bond[27]. This effectively stabilizes the trimeric Env oligomeric state and also prevents the possibility of gp120 shedding. These modifications do not impact upon previously characterised bnAb binding and do not influence other post translational modifications such as N-linked glycosylation and therefore demonstrate the ability to configure Env post-translational modifications for superior immunogen design[27].
Glycan engineering of germline targeting immunogens
In addition to their role in protein folding, the high density of N-linked glycans is thought to be driven by immune selection whereby glycans mask underlying conserved protein epitopes. This is evidenced by their accumulation during longitudinal infection and depletion during collapse of an effective adaptive immune response in late infection[38–42]. The high density of glycans means that the evolution of the glycan shield can be understood as holes being formed and being filled rather than there being a continuum of options for glycan locations[43]. This is less so in the variable loops where there is very low conservation of glycan positions (Figure 1C and 1D)[44]. One interesting consequence of the trimeric structure of the viral spike is the role of conserved glycans at the protomer interface. Presumably the interfacial glycans act to obscure important surface features and may help stabilize the trimeric structure[45]. The highly conserved N-glycan sites across the surface of Env also form epitopes for a range of bnAbs and with the characterisation of one such bnAb that is able to recognise the highly conserved N262 and N448 glycans the entire surface of Env forms bnAb epitopes[46,47].
The conservation of potential N-linked glycan sites across the envelope spike far exceeds other regions of the envelope spike and present a robust platform for immunogen design[47,48]. In addition, the high density of glycosylation and the trimeric architecture places steric restrictions on glycan processing which drives the formation of a population of under-processed oligomannose-type glycans[9,49–53]. However, the extensive heterogeneity inherent to N-linked glycan processing means that these steric constraints alone are not entirely sufficient for native-like glycosylation[49].
The producer cell of the Env spike, either viral or recombinant, can lead to changes in glycosylation that may impact on bnAb binding. In one extreme, when the glycans of macrophage-derived HIV particles are compared to those from peripheral blood mononuclear cells (PBMCs) there was a significant shift in the composition of the population of complex-type glycans with the macrophage-derived glycans exhibiting large polylactosamine structures[54]. Importantly, the viruses exhibited different sensitivities to antibody neutralization and this may be attributed to glycosylation.
Glycan heterogeneity is an important parameter when assessing recombinant Env spike mimetics. Such heterogeneity has been shown to contribute to partial neutralization by glycan-dependent bnAbs [55–57]. Encouragingly, trimeric SOSIPs exhibit native-like levels of oligomannose-type glycans. However, there will be potentially important variations in glycan structure between recombinant immunogens and their viral counterparts. For example, between PBMC and HEK293T derived gp120 there are subtle differences such as the sialic acid linkages of complex type glycans, with PBMCs presenting a larger population of α2,6 linkages. Such differences between viral and recombinant glycosylation may result in diminished presentation of potential glycan epitopes to the immune system when recombinant constructs are used as immunogens[58]. Despite this caveat, promising immunogens are being developed that exploit viral glycosylation.
Despite the broad neutralization and high affinities of bnAbs to the viral spike, few patients develop bnAb lineages before the genetic diversity of the viral spike overwhelm the immune system. The original goal of mimicking the natively glycosylated viral spike has produced strong autologous responses but have not managed to elicit broadly neutralizing antibodies in macaques[17]. The focus of immunogen design has therefore shifted and now focuses on manipulating the post-translational modifications of Env to guide the immune system to mature the rare B cell lineages of broadly neutralizing antibodies. This approach has been greatly stimulated by the observation that patients that have eventually developed a potent bnAb response often have virus missing key glycan sites[59]. Many investigators are now examining immunogens displaying glycan holes[28–31,60,61].
One such platform is the glycan depleted trimer, termed BG505 SOSIP.v4.1-GT1 which has been designed to trigger B cells corresponding to germline PGT121 and germline VRC01. This trimer lacks 15 glycans across the viral spike and initiates antibody responses in knock-in mice expressing the predicted germlines for these bnAbs[62]. In addition, a comparative glycan analysis revealed that the remaining oligomannose-type glycans were largely unaffected by the glycan deletions with changes in their mannose trimming localised to regions proximal to a depleted glycan[63]. The overall integrity of the mannose patch is a promising observation and broadens the possibilities of using extensive glycan holes as a design feature of new immunogens.
It is also possible to harness the microheterogeneity of conserved glycan sites to present glycan epitopes that result in the proliferation of the rare B-cell lineages of bnAbs. In trimers derived from env sequences from a prolonged infection such as the clade A BG505 sequence used for BG505 SOSIP.664 the apex sites N160 and N156 display oligomannose glycans[50–52]. Andrabi et al. demonstrated that bnAb precursors require sialic acid to bind to the apex viral spike and that Env sequences derived from early infection appear to have sialic acid glycans at N160 and N156[64]. Although the bnAb precursors require sialic acid the resultant mature bnAbs are able to bind to trimers containing apex sites that are well defined as oligomannose. These observations teach us that glycan heterogeneity can evolve during the course of infection and that this feature can potentially be exploited in vaccine design.
Optimising and bypassing furin cleavage
Another critical post-translational modification of Env is furin cleavage of the gp160 pro-protein. Furin cleavage is thought to occur in the trans Golgi network and is an essential step in the formation of functional correctly folded trimers. Furin cleaves between the gp120 and gp41 subunits of the gp160 polypeptide, recognising the amino acid motif Arg-X-(Arg/Lys)-Arg[65]. Negative stain electron microscopy of uncleaved trimers, which have been explored as viral spike mimetics, revealed that they do not naturally adopt the classical trimeric architecture[58,66]. The perturbation of the structural integrity resulting from a lack of furin cleavage also has implications for the post-translational modifications of candidate immunogens. Analysis of N-linked glycosylation on a global and a site-specific level on a number of uncleaved structures described an elevation in glycan processing, most likely resulting from aberrant trimerization[9,67]. As broadly neutralizing antibody epitopes frequently contain N-linked glycans the aberrant glycosylation resulting from a lack of furin cleavage will have knock-on effects on the antigenicity of those trimers. Furthermore, the destabilization of the quaternary structure ablates the binding of quaternary-specific bnAbs such as PGT151 and also reduces the affinity of other quaternary specific bnAbs such as PGT145[68,69].
The requirement for furin cleavage for correct assembly is an important feature in the expression of BG505 SOSIP.664 as the low levels of endogenous furin result in large populations of uncleaved trimers. Binley et al. circumvented this problem by co-transfecting plasmid containing the furin gene concomitantly with BG505 SOSIP.664. In addition, it is important to optimise the protease cleavage step as much as possible. The majority of HIV strains present a furin cleavage site consisting of a REKR motif. A panel of mutations introduced to this region demonstrated that this is not the optimal motif for furin mediated Env glycoprotein cleavage. By replacing the REKR motif with 6 arginine residues the proteolytic separation of gp120 and gp41 is greatly improved[70]. By modifying the amino acid sequence in this way, it is possible to manipulate post-translational protease cleavage to allow for a larger amount of native-like material. Impressively, efficient furin cleavage has also now been achieved in production of clinical grade BG505 SOSIP.664 using a stable CHO cell line containing the target SOSIP, possessing an optimised furin cleavage site, and also the gene encoding furin[71].
As well using recombinant glycoproteins as immunogens, a further strategy currently under investigation to boost the neutralizing antibody titre is to use DNA-based approaches. For DNA vaccines, the env gene is administered and the trimers are expressed by the host. In order to utilise this approach, it is favourable to bypass the furin cleavage stage as it removes the requirement of co-transfection with furin. Although previously defined uncleaved trimers have produced large populations of misfolded trimers, several constructs now exist that are able to form native-like trimers without the need for furin cleavage. By replacing the furin cleavage site with a flexible linker it is possible to generate native-like trimers. With additional stabilizing mutations native flexibly linked (NFL) trimer display native-like bnAb binding and analogous glycosylation to BG505 SOSIP.664[6,30,72]. Using in silico methods Kong et al. redesigned the HR1 loop to generate uncleaved trimers that were stabilized in the pre-fusion conformation (UFO) and present trimers with greater stability that the equivalent SOSIP construct[73]. The ability to redesign fundamental post-translational modifications with little impact on the overall glycosylation and topology of the envelope spike further highlights the robustness of the glycan shield as a target for bnAb elicitation by immunization.
Perspective
As immunogen strategies continue to move towards activating precursor B cells, innovations are increasingly exploiting or bypassing post-translational modifications of the envelope glycoprotein. These advances may well stimulate developments of efficacious vaccines against a much wider range of pathogens where classical vaccine design strategies have proven ineffective.
Acknowledgments
M.C. is supported by the Scripps CHAVI-ID (1UM1AI100663), the International AIDS Vaccine Initiative (IAVI) Neutralizing Antibody Center CAVD grant (Glycan characterization and Outer Domain glycoform design; agreement 1981) and IAVI (VxPDC agreement 2402), and R.W.S. by NIH HIVRAD grant P01 AI110657. This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 681137. R.W.S. is a recipient of a Vidi grant from the Netherlands Organization for Scientific Research (NWO) and a Starting Investigator Grant from the European Research Council (ERC-StG-2011–280829-SHEV). K.J.D. was funded by the Medical Research Council (MRC) (MR/K024426/1).
References
- 1.Barouch DH, Stephenson KE, Borducchi EN, Smith K, Stanley K, McNally AG, et al. Protective efficacy of a global HIV-1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell. 2013;155:531–539. doi: 10.1016/j.cell.2013.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung AW, Kumar MP, Arnold KB, Yu WH, Schoen MK, Dunphy LJ, et al. Dissecting Polyclonal Vaccine-Induced Humoral Immunity against HIV Using Systems Serology. Cell. 2015;163:988–998. doi: 10.1016/j.cell.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, et al. Vaccination with ALVAC and AIDSVAX to Prevent HIV-1 Infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 4.Chuang G-Y, Geng H, Pancera M, Xu K, Cheng C, Acharya P, et al. Structure-Based Design of a Soluble Prefusion-Closed HIV-1 Env Trimer with Reduced CD4 Affinity and Improved Immunogenicity. J Virol. 2017;91:e02268–16. doi: 10.1128/JVI.02268-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanders RW, Derking R, Cupo A, Julien J, Yasmeen A, De N, et al. A Next-Generation Cleaved, Soluble HIV-1 Env Trimer, BG505 SOSIP. 664 gp140, Expresses Multiple Epitopes for Broadly Neutralizing but Not Non-Neutralizing Antibodies. 2013;9 doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, et al. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep. 2015;11:539–50. doi: 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulp DW, Steichen JM, Pauthner M, Hu X, Schiffner T, Liguori A, et al. Structure-based design of native-like HIV-1 envelope trimers to silence non-neutralizing epitopes and eliminate CD4 binding. Nat Commun. 2017;8:1655. doi: 10.1038/s41467-017-01549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behrens A-J, Harvey DJ, Milne E, Cupo A, Kumar A, Zitzmann N, et al. Molecular architecture of the cleavage-dependent mannose patch on a soluble HIV-1 envelope glycoprotein trimer. J Virol. 2016;44 doi: 10.1128/JVI.01894-16. JVI.01894-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behrens A, Harvey DJ, Milne E, Cupo A, Kumar A, Zitzmann N, et al. Molecular Architecture of the Cleavage- Dependent Mannose Patch on a Soluble HIV-1 Envelope Glycoprotein Trimer. J Virol. 2017;91:1–16. doi: 10.1128/JVI.01894-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, et al. A Next-Generation Cleaved, Soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, Expresses Multiple Epitopes for Broadly Neutralizing but Not Non-Neutralizing Antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart-Jones GBE, Soto C, Lemmin T, Chuang G-Y, Druz A, Kong R, et al. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell. 2016;165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JH, de Val N, Lyumkis D, Ward AB. Model Building and Refinement of a Natively Glycosylated HIV-1 Env Protein by High-Resolution Cryoelectron Microscopy. Structure. 2015;23:1943–1951. doi: 10.1016/j.str.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gristick HB, von Boehmer L, West AP, Jr, Schamber M, Gazumyan A, Golijanin J, et al. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat Struct Mol Biol. 2016;23:906–915. doi: 10.1038/nsmb.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JH, Ozorowski G, Ward AB. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043–8. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crispin M, Ward AB, Wilson IA. Structure and Immune Recognition of the HIV Glycan Shield. Annu Rev Biophys. 2018;47 doi: 10.1146/annurev-biophys-060414-034156. annurev-biophys-060414-034156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torrents de la Peña A, de Taeye SW, Sliepen K, LaBranche CC, Burger JA, Schermer EE, et al. Immunogenicity in rabbits of SOSIP trimers from clades A, B and C, given individually, sequentially or in combinations. J Virol. 2018 doi: 10.1128/JVI.01957-17. JVI.01957-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pauthner M, Havenar-Daughton C, Sok D, Nkolola JP, Bastidas R, Boopathy AV, et al. Elicitation of Robust Tier 2 Neutralizing Antibody Responses in Nonhuman Primates by HIV Envelope Trimer Immunization Using Optimized Approaches. Immunity. 2017;46:1073–1088. e6. doi: 10.1016/j.immuni.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klasse PJ, LaBranche CC, Ketas TJ, Ozorowski G, Cupo A, Pugach P, et al. Sequential and Simultaneous Immunization of Rabbits with HIV-1 Envelope Glycoprotein SOSIP.664 Trimers from Clades A, B and C. PLOS Pathog. 2016;12:e1005864. doi: 10.1371/journal.ppat.1005864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burton DR, Hangartner L. Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annu Rev Immunol. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 21.Hessell AJ, Rakasz EG, Tehrani DM, Huber M, Weisgrau KL, Landucci G, et al. Broadly Neutralizing Monoclonal Antibodies 2F5 and 4E10 Directed against the Human Immunodeficiency Virus Type 1 gp41 Membrane-Proximal External Region Protect against Mucosal Challenge by Simian-Human Immunodeficiency Virus SHIVBa-L. J Virol. 2010;84:1302–1313. doi: 10.1128/JVI.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shingai M, Donau OK, Plishka RJ, Buckler-White A, Mascola JR, Nabel GJ, et al. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med. 2014;211:2061–74. doi: 10.1084/jem.20132494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Julg B, Liu P-T, Wagh K, Fischer WM, Abbink P, Mercado NB, et al. Protection against a mixed SHIV challenge by a broadly neutralizing antibody cocktail. Sci Transl Med. 2017;9:eaao4235. doi: 10.1126/scitranslmed.aao4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Julg B, Tartaglia LJ, Keele BF, Wagh K, Pegu A, Sok D, et al. Broadly neutralizing antibodies targeting the HIV-1 envelope V2 apex confer protection against a clade C SHIV challenge. Sci Transl Med. 2017;9:eaal1321. doi: 10.1126/scitranslmed.aal1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shibata R, Igarashi T, Haigwood N, Buckler–White A, Ogert R, Ross W, et al. Neutralizing antibody directed against the HIV–1 envelope glycoprotein can completely block HIV–1/SIV chimeric virus infections of macaque monkeys. Nat Med. 1999;5:204–210. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- 26.Sanders RW, Moore JP. Native-like Env trimers as a platform for HIV-1 vaccine design. Immunol Rev. 2017;275:161–182. doi: 10.1111/imr.12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torrents de la Peña A, Julien JP, de Taeye SW, Garces F, Guttman M, Ozorowski G, et al. Improving the Immunogenicity of Native-like HIV-1 Envelope Trimers by Hyperstabilization. Cell Rep. 2017;20:1805–1817. doi: 10.1016/j.celrep.2017.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74:627–43. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou T, Doria-Rose NA, Cheng C, Stewart-Jones GBE, Chuang G-Y, Chambers M, et al. Quantification of the Impact of the HIV-1-Glycan Shield on Antibody Elicitation. Cell Rep. 2017;19:719–732. doi: 10.1016/j.celrep.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubrovskaya V, Guenaga J, de Val N, Wilson R, Feng Y, Movsesyan A, et al. Targeted N-glycan deletion at the receptor-binding site retains HIV Env NFL trimer integrity and accelerates the elicited antibody response. PLOS Pathog. 2017;13:e1006614. doi: 10.1371/journal.ppat.1006614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoy LE, van Gils MJ, Ozorowski G, Messmer T, Briney B, Voss JE, et al. Holes in the Glycan Shield of the Native HIV Envelope Are a Target of Trimer-Elicited Neutralizing Antibodies. Cell Rep. 2016;16:2327–2338. doi: 10.1016/j.celrep.2016.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stolf BS, Smyrnias I, Lopes LR, Vendramin A, Goto H, Laurindo FRM, et al. Protein disulfide isomerase and host-pathogen interaction. ScientificWorldJournal. 2011;11:1749–61. doi: 10.1100/2011/289182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Land A, Zonneveld D, Braakman I. Folding of HIV-1 envelope glycoprotein involves extensive isomerization of disulfide bonds and conformation-dependent leader peptide cleavage. FASEB J. 2003;17:1058–67. doi: 10.1096/fj.02-0811com. [DOI] [PubMed] [Google Scholar]
- 34.Go EP, Hua D, Desaire H. Glycosylation and disulfide bond analysis of transiently and stably expressed clade C HIV-1 gp140 trimers in 293T cells identifies disulfide heterogeneity present in both proteins and differences in O-linked glycosylation. J Proteome Res. 2014;13:4012–27. doi: 10.1021/pr5003643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallina A, Hanley TM, Mandel R, Trahey M, Broder CC, Viglianti GA, et al. Inhibitors of protein-disulfide isomerase prevent cleavage of disulfide bonds in receptor-bound glycoprotein 120 and prevent HIV-1 entry. J Biol Chem. 2002;277:50579–88. doi: 10.1074/jbc.M204547200. [DOI] [PubMed] [Google Scholar]
- 36.Go EP, Cupo A, Ringe R, Pugach P, Moore JP, Desaire H. Native Conformation and Canonical Disulfide Bond Formation Are Interlinked Properties of HIV-1 Env Glycoproteins. J Virol. 2016;90:2884–2894. doi: 10.1128/JVI.01953-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science (80- ) 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 39.Reitter JN, Means RE, Desrosiers RC. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 40.Coss KP, Vasiljevic S, Pritchard LK, Krumm SA, Glaze M, Madzorera S, et al. HIV-1 Glycan Density Drives the Persistence of the Mannose Patch within an Infected Individual. J Virol. 2016;90:11132–11144. doi: 10.1128/JVI.01542-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blay WM, Gnanakaran S, Foley B, Doria-Rose NA, Korber BT, Haigwood NL. Consistent patterns of change during the divergence of human immunodeficiency virus type 1 envelope from that of the inoculated virus in simian/human immunodeficiency virus-infected macaques. J Virol. 2006;80:999–1014. doi: 10.1128/JVI.80.2.999-1014.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borggren M, Repits J, Sterjovski J, Uchtenhagen H, Churchill MJ, Karlsson A, et al. Increased sensitivity to broadly neutralizing antibodies of end-stage disease R5 HIV-1 correlates with evolution in Env glycosylation and charge. PLoS One. 2011;6:e20135. doi: 10.1371/journal.pone.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore PL, Gray ES, Wibmer CK, Bhiman JN, Nonyane M, Sheward DJ, et al. Evolution of an HIV glycan–dependent broadly neutralizing antibody epitope through immune escape. Nat Med. 2012;18:1688–1692. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 2014;515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Auwerx J, François KO, Covens K, Van Laethem K, Balzarini J. Glycan deletions in the HIV-1 gp120 V1/V2 domain compromise viral infectivity, sensitize the mutant virus strains to carbohydrate-binding agents and represent a specific target for therapeutic intervention. Virology. 2008;382:10–19. doi: 10.1016/j.virol.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 46.Zhou T, Zheng A, Baxa U, Chuang GY, Georgiev IS, Kong R, et al. A Neutralizing Antibody Recognizing Primarily N-linked Glycan Targets the Silent Face of the HIV Envelope. Immunity. 2018;48:500–513. e6. doi: 10.1016/j.immuni.2018.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward AB, Wilson IA. The HIV-1 envelope glycoprotein structure: nailing down a moving target. Immunol Rev. 2017;275:21–32. doi: 10.1111/imr.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Travers SA. Conservation, Compensation, and Evolution of N-Linked Glycans in the HIV-1 Group M Subtypes and Circulating Recombinant Forms. ISRN AIDS. 2012;2012:823605. doi: 10.5402/2012/823605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Behrens A-J, Crispin M. Structural principles controlling HIV envelope glycosylation. Curr Opin Struct Biol. 2017;44:125–133. doi: 10.1016/j.sbi.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao L, Diedrich JK, Kulp DW, Pauthner M, He L, Park S-KR, et al. Global site-specific N-glycosylation analysis of HIV envelope glycoprotein. Nat Commun. 2017;8:14954. doi: 10.1038/ncomms14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Panico M, Bouché L, Binet D, O’Connor M-J, Rahman D, Pang P-C, et al. Mapping the complete glycoproteome of virion-derived HIV-1 gp120 provides insights into broadly neutralizing antibody binding. Sci Rep. 2016;6:32956. doi: 10.1038/srep32956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Behrens AJ, Vasiljevic S, Pritchard LK, Harvey DJ, Andev RS, Krumm SA, et al. Composition and Antigenic Effects of Individual Glycan Sites of a Trimeric HIV-1 Envelope Glycoprotein. Cell Rep. 2016;14:2695–2706. doi: 10.1016/j.celrep.2016.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pritchard LK, Vasiljevic S, Ozorowski G, Seabright GE, Cupo A, Ringe R, et al. Structural Constraints Determine the Glycosylation of HIV-1 Envelope Trimers. Cell Rep. 2015;11:1604–1613. doi: 10.1016/j.celrep.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willey RL, Shibata R, Freed EO, Cho MW, Martin MA. Differential Glycosylation, Virion Incorporation, and Sensitivity to Neutralizing Antibodies of Human Immunodeficiency Virus Type 1 Envelope Produced from Infected Primary T-Lymphocyte and Macrophage Cultures. J Virol. 1996;70:6431–6436. doi: 10.1128/jvi.70.9.6431-6436.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pritchard LK, Spencer DIR, Royle L, Vasiljevic S, Krumm SA, Doores KJ, et al. Glycan Microheterogeneity at the PGT135 Antibody Recognition Site on HIV-1 gp120 Reveals a Molecular Mechanism for Neutralization Resistance. J Virol. 2015;89:6952–9. doi: 10.1128/JVI.00230-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCoy LE, Falkowska E, Doores KJ, Le K, Sok D, van Gils MJ, et al. Incomplete Neutralization and Deviation from Sigmoidal Neutralization Curves for HIV Broadly Neutralizing Monoclonal Antibodies. PLoS Pathog. 2015;11:1–19. doi: 10.1371/journal.ppat.1005110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Katie, Doores J, Burton DR. Variable Loop Glycan Dependency of the Broad and Potent HIV-1-Neutralizing Antibodies PG9 and PG16. J Virol. 2010;84:10510–10521. doi: 10.1128/JVI.00552-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pritchard LK, Harvey DJ, Bonomelli C, Crispin M, Doores KJ. Cell- and Protein-Directed Glycosylation of Native Cleaved HIV-1 Envelope. J Virol. 2015;89:8932–8944. doi: 10.1128/JVI.01190-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crooks ET, Tong T, Chakrabarti B, Narayan K, Georgiev IS, Menis S, et al. Vaccine-Elicited Tier 2 HIV-1 Neutralizing Antibodies Bind to Quaternary Epitopes Involving Glycan-Deficient Patches Proximal to the CD4 Binding Site. PLoS Pathog. 2015;11:e1004932. doi: 10.1371/journal.ppat.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Voss JE, Andrabi R, McCoy LE, de Val N, Fuller RP, Messmer T, et al. Elicitation of Neutralizing Antibodies Targeting the V2 Apex of the HIV Envelope Trimer in a Wild-Type Animal Model. Cell Rep. 2017;21:222–235. doi: 10.1016/j.celrep.2017.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wibmer CK, Gorman J, Anthony CS, Mkhize NN, Druz A, York T, et al. Structure of an N276-Dependent HIV-1 Neutralizing Antibody Targeting a Rare V5 Glycan Hole Adjacent to the CD4 Binding Site. J Virol. 2016;90:10220–10235. doi: 10.1128/JVI.01357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Medina-Ramírez M, Garces F, Escolano A, Skog P, de Taeye SW, Del Moral-Sanchez I, et al. Design and crystal structure of a native-like HIV-1 envelope trimer that engages multiple broadly neutralizing antibody precursors in vivo. J Exp Med. 2017;214:2573–2590. doi: 10.1084/jem.20161160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Behrens AJ, Kumar A, Medina-Ramirez M, Cupo A, Marshall K, Cruz Portillo VM, et al. Integrity of Glycosylation Processing of a Glycan-Depleted Trimeric HIV-1 Immunogen Targeting Key B-Cell Lineages. J Proteome Res. 2018;17:987–999. doi: 10.1021/acs.jproteome.7b00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andrabi R, Su C-Y, Liang C-H, Shivatare SS, Briney B, Voss JE, et al. Glycans Function as Anchors for Antibodies and Help Drive HIV Broadly Neutralizing Antibody Development. Immunity. 2018;47:524–537. e3. doi: 10.1016/j.immuni.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Molloy SS, Bresnahan PA, Leppla SH, Klimpel KR, Thomas G. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J Biol Chem. 1992;267:16396–16402. [PubMed] [Google Scholar]
- 66.Ringe RP, Sanders RW, Yasmeen A, Kim HJ, Lee JH, Cupo A, et al. Cleavage strongly influences whether soluble HIV-1 envelope glycoprotein trimers adopt a native-like conformation. Proc Natl Acad Sci U S A. 2013;110:18256–61. doi: 10.1073/pnas.1314351110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pritchard LK, Vasiljevic S, Ozorowski G, Seabright GE, Cupo A, Ringe R, et al. Structural Constraints Determine the Glycosylation of HIV-1 Envelope Trimers. Cell Rep. 2015;11:1604–1613. doi: 10.1016/j.celrep.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, et al. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Binley JM, Sanders RW, Master A, Cayanan CS, Wiley CL, Schiffner L, et al. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 2002;76:2606–16. doi: 10.1128/JVI.76.6.2606-2616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dey AK, Cupo A, Ozorowski G, Sharma VK, Behrens A-J, Go EP, et al. cGMP production and analysis of BG505 SOSIP.664, an extensively glycosylated, trimeric HIV-1 envelope glycoprotein vaccine candidate. Biotechnol Bioeng. 2017 doi: 10.1002/bit.26498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guenaga J, Dubrovskaya V, de Val N, Sharma SK, Carrette B, Ward AB, et al. Structure-Guided Redesign Increases the Propensity of HIV Env To Generate Highly Stable Soluble Trimers. J Virol. 2015;90:2806–17. doi: 10.1128/JVI.02652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kong L, He L, de Val N, Vora N, Morris CD, Azadnia P, et al. Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability. Nat Commun. 2016;7:12040. doi: 10.1038/ncomms12040. [DOI] [PMC free article] [PubMed] [Google Scholar]