Abstract
The formation of C-C bonds by using CoA independent acyltransferases may have significant impact for novel methods for biotechnology. We report the identification of Pseudomonas strains with CoA-independent acyltransferase activity as well as the heterologous expression of the enzyme in E. coli. The cloning strategies and selected expression studies are discussed. The recombinant acyltransferases were characterized with regard to thermal and storage stability, pH,- and co-solvent tolerance. Moreover, the impact of bivalent metals, inhibitors, and other additives was tested. Careful selection of expression and working conditions led to obtain recombinant acyltransferase form Pseudomonas protegens with up to 11 U mL−1 activity.
Electronic supplementary material
The online version of this article (10.1007/s00253-018-9052-z) contains supplementary material, which is available to authorized users.
Keywords: Acyltransferase; 2,4-diacetylphloroglucinol; Pseudomonas protegens; Friedel-Crafts reaction
Introduction
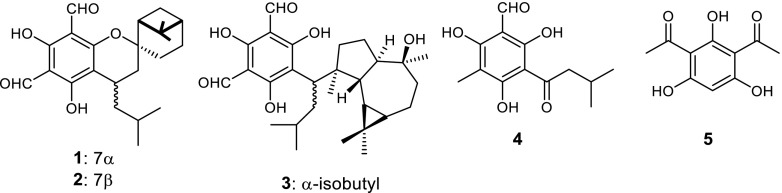
The phenolic compound phloroglucinol (PG, 1,3,5-trihydoxybenzene) is a polyketide and represents the key scaffold of around 700 natural products isolated from different natural sources including microorganisms, plants, and marine (Singh and Bharate 2006, Singh et al. 2010). This includes halogenated, prenylated or acylated phloroglucinols, phloroglucinol glycosides, and cyclic polyketides or terpene-adducts. Those natural compounds exhibit a broad spectrum of antibacterial, antiviral, antifungal, antihelminthic, phytotoxic, and antioxidant properties (Tada et al. 1990; Bae 2011; Mathekga et al. 2000; Artan et al. 2008; Veena et al. 2016). For instance, robustadials A and B are known for their in vivo antimalarial activity (1–2, Fig. 1; Xu et al. 1984), macrocarpal B possesses potent HIV-RTase inhibitory activity (3, Fig. 1; Nishizawa et al. 1992), and gerandinol (4, Fig. 1) shows Epstein-Barr virus inhibitory activity (Takasaki et al. 1990).
Fig. 1.
Biologically active phloroglucinol derivatives
Among acylated phloroglucinols, the bis-acetylated compound 2,4-diacetylphloroglucinol (5; DAPG, Fig. 1) is of special interests due to its strong antibiotic properties. DAPG is a secondary metabolite excreted by several plant-associated Pseudomonas sp. and Pseudomonas fluorescens ssp. to protect the rhizosphere from soil-borne pathogens (Dowling and O'Gara 1994). The biosynthesis of DAPG is regulated by the phlACBDEFGHI gene cluster, conserved among all DAPG-producing Pseudomonas strains (Keel et al. 1996; Moynihan et al. 2009), which is divided into regulatory genes phlEFGHI and the biosynthetic operon phlACBD.
In addition to complex genetic regulations, several biotic and abiotic factors were found to modulate DAPG production as well (Notz et al. 2001; Matano et al. 2010; Paulin et al. 2009). Among the proteins which are not directly involved in the biosynthesis, phlE encodes a putative permease (Abbas et al. 2004) and phlG a hydrolase (Bottiglieri and Keel 2006; Saitou et al. 2012). PhlF and phlH are transcriptional regulators (Schnider-Keel et al. 2000) and phlI is a so-far uncharacterized protein. Among the biosynthetic genes, phlD encodes a type-III polyketide synthase which is responsible for the biosynthesis of the DAPG-precursor phloroglucinol (PG) (Achkar et al. 2005; Zha et al. 2006; Cao and Xian 2011; Yang and Cao 2012). Finally, the operon phlACB encodes an acetyl-CoA independent acyltransferase (ATase), which catalyzes the acetylation of PG leading to the target polyketide DAPG (Shanahan et al. 1993; Bangera and Thomashow 1999). Expression of the entire phlACB operon is essential to obtain functional ATase, since individual expression of phlA, phlC, and phlB and subsequent incubation of the three individual proteins did not show any activity towards disproportionation of the natural substrate monoacylphloroglucinol (MAPG) (Bangera and Thomashow 1999; Achkar et al. 2005; Hayashi et al. 2012). This leads to the assumption that the ATase exists as a multienzyme complex, what was furthermore confirmed by the fact that mutations in any of the genes resulted in a loss of catalytic activity (Bangera and Thomashow 1999; Kidarsa et al. 2011). According to literature, biocatalytic applications were limited to phlD which was employed for the in-vivo production of PG in either E. coli or Pseudomonas sp. under controlled conditions in bioreactors (Cao et al. 2011; Banotai et al. 2012; Frost 2012; Rao et al. 2013).
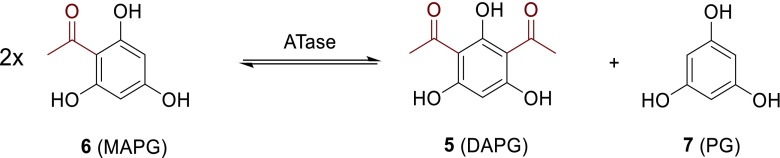
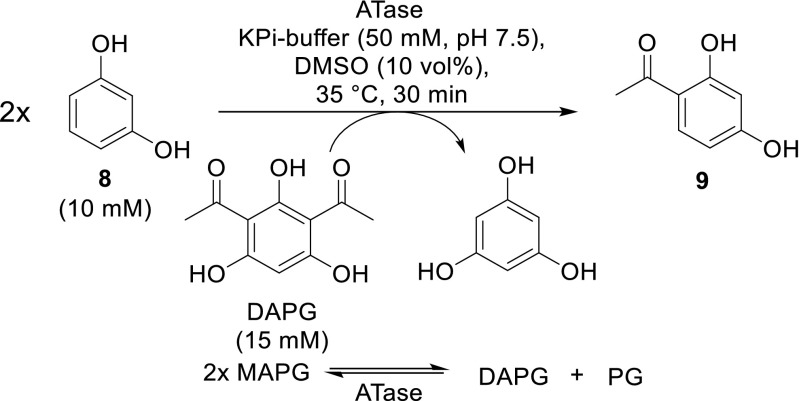
Very recently, a multi-component acyltransferase (ATase) originating from the bacterium Pseudomonas sp. YGJ3 was identified to catalyze the reversible disproportionation of two molecules of MAPG (6, Fig. 2) into one molecule of PG (7) and DAPG (5) in the forward reaction (Hayashi et al. 2012; Yang and Cao 2012; Almario et al. 2017). Our previous report proved that a multi-component ATase from Pseudomonas protegens catalyzes transfer of acyl moieties, not only from natural but also from non-natural donor substrates, to the aromatic ring of a phenolic acceptor substrate by forming a new C-C bond in a Friedel-Crafts-type acylation reaction (Schmidt et al. 2017). In this work, we report on the cloning of the key biosynthetic operon phlACB, into a bacterial expression vector to produce the ATase in E. coli and developed optimal conditions for robust synthesis of this enzyme.
Fig. 2.
Natural reaction catalyzed by the acyltransferase (ATase) involved in the biosynthesis of DAPG
Materials and methods
General information
All starting materials in this study were obtained from commercial suppliers (Sigma- Aldrich, Alfa Aesar, or TCI-Chemicals) and used as received unless stated otherwise. 1,3-Diacetyl-2,4,6-trihydroxy benzene (5, DAPG) was chemically synthesized as previously described (Sato et al. 2005; Khazaei et al. 2010; Mudududdla et al. 2012). pH-measurements were carried out on a pH Meter (Hanna Instruments, HI2211 pH/ORP Meter), equipped with a conventional Ag/AgCl pH-electrode (SI-Analytics, BlueLine 16 pH). Pseudomonas protegens DSM 19095 and Pseudomonas brassicacearum DSM 13227 were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ). Pseudomonas fluorescens Pf-5 was obtained from the American Type Culture Collection (ATCC BAA-477). gBlocks® gene fragments, primers for polymerase-chain reactions (PCR) were purchased from IDT and Eurofins MWG Operon. The expression vector pASK-IBA3plus was purchased from IBA©-Solutions for Lifesciences. Restriction enzymes and the PureLink® Genomic DNA Minikit were obtained from ThermoFisher Scientific. Chemically competent E. coli DH5α and E. coli BL21 (DE3) cells and the Gibson Assembly® master mix were purchased from New England Biolabs. Conversions for product 9 were determined by HPLC (Shimadzu-prominence liquid chromatograph, SPD-M20A diode array detector), equipped with a Phenomenex Luna® 5 μ C18 (2) 100A (250 × 4.6 mm) column. Gradient elution with H2O and MeCN (+TFA, 0.1 vol%) was applied: 0–15% MeCN (0–5 min), 15–60% MeCN (5–22 min), 60–100% MeCN (22–25 min), 100–0% MeCN (25–30 min), flow rate = 1 mL min−1, sample vol. = 2 μL, λ = 280 nm. GC-MS spectra were recorded with an Agilent 7890A GC-system (Agilent 5975C mass selective detector) equipped with a HP-5 MS column (30 m × 0.25 mm × 0.25 μm); injector 250 °C, constant flow 0.7 mL; carrier gas = He. Temperature program 100 °C (hold 0.5 min) 100 to 300 °C (10 °C min−1), 300 °C (hold 2 min). 1H- and 13C-NMR spectra were recorded at 20 °C on a 300 Bruker NMR unit; chemical shifts are given in ppm relative to Me4Si (1H: Me4Si = 0.0 ppm) or relative to the resonance of the solvent (1H: acetone-d6 = 2.05 ppm; 13C: acetone-d6 = 29.84 and 206.6 ppm). TLC was carried out with pre-coated aluminum sheets (TLC Silica gel 60 F254, Merck) with detection by UV (254 nm) and/or by staining with cinnamaldehyde/HCl solution [abs. EtOH (72.2 vol%), conc. HCl (3.6 vol%), trans-cinnamaldehyde (3.6 vol%)]. Shake flask cultivation, expression of the wild-type, and recombinant ATase was performed as described previously (Schmidt et al. 2017).
Shake flask cultivation of the Pseudomonas wildtypes
For the cultivation of the Pseudomonas wildtypes, P. protegens and P. brassicacearum, a pre-culture (100 mL) containing M1-media (peptone 5 g L−1, meat-extract 3 g L−1) was inoculated with a glycerol stock (250 μL). The pre-culture was grown at 28 °C (P. protegens) or 30 °C (P. brassicacearum) for 3 days and shaken with 120 rpm. The main culture (3 × 330 mL) in baffled shake flasks (1 L) was inoculated with the pre-culture (10 mL), and incubation was continued for another 3 days under the given conditions. The cells were harvested by centrifugation (10 min, 8000 rpm) and washed with potassium phosphate buffer (50 mM, pH 7.5) prior to lyophilization.
Plasmid construction
The phlACB operon from P. protegens DSM19095 and P. brassicacearum DSM13227 was amplified from the genomic DNA using primer sequences which were identified in a BLAST-search (Supporting Information). The phlACB open-reading frames (≈ 2770 bp) of P. protegens DSM19095 and P. brassicacearum DSM13227 were identified from a multiple sequence alignment using the phl gene cluster sequence phlACBDEFGHI of Pseudomonas sp. YGJ3 as template (Noyori 2009). Since P. protegens DSM19095 shares 99% sequence identity to the reference strain Pseudomonas sp. YGJ3, primer sequences for the amplification of phlACB were taken from literature without further modifications (Hayashi et al. 2012). Whereas, P. brassicacearum DSM13227 was unavailable in databases, P. brassicacearum NFM241 was chosen as a best substitute with 80% sequence identity. A second sequence alignment indicated 99% identity between phl genes of P. brassicacearum NFM241 and P. fluorescens J2. For this reason, the phlACB genes of P. fluorescens J2 were used to design the primers for P. brassicacearum DSM13227. The isolated WT-ATase encoding genes were subsequently cloned into pASK-IBA3 (tet-promoter) to obtain the plasmids pEG330 and pEG331.
Construction of recombinant Pseudomonas protegens
To construct the recombinant PpATaseCH, the ATase encoding open-reading frames phlA, phlC, and phlB of P. protegens were codon-optimized by manually matching the codon-frequency of the Pseudomonas wild-type with E. coli. For this purpose, codon-usage tables for Escherichia coli B and Pseudomonas fluorescens were obtained from the Kazusa-database (http://www.kazusa.or.jp/codon/). Ribosomal binding sites suitable for E. coli were introduced upstream of each start codon of every individual phl gene. The optimized phl genes were obtained as gene fragments (gBlocks©) and assembled together with the double-digested pASKIBA3plus backbone (EcoRI/HindIII) by Gibson cloning (Gibson Assembly® master mix) and subsequent overlap extension-PCR (OE-PCR). The resulting expression vector carries the E. coli codon-optimized ATase encoding genes phlACB under the control of the PTet promoter.
Shake flask cultivation and expression of the recombinant ATases
The expression plasmids (pEG330-PbATaseWT, pEG331-PpATaseWT or pEG332-PpATaseCH) were chemically transformed into E. coli BL21 (DE3) host cells. A single colony, picked from an agar-plate (LB/ampicilin, 100 μg mL−1), was used for inoculation of an overnight-culture (10 mL, LB/Amp, 100 μg mL−1) which was grown at 37 °C and 135 rpm for 15 h. The main culture (1 L) containing LB/Amp (100 μg mL−1) was then inoculated with the overnight culture (10 mL) and shaken in a non-baffled flask (5 L) at 37 °C and 140 rpm until the OD600 reached 0.7. The cells were induced with anhydrotetracycline (AHTC, 200 μg L−1), and protein expression was continued for 21 h at 30 °C (PpATaseWT or PpATaseCH) or 6 h at 25 °C (PbATaseWT). The cells were harvested by centrifugation (15 min, 8000 rpm, 12,028×g), washed with potassium phosphate buffer (50 mM, pH 7.5), and resuspended again in the same buffer (7 mL buffer to 1 g wet cells). The suspension was disrupted by ultrasonication (40% amplitude, 8 min, pulse 1 s, pause 4 s). After centrifugation (30 min, 14,000 rpm, 23,519×g), the cell-free extract was analyzed by SDS-PAGE, shock-frozen in liquid nitrogen and stored at − 20 °C until direct use for biotransformations. Initial rates and protein concentrations were measured to determine the batch activity.
Acylation of resorcinol with DAPG - general procedure
Resorcinol (8, 1.1 mg, 0.01 mmol, 10 mM final concentration) was dissolved in KPi-buffer (50 mM, pH 7.5), and the mixture was preheated to 35 °C for 10 min. Cell-free E. coli extract containing the recombinant ATase (50 μL or vol. ≡ 20 mg lyophilized cells) was added, and the reaction was started by addition of DAPG (3.15 mg, 0.015 mmol, 15 mM final concentration) dissolved in DMSO (100 μL). DMSO was required to improve the solubility of the donor in buffer. The final reaction mixture (1 mL, 10 vol% DMSO) was shaken for 18 h at 35 °C and 550 rpm in an Eppendorf benchtop shaker. The reaction was aborted by addition of HPLC-grade MeCN (1 mL) and vigorous shaking. The precipitated protein was removed by centrifugation (7 min, 14,000 rpm, 18,407×g) and the supernatant (800 μL) was transferred to an Eppendorf tube and left standing for another 40 min. Any precipitated protein was removed by centrifugation (10 min, 14,000 rpm, 18,407×g), and the supernatant was directly subjected to HPLC for determination of conversions.
Activity assay
ATase-batch activities were measured on a Thermo Scientific Genesys 10 UV Scanning UV/Vis spectrophotometer according to a modified procedure (Hayashi et al. 2012) from literature. When following the disproportionation of MAPG into DAPG and PG spectrophotometrically (Fig. 2), an increase of absorption is recorded due to the formation of DAPG (ε = 205.6 M−1 cm−1, λ = 395 nm). One unit of activity was defined as the amount of ATase that catalyzed the formation of 1 μmol of DAPG per minute under the following conditions: potassium phosphate buffer (800 μL, 50 mM, pH 7.5) and MAPG (2.0 μmol, 100 μL of a 20 mM stock solution prepared in DMSO) were added to a cuvette and preheated to 35 °C. The reaction (1 mL total volume, 10 vol% DMSO) was started by the addition of the enzyme-containing cell-free extract (100 μL ≡ 14.3 mg wet cells). The reaction was followed for 3 min. All reactions were performed as a duplicate. As a negative control, a sample with cell-free extract of E. coli without plasmid (empty host cells) was run. The protein concentration (Bradford) was measured [ε = 0.083 mL mg−1 cm−1, λ = 595 nm], and specific activities were determined as units per mg cell-free extract or as units per mg purified protein. PG does not absorb at this wavelength.
Nucleotide sequences used in this study have been deposited at NCBI.
PbATaseWT (wild-type nucleotide phlACB gene sequence, P. brassicacearum DSM 13227, Genbank accession no.: KY173354).
PpATaseWT (wild-type nucleotide phlACB gene sequence, P. protegens DSM 19095, Genbank accession no.: CP003190.1).
PpATaseCH (codon-optimized phlACB gene sequence, P. protegens DSM 19095, Genbank accession no.: KY173355).
Results
Screening of Pseudomonas wild type strains
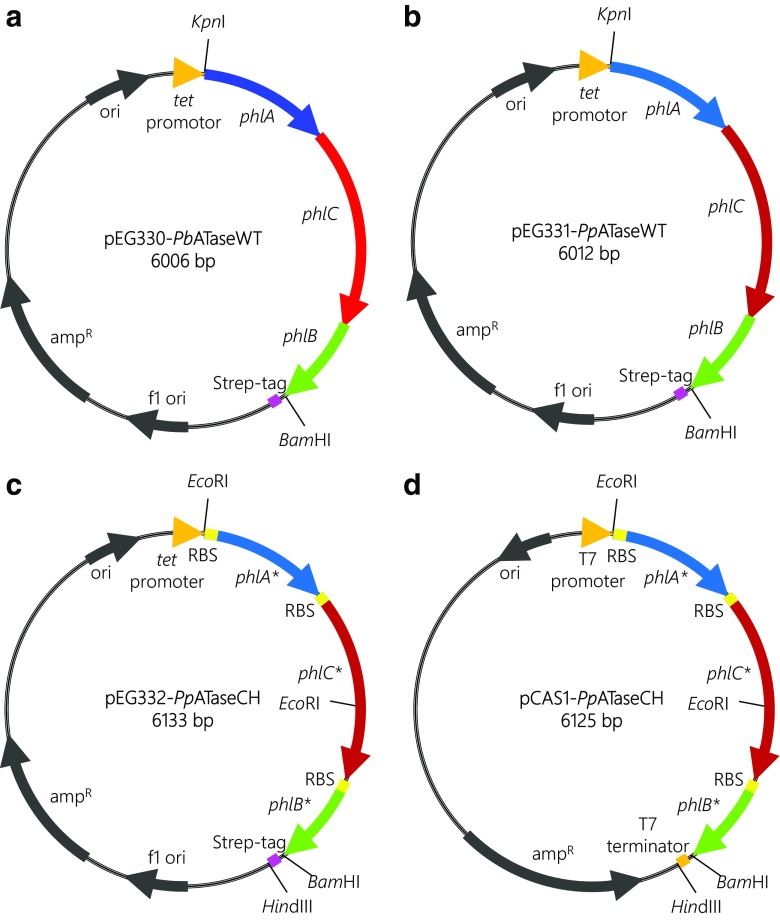
Freeze-dried cell preparations of the respective Pseudomonas wild-type were screened for ATase activity. For this purpose, 32 strains from the in-house culture collection (Table S1, Fig. S1) were qualitatively assayed for the acetylation and deacetylation of MAPG. Additionally, the reverse direction, the acetylation of PG by DAPG, was tested. Among the investigated Pseudomonas wild-type strains, three strains were found to be active, namely P. brassicacearum DSM13227, P. fluorescens Pf-5 ATCC BAA-477, and P. protegens DSM19095. The phlACB operon from P. protegens DSM19095 and P. brassicacearum DSM13227 was amplified from the genomic DNA using primer sequences which were identified in a BLAST-search. The isolated WT-ATase encoding genes were subsequently cloned into pASK-IBA3 (tet-promoter) to obtain the plasmids pEG330 (PbATaseWT, Fig. 3) and pEG331 (PpATaseWT, Fig. 3b).
Fig. 3.
Plasmids harboring a the WT-derived ATase genes phlACB from P. brassicacearum (PbATaseWT, pEG330), b the WT-derived ATase genes phlACB from P. protegens (PpATaseWT, pEG330), c the codon-harmonized phlACB* genes and ribosmomal binding sites (RBS), optimized ATase from P. protegens (PpATaseCH, pEG332), tet-promoter regulated, d optimized ATase from P. protegens (PpATaseCH, pCAS1), T7-promoter regulated
Expression of the wild-type acyltransferases
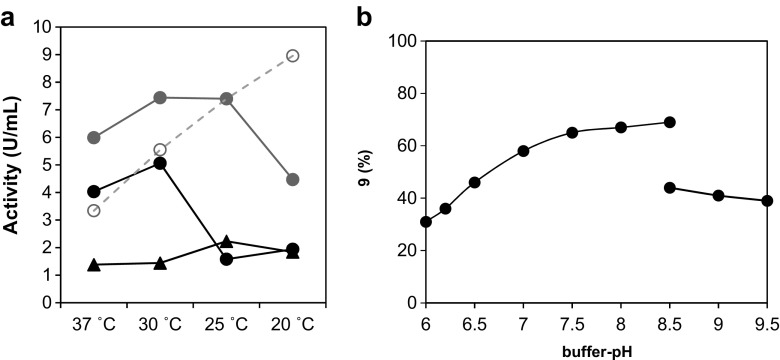
In order to establish the optimum expression conditions required to obtain soluble and active protein, parameters such as temperature, culture media, and time, were investigated. Both WT-ATases were expressed in E. coli BL21 (DE3) host cells, and ATase activities were measured spectrophotometrically, following the disproportionation of MAPG into DAPG and PG. The activity of the PpATaseWT was highest when expression was performed at 30 °C with a rapid decrease in activity below 30 °C, whereas 25 °C was the optimum expression temperature for the PbATaseWT (Fig. 4a).
Fig. 4.
a Batch activities (units per mL cell-free extract) of PpATaseWT (black circles), PbATaseWT (black triangles) after expression in LB-media for 21 h in the absence of ZnCl2, and PpATaseCH in the presence (open gray circles) and absence of ZnCl2 (filled gray circles) at varied temperatures. b pH-study of the PpATaseCH. Assay conditions: Lyophilized cells of E. coli containing the recombinant PpATaseCH (20 mg), KPi-buffer (50 mM) for pH 6.0–8.5 or CHES-buffer (50 mM) for pH 8.5–9.5, resorcinol (8, 10 mM), DAPG (15 mM), DMSO (10 vol%), 35 °C, 30 min, 750 rpm
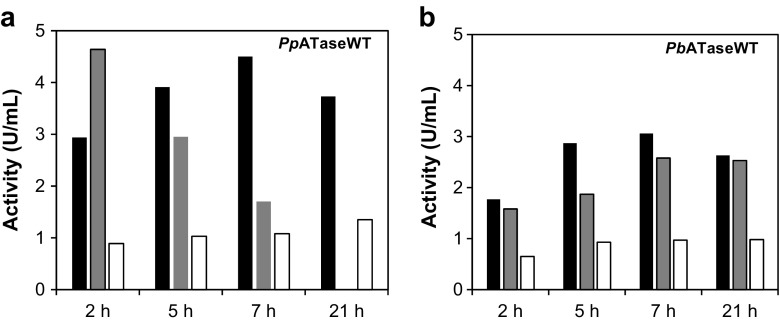
In the next step, the effect of different growth media on the expression of the WT-ATases was studied at various time frames. LB-, TB-, or YENB-broth media were tested at previously determined temperatures, thus 30 °C for PpATaseWT and 25 °C for PbATaseWT. Results are shown in Fig. 5. Activities after expression of the PpATaseWT in TB-media linearly decreased with increasing expression time. Even though a 2 h expression in TB gave the best results in terms of protein activity, cell densities were too low to provide enough protein after such a short expression time. The use of chloride-free YENB-media suffered from low activities for both enzymes.
Fig. 5.
a Activities of PpATaseWT and b PbATaseWT (units per mL cell-free extract) after expression at different times in different culture media: LB-media (black column), TB-media (gray column), YENB-media (white column). The activity of the PpATaseWT in TB after 21 h was not determined
Cloning strategy for PpATaseCH
To obtain a higher and more balanced expression of the three subunits of the ATase encoding genes in E. coli, phlACB from P. protegens (Almario et al. 2017) were codon-harmonized and ribosomal binding sites (RBS) were introduced at the 5′-end of each phl gene. For this purpose, the optimized phl* genes were assembled by Gibson cloning (Smith and March 2007; Turner and O'reilly 2013) and overlap-extension PCR (OExPCR) (Fessner and Anthonsen 2009; Patel 2006; Bommarius and Riebel-Bommarius 2004) to obtain the tet-promoter regulated plasmid pEG332 (PpATaseCH, Fig. 3c). Alternatively, the phl* genes were cloned into the T7-promoter-regulated vector pCAS1 to obtain the construct pCAS1- PpATaseCH (Fig. 3d). Expression conditions optimized for the related PpATaseWT served as a starting point for the expression of the codon-harmonized PpATaseCH (pEG332), i.e., E. coli BL21, LB-media, 30 °C, 21 h. The cell-free extracts produced by codon-harmonized construct PpATaseCH displayed twofold higher activities towards disproportionation of MAPG compared to analogous preparations derived from wild-type sequences. According to SDS-PAGE, all phl genes of the codon-harmonized ATase were clearly overexpressed in high amounts and soluble form, in contrast to the WT-ATase, which was not clearly detectable at the same conditions (see Supporting Information). The expression of the PpATaseCH was performed efficiently under control of the PTet promoter, whereas, the expression under regulation of the strong T7-promoter (pCAS1-construct) did not lead to functional ATase.
Influence of Zn2+ on the expression
Since the impact of Zn2+ on the functional expression of the ATase was of particular interest, the influence of ZnCl2 (1 mM) and temperature was studied in detail (Fig. 4a). Batch activities determined for PpATaseWT (Fig. 4a, black circles) and PpATaseCH (Fig. 4a, open gray circles) after expression with ZnCl2 were compared to the expression without additives (Fig. 4a, gray-filled circles). The expression level of PpATasCH was enhanced in the presence of Zn2+ as deduced from the SDS-page (Supporting Information, SDS-gel); however, the obtained activities in the cell-free extract at 30 °C were 1.5 times lower compared to the expression in the absence of ZnCl2 (Fig. 4a, filled vs. open gray circles). Even more interesting was the effect of Zn2+ at various temperatures. Again, Zn2+ led to slightly enhanced visible expression, whereas the activity linearly increased with decreasing temperature, thereby reaching the highest activity at 20 °C.
Cultivation scale and size of shake flasks
To evaluate the possible influence of the cultivation scale and the size of the shaking flasks on the measured ATase activities, total protein concentration and enzyme specific activity were determined after cultivation in various size of baffled and non-baffled flasks. Although there are variations (Table 1, entry 4) for related conditions, the activity obtained expressed as U/mL was comparable (Entries 1–3).
Table 1.
Activities depending on the scale of the shake flask cultivation and the flask size
| Entry | Flask size (L) | Medium/flask (L) | Total protein (mg/mL) | U/mL | U/mg |
|---|---|---|---|---|---|
| 1 | 0.3[a] | 0.006 | 20.03 | 7.44 | 0.49 |
| 2 | 1.0[a] | 0.33 | 15.25 | 8.90 | 0.58 |
| 3 | 5.0 | 1.0 | 14.85 | 8.2 | 0.55 |
| 4 | 5.0 | 1.0 | 8.95 | 10.95 | 1.22 |
Cultivation conditions: LB-media, 200 μg L−1 AHTC, 30 °C, 21 h, 140 rpm
[a]Baffled flask
pH-study and co-solvent tolerance
For the evaluation of pH, the model substrate was changed to resorcinol (Fig. 6), since we were interested to apply this enzyme for various acyl donors and acceptors (Schmidt et al. 2017). Thus, the ATase activity in the following studies was evaluated based on the formation of the C-acetyl product 9 arising from the bioacetylation of model substrate 8 by DAPG. Alternatively to DAPG, also MAPG may be used as an acyl donor. However, bioacylation of 8 performed with MAPG resulted in 13% product yield within 30 min, while using DAPG gave 65% in the same conditions (KPi-buffer pH 7.5, 35 °C, 750 rpm). Following the disproportionation of MAPG into DAPG and PG on 10 mM scale resulted in 59% conversion.
Fig. 6.
The bioacetylation of resorcinol (8) by DAPG served as a new model system to characterize the ATases
The pH-optimum was determined based on the ATase catalyzed formation of 9 upon bioacetylation of 8 with DAPG at varied pH-values (Fig. 4b). pH-values, from slightly acidic (pH 6.0) to alkaline (pH 9.5) were investigated using two buffer salts, namely KPi-buffer (50 mM) for pH 6.0–8.5 and CHES-buffer (50 mM) for pH 8.5–9.5. The PpATaseCH showed the highest activity at slightly alkaline pH, as product formation was most efficient when reactions were carried out between pH 7.5 and 8.5 KPi-buffer.
Besides temperature and pH, the application of co-solvent was considered. Different water-immiscible (toluene, cyclohexane), moderately water-miscible (MTBE, DIPE, Et2O, EtOAc), aprotic water-miscible (DMSO, DMF, THF, 1,4-dioxane, acetone, MeCN), and protic water-miscible (MeOH, EtOH, glycerol, ethylene glycole) solvents were tested for the bioacetylation of model substrate 8 either at 5 vol% (Fig. S11, black columns) or 20 vol% (Fig. S11, gray columns), and the compatibility with PpATaseCH was determined based on the formation of C-acetyl product 9. It is worth to note that only toluene, cyclohexane, and MTBE formed a biphasic system, whereas the other solvents were completely miscible under the given conditions. Most of the investigated co-solvents were well tolerated by the enzyme at both tested concentrations, although no substantial improvements were observed compared to the reaction without solvent (Fig. S11, gray dashed line). Weakly water-miscible ethers, such as MTBE, DIPE or diethyl ether, and the water-miscible aprotic solvent DMSO, seemed to be slightly preferred, because the formation of 9 went almost equally well at 5 and 20 vol% concentration, respectively. Larger differences with respect to conversion and solvent concentration were obtained with the water-miscible ones. Especially THF and, at higher concentrations, also DMF as well as several other protic and aprotic co-solvents turned out to be rather detrimental to the ATase.
Storage and thermo-stability of different types of enzyme preparation
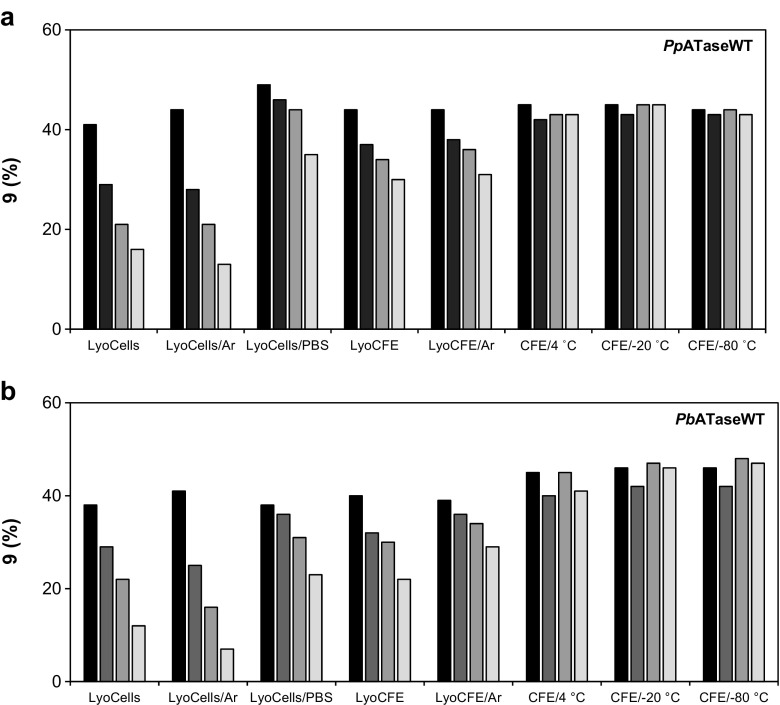
To learn about the storage stability of the recombinant ATases, batch activities of different types of enzyme preparations under various conditions were monitored over 9 weeks (Fig. 7). To test the residual activity of the enzyme batches, the bioacetylation of 8 with DAPG was performed after 0, 14, 35, or 63 days of storage. The impact of lyophilization and storage temperature was evaluated. Additionally, the sensitivity to molecular oxygen was tested by storing the preparation under inert atmosphere (argon). For instance, freeze-dried cells prepared in KPi-buffer lost almost their entire activity after being stored for 9 weeks at 4 °C; inert storage under argon did not help to retain activity (Fig. 7a and b, rows 1 and 2). However, if cells were treated in salt-rich PBS-buffer instead of KPi-buffer prior to lyophilization, the activity loss was significantly slower, indicating that high salt concentrations contribute to the stabilization of the enzyme (Fig. 7, row 3). If the ATase-containing cell-free extract was lyophilized and stored at 4 °C, activity was lost as well. However, the degradation was much slower than observed for the lyophilized cells (Fig. 7, rows 4–5 vs. rows 1–2). Interestingly, when the liquid cell-free extract was frozen or simply stored at 4 °C instead of lyophilized, the entire batch activity was retained for at least 9 weeks. Preparations stored at + 4 °C, − 20 or − 80 °C were equally active (Fig. 7, rows 6–8) leading to 9 with 44–45% product yield.
Fig. 7.
Studies on the storage stability of different types of enzyme preparations a PpATaseWT and b PbATaseWT. The conversions were measured after 0 days (black columns), 14 days (dark gray columns), 35 days (pale gray column), and 63 days (white column) after preparing the catalyst. Assay conditions: Cell preparation of the respective recombinant ATase (20 mg or vol. ≡ 20 mg lyophilisate), KPi-buffer (50 mM, pH 7.5), resorcinol (8, 10 mM), DAPG (15 mM), DMSO (10 vol%), 35 °C, 30 min, 750 rpm; LyoCells, (lyophilized cells, KPi-buffer, 4 °C); LyoCells/Ar, (lyophilized cells, inert storage, KPi-buffer, 4 °C); LyoCells/PBS (Lyophilized cells, PBS, 50 Mm, pH 7.5, 4 °C), LyoCFE, (lyophilized cell-free extract, KPi-buffer, 4 °C); LyoCFE/Ar, (lyophilized cell-free extract, inert storage, KPi-buffer, 4 °C); CFE/4 °C, (liquid cell-free extract, KPi-buffer, 4 °C); CFE/− 20 °C, (liquid cell-free extract, KPi-buffer, − 20 °C); CFE/− 80 °C, (liquid cell-free extract, KPi-buffer, − 80 °C)
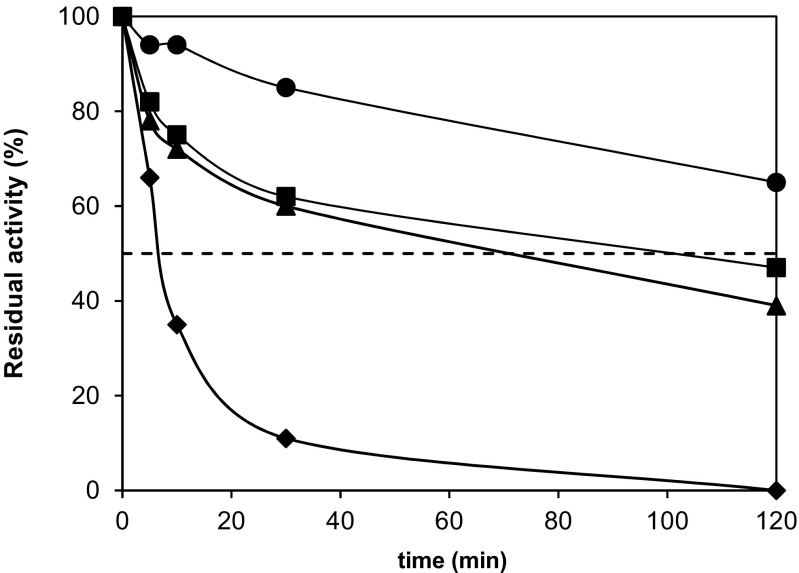
Tolerance to an increased reaction temperature is a crucial parameter having significant effects on both, the enzymatic activity and its stability. The thermostability of PpATaseCH was determined by heat-treatment of the ATase-containing free extract assay at various temperatures. When incubating the ATase at 35 °C, the residual activity after 2 h was 68% (Fig. 8, circle); however, at higher temperatures, such as 50 °C (Fig. 8, square) or 60 °C (Fig. 8, triangle), the enzyme lost half of its activity within 70 min. Prolonging the incubation time at these temperatures up to 2 h resulted in residual activities of approximately 40%. At even higher temperatures, severe heat-deactivation was noticed. For instance, at 70 °C (Fig. 8, diamond) the half-life was less than 10 min and consequently, after 2 h the enzyme was not active anymore.
Fig. 8.
Thermostability of PpATaseCH. The residual activity was determined after heat-treating the ATase-containing cell-free extract at 35 °C (circle), 50 °C (square), 60 °C (triangle), or 70 °C (diamond) for up to 120 min. Assay conditions: Cell-free E. coli extract containing the recombinant PpATaseCH (50 μL), KPi-buffer (50 mM, pH 7.5), resorcinol (8, 10 mM), DAPG (15 mM), DMSO (10 vol%), 35 °C, 30 min, 750 rpm. Activities in % were related to an untreated control sample (= 100%)
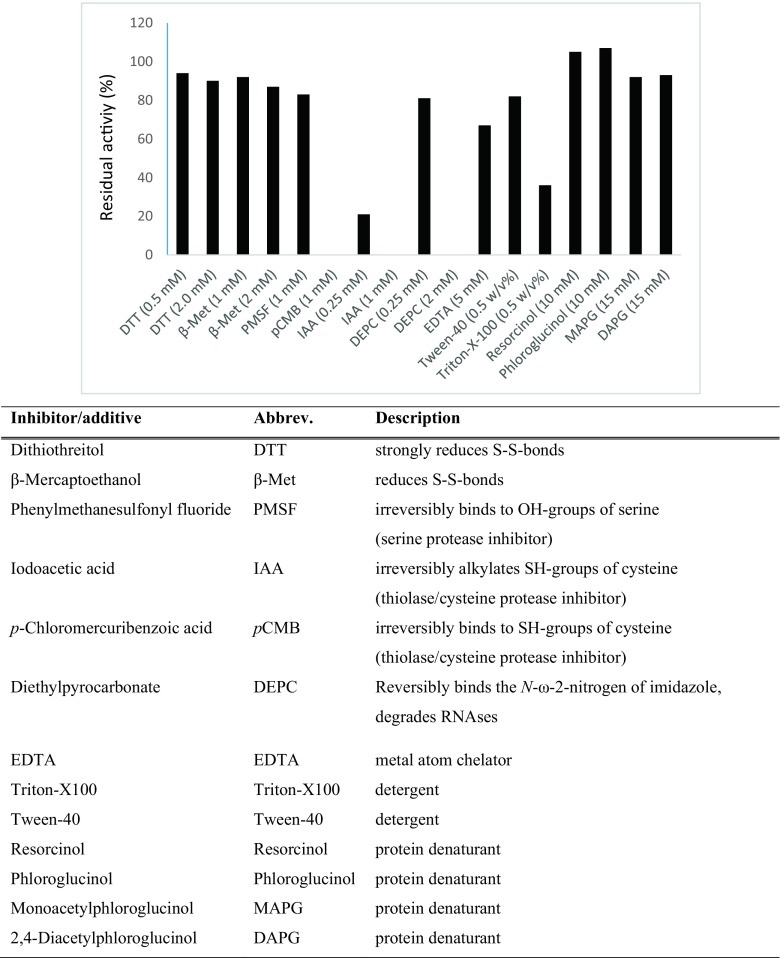
Inhibitors and additives
The influence of commonly used inhibitors was studied in order to gain more information about the ATase (Fig. 9). For testing a small aliquot of cell-free E. coli extract containing the recombinant ATase (50 μL) was pretreated with the respective inhibitor/additive for 40 min at 28 °C: dithiothreitol (DTT, 0.5 or 2 mM), 2-mercaptoethanol (β-Met, 1 or 2 mM), phenylmethanesulfonyl fluoride (PMSF, 1 mM), iodoacetic acid (IAA, 1 or 2 mM), p-chloromecuribenzoic acid (pCMB, 1 mM), diethylpyrocarbonate (DEPC, 2 or 3 mM), EDTA (5 mM), Triton-X100 (0.5 w/v%), or Tween-40 (0.5 w/v%). All studied inhibitors affected the ATase activity, leading in general to a loss of activity (Fig. 9). After treatment with pCMP or IAA, which are known for their ability to covalently bind to thiol moieties, the enzyme activity was abolished. Furthermore DEPC, which is known to modify histidine residues also led to a loss of conversion at 2 mM concentration. DTT, β-Met, PMSF, and EDTA led to slightly reduced activities. Furthermore, the enzyme turned out to be sensitive towards detergents (0.5% w/v, Triton-X100 and Tween-40).
Fig. 9.
The influence of inhibitors and additives on PpATaseCH. The residual activity was determined after pre-treating the ATase-containing cell-free extract at 28 °C for 40 min with the respective additive. Assay conditions: Cell-free E. coli extract containing the recombinant PpATaseCH (50 μL), KPi-buffer (50 mM, pH 7.5), resorcinol (8, 10 mM), DAPG (15 mM), DMSO (10 vol%), 35 °C, 30 min, 750 rpm. Activities in % were related to an untreated control sample (= 100%)
Additionally, the enzyme was incubated with resorcinol (10 mM), phloroglucinol (10 mM), DAPG (15 mM), and MAPG (15 mM) prior to biotransformation. In case of resorcinol and phloroglucinol, a slight increase in residual activity was observed (105 and 107% respectively, while for MAPG and DAPG, a slight decrease was noticed, 92–93% (Fig. 9).
Enzyme activity was also investigated in the presence of the chloride salts of Ca2+, Mg2+, Zn2+, Cu2+, Co2+, Mn2+, Sr2+, or Ni2+ at 5 or 8 mM concentration and compared to a control reaction in the absence of the metal (Fig. S9). To avoid precipitation of the metal with a buffer component (e.g., phosphate), all reactions were performed in HEPES-buffer (50 mM, pH 7.5) using solutions and ATase containing cell-free extract prepared in the same buffer. Most of the bivalent metals were tolerated by the ATase without having noteworthy effects on the enzyme. Only Cu2+ led to significantly reduced conversions.
Discussion
Three Pseudomonas strains, P. protegens DSM19095, P. brassicacearum DSM13227, and P. fluorescens Pf-5 ATCC BAA-477 with acyltransferase activity were identified to catalyze the reversible disproportionation of two molecules of natural substrate MAPG into PG and DAPG. In contrast to other acyltransferases, the ATase involved in the biosynthesis of DAPG is independent of the coenzyme A (CoA) cofactor (Hayashi et al. 2012). No product was detected in the absence of the cell preparations, indicating that product formation can be assigned to the acyltransferase activity present in the mentioned Pseudomonas strains. As P. fluorescens Pf-5 was the least active strain under the conditions employed, subsequent investigations of the substrate spectrum were exclusively performed with P. brassicacearum and P. protegens. Although higher activities were obtained with the recombinant enzyme from P. protegens (PpATaseWT) compared to P. brassicacearum (PbATaseWT), optimum expression conditions were established for both enzyme preparations.
In order to obtain soluble and active protein, parameters such as temperature, culture media, and time were investigated. Major differences in terms of functional expression of both ATases were noticed. Equally important was the type of the expression medium. LB-media turned out to be the best compromise between cell densities and protein activity. A time period of 21 h was chosen as an optimum for the expression of the PpATaseWT and 6–7 h for the PbATaseWT in LB-media.
For expression of the heterotrimeric enzyme in E. coli, it was shown that the introduction of ribosomal binding site upstream of the phl coding sequences as well as the codon-optimization significantly improved the expression of the ATase in E. coli. Furthermore, it is worth mentioning that almost no inclusion bodies were formed under these conditions indicating that the proteins PhlA, PhlC, and PhlB were expressed in a soluble form. Despite the high-level expression observed for T7-promoter, formation of inclusion bodies was observed at any investigated temperature. From these observations, it can be deduced that a weaker promoter, such as the tet-promoter in pEG332 is much better suited for the overexpression of the ATase than the strong T7-system in pCAS1.
During optimization, it was noticed that the size and shape of the shaking flasks did not significantly influence the ATase expression, although significant variation between experiments were observed (Table 1, entry 3, 4). Optimum expression conditions can be summarized as follows: E. coli BL21 worked best in LB-media, non-baffled shake flasks (5 L) with 200 μg L−1 AHTC at 30 °C and 140 rpm. Optimization of the expression conditions, as well as codon-optimization, let to significantly improved enzyme activities compared to PpATaseWT (Fig. 4a, black vs. gray circles).
The type of cell/enzyme treatment had also a significant impact on the activity of the catalyst preparation, while the storage temperature seemed to play a minor role. Inert storage did not prevent from activity loss, which leads to the assumption that oxygen is not involved in the observed degradation. Both enzyme preparations, PpATaseWT, and PbATaseWT showed similar behavior upon long-term storage. Finally, after proving the negative effect of lyophilization, the enzymes were subsequently stored at − 20 °C as a cell-free extract or as PBS-treated lyophilized cells.
It is worth mentioning, that the enzyme is not only sensitive to lyophilization, but also to increased temperatures. From thermostability studies, it can be concluded, that ATase is temperature-sensitive, and 35 °C seems to be the best compromise between stability and activity.
The PpATaseCH showed the highest activity at slightly alkaline pH, as product formation was most efficient when reactions were carried out between pH 7.5 and 8.5 KPi-buffer (Fig. 4b). Alkaline pH resulted in lower enzyme activity; however, it has to be mentioned that CHES-buffer was obviously less suitable for the enzyme because overall conversions were considerably lower compared to reactions performed in KPi-buffer. Taking into account that pH 8.0 constitutes the upper operational limit of the KPi buffer salt, pH 7.5 was used as best pH for the substrates investigated here.
High co-solvent tolerance is particularly useful if the solubility and thus the availability of hydrophobic substrates in the reaction medium has to be increased. Furthermore, product separation and work-up procedures can be significantly facilitated by integrating a suitable co-solvent into the reaction system. After mapping the general co-solvent tolerance of the ATase, potential solubility issues may be addressed by implementing one of the tolerated solvents, preferably MTBE, diethyl ether, or EtOAc.
Small chemical compounds may affect the enzymatic activity, either by specifically interacting with distinct amino acids such as active-site residues or by influencing the overall structure. Some of tested compounds react with SH-groups (DTT, β-Met, IAA, pCMB), OH-moieties (PMSF) or NH-groups (DEPC). After treatment with pCMP or IAA, which are known for their ability to covalently bind to thiol moieties, the enzyme activity was abolished. This may support the assumption that cysteine (most likely cysteine 88) is involved in the acyl transfer. Furthermore, the enzyme turned out to be sensitive towards detergents (0.5% w/v, Triton-X100, and Tween-40). Detergents tend to alter the quaternary and tertiary structure of proteins, and may even lead to denaturation. At this point, it has to be mentioned that none of the investigated reagents are ATase-specific inhibitors; thus, the influence on other structural elements besides active-site residues might be considered as well. Nevertheless, this study allows a rough estimation of possible hot-spots, responsible for the observed catalytic action of the ATase.
Although phenols are known for their denaturating properties, resorcinol or phloroglucinol did not cause any loss of activity but actually improved activity, while MAPG and DAPG decreased activity only slightly (92–93%, Fig. 9); thus, apparently these reagents had no significant denaturing effect under the conditions investigated.
The effects of bivalent metals on the stability and activity of proteins is well studied and generally depends on the ionic radii of the metal and the coordination geometries it can adopt (Yang et al. 2003; Coolbear et al. 1992). Neutral impact of investigated bivalent alkaline-earth and transition metals on the biotransformation confirmed, that ATase catalyzes C-acyl transfer reactions independently of a metal cofactor. Only Cu2+ led to significantly reduced conversions, indicating enzyme inhibition.
In summary, identification of suitable expression conditions using E. coli BL21 cells, optimum temperature (30 °C for PpATaseWT and 25 °C for PbATaseWT), LB-medium, expression time (21 h for PpATaseWT and 7 h for PbATaseWT), and cultivation in 5-L non-baffled flask led to catalyst preparations of highest activity. We identified KPi-buffer pH 7.5 as the most suitable reaction medium, while the addition of co-solvents and bivalent metals had no notable influence on the performance of the ATase. Introduction of ribosomal binding sites upstream of the phl genes and the codon-optimization significantly improved the expression of the ATase in E. coli compared to the wild-type genes.
Electronic supplementary material
(PDF 1431 kb)
Funding information
Open access funding provided by Austrian Science Fund (FWF). This study was financed by the Austrian FFG, BMWFJ, BMVIT, SFG, Standortagentur Tirol, and ZIT through the Austrian FFG-COMET- Funding Program. AZD is financed by the Austrian Science Fund (FWF) Lise Meitner Fellowship grant M 2172-B21.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants performed by any of the authors.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s00253-018-9052-z) contains supplementary material, which is available to authorized users.
References
- Abbas A, McGuire JE, Crowley D, Baysse C, Dow M, O'Gara F. The putative permease PhlE of Pseudomonas fluorescens F113 has a role in 2, 4-diacetylphloroglucinol resistance and in general stress tolerance. Microbiology. 2004;150(7):2443–2450. doi: 10.1099/mic.0.27033-0. [DOI] [PubMed] [Google Scholar]
- Achkar J, Xian M, Zhao H, Frost JW. Biosynthesis of phloroglucinol. J Am Chem Soc. 2005;127(15):5332–5333. doi: 10.1021/ja042340g. [DOI] [PubMed] [Google Scholar]
- Almario J, Bruto M, Vacheron J, Prigent-Combaret C, Moënne-Loccoz Y, Muller D. Distribution of 2, 4-Diacetylphloroglucinol biosynthetic genes among the Pseudomonas spp. reveals unexpected polyphyletism. Front Microbiol. 2017;8:1218. doi: 10.3389/fmicb.2017.01218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artan M, Li Y, Karadeniz F, Lee SH, Kim MM, Kim SK. Anti-HIV-1 activity of phloroglucinol derivative, 6, 6′-bieckol, from Ecklonia cava. Bioorg Med Chem. 2008;16(17):7921–7926. doi: 10.1016/j.bmc.2008.07.078. [DOI] [PubMed] [Google Scholar]
- Bae JS. Antithrombotic and profibrinolytic activities of phloroglucinol. Food Chem Toxicol. 2011;49(7):1572–1577. doi: 10.1016/j.fct.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Bangera MG, Thomashow LS. Identification and characterization of a gene cluster for synthesis of the polyketide antibiotic 2, 4-diacetylphloroglucinol from Pseudomonas fluorescens Q2–87. J Bacteriol. 1999;181(10):3155–3163. doi: 10.1128/jb.181.10.3155-3163.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banotai C, Frost J, Jancauskas J, Lau MK (2012) Lehner B (Draths Coporations) WO2012003461 A2
- Bommarius AS, Riebel-Bommarius BR. Biocatalysis. Weinheim: Wiley-VCH; 2004. [Google Scholar]
- Bottiglieri M, Keel C. Characterization of PhlG, a hydrolase that specifically degrades the antifungal compound 2, 4-diacetylphloroglucinol in the biocontrol agent Pseudomonas fluorescens CHA0. Appl Environ Microbiol. 2006;72(1):418–427. doi: 10.1128/AEM.72.1.418-427.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Xian M. Production of phloroglucinol by Escherichia coli using a stationary-phase promoter. Biotechnol Lett. 2011;33(9):1853–1858. doi: 10.1007/s10529-011-0638-0. [DOI] [PubMed] [Google Scholar]
- Cao Y, Jiang X, Zhang R, Xian M. Improved phloroglucinol production by metabolically engineered Escherichia coli. Appl Microbiol Biotechnol. 2011;91(6):1545–1552. doi: 10.1007/s00253-011-3304-5. [DOI] [PubMed] [Google Scholar]
- Coolbear T, Whittaker JM, Daniel RM. The effect of metal ions on the activity and thermostability of the extracellular proteinase from a thermophilic Bacillus, strain EA. 1. Biochem J. 1992;287(2):367–374. doi: 10.1042/bj2870367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling DN, O'Gara F. Metabolites of Pseudomonas involved in the biocontrol of plant disease. Trends Biotechnol. 1994;12(4):133–141. doi: 10.1016/0167-7799(94)90091-4. [DOI] [Google Scholar]
- Fessner WD, Anthonsen T (2009) Modern biocatalysis: stereoselective and environmentally friendly reactions. Wiley-VCH, Weinheim
- Frost JW (2012) US8329445 B2
- Hayashi A, Saitou H, Mori T, Matano I, Sugisaki H, Maruyama K. Molecular and catalytic properties of monoacetylphloroglucinol acetyltransferase from Pseudomonas sp. YGJ3. Biosci Biotechnol Biochem. 2012;76(3):559–566. doi: 10.1271/bbb.110860. [DOI] [PubMed] [Google Scholar]
- Keel C, Weller DM, Natsch A, Défago G, Cook RJ, Thomashow LS. Conservation of the 2, 4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl Environ Microbiol. 1996;62(2):552–563. doi: 10.1128/aem.62.2.552-563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazaei A, Rostami A, Mantashlo F. p-Toluenesulfonyl chloride as a new and effective catalyst for acetylation and formylation of hydroxyl compounds under mild conditions. Chin Chem Lett. 2010;21(12):1430–1434. doi: 10.1016/j.cclet.2010.05.025. [DOI] [Google Scholar]
- Kidarsa TA, Goebel NC, Zabriskie TM, Loper JE. Phloroglucinol mediates cross-talk between the pyoluteorin and 2, 4-diacetylphloroglucinol biosynthetic pathways in Pseudomonas fluorescens Pf-5. Mol Microbiol. 2011;81(2):395–414. doi: 10.1111/j.1365-2958.2011.07697.x. [DOI] [PubMed] [Google Scholar]
- Matano I, Tsunekawa M, Shimizu S, Tanaka I, Mitsukura K, Maruyama K. The chloride ion is an environmental factor affecting the biosynthesis of pyoluteorin and 2, 4-diacetylphloroglucinol in Pseudomonas sp. YGJ3. Biosci Biotechnol Biochem. 2010;74(2):427–429. doi: 10.1271/bbb.90664. [DOI] [PubMed] [Google Scholar]
- Mathekga AD, Meyer JM, Horn MM, Drewes SE. An acylated phloroglucinol with antimicrobial properties from Helichrysum caespititium. Phytochemistry. 2000;53(1):93–96. doi: 10.1016/S0031-9422(99)00424-0. [DOI] [PubMed] [Google Scholar]
- Moynihan JA, Morrissey JP, Coppoolse ER, Stiekema WJ, O'Gara F, Boyd EF. Evolutionary history of the phl gene cluster in the plant-associated bacterium Pseudomonas fluorescens. Appl Environ Microbiol. 2009;75(7):2122–2131. doi: 10.1128/AEM.02052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudududdla R, Jain SK, Bharate JB, Gupta AP, Singh B, Vishwakarma RA, Bharate SB. Ortho-Amidoalkylation of phenols via tandem one-pot approach involving oxazine intermediate. J Org Chem. 2012;77(19):8821–8827. doi: 10.1021/jo3017132. [DOI] [PubMed] [Google Scholar]
- Nishizawa M, Emura M, Kan Y, Yamada H, Ogawa K, Hamanaka N. Macrocarpals: HIV-RTase inhibitors of Eucalyptus globulus. Tetrahedron Lett. 1992;33(21):2983–2986. doi: 10.1016/S0040-4039(00)79578-5. [DOI] [Google Scholar]
- Notz R, Maurhofer M, Schnider-Keel U, Duffy B, Haas D, Défago G. Biotic factors affecting expression of the 2, 4-diacetylphloroglucinol biosynthesis gene phlA in Pseudomonas fluorescens biocontrol strain CHA0 in the rhizosphere. Phytopathology. 2001;91(9):873–881. doi: 10.1094/PHYTO.2001.91.9.873. [DOI] [PubMed] [Google Scholar]
- Noyori R. Synthesizing our future. Nat Chem. 2009;1(1):5–6. doi: 10.1038/nchem.143. [DOI] [PubMed] [Google Scholar]
- Patel RN. Biocatalysis in the pharmaceutical and biotechnology industries. New York: CRC Press; 2006. [Google Scholar]
- Paulin MM, Novinscak A, St-Arnaud M, Goyer C, DeCoste NJ, Prive JP, Owen J, Filion M. Transcriptional activity of antifungal metabolite-encoding genes phlD and hcnBC in Pseudomonas spp. using qRT-PCR. FEMS Microbiol Ecol. 2009;68(2):212–222. doi: 10.1111/j.1574-6941.2009.00669.x. [DOI] [PubMed] [Google Scholar]
- Rao G, Lee JK, Zhao H. Directed evolution of phloroglucinol synthase PhlD with increased stability for phloroglucinol production. Appl Microbiol Biotechnol. 2013;97(13):5861–5867. doi: 10.1007/s00253-013-4713-4. [DOI] [PubMed] [Google Scholar]
- Saitou H, Watanabe M, Maruyama K. Molecular and catalytic properties of 2, 4-Diacetylphloroglucinol hydrolase (PhlG) from Pseudomonas sp. YGJ3. Biosci Biotechnol Biochem. 2012;76(6):1239–1241. doi: 10.1271/bbb.120054. [DOI] [PubMed] [Google Scholar]
- Sato S, Kusakari T, Suda T, Kasai T, Kumazawa T, Onodera JI, Obara H. Efficient synthesis of analogs of safflower yellow B, carthamin, and its precursor: two yellow and one red dimeric pigments in safflower petals. Tetrahedron. 2005;61(40):9630–9636. doi: 10.1016/j.tet.2005.07.080. [DOI] [Google Scholar]
- Schmidt NG, Pavkov-Keller T, Richter N, Wiltschi B, Gruber K, Kroutil W. Biocatalytic Friedel–crafts acylation and fries reaction. Angew Chem Int Ed. 2017;56(26):7615–7619. doi: 10.1002/anie.201703270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnider-Keel U, Seematter A, Maurhofer M, Blumer C, Duffy B, Gigot-Bonnefoy C, Reimmann C, Notz R, Défago G, Haas D, Keel C. Autoinduction of 2, 4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J Bacteriol. 2000;182(5):1215–1225. doi: 10.1128/JB.182.5.1215-1225.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan P, Glennon JD, Crowley JJ, Donnelly DF, O'Gara F. Liquid chromatographic assay of microbially derived phloroglucinol antibiotics for establishing the biosynthetic route to production, and the factors affecting their regulation. Anal Chim Acta. 1993;272(2):271–277. doi: 10.1016/0003-2670(93)80579-A. [DOI] [Google Scholar]
- Singh IP, Bharate SB. Phloroglucinol compounds of natural origin. Nat Prod Rep. 2006;23(4):558–591. doi: 10.1039/b600518g. [DOI] [PubMed] [Google Scholar]
- Singh IP, Sidana J, Bharate SB, Foley WJ. Phloroglucinol compounds of natural origin: synthetic aspects. Nat Prod Rep. 2010;27(3):393–416. doi: 10.1039/b914364p. [DOI] [PubMed] [Google Scholar]
- Smith MB, March J. March’s Advanced Organic Chemistry. ed. 6. New Jersey: Wiley; 2007. [Google Scholar]
- Tada M, Takakuwa T, Nagai M, Yoshii T. Antiviral and antimicrobial activity of 2, 4-Diacylphloroglucinols, 2-Acylcyclohexane-1, 3-diones and 2-Carboxamidocyclo-hexane-1, 3-diones. Agric Biol Chem. 1990;54(11):3061–3063. [Google Scholar]
- Takasaki M, Konoshima T, Fujitani K, Yoshida S, Nishimura H, Tokuda H, Nishino H, Iwashima A, Kozuka M. Inhibitors of skin-tumor promotion. VIII: inhibitory effects of Euglobals and their related compounds on Epstein-Barr virus activation.(1) Chem Pharm Bull. 1990;38(10):2737–2739. doi: 10.1248/cpb.38.2737. [DOI] [PubMed] [Google Scholar]
- Turner NJ, O'Reilly E. Biocatalytic retrosynthesis. Nat Chem Biol. 2013;9(5):285–288. doi: 10.1038/nchembio.1235. [DOI] [PubMed] [Google Scholar]
- Veena VK, Kennedy K, Lakshmi P, Krishna R, Sakthivel N. Anti-leukemic, anti-lung, and anti-breast cancer potential of the microbial polyketide 2, 4-diacetylphloroglucinol (DAPG) and its interaction with the metastatic proteins than the antiapoptotic Bcl-2 proteins. Mol Cell Biochem. 2016;414(1–2):47–56. doi: 10.1007/s11010-016-2657-6. [DOI] [PubMed] [Google Scholar]
- Xu R, Snyder JK, Nakanishi K. Robustadials A and B from Eucalyptus robusta. J Am Chem Soc. 1984;106(3):734–736. doi: 10.1021/ja00315a047. [DOI] [Google Scholar]
- Yang F, Cao Y. Biosynthesis of phloroglucinol compounds in microorganisms. Appl Microbiol Biotechnol. 2012;93(2):487–495. doi: 10.1007/s00253-011-3712-6. [DOI] [PubMed] [Google Scholar]
- Yang Q, Xu J, Li M, Lei X, An L. High-level expression of a soluble snake venom enzyme, gloshedobin, in E. coli in the presence of metal ions. Biotechnol Lett. 2003;25(8):607–610. doi: 10.1023/A:1023067626846. [DOI] [PubMed] [Google Scholar]
- Zha W, Rubin-Pitel SB, Zhao H. Characterization of the substrate specificity of PhlD, a type III polyketide synthase from Pseudomonas fluorescens. J Biol Chem. 2006;281(42):32036–32047. doi: 10.1074/jbc.M606500200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1431 kb)