ABSTRACT
Streptococcus suis has received increasing attention for its involvement in severe human infections worldwide as well as in multidrug resistance. Two-component signaling systems (TCSSs) play important roles in bacterial adaptation to various environmental stimuli. In this study, we identified a novel TCSS located in S. suis serotype 2 (SS2), designated VraSRSS, which is involved in bacterial pathogenicity and susceptibility to antimicrobials. Our data demonstrated that the yvqFSS gene, located upstream of vraSRSS, shared the same promoter with the TCSS genes, which was directly regulated by VraSRSS, as shown in electrophoretic mobility shift assays. Notably, YvqFSS and VraSRSS constitute a novel multidrug resistance module of SS2 that participates in resistance to certain groups of antimicrobials. Further analyses showed that VraSRSS inactivation significantly attenuated bacterial virulence in animal models, which, coupled with the significant activation of VraSRSS expression observed in host blood, strongly suggested that VraSRSS is an important regulator of SS2 pathogenicity. Indeed, RNA-sequencing analyses identified 106 genes that were differentially expressed between the wild-type and ΔvraSRSS strains, including genes involved in capsular polysaccharide (CPS) biosynthesis. Subsequent studies confirmed that VraSRSS indirectly regulated the transcription of CPS gene clusters and, thus, controlled the CPS thickness shown by transmission electron microscopy. Decreased CPS biosynthesis caused by vraSRSS deletion subsequently increased bacterial adhesion to epithelial cells and attenuated antiphagocytosis against macrophages, which partially clarified the pathogenic mechanism mediated by VraSRSS. Taken together, our data suggest that the novel TCSS, VraSRSS, plays critical roles for multidrug resistance and full virulence in SS2.
KEYWORDS: Streptococcus suis, capsular biosynthesis, multidrug resistance, two-component signaling system, virulence
INTRODUCTION
Streptococcus suis is an important swine pathogen responsible for a wide range of diseases, including meningitis, septicemia, endocarditis, arthritis, and sudden death (1). Thirty-three reference serotypes have been identified based on capsular polysaccharide (CPS) (2), which is recognized as an important virulence factor in S. suis (3, 4). S. suis serotype 2 (SS2) is considered one of the most prevalent serotypes worldwide and an increasingly important zoonotic pathogen for humans (1). Two large-scale outbreaks of SS2 in humans with severe clinical symptoms emerged in 1998 and 2005 in China, during which many cases exhibited streptococcal toxic shock syndrome (5).
Two-component signaling systems (TCSSs), comprised of a membrane-bound sensor histidine kinase (HK) and a cytoplasmic response regulator (RR), are thought to play key roles in bacterial colonization and virulence (6, 7). The C-terminal catalytic and ATP-binding (CA) domain in HK protein phosphorylates the histidine residue of the dimerization and histidine phosphotransfer (DHp) domain when encountering certain external signals. Subsequently, the phosphoryl group is transferred to the receiver (REC) domain in the RR protein, and the helix-turn-helix (HTH) domain subsequently binds the target DNA promoters to induce or repress the expression of downstream genes (8, 9). The genome of SS2 encodes at least 13 TCSSs (10), among which RevS, Salk/SalR, CovR, CiaRH, Ihk/Irr, VirR/VirS, NisK/NisR, and 1910HK/RR have been reported to regulate the expression of key virulence factors or alter the bacterial metabolism, thereby contributing to the virulence of SS2 (11–18). RevS was the first TCSS in SS2 that was found to be involved in the pathogenesis of infected piglets (11), and CovR was identified as a global virulence regulator by repressing several virulence factors, such as CPS, sortase A, DNase streptodornase, laminin-binding protein, and hemolysin (13). SalK/SalR is located in the 89K pathogenicity island and was previously found to be required for the overall virulence of Chinese isolates of highly pathogenic SS2 (12). Ihk/Irr and VirR/VirS were shown to contribute to the virulence of SS2 by altering the bacterial cell metabolism (15, 16). However, it remains unclear which underlying mechanisms are employed by CiaRH, NisK/NisR, and 1910HK/RR to contribute to SS2 virulence (14, 17, 18).
To date, TCSSs implicated in antimicrobial resistance have not been reported in SS2. VraSR, a well-known TCSS, was identified to interact with its upstream gene, yvqF, to confer resistance to diverse cell wall-targeting antibiotics in Staphylococcus aureus (19, 20) and shares high sequence identity with one of the predicted TCSSs (ZY05719_02080/ZY05719_02085) in the virulent SS2 strain ZY05719. Four genes (orf1, yvqF, vraS, and vraR) form an autoregulatory operon in S. aureus, among which orf1 played no observable role, whereas the gene products YvqF and VraSR acted together to recognize members of cell wall-targeting antibiotics; thus, these proteins were proposed to function as a three-component system (20). ZY05719_02075, encoded by the upstream gene of zy05719_02080/zy05719_02085, was predicted to be a transmembrane protein but only shared 26% amino acid identity with the YvqF protein of S. aureus. It will be worthwhile to determine whether this VraSR homolog from S. aureus or other TCSSs regulate the multidrug resistance in SS2. With the massive use of multiple antimicrobial agents for S. suis prophylaxis and therapy, the emergence of resistance to tetracyclines, macrolides, β-lactams, and aminoglycosides has been frequently reported in S. suis worldwide (21–25). Indeed, antibiotic resistance determinants for tetracyclines, aminoglycosides, β-lactams, quinolones, and macrolides have been sequentially identified in S. suis, as have several integrative and conjugative elements (ICEs) carrying these resistance determinants (24, 26–33). Additionally, a multidrug gene, cfr, conferring resistance to diverse antimicrobials, including phenicols, lincosamides, oxazolidinones, pleuromutilins, streptogramin A, and macrolides, was detected on the pStrcfr plasmid in S. suis (34). Although these related genes can clarify the underlying mechanism for most antibiotic resistance in S. suis, the resistance mechanisms to the cell wall-targeting antimicrobials have remained unclear for this Gram-positive coccus.
In this study, we redesignated the ZY05719_02075 and ZY05719_02080/ZY05719_02085 proteins YvqFSS and VraSRSS, respectively. Our results demonstrated that VraSRSS contributes to multidrug resistance in SS2 by directly regulating the accessory protein YvqFSS. In addition, we found that the inactivation of VraSRSS significantly attenuates the virulence of SS2. Further RNA-sequencing (RNA-Seq) analyses identified 52 genes in the ΔvraSRSS mutant that were markedly downregulated compared with that of the wild-type (WT) strain. Of note, more than 15 downregulated genes are known to be involved in the biosynthesis of CPS. Transmission electron microscopy and capsule staining confirmed that the ΔvraSRSS strain indeed reduced the CPS thickness significantly. These results suggest that VraSRSS is a novel TCSS contributing to antimicrobial drug susceptibility and the full virulence of SS2.
RESULTS
Identification of a potential TCSS in SS2.
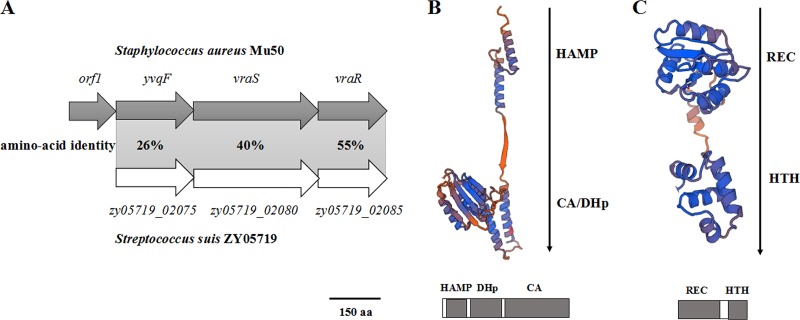
Homology analysis of the ZY05719_02080 and ZY05719_02085 proteins revealed that their amino acid sequences shared 40% and 55% identity with the VraS and VraR proteins of S. aureus (Fig. 1A) (19), respectively, suggesting that they also constitute a functional TCSS in S. suis. Domains and three-dimensional structures of ZY05719_02080 and ZY05719_02085 proteins were predicted using the SWISS-MODEL online server (https://www.swissmodel.expasy.org/). ZY05719_02080 was predicted to be a typical sensor kinase harboring a DHp domain, CA domain (Fig. 1B), and HAMP domain that may mediate transmembrane signal transduction. As shown in Fig. 1C, a REC domain catalyzing phosphoryl and an HTH domain used to conduct DNA binding were predicted to be present in ZY05719_02085, and a similar domain architecture is commonly found in most RRs. Such observations are consistent with the characteristics of a TCSS. Additionally, DNA sequence analyses were performed using the BLAST search service in NCBI, and zy05719_02080 and zy05719_02085 did not show any homology to any characterized HK or RR genes in Streptococcus species, suggesting that ZY05719_02080/ZY05719_02085 is an unknown TCSS in S. suis; thus, here it was redesignated VraSSS/VraRSS.
FIG 1.
Identification of a novel TCSS in SS2 strain ZY05719. (A) Homology analyses of the ZY05719_02075, ZY05719_02080, and ZY05719_02085 proteins by the protein BLAST algorithm. The identities of the amino acid sequences encoded by each gene are shown between SS2 strain ZY05719 and S. aureus strain Mu50. (B) Predicted three-dimensional structure and conserved domains of the ZY05719_02080 protein, as determined using the SWISS-MODEL online server. The HAMP domain that potentially mediates transmembrane signal transduction, the DHp domain containing a conserved histidine residue, and the CA domain were identified by sequence alignment and are labeled accordingly. (C) Three-dimensional structure and conserved domains of the ZY05719_02085 protein, predicted using the SWISS-MODEL online server. A REC domain catalyzing phosphoryl and an HTH domain used for DNA binding were identified by sequence alignment and are labeled accordingly.
The TCSS vraSRSS gene and the upstream zy05719_02075 gene constituted a novel multidrug resistance module in SS2.
It should be noted that the protein encoded by zy05719_02075, located upstream of the vraSSS gene, shows 26% sequence identity to the YvqF protein in S. aureus (Fig. 1A), which interacted with VraSR to contribute to resistance against cell wall-targeting antimicrobials (20). To assess whether ZY05719_02075 also functions in the resistance to antimicrobials, we examined the susceptibility profile of the Δzy05719_02075 strain against selected antimicrobial agents in vitro. As shown in Table 1, the Δzy05719_02075 strain was more sensitive than the WT strain to a wide range of antimicrobials, including ampicillin, amikacin, ceftriaxone, gentamicin, kanamycin, penicillin, streptomycin, vancomycin, bacitracin, colistin, polymyxin B, and nisin, and this deficiency was completely restored by complementation. These results indicated that zy05719_02075 functioned as a novel multidrug resistance gene in S. suis and showed a broader spectrum of antimicrobial resistance than yvqF of S. aureus; thus, here zy05719_02075 was redesignated yvqFSS.
TABLE 1.
Sensitivity of the strains to antimicrobials
| Antimicrobial | MIC (μg/ml) |
Classification | ||||
|---|---|---|---|---|---|---|
| ZY05719 | ΔvraSRSS | CΔvraSRSS | ΔyvqFSS | CΔyvqFSS | ||
| Amikacin | 32 | 4 | 32 | 4 | 16 | Aminoglycosides |
| Gentamicin | 4 | 0.5 | 4 | 1 | 4 | Aminoglycosides |
| Kanamycin | 16 | 2 | 16 | 4 | 8 | Aminoglycosides |
| Streptomycin | 128 | 32 | 128 | 64 | 128 | Aminoglycosides |
| Ampicillin | 1/8 | ≤1/32 | 1/8 | 1/16 | 1/8 | β-Lactams |
| Penicillin | 1/32 | ≤1/64 | 1/32 | ≤1/64 | 1/32 | β-Lactams |
| Ceftriaxone | 4 | 0.5 | 4 | 2 | 4 | β-Lactams |
| Vancomycin | 0.5 | 0.25 | 0.5 | 0.5 | 0.5 | Peptides |
| Bacitracin | 8 | 0.5 | 4 | 0.5 | 4 | Peptides |
| Colistin | 256 | 32 | 256 | 128 | 256 | Peptides |
| Polymyxin B | 128 | 16 | 64 | 64 | 128 | Peptides |
| Nisin | 256 | 64 | 256 | 128 | 256 | Peptides |
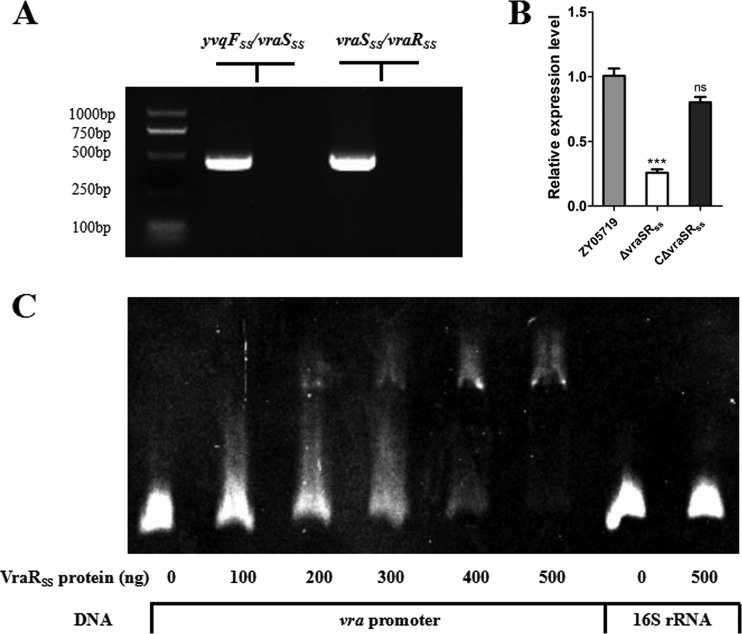
We then asked whether the multidrug resistance associated with YvqFSS was dependent on the regulation by VraSRSS and constructed a vraSRSS mutant strain. Subsequent study showed that the MICs of all antimicrobials for the ΔvraSRSS strain were close to that of the ΔyvqFSS strain and were significantly lower than that for the WT strain. Complementation restored resistance to these antimicrobials to levels similar to that of the WT strain, which further confirmed the susceptibility phenotype of vraSRSS deletion. In addition, we conducted quantitative real-time PCR (qRT-PCR) assays, which verified that the yvqFSS deletion did not affect expression of the downstream gene, vraSRSS (see Fig. S1 in the supplemental material). These results suggested that VraSRSS regulates the transcription of yvqFSS to control multidrug resistance in SS2.
VraSRSS directly regulated the expression of yvqFSS.
The closely encoding neighborhood between yvqFSS and vraSRSS, coupled with the mechanism of VraR in autoregulation (20), prompted us to consider that these three genes are controlled by one promoter. Indeed, prediction by the BProm program SoftBerry followed by RT-PCR analysis indicated that yvqFSS-vraSRSS formed a single operon (Fig. 2A) and thus shared one promoter. Furthermore, vraSRSS deletion significantly decreased the expression of yvqFSS (Fig. 2B), while reintroducing vraSRSS into the mutant recovered yvqFSS expression to WT levels (Fig. 2B), suggesting that VraSRSS indeed regulated yvqFSS expression in SS2. To determine the mechanisms of VraSRSS-mediated transcriptional regulation of the yvqFSS gene, we analyzed the upstream region of yvqFSS to identify a potential promoter region for the regulator-binding assay. Electrophoretic mobility shift assay (EMSA) showed that the VraRSS protein could bind to the upstream region of this operon but not the control fragment from 16S rRNA (Fig. 2C), which indicated that VraRSS directly regulated the expression of yvqFSS and the downstream TCSS genes, thus controlling multidrug resistance in the SS2 strain ZY05719.
FIG 2.
VraSRSS directly regulated the expression of YvqFSS and itself in SS2. (A) The yvqFSS, vraSSS, and vraRSS genes formed an operon, as determined by RT-PCR. An RNA sample that was not reverse transcribed served as a negative control. (B) Expression levels of yvqFSS in the WT, ΔvraSRSS, and complemented strains were measured by qRT-PCR. qRT-PCR expression values are shown as the means plus standard deviations (error bars) from at least three independent experiments. The unpaired two-tailed Student's t test was used for statistical analysis (ns, P > 0.05; ***, P < 0.001). (C) An EMSA showing VraRSS binding to the promoter region of the yvqFSS operon. The purified VraRSS fusion protein was added to each reaction mixture at different concentrations. DNA probes containing the yvqFSS operon promoter region were used at 80 ng per reaction mixture, and fragments amplified from 16S rRNA served as a negative control.
VraSRSS is required for the full virulence of SS2.
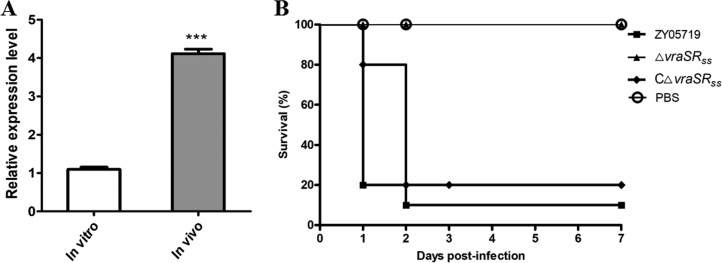
TCSSs enable bacteria to sense, respond, and adapt to a wide range of environments, stressors, and growth conditions (6, 7). Many TCSSs have been reported to respond to host stimulation, thereby contributing to the pathogenic process. Our results showed that vraSRSS transcription significantly increased in vivo by more than 4.1-fold compared to its expression level in vitro (Fig. 3A), suggesting that VraRSS regulates pathogenicity in SS2. To verify this possibility, an animal challenge test was performed using BALB/c mice injected with 5 × 108 CFU of different bacterial strains. We found that the mice infected by the ΔvraSRSS strain showed a significantly higher survival rate (100%), with or without slight clinical signs, compared with the survival of mice infected by the WT strain (10%), which showed acute clinical signs, such as shivering, rough hair coat, and depression. Although the BALB/c mouse model has been widely used for S. suis virulence studies, the model remains controversial (35–38). Thus, we then employed a zebrafish model to repeat the S. suis infection assays, as it has been identified as a convincing model for evaluating virulence with Streptococcus spp. (39, 40). Similarly, the 50% lethal dose (LD50) value of the ΔvraSRSS strain (6.84 × 106 CFU/fish) was significantly higher than that of the WT strain (3.09 × 105 CFU/fish) in the zebrafish infection model (Table S1). These observations, coupled with the complete restoration of virulence by complementation to the WT strain levels in both mouse and zebrafish models, indicated that the virulence of SS2 was significantly attenuated by VraSRSS inactivation.
FIG 3.
VraSRSS contributed to the full virulence of SS2. (A) Levels of vraSRSS gene expression during THB culture (in vitro) and animal infection (mouse blood, in vivo) were analyzed by qRT-PCR. The unpaired two-tailed Student's t test was used for statistical analysis (***, P < 0.001). (B) Deletion of vraSRSS resulted in significant attenuation of mortality in SS2 infection. Randomized groups of 10 SPF BALB/c mice were challenged with different bacterial strains at a dose of 5 × 108 CFU/mouse. Another 10 SPF BALB/c mice injected with PBS served as negative controls. The survival rates were monitored for 7 days after challenge. Significant differences in survival between different groups were analyzed by log-rank (Mantel-Cox) test (P < 0.05).
Identification of VraSRSS-regulated genes related to virulence via transcriptional analysis.
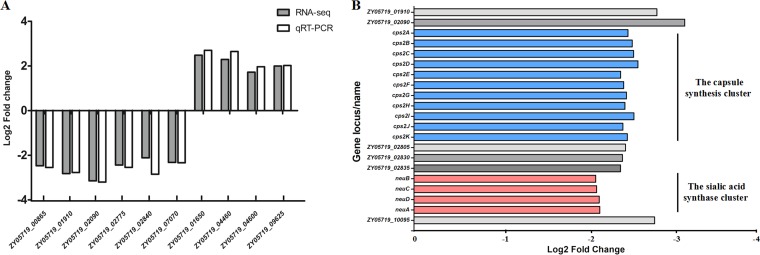
RNA-Seq analysis was performed to further explore the underlying mechanism whereby VraSRSS regulates the pathogenicity of SS2. The results showed that 54 genes were significantly upregulated and 52 were significantly downregulated in the ΔvraSRSS strain compared to the levels for the WT strain (Table S2). To confirm the changes in RNA transcription levels, qRT-PCR was performed to verify the expression profile of 10 selected genes in the WT and ΔvraSRSS strains. The relative expression levels of six downregulated genes and four upregulated genes identified by RNA-Seq were consistent in the qRT-PCR assays, which demonstrated the reliability of the RNA-Seq transcriptional data (Fig. 4A). These regulated genes can be classified into different functional categories, including ABC transporters, protein folding, and capsular synthesis. Although the expression levels of several well-known virulence factors, like fibronectin and fibrinogen-binding protein (FBPS), muramidase-released protein (MRP), and extracellular protein factor (EF), were unchanged, the expression level of gene clusters driving CPS biosynthesis were significantly decreased in the ΔvraSRSS strain (Fig. 4B), suggesting that VraSRSS regulates virulence by controlling CPS biosynthesis in SS2.
FIG 4.
Identification of VraSRSS-regulated genes via transcriptional analysis. (A) Comparison of gene regulation by RNA-Seq analysis or qRT-PCR. qRT-PCR was used to validate the expression level changes of 10 selected genes, including 4 upregulated genes and 6 downregulated genes, identified by RNA-Seq analysis. Each sample was run in triplicate, and the reference gene parC was detected as a control. Relative fold changes in expression were calculated using the 2−ΔΔCT method. (B) The VraSRSS-regulated genes identified by transcriptional analysis were involved in CPS biosynthesis. The expression levels of CPS biosynthesis-related genes were significantly lower in the ΔvraSRSS strain.
VraRSS indirectly regulated the expression of gene clusters driving CPS biosynthesis in SS2.
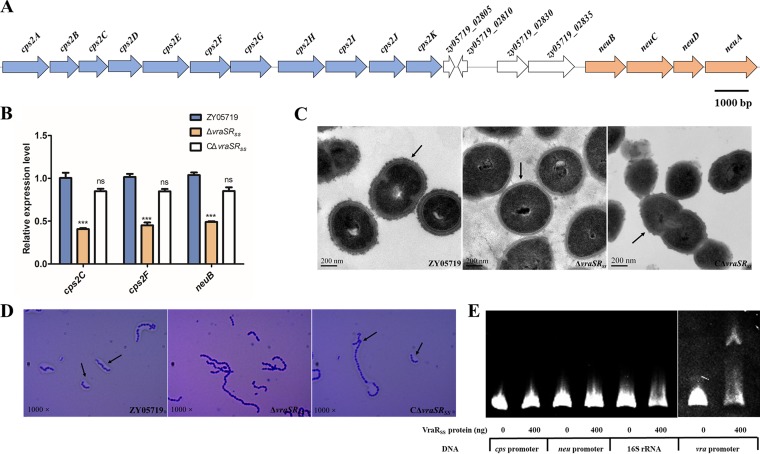
CPS has been proven to be a critical virulence factor in SS2, and the N-acetylneuraminic acid (sialic acid) linked to the end of the CPS chain also played a role in SS2 virulence (3, 4, 41–43). Our RNA-Seq data revealed that both the capsule synthesis cluster (cps2A-K) and the sialic acid synthase cluster (neuBCDA) were significantly downregulated in the ΔvraSRSS compared to the WT strain (Fig. 4B and 5A). qPCR results confirmed similar decreases in the gene expression levels of cps2C, cps2F, and neuB in the ΔvraSRSS strain, while complementation completely restored transcription-level deficiencies caused by the vraSRSS deletion (Fig. 5B). To confirm that CPS biosynthesis was attenuated in ΔvraSRSS, transmission electron microscopy (TEM) was performed to visualize the colony morphology. As shown in Fig. 5C, inactivation of VraSRSS caused a significant decrease of capsule thickness compared to that in the WT strain, which generally matched the transcriptomics and qPCR assay results. This effect on capsule formation was confirmed at the population level by crystal violet staining (Fig. 5D). As expected, complementation nearly restored the capsule thickness to that of the WT strain. We also conducted EMSAs to examine whether VraRSS directly regulated expression of the cps and neu cluster. The promoter of the vra operon and the fragments amplified from the 16S rRNA gene served as controls. As shown in Fig. 5E, neither the promoter region of the cps cluster nor that of the neu cluster was shifted by the recombinant VraRSS protein, and only the upstream region of the yvqFSS operon (positive control) was clearly bound. These results suggested that VraSRSS controlled capsular biosynthesis via indirect regulation to contribute to the full virulence of SS2. Future work will need to be performed to identify the downstream factor(s) of VraSRSS and its regulatory mechanism for the cps and neu clusters.
FIG 5.
VraSRSS indirectly regulated expression of the CPS biosynthesis gene cluster in SS2. (A) Schematic diagram of the genetic organization of the CPS synthesis and the sialic acid synthase clusters. The arrows indicate the direction of transcription. (B) Expression levels of cps2C, cps2F, and neuB in ZY05719, ΔvraSRSS, and CΔvraSRSS strains were measured by qRT-PCR. The unpaired two-tailed Student's t test was used for statistical analysis (ns, P > 0.05; ***, P < 0.001). (C) Transmission electron micrographs of the ZY05719, ΔvraSRSS, and CΔvraSRSS strains. The scale bars indicate the magnification size. (D) Photomicrographs of the capsules of the ZY05719, ΔvraSRSS, and CΔvraSRSS strains stained with crystal violet (1,000×). (E) VraRSS indirectly regulated the CPS biosynthesis gene cluster. Either the cps or neu promoter region could be shifted by the recombinant VraRSS protein in EMSAs. DNA probes containing the yvqFSS operon promoter region were used as a positive control, and fragments amplified from 16S rRNA served as a negative control.
Downregulation of CPS biosynthesis caused by vraSRSS deletion is responsible for the increased adhesion to bEnd.3 cells and attenuated antiphagocytosis against RAW 264.7 cells.
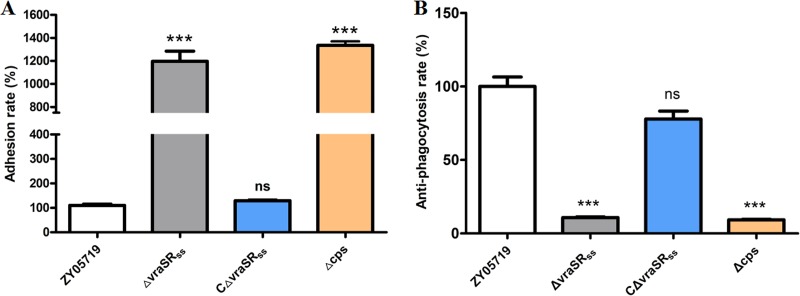
Adhesion to the host cell surface and antiphagocytosis against macrophages are considered essential steps in the pathogenesis of bacterial infections (44). Previous studies have confirmed that loss of capsule in SS2 could impair antiphagocytosis against macrophages and increase adhesion to epithelial cells (45, 46). Here, the WT, a ΔvraSRSS complementary strain, and a capsular deficient mutant strain (Δcps) were tested to assess whether VraSRSS inactivation would cause similar effects. As shown in Fig. 6, both the vraSRSS and cps mutant strains displayed significantly increased adhesion to bEnd.3 cells (P < 0.001) but significantly attenuated antiphagocytosis against RAW264.7 cells versus the WT strain. Notably, the vraSRSS-complemented strain showed complete restoration of the above-described changes to levels similar to that of the WT strain. These results further confirmed that VraSRSS could control capsular biosynthesis to be involved in antiphagocytosis and host cell adhesion, thereby playing key roles in the full virulence of SS2.
FIG 6.
VraSRSS is involved in SS2 adhesion to bEnd.3 cells and resisting phagocytosis by RAW 264.7 cells. (A) Effect of the vraSRSS mutation on the ability of SS2 to adhere to bEnd.3 cells. The adhesion rate of the ZY05719 strain was significantly lower than that of the ΔvraSRSS and Δcps strains. The vraSRSS-complemented strain showed adhesion restored to the level of the WT strain. (B) Effect of the vraSRSS deletion on the ability of SS2 to resist phagocytosis by RAW 264.7 cells. The antiphagocytosis rate of the ZY05719 strain was significantly higher than that of the ΔvraSRSS and Δcps strains. The vraSRSS-complemented strain showed an ability restored to the level of the WT strain. The data are shown as the means and standard deviations of the results from three independent experiments performed in triplicate. Two-tailed unpaired Student's t tests were used for statistical analysis (***, P < 0.001).
DISCUSSION
TCSSs are comprised of a membrane-bound HK protein and a cytoplasmic RR protein, which represent one of the major mechanisms used by bacterial cells to adapt to harsh environmental stimuli, such as pH, temperature, and antimicrobial drugs (7, 47). VraSR, a TCSS identified in S. aureus, was initially characterized as the master regulator of vancomycin resistance that functions by interacting with the membrane-anchored protein YvqF, and it was subsequently demonstrated to respond to various cell wall-targeting antimicrobials, including vancomycin, β-lactams, and certain cationic peptides (19, 20, 48). VraSRSS is a homolog carried by S. suis that shares ∼50% sequence identity with the VraSR of S. aureus. The upstream gene yvqFSS also encodes a membrane-anchored protein but shares very low homology with the YvqF protein of S. aureus.
Our results showed that VraSRSS inactivation caused a direct and significant downregulation of the yvqFSS gene and confirmed that either yvqFSS or vraSRSS deletion significantly increased the susceptibility to diverse antimicrobial agents. Notably, YvqFSS and VraSRSS showed a broader spectrum than their homologs in S. aureus in terms of multidrug resistance, which are classified based on differing chemical structures and compositions, including aminoglycosides (amikacin, gentamicin, kanamycin, and streptomycin), β-lactams (ampicillin, penicillin, and ceftriaxone), and peptides (vancomycin, bacitracin, colistin, polymyxin B, and nisin). As reported previously, VraSR in S. aureus responds to cell wall-targeting antibiotics but not aminoglycoside antimicrobials (20). The β-lactam- and peptide-based antimicrobials target the cell wall to kill bacteria, while the aminoglycosides employ a different antibacterial mechanism by inhibiting protein synthesis (49). Although a previous study demonstrated that S. suis resistance to β-lactams occurs due to the alteration of penicillin-binding proteins (32), data generated for VraSRSS and the accessory YvqFSS in this study reveal a novel mechanism of this antibiotic resistance. Perhaps the membrane-anchored protein YvqFSS was involved in the cell wall biosynthesis pathway in S. suis, and the decreased production of YvqFSS might damage the integrity of both cell membrane and cell wall, thus enhancing the permeability of bacterial cells to aminoglycosides and increasing susceptibility to this class of antimicrobials. Unlike the plasmid-located multidrug resistance gene cfr, the chromosomal yvqFSS gene is not easily transferred during interbacterial communication and is directly regulated by the TCSS VraSRSS to more precisely respond to antibiotics. Recently, TCSSs have become increasingly recognized as potential targets of adjuvant antibiotic therapies, because their regulation of virulence and responses to cell envelope stress could affect the proliferation of bacterial pathogens in the host (50). For example, walkmycin B and waldiomycin showed antibiotic activity against Bacillus subtilis and S. aureus by inhibiting HK protein WalK autophosphorylation (51, 52). Therefore, an inhibitor of VraSRSS could reduce the mortality associated with S. suis infection.
The acquisition of vancomycin resistance in S. aureus is often accompanied by reduced virulence. A recent study found that VraSR directly regulated the function of the Agr quorum-sensing system and, thus, inhibited virulence in S. aureus (53). However, our data showed that vraSRSS expression was significantly higher in vivo than in vitro, which prompted us to consider its potential roles in pathogenesis. Indeed, the deletion of the vraSRSS gene significantly attenuated the virulence of SS2 in both the mouse and zebrafish models, suggesting that VraSRSS is an important TCSS in mediating responses to host stimulation and for the expression of many virulence factors in vivo. RNA-Seq analyses identified 106 genes in the ΔvraSRSS strain with significantly different transcription levels compared with those in the WT strain, which helped elucidate the underlying regulatory mechanism of VraSRSS in SS2 strain ZY05719. Of these 106 genes, more than 15 downregulated genes were from the same clusters and related to capsular biosynthesis. It is well known that CPS are key virulence factors in SS2 and play critical roles in antiphagocytic processes against host macrophage phagocytosis and killing (3, 4). Further study showed that vraSRSS deletion caused the results consistent with CPS deficiency in increasing bacterial adhesion to epithelial cells and attenuating antiphagocytosis against macrophages, which partially explained the attenuated bacterial pathogenicity in animal models.
To further examine the process whereby VraSRSS regulates CPS biosynthesis, we visualized the bacterial morphology by TEM and capsule staining. The results confirmed that VraSRSS inactivation significantly decreased the capsule thickness in SS2 compared to that in the WT strain, which was consistent with the transcriptional data and the attenuated virulence observed in animal challenge assays. In addition, our EMSA showed that VraRSS did not bind the cps and nue promoter regions from the SS2 capsule biosynthesis cluster. This finding, coupled with the qRT-PCR results displaying a significant downregulation of clustered genes related to CPS expression in ΔvraSRSS compared to the WT and complementary strains, suggested that TCSS VraSRSS indirectly regulates CPS biosynthesis.
Environmental signals can also modulate the expression of virulence factors, and S. suis growth in Todd-Hewitt broth (THB) exhibits a weaker invasion ability than the growth in culture medium with fewer nutrients (46). Not only transcriptional regulation of the cps gene cluster but also the availability of glucose or other carbohydrates affects CPS biosynthesis (54). Our RNA-Seq results highlighted several sugar transporter genes (zy05719_10085 to zy05719_10100) that were significantly downregulated in the ΔvraSRSS strain, suggesting that VraSRSS controlled CPS biosynthesis by regulating carbohydrate acquisition in SS2 cells. In addition, given that a previous report showed that the global virulence regulator CovR could repress CPS biosynthesis (13), we compared the transcriptome data between ΔvraSRSS and ΔcovR strains to clarify the potential correlation between both TCSSs. Unlike VraSRSS, which upregulated transcription of the whole CPS biosynthesis cluster (cps2A-K and neuBCDA), CovR downregulated the transcription of only two genes (cps2C and neuA) in the cluster (13), suggesting that these TCSSs have regulated CPS biosynthesis through different mechanisms. Notably, VraSRSS are activated during systemic infection (Fig. 3), which may help relieve the repression of CovR on cps2C and neuA for CPS biosynthesis, thus increasing the bacterial fitness in host bloodstream for full virulence.
In summary, a novel TCSS (VraSRSS) associated with multidrug resistance and virulence was identified in SS2. We demonstrated that VraSRSS directly regulated the expression of the membrane-anchored protein encoded by yvqFSS to mediate multidrug resistance and contributed to the full virulence of SS2 by indirectly regulating CPS biosynthesis. However, the external signal that activates HK and the underlying mechanism of VraSRSS in regulating CPS biosynthesis remain unknown and require further exploration in our future work.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and plasmids.
The SS2 strain ZY05719 was originally isolated from a diseased pig during an outbreak in the Sichuan Province of China. The strain was grown at 37°C in Todd-Hewitt broth (THB; Becton-Dickinson, USA) or Todd-Hewitt broth agar (THA) and harvested at the mid-exponential growth phase (optical density at 600 nm [OD600] of 0.6). Escherichia coli strains DH5α and BL21(DE3) were grown on Luria-Bertani (LB) agar plates or in LB broth at 37°C. When antibiotics (Sigma-Aldrich) were required, the following concentrations were added to the medium: for S. suis, spectinomycin (Spc) at 100 μg/ml; for E. coli, Spc at 50 μg/ml and kanamycin (Kan) at 50 μg/ml. All strains and plasmids used in this study are listed in Table S3 in the supplemental material.
Recombinant DNA techniques.
The pSET4S vector was used to construct the target gene deletion mutants as previously described (55). The upstream and downstream regions of target genes were amplified from genomic DNA of the ZY05719 strain and fused together by overlap extension PCR. After digestion with respective endonucleases, the fusion fragments were inserted into pSET4S vector with the same endonucleases sites. ZY05719 competent cells were prepared and electrotransformed with the recombinant vector, after which the resultant strains were selected at 28°C in Spc and then grown at 37°C without Spc. The deletion of target genes was verified by PCR and sequencing. To construct the complemented strains, the target genes (including their putative promoter sequences) were amplified from the ZY05719 strain and independently inserted into the pSET2 plasmid. The recombinant pSET2 plasmids then were electroporated into mutant competent cells.
To express the recombinant VraRSS protein, the coding region of vraRSS was amplified from genomic DNA and inserted into the pET28a vector. After confirming the DNA sequence, the recombinant pET28a-vraRSS vector was transformed into BL21(DE3) cells. The sequences of all primers used in this study are shown in Table S4.
Bioinformatics analysis.
DNAStar Lasergene 7 software and the BLAST program (available from the NCBI website, http://blast.ncbi.nlm.nih.gov/) were used to analyze the DNA sequences. The three-dimensional structures of VraSSS and VraRSS were predicted using the SWISS-MODEL online server (https://www.swissmodel.expasy.org/) (56–59). The BProm program (SoftBerry) was used to predict the promoter region.
Antimicrobial susceptibility testing.
The MIC of an antimicrobial substance was defined as the lowest concentration resulting in no detectable bacterial growth. The MICs of antimicrobials (Sigma-Aldrich) were determined by performing the standardized microdilution assay in a 96-well microtiter plate containing 100 μl THB medium in each well. The strains were grown to mid-exponential growth phase, diluted to an OD600 of 0.05, and used to inoculate wells containing THB with standard 2-fold (vol/vol) dilutions of the antimicrobial of interest. Plates were incubated at 37°C for 24 h. Each assay was repeated independently three times.
EMSAs.
EMSAs were performed to analyze the binding of VraRSS to the DNA probe. The recombinant VraRSS protein was expressed in BL21 cells from the pET28a-vraRSS vector and purified using nickel-nitrilotriacetic acid spin columns (GE Healthcare). The DNA probes were obtained by PCR amplification and purified using a gel extraction kit (TaKaRa). The negative-control probe was amplified from 16S rRNA. Increasing amounts of native purified VraRSS protein (0 to 500 ng) were added to the DNA probe (80 ng) in binding buffer (10 mM Tris base, 50 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 0.05% Nonidet P-40, 2.5% glycerol, pH 7.5). The reaction mixtures were incubated at 37°C for 30 min and then loaded onto a 6% polyacrylamide gel in 0.5× TBE buffer (44.5 mM Tris base, 44.5 mM boric acid, 1 mM EDTA; pH 7.5) at 200 V for 45 min. After electrophoresis, the gel was stained in 0.5× TBE buffer with ethidium bromide for 20 min.
Animal tests.
To investigate the survival curves of mice, 10 specific-pathogen-free (SPF) BALB/c mice (female, 6 weeks old) were challenged by intraperitoneal injection with the strains of interest at a dose of 5 × 108 CFU/mouse. Another 10 SPF BALB/c mice were injected with an equal volume of sterile phosphate-buffered saline (PBS) as a negative control. Clinical signs and survival rates were monitored for 7 days after challenge.
In addition, to calculate the LD50 values, 15 zebrafish per group were challenged by intraperitoneal injection with 20 μl of 10-fold-diluted bacteria, ranging from 5 × 104 to 5 × 107 total CFU/fish. Another group was injected with an equal volume of sterile PBS as a negative control. Mortality was monitored for 7 days postchallenge. This experiment was repeated three times. LD50 values were calculated using the method of Reed and Muench (60).
All animal experiments were conducted in the Laboratory Animal Center of the Nanjing Agricultural University with the approval of the Laboratory Animal Monitoring Committee of Jiangsu Province.
RNA isolation, transcriptome sequencing, RT-PCR, and qRT-PCR.
RNAs from the ZY05719 and ΔvraSRSS strains were extracted using TRIzol (TaKaRa) according to the manufacturer's instructions. After removing contaminating DNA, the extracted RNAs were sent to Novogene (Tianjin, China) for RNA-Seq analysis. Each strain was analyzed using three biological replicates. The sequencing library was constructed and sequenced using the Illumina HiSeq 2000 platform, as previously described (61). To control the false discovery rate for the transcriptome data, comparisons with estimated fold changes of at least 3 (log2 fold changes of at least 1.6) and q values of ≤0.05 were declared significant.
RT-PCR and qRT-PCR were performed to validate the RNA transcription levels determined by RNA-Seq. Total RNAs were isolated from strains cultured in THB and from the infected blood of mice. After extraction as described above, total RNAs were reverse transcribed by following the recommended protocol of the HiScript II first-strand cDNA synthesis kit (Vazyme). The QuantStudio 6 Flex real-time PCR system (Thermo Fisher Scientific) and SYBR Premix Ex Taq (TaKaRa) were used per the manufacturers' instructions. The sequences of primers used for qRT-PCR are shown in Table S4, and the housekeeping gene (parC) was used as a control (62). Each sample procedure was repeated three times. Relative fold changes in expression were calculated using the 2−ΔΔCT method (63).
For the cotranscription test, total RNAs were extracted and reverse transcribed as described above. Primers were designed to span the open reading frames (ORFs) yvqFSS and vraSSS, as well as vraSSS and vraRSS. RNA was purified and reverse transcribed to prepare cDNA. One microgram of total RNA samples (without reverse transcription) served as a control to confirm that the samples were free of contaminating DNA. The RNA and matching cDNA samples then were used as PCR templates.
TEM and capsule staining.
The colony morphology was analyzed by TEM. Briefly, bacteria growing in mid-exponential phase were harvested by centrifugation at 5,000 × g for 10 min and fixed in 2.5% glutaraldehyde for more than 2 h. The samples were dehydrated in propylene oxide for 10 min, embedded in epoxy resin, and examined using a Hitachi H-7650 system (Hitachi) according to the manufacturer's instructions. This experiment was performed independently three times.
To visualize the bacterial capsules at the population level, capsule staining was performed as previously described (64). A drop of bacterial culture was placed on a microscope slide and spread in a thin film using another slide. After gentle flame fixation, the smear was stained with 0.1% crystal violet and gently heated. Subsequently, the dye was washed with 20% copper sulfate. The slide was air dried and examined immediately under oil immersion.
Cell experiments.
Mouse brain microvascular endothelial cells (bEnd.3) were used to perform adhesion assays, and murine macrophage-like RAW264.7 cells were used to study antiphagocytosis of SS2. These cells were cultured in Dulbecco's modified Eagle medium (DMEM), supplemented with 10% fetal bovine serum (FBS), and maintained at 37°C with 5% CO2. The strains were grown to mid-exponential growth phase (OD600 of 0.6) and washed twice in PBS.
Adhesion assays were performed as previously reported (65). The bEnd.3 cells were cultured in 24-well cell plates and washed three times with PBS. The bacteria were suspended in DMEM without antibiotics to a density of 1 × 107 CFU/ml. After infecting the cells in each well with 1 ml of the bacterial suspension, the plates were centrifuged at 800 × g for 15 min and incubated at 37°C for 90 min. Subsequently, the infected cells were washed five times and trypsinized at 37°C for 10 min. Serial dilutions of the cell lysate were plated onto THA, and the plates were incubated overnight at 37°C. Each assay was repeated independently three times.
For the antiphagocytosis assay, the strains were incubated with RAW264.7 macrophages at a bacterium-to-cell ratio of 100:1. After coculturing for 1.5 h, the infected cells were washed three times with PBS and incubated in DMEM containing gentamicin (100 μg/ml) and penicillin (5 μg/ml). After coculturing for another 1.5 h, the macrophages were washed three times and lysed with water. Serial dilutions of the cell lysate were plated onto THA. Each assay was repeated independently three times.
Data analysis.
All experiments were performed at least three times, and the data were analyzed using an unpaired two-tailed Student's t test, or the log-rank (Mantel-Cox) test, with the GraphPad software package. For all tests, a P value of <0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Special Fund for Public Welfare Industry of Chinese Ministry of Agriculture (no. 201303041), Shanghai Agriculture Applied Technology Development Program (no. G2016060201), and the project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
We have no competing financial interests to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00096-18.
REFERENCES
- 1.Fittipaldi N, Segura M, Grenier D, Gottschalk M. 2012. Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol 7:259–279. doi: 10.2217/fmb.11.149. [DOI] [PubMed] [Google Scholar]
- 2.Palmieri C, Varaldo PE, Facinelli B. 2011. Streptococcus suis, an emerging drug-resistant animal and human pathogen. Front Microbiol 2:235. doi: 10.3389/fmicb.2011.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith HE, Damman M, van der Velde J, Wagenaar F, Wisselink HJ, Stockhofe-Zurwieden N, Smits MA. 1999. Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect Immun 67:1750–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Segura M, Gottschalk M, Olivier M. 2004. Encapsulated Streptococcus suis inhibits activation of signaling pathways involved in phagocytosis. Infect Immun 72:5322–5330. doi: 10.1128/IAI.72.9.5322-5330.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng Y, Zhang H, Ma Y, Gao GF. 2010. Uncovering newly emerging variants of Streptococcus suis, an important zoonotic agent. Trends Microbiol 18:124–131. doi: 10.1016/j.tim.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 6.West AH, Stock AM. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci 26:369–376. doi: 10.1016/S0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen CT, Park SS, Rhee DK. 2015. Stress responses in Streptococcus species and their effects on the host. J Microbiol 53:741–749. doi: 10.1007/s12275-015-5432-6. [DOI] [PubMed] [Google Scholar]
- 8.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 9.Marina A, Waldburger CD, Hendrickson WA. 2005. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J 24:4247–4259. doi: 10.1038/sj.emboj.7600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C, Tang J, Dong W, Wang C, Feng Y, Wang J, Zheng F, Pan X, Liu D, Li M, Song Y, Zhu X, Sun H, Feng T, Guo Z, Ju A, Ge J, Dong Y, Sun W, Jiang Y, Wang J, Yan J, Yang H, Wang X, Gao GF, Yang R, Wang J, Yu J. 2007. A glimpse of streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PLoS One 2:e315. doi: 10.1371/journal.pone.0000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Greeff A, Buys H, van Alphen L, Smith HE. 2002. Response regulator important in pathogenesis of Streptococcus suis serotype 2. Microb Pathog 33:185–192. doi: 10.1006/mpat.2002.0526. [DOI] [PubMed] [Google Scholar]
- 12.Li M, Wang C, Feng Y, Pan X, Cheng G, Wang J, Ge J, Zheng F, Cao M, Dong Y, Liu D, Wang J, Lin Y, Du H, Gao GF, Wang X, Hu F, Tang J. 2008. SalK/SalR, a two-component signal transduction system, is essential for full virulence of highly invasive Streptococcus suis serotype 2. PLoS One 3:e2080. doi: 10.1371/journal.pone.0002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan X, Ge J, Li M, Wu B, Wang C, Wang J, Feng Y, Yin Z, Zheng F, Cheng G, Sun W, Ji H, Hu D, Shi P, Feng X, Hao X, Dong R, Hu F, Tang J. 2009. The orphan response regulator CovR: a globally negative modulator of virulence in Streptococcus suis serotype 2. J Bacteriol 191:2601–2612. doi: 10.1128/JB.01309-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Tan C, Zhou Y, Fu S, Hu L, Hu J, Chen H, Bei W. 2011. The two-component regulatory system CiaRH contributes to the virulence of Streptococcus suis 2. Vet Microbiol 148:99–104. doi: 10.1016/j.vetmic.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Han H, Liu C, Wang Q, Xuan C, Zheng B, Tang J, Yan J, Zhang J, Li M, Cheng H, Lu G, Gao GF. 2012. The two-component system Ihk/Irr contributes to the virulence of Streptococcus suis serotype 2 strain 05ZYH33 through alteration of the bacterial cell metabolism. Microbiology 158:1852–1866. doi: 10.1099/mic.0.057448-0. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Shen X, Zhao Y, Wang M, Zhong Q, Chen T, Hu F, Li M. 2012. Identification and proteome analysis of the two-component VirR/VirS system in epidemic Streptococcus suis serotype 2. FEMS Microbiol Lett 333:160–168. doi: 10.1111/j.1574-6968.2012.02611.x. [DOI] [PubMed] [Google Scholar]
- 17.Xu J, Fu S, Liu M, Xu Q, Bei W, Chen H, Tan C. 2014. The two-component system NisK/NisR contributes to the virulence of Streptococcus suis serotype 2. Microbiol Res 169:541–546. doi: 10.1016/j.micres.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Yuan F, Tan C, Liu Z, Yang K, Zhou D, Liu W, Duan Z, Guo R, Chen H, Tian Y, Bei W. 2017. The 1910HK/RR two-component system is essential for the virulence of Streptococcus suis serotype 2. Microb Pathog 104:137–145. doi: 10.1016/j.micpath.2016.12.026. [DOI] [PubMed] [Google Scholar]
- 19.Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol 49:807–821. doi: 10.1046/j.1365-2958.2003.03599.x. [DOI] [PubMed] [Google Scholar]
- 20.Boyle-Vavra S, Yin S, Jo DS, Montgomery CP, Daum RS. 2013. VraT/YvqF is required for methicillin resistance and activation of the VraSR regulon in Staphylococcus aureus. Antimicrob Agents Chemother 57:83–95. doi: 10.1128/AAC.01651-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wisselink HJ, Veldman KT, Van den Eede C, Salmon SA, Mevius DJ. 2006. Quantitative susceptibility of Streptococcus suis strains isolated from diseased pigs in seven European countries to antimicrobial agents licensed in veterinary medicine. Vet Microbiol 113:73–82. doi: 10.1016/j.vetmic.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 22.Zhang C, Ning Y, Zhang Z, Song L, Qiu H, Gao H. 2008. In vitro antimicrobial susceptibility of Streptococcus suis strains isolated from clinically healthy sows in China. Vet Microbiol 131:386–392. doi: 10.1016/j.vetmic.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 23.Chu YW, Cheung TK, Chu MY, Tsang VY, Fung JT, Kam KM, Lo JY. 2009. Resistance to tetracycline, erythromycin and clindamycin in Streptococcus suis serotype 2 in Hong Kong. Int J Antimicrob Agents 34:181–182. doi: 10.1016/j.ijantimicag.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 24.Princivalli MS, Palmieri C, Magi G, Vignaroli C, Manzin A, Camporese A, Barocci S, Magistrali C, Facinelli B. 2009. Genetic diversity of Streptococcus suis clinical isolates from pigs and humans in Italy (2003-2007). Euro Surveill 14:19310. doi: 10.2807/ese.14.33.19310-en. [DOI] [PubMed] [Google Scholar]
- 25.Hoa NT, Chieu TT, Nghia HD, Mai NT, Anh PH, Wolbers M, Baker S, Campbell JI, Chau NV, Hien TT, Farrar J, Schultsz C. 2011. The antimicrobial resistance patterns and associated determinants in Streptococcus suis isolated from humans in southern Vietnam, 1997-2008. BMC Infect Dis 11:6. doi: 10.1186/1471-2334-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wasteson Y, Hoie S, Roberts MC. 1994. Characterization of antibiotic resistance in Streptococcus suis. Vet Microbiol 41:41–49. doi: 10.1016/0378-1135(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 27.Martel A, Baele M, Devriese LA, Goossens H, Wisselink HJ, Decostere A, Haesebrouck F. 2001. Prevalence and mechanism of resistance against macrolides and lincosamides in Streptococcus suis isolates. Vet Microbiol 83:287–297. doi: 10.1016/S0378-1135(01)00426-6. [DOI] [PubMed] [Google Scholar]
- 28.Escudero JA, San Millan A, Catalan A, de la Campa AG, Rivero E, Lopez G, Dominguez L, Moreno MA, Gonzalez-Zorn B. 2007. First characterization of fluoroquinolone resistance in Streptococcus suis. Antimicrob Agents Chemother 51:777–782. doi: 10.1128/AAC.00972-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holden MT, Hauser H, Sanders M, Ngo TH, Cherevach I, Cronin A, Goodhead I, Mungall K, Quail MA, Price C, Rabbinowitsch E, Sharp S, Croucher NJ, Chieu TB, Mai NT, Diep TS, Chinh NT, Kehoe M, Leigh JA, Ward PN, Dowson CG, Whatmore AM, Chanter N, Iversen P, Gottschalk M, Slater JD, Smith HE, Spratt BG, Xu J, Ye C, Bentley S, Barrell BG, Schultsz C, Maskell DJ, Parkhill J. 2009. Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS One 4:e6072. doi: 10.1371/journal.pone.0006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chander Y, Oliveira SR, Goyal SM. 2011. Identification of the tet(B) resistance gene in Streptococcus suis. Vet J 189:359–360. doi: 10.1016/j.tvjl.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 31.Palmieri C, Princivalli MS, Brenciani A, Varaldo PE, Facinelli B. 2011. Different genetic elements carrying the tet(W) gene in two human clinical isolates of Streptococcus suis. Antimicrob Agents Chemother 55:631–636. doi: 10.1128/AAC.00965-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu D, Zhang F, Zhang H, Hao L, Gong X, Geng M, Cao M, Zheng F, Zhu J, Pan X, Tang J, Feng Y, Wang C. 2014. The beta-galactosidase (BgaC) of the zoonotic pathogen Streptococcus suis is a surface protein without the involvement of bacterial virulence. Sci Rep 4:4140. doi: 10.1038/srep04140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang J, Ma J, Shang K, Hu X, Liang Y, Li D, Wu Z, Dai L, Chen L, Wang L. 2016. Evolution and diversity of the antimicrobial resistance associated mobilome in Streptococcus suis: a probable mobile genetic elements reservoir for other streptococci. Front Cell Infect Microbiol 6:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Li D, Song L, Liu Y, He T, Liu H, Wu C, Schwarz S, Shen J. 2013. First report of the multiresistance gene cfr in Streptococcus suis. Antimicrob Agents Chemother 57:4061–4063. doi: 10.1128/AAC.00713-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams AE, Blakemore WF, Alexander TJ. 1988. A murine model of Streptococcus suis type 2 meningitis in the pig. Res Vet Sci 45:394–399. [PubMed] [Google Scholar]
- 36.Vecht U, Stockhofe-Zurwieden N, Tetenburg BJ, Wisselink HJ, Smith HE. 1997. Virulence of Streptococcus suis type 2 for mice and pigs appeared host-specific. Vet Microbiol 58:53–60. doi: 10.1016/S0378-1135(97)00131-4. [DOI] [PubMed] [Google Scholar]
- 37.Dominguez-Punaro MC, Segura M, Plante MM, Lacouture S, Rivest S, Gottschalk M. 2007. Streptococcus suis serotype 2, an important swine and human pathogen, induces strong systemic and cerebral inflammatory responses in a mouse model of infection. J Immunol 179:1842–1854. doi: 10.4049/jimmunol.179.3.1842. [DOI] [PubMed] [Google Scholar]
- 38.Seitz M, Beineke A, Seele J, Fulde M, Valentin-Weigand P, Baums CG. 2012. A novel intranasal mouse model for mucosal colonization by Streptococcus suis serotype 2. J Med Microbiol 61:1311–1318. doi: 10.1099/jmm.0.043885-0. [DOI] [PubMed] [Google Scholar]
- 39.Wu Z, Zhang W, Lu Y, Lu C. 2010. Transcriptome profiling of zebrafish infected with Streptococcus suis. Microb Pathog 48:178–187. doi: 10.1016/j.micpath.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 40.Neely MN, Pfeifer JD, Caparon M. 2002. Streptococcus-zebrafish model of bacterial pathogenesis. Infect Immun 70:3904–3914. doi: 10.1128/IAI.70.7.3904-3914.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elliott SD, Tai JY. 1978. The type-specific polysaccharides of Streptococcus suis. J Exp Med 148:1699–1704. doi: 10.1084/jem.148.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson TL, Jeffers J, Rapp-Gabrielson VJ, Martin S, Klein LK, Lowery DE, Fuller TE. 2007. A novel signature-tagged mutagenesis system for Streptococcus suis serotype 2. Vet Microbiol 122:135–145. doi: 10.1016/j.vetmic.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 43.Van Calsteren MR, Gagnon F, Lacouture S, Fittipaldi N, Gottschalk M. 2010. Structure determination of Streptococcus suis serotype 2 capsular polysaccharide. Biochem Cell Biol 88:513–525. doi: 10.1139/O09-170. [DOI] [PubMed] [Google Scholar]
- 44.Tamura GS, Kuypers JM, Smith S, Raff H, Rubens CE. 1994. Adherence of group B streptococci to cultured epithelial cells: roles of environmental factors and bacterial surface components. Infect Immun 62:2450–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lalonde M, Segura M, Lacouture S, Gottschalk M. 2000. Interactions between Streptococcus suis serotype 2 and different epithelial cell lines. Microbiology 146(Part 8):1913–1921. doi: 10.1099/00221287-146-8-1913. [DOI] [PubMed] [Google Scholar]
- 46.Benga L, Goethe R, Rohde M, Valentin-Weigand P. 2004. Non-encapsulated strains reveal novel insights in invasion and survival of Streptococcus suis in epithelial cells. Cell Microbiol 6:867–881. doi: 10.1111/j.1462-5822.2004.00409.x. [DOI] [PubMed] [Google Scholar]
- 47.Stephenson K, Hoch JA. 2002. Two-component and phosphorelay signal-transduction systems as therapeutic targets. Curr Opin Pharmacol 2:507–512. doi: 10.1016/S1471-4892(02)00194-7. [DOI] [PubMed] [Google Scholar]
- 48.McCallum N, Meier PS, Heusser R, Berger-Bachi B. 2011. Mutational analyses of open reading frames within the vraSR operon and their roles in the cell wall stress response of Staphylococcus aureus. Antimicrob Agents Chemother 55:1391–1402. doi: 10.1128/AAC.01213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vakulenko SB, Mobashery S. 2003. Versatility of aminoglycosides and prospects for their future. Clin Microbiol Rev 16:430–450. doi: 10.1128/CMR.16.3.430-450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tiwari S, Jamal SB, Hassan SS, Carvalho P, Almeida S, Barh D, Ghosh P, Silva A, Castro TLP, Azevedo V. 2017. Two-component signal transduction systems of pathogenic bacteria as targets for antimicrobial therapy: an overview. Front Microbiol 8:1878. doi: 10.3389/fmicb.2017.01878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okada A, Igarashi M, Okajima T, Kinoshita N, Umekita M, Sawa R, Inoue K, Watanabe T, Doi A, Martin A, Quinn J, Nishimura Y, Utsumi R. 2010. Walkmycin B targets WalK (YycG), a histidine kinase essential for bacterial cell growth. J Antibiot (Tokyo) 63:89-94. doi: 10.1038/ja.2009.128. [DOI] [PubMed] [Google Scholar]
- 52.Eguchi Y, Okajima T, Tochio N, Inukai Y, Shimizu R, Ueda S, Shinya S, Kigawa T, Fukamizo T, Igarashi M, Utsumi R. 2017. Angucycline antibiotic waldiomycin recognizes common structural motif conserved in bacterial histidine kinases. J Antibiot (Tokyo) 70:251–258. doi: 10.1038/ja.2016.151. [DOI] [PubMed] [Google Scholar]
- 53.Dai Y, Chang W, Zhao C, Peng J, Xu L, Lu H, Zhou S, Ma X. 2017. VraR binding to the promoter region of agr inhibits its function in vancomycin-intermediate Staphylococcus aureus (VISA) and heterogeneous VISA. Antimicrob Agents Chemother 61:e02740-16. doi: 10.1128/AAC.02740-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willenborg J, Fulde M, de Greeff A, Rohde M, Smith HE, Valentin-Weigand P, Goethe R. 2011. Role of glucose and CcpA in capsule expression and virulence of Streptococcus suis. Microbiology 157:1823–1833. doi: 10.1099/mic.0.046417-0. [DOI] [PubMed] [Google Scholar]
- 55.Takamatsu D, Osaki M, Sekizaki T. 2001. Thermosensitive suicide vectors for gene replacement in Streptococcus suis. Plasmid 46:140–148. doi: 10.1006/plas.2001.1532. [DOI] [PubMed] [Google Scholar]
- 56.Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 57.Guex N, Peitsch MC, Schwede T. 2009. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30(Suppl 1):S162–S173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 58.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. 2009. The SWISS-MODEL repository and associated resources. Nucleic Acids Res 37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. [Google Scholar]
- 61.Wilhelm BT, Landry JR. 2009. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods 48:249–257. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 62.Wu Z, Wu C, Shao J, Zhu Z, Wang W, Zhang W, Tang M, Pei N, Fan H, Li J, Yao H, Gu H, Xu X, Lu C. 2014. The Streptococcus suis transcriptional landscape reveals adaptation mechanisms in pig blood and cerebrospinal fluid. RNA 20:882–898. doi: 10.1261/rna.041822.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 64.Hiss PH. 1905. A contribution to the physiological differentiation of pneumococcus and streptococcus, and to methods of staining capsules. J Exp Med 6:317–345. doi: 10.1084/jem.6.4-6.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vanier G, Segura M, Friedl P, Lacouture S, Gottschalk M. 2004. Invasion of porcine brain microvascular endothelial cells by Streptococcus suis serotype 2. Infect Immun 72:1441–1449. doi: 10.1128/IAI.72.3.1441-1449.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.