ABSTRACT
The morbidity and mortality resulting from acute gastroenteritis and associated chronic sequelae represent a substantial burden on health care systems worldwide. Few studies have investigated changes in the gut microbiome following an episode of acute gastroenteritis. By using nondirected 16S rRNA gene amplicon sequencing, the fecal microbiota of 475 patients with acute gastroenteritis was examined. Patient age was correlated with the overall microbial composition, with a decrease in the abundance of Faecalibacterium being observed in older patients. We observed the emergence of a potential Escherichia-Shigella-dominated enterotype in a subset of patients, and this enterotype was predicted to be more proinflammatory than the other common enterotypes, with the latter being dominated by Bacteroides or Faecalibacterium. The increased abundance of Escherichia-Shigella did not appear to be associated with infection with an agent of a similar sequence similarity. Stool color and consistency were associated with the diversity and composition of the microbiome, with deviations from the norm (not brown and solid) showing increases in the abundances of bacteria such as Escherichia-Shigella and Veillonella. Analysis of enriched outliers within the data identified a range of genera previously associated with gastrointestinal diseases, including Treponema, Proteus, Capnocytophaga, Arcobacter, Campylobacter, Haemophilus, Aeromonas, and Pseudomonas. Our data represent the first in-depth analysis of gut microbiota in acute gastroenteritis. Phenotypic changes in stool color and consistency were associated with specific changes in the microbiota. Enriched bacterial taxa were detected in cases where no causative agent was identified by using routine diagnostic tests, suggesting that in the future, microbiome analyses may be utilized to improve diagnostics.
KEYWORDS: microbiota, acute gastroenteritis, enterotype, Escherichia coli, pathogenesis, gut
INTRODUCTION
Acute infectious gastroenteritis results when an enteric pathogen, pathobiont, or toxin breaches the intestinal epithelial layer, leading to inflammation and a disruption of normal functions. The most common route of infection is the ingestion of contaminated food or water. Although often preventable and treatable, gastroenteritis is one of leading causes of death in children under 5 years of age (1). The majority of gastroenteritis-associated deaths can be attributed to severe dehydration and/or sepsis (1).
While in many cases, the clinical symptoms associated with gastroenteritis are acute or transient, the disease can be associated with a range of chronic sequelae, including reactive arthritis, irritable bowel syndrome, and Guillain-Barré syndrome (2). In particular, a lasting impact of gastroenteritis on intestinal physiology is that it appears to benefit atypical microbial species, which can lead to a disruption of the gut microbiome (3). Such changes in the microbiome have been shown to persist long after the clearance of infection and have been associated with an increased risk of developing chronic sequelae (4).
While current standard laboratory diagnostics are capable of identifying the causative agent, for a substantial number of patients, a specific pathogen is not identified. To date, a limited number of studies have characterized the changes in the gut microbiome following an episode of gastroenteritis. Screening of the gut microbiome of gastroenteritis patients not only has the potential to identify putative infectious agents in samples where no agent is identified but also can reveal shifts in the composition of the microbiome related to infection with specific gastrointestinal pathogens. The fecal microbiomes of 475 gastroenteritis patients were investigated, and the results were validated in a replicate analysis of 23 patients and with data from four cohorts of healthy individuals (n = 688 samples from 590 individuals).
RESULTS
Faecalibacterium abundance contributes to differences in the gut microbiome across age.
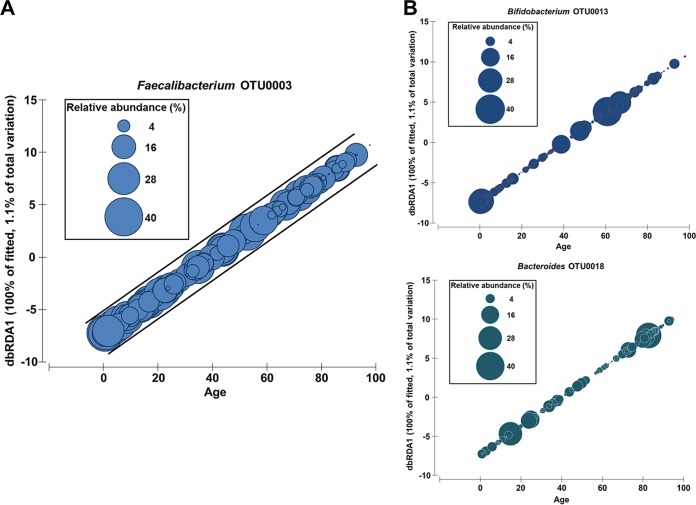
Patient age but not gender was found to be significantly correlated with the composition of the gut microbiota in patients (pseudo-F = 3.8; P = 0.001; df = 354). A subsequent analysis of the relationship of patient age with individual microbial taxa identified a significant decrease in the relative abundance of Faecalibacterium (pseudo-F = 19.5; P = 0.001; df = 354) with age (Fig. 1A). Other taxa found to be significantly associated with age were Bifidobacterium operational taxonomic unit 0013 (OTU0013) (pseudo-F = 7.9; P = 0.006; df = 354) and Bacteroides OTU0018 (pseudo-F = 4.2; P = 0.031; df = 354); however, unlike Faecalibacterium, their relationships appeared to be bimodal (Fig. 1B).
FIG 1.
Relationship between the gut microbiota and patient age. (A) Faecalibacterium OTU0003 was significantly correlated with age, showing a decrease in the relative abundance with increasing age. Graphs are distance-based redundancy analysis (dbRDA) plots of coordinate scores against age. The sizes of the circles represent the relative abundances (4 to 40%) of the OTUs of interest within each patient. DistLM analysis between the Euclidean distance matrix of patient ages and relative abundances of microbial taxa was performed. (B) Bifidobacterium OTU0013 and Bacteroides OTU0018 were also found to be significantly correlated with age. The sizes of the circles represent the relative abundances (percent) of the OTUs of interest within each patient.
Potential emergence of an enterotype in acute gastroenteritis defined by Escherichia-Shigella abundance.
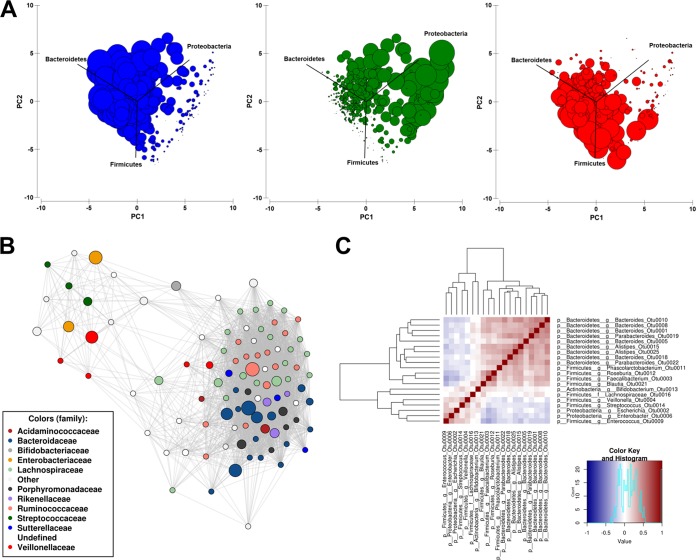
Patient microbiomes clustered into three putative enterotypes, dominated by Bacteroidetes (Bacteroides), Proteobacteria (Escherichia-Shigella), or Firmicutes (Faecalibacterium) (Fig. 2A; see also Fig. S1A in the supplemental material). A network analysis taking into account an ensemble of Pearson and Spearman correlations across microbial taxa showed clustering of common members of the gut microbiota belonging to the Bacteroidetes and Firmicutes (Fig. 2B). Each of the networks arising from Pearson and Spearman correlations showed similar relationships (Fig. S1B). In line with the presence of a third enterotype, Enterobacteriaceae, Streptococcaceae, and Veillonellaceae formed a distinct cluster within the network (Fig. 2B and Fig. S1B). The exclusion relationship between the Bacteroidetes and Firmicutes and the Enterobacteriaceae was confirmed through a heat map of the correlations (Fig. 2C). The absence of the Escherichia-Shigella enterotype was confirmed in four cohorts of healthy individuals from Australia, France, Chile, and Japan (n = 688 samples from 590 individuals) (Fig. S2).
FIG 2.
Composition of the gut microbiota in cases of acute gastroenteritis. (A) Principal-component analysis of square-root-transformed relative abundances showing samples clustering into three possible enterotypes, dominated by Bacteroidetes (Bacteroides OTU0001) (blue), Proteobacteria (Escherichia-Shigella OTU0002) (green), or Firmicutes (Faecalibacterium OTU0003) (red). The sizes of the circles represent the relative abundances (percent) of OTUs within each patient. (B) Network analysis (ensemble between Spearman and Pearson correlations) showing Bacteroidaceae, Rikenellaceae, Lachnospiraceae, and Ruminococcaceae forming a distinct cluster that can be differentiated from a cluster of Enterobacteriaceae, Veillonellaceae, and Streptococcaceae. (C) Heat map of correlations across the top 20 OTUs confirms the relationships observed in the network analysis.
Dirichlet multinomial mixtures were employed to confirm the classification of enterotypes in our data set using an unbiased unsupervised clustering method (Fig. S3). The analysis partitioned the patient data into two groups dominated by Bacteroides or Escherichia-Shigella. Partition 1 had an average relative abundance of Bacteroides OTU0001 of 21.4%, an average relative abundance of Escherichia-Shigella OTU0002 of 28.8%, and an average relative abundance of Faecalibacterium OTU0003 of 44.0%, while partition 2 contained Bacteroides OTU0001 at an average relative abundance of 44.5% (2-fold higher), Escherichia-Shigella OTU0002 at an average relative abundance of 7.3% (3.9-fold lower), and Faecalibacterium OTU0003 at an average relative abundance of 43.6% (0.98-fold). A total of 88% of samples classified into the Escherichia-Shigella-dominated enterotype were classified into partition 1.
Stool color and consistency appear to be phenotypic markers of the Escherichia-Shigella enterotype.
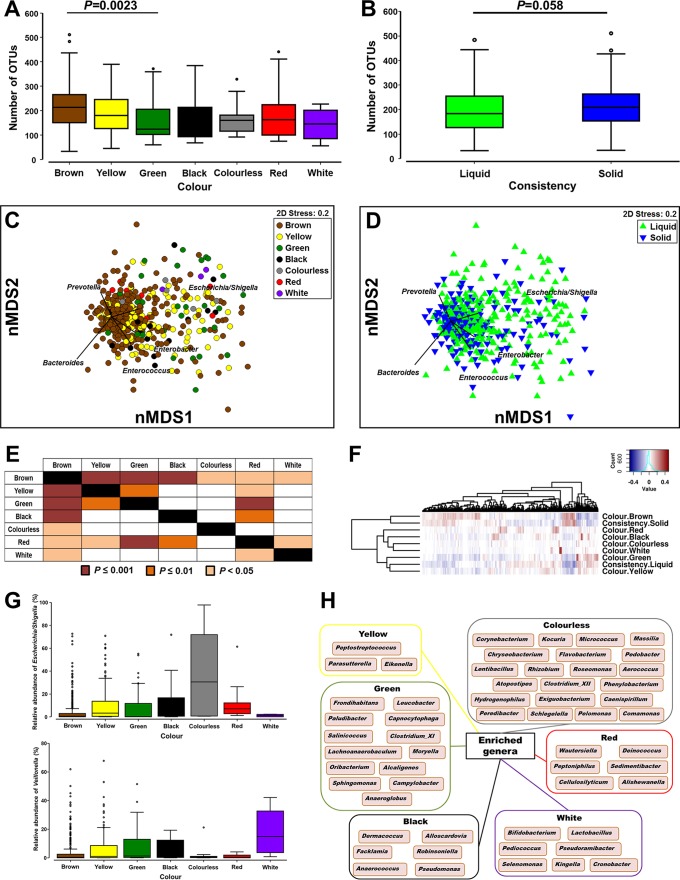
The color and consistency of stool samples were significantly associated with α-diversity measures. There was a nonsignificant drop in species richness (number of OTUs) when stools of other colors were compared to brown stools, except for green stools, which were significantly different from brown stools (Fig. 3A). A range of differences was also identified across evenness and Shannon's diversity values (see Fig. S4A and S4B in the supplemental material). Furthermore, a nonsignificant drop in the number of OTUs was identified in liquid stool compared to solid stool (Fig. 3B). No differences in evenness or Shannon's diversity values were identified across consistency (Fig. S4C and S4D).
FIG 3.
Microbial diversity across colors and consistencies of fecal samples. (A and B) Changes in species richness (number of OTUs) across stool color (A) and consistency (B). (C and D) Nonmetric multidimensional scaling plots on Bray-Curtis resemblances of square-root-transformed relative abundances across stool color (C) and consistency (D). (E) Results of PERMANOVA following Bray-Curtis resemblance analysis of square-root-transformed relative abundances across colors of fecal samples. (F) Analysis of hierarchal clustering across different variables within sample colors and consistencies. (G) Box plot of relative abundances (percent) of Escherichia-Shigella and Veillonella across colors of samples. (H) Microbial genera found to be enriched by LEfSe analysis and unique for stool of a specific color following comparison with brown fecal samples.
The global microbial composition was also impacted by the color and consistency of stool samples. Nonmetric multidimensional scaling (nMDS) plots showed that stool of colors other than brown shifted the microbial composition toward an increased relative abundance of Escherichia-Shigella (Fig. 3C). A similar pattern was observed for liquid stool compared to solid stool (Fig. 3D). Differences in microbial compositions were confirmed by using permutational multivariate analysis of variance (PERMANOVA). Brown stools were found to be significantly different from stools of all other colors with respect to bacterial content (Fig. 3E). Furthermore, red stools were also found to be significantly different from other stools of colors except for colorless stools (Fig. 3E). Compared to solid stool samples, liquid stool samples were found to be not only significantly different (t = 3.85 and P = 0.0001 for phylum; t = 2.22 and P = 0.0001 for OTU) but also more dispersed in content (t = 3.99 and P = 0.0001 for phylum; t = 2.01 and P = 0.063 for OTU). Hierarchical clustering analysis across different variables within sample color and consistency showed that the microbiomes in brown and solid stools clustered together and away from those of liquid stools and stools of other colors (Fig. 3F). These findings were supported by hierarchal clustering analyses at the individual OTU and sample levels (Fig. S4E and S4F).
Krona plots of the relative abundances of microbial taxa confirmed the expansion of Proteobacteria in stool samples of all colors compared to brown stool as well as in liquid stool compared to solid stool (Fig. S5). This expansion, most strikingly seen in colorless stool (Fig. 3G), was due mostly to an increase in the abundance of Escherichia-Shigella and was accompanied by a drop in the abundances of Bacteroides and Faecalibacterium (Fig. S6). Of interest, Veillonella, previously identified to be correlated with Escherichia-Shigella (Fig. 2B), was consistently enriched in white stool samples (Fig. 3G). Linear discriminant-analysis effect size (LEfSe) analysis was also performed to identify additional microbial taxa that were significantly different between stools of each color and brown stool, and genera uniquely enriched in stools of each color relative to brown stool are shown in Fig. 3H.
Predicted enrichment of virulence-associated functions in the Escherichia-Shigella enterotype.
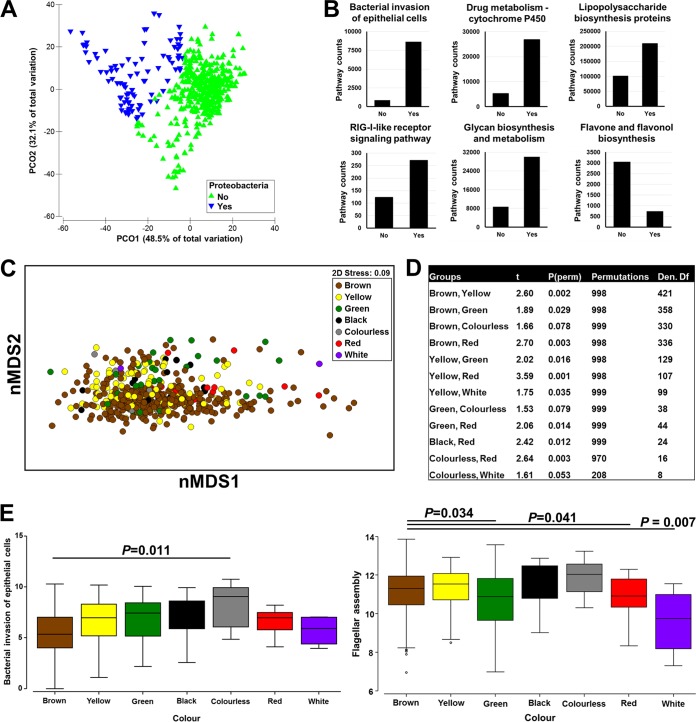
Metabolic differences arising from changes in the microbiome were predicted by using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt), and the nearest-sequenced-taxon index (NSTI) was calculated for these predictions. The average NSTI score for all samples was 0.054 ± 0.001.
Samples were separated into two groups according to their levels of Proteobacteria (Fig. 4A). A relative abundance of 25%, which corresponded to the mean plus 0.5 standard deviations, was used as a cutoff. As expected, this showed that the two groups differed significantly at the phylum (t = 15.2; P = 0.001; number of permutations = 998; df = 491) and OTU (t = 5.8; P = 0.001; number of permutations = 999) levels as well as in predicted KEGG pathways (t = 5.3; P = 0.001; number of permutations = 998). Microbial taxa and pathways that were found to be significantly different between the two groups by LEfSe analysis are shown in Tables S1 and S2 in the supplemental material. Six pathways showed large fold changes in abundances between the two groups, including bacterial invasion of epithelial cells, drug metabolism-cytochrome P450, lipopolysaccharide biosynthesis proteins, the RIG-I-like receptor signaling pathway, and glycan biosynthesis and metabolism, which were increased in the Proteobacteria-enriched group, as well as flavone and flavonol biosynthesis, which was decreased (Fig. 4B). In support of the impact of Escherichia-Shigella, shifts in the predicted metabolic contributions were identified between brown stool and stools of other colors (Fig. 4C to E).
FIG 4.
Predicted bacterial metabolic contributions. (A) Principal-coordinate analysis of the Bray-Curtis resemblance matrix of square-root-transformed relative abundances. Samples were separated into two groups according to their levels of Proteobacteria, with a relative abundance of 25% being used as a cutoff. (B) KEGG pathways (level 3) significantly different between the two groups (Yes, high abundance of Proteobacteria; No, low abundance of Proteobacteria). Only pathways that had relatively high counts and a high fold change were plotted. (C) nMDS plot of log-transformed pathway (KEGG level 3) counts across colors of fecal samples. (D) PERMANOVA following Bray-Curtis resemblance of log-transformed pathway (KEGG level 3) counts. Only comparisons that were significant or close to significant are shown. (E) Box plot of PICRUSt counts across colors of samples. The two pathways “bacterial invasion of epithelial cells” and “flagellar assembly” are shown. P values were generated by analysis of variance with Tukey's multiple-comparison test.
Analysis of the possible origins of the Escherichia-Shigella enterotype.
To understand the possible origins of the Escherichia-Shigella enterotype, the frequency and identity of known pathogens in samples belonging to this enterotype (n = 87 samples) were examined. No infectious agent was identified in 77/87 samples by using routine diagnostics. Three of the 10 remaining samples were infected with Gammaproteobacteria with high sequence similarity to Escherichia-Shigella: Shigella sonnei (n = 2) and Salmonella (n = 1). The other seven samples had a diverse array of infectious agents, including Clostridium difficile (n = 2), rotavirus group A (n = 1), Giardia (n = 1), astrovirus (n = 1), adenovirus serotype 40/41 (n = 1), and Campylobacter (n = 1), suggesting that the dominance of Escherichia-Shigella in this enterotype is not necessarily the causative agent itself but a consequence of infection.
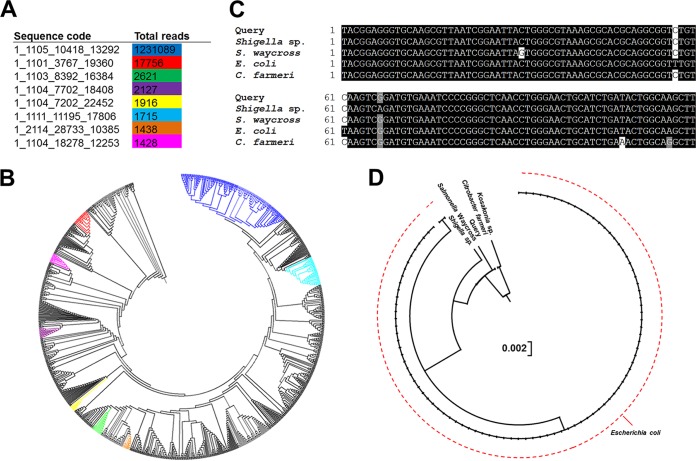
Individual sequences corresponding to Escherichia-Shigella were then examined further (classified at 100% similarity) to differentiate between those potentially associated with disease and those arising from the resident microbiota. Of the eight unique sequences that had a reasonable presence in the cohort (i.e., total read count of >1,000) (Fig. 5A), the second most abundant sequence clustered farthest from the most common sequence (Fig. 5B). Upon further analysis, this sequence was found to cluster closer to Salmonella within the Enterobacteriaceae (Fig. 5C and D) and was present at a relatively high abundance in 3/6 of the confirmed cases of Salmonella infection in this cohort.
FIG 5.
Analysis of sequences belonging to Escherichia-Shigella. (A) Total numbers of reads of the top eight most abundant unique sequences classified as Escherichia-Shigella following classification at 100% similarity and not 97% similarity. Only sequences with >1,000 reads are included. Sequence codes correspond to the sequence names in the raw data, given that these are unique sequences. (B) Phylogenetic analysis (Clear-cut) of all unique sequences classified as Escherichia-Shigella and branch locations of the top eight sequences within the tree. The color coding in the tree corresponds to the colors in the table in panel A. (C) Sequence alignment of the second most abundant Escherichia-Shigella sequence (1_1101_3767_19360) (red) with the sequences of other microbial taxa within the Gammaproteobacteria. (D) Phylogenetic analysis of the second most abundant Escherichia-Shigella sequence (1_1101_3767_19360) relative to other microbial taxa within the Gammaproteobacteria. Salmonella Waycross, Salmonella enterica serovar Waycross.
Specific infectious agents induce distinct shifts in the gut microbiota.
Microbial compositions differed significantly at the OTU level (t = 1.8; P = 0.001; number of permutations = 998; df = 70) between confirmed bacterial (n = 45) and viral (n = 27) infections. Specifically, significant differences in microbial compositions were observed between cases of C. difficile (n = 29) and norovirus genotype II (n = 11) infections (t = 2.0; P = 0.001; number of permutations = 999; df = 38) and between cases of C. difficile and rotavirus group A (n = 11) infections (t = 1.6; P = 0.002; number of permutations = 998; df = 38) but not between norovirus genotype II and rotavirus group A (t = 1.0; P = 0.28; number of permutations = 999; df = 20). No other comparisons were performed, as the numbers of confirmed cases of infections by other pathogens were <10 and were not considered powered enough. The identification of bacterial taxa that were differentially abundant between cases of viral and bacterial infections showed that Bacteroides OTUs were enriched in cases of viral infection and that Fusobacterium and Enterobacter OTUs were enriched in cases of bacterial infection (see Table S3 in the supplemental material).
Identification of outlier bacterial taxa within the microbiome of patients with acute gastroenteritis.
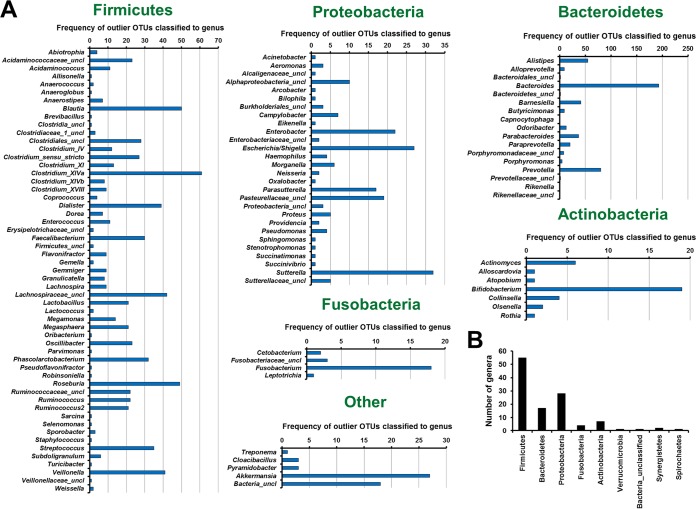
A strict analysis was developed to identify outlier bacterial taxa in samples where no known agent was identified by using routine diagnostics. OTU abundances within individual samples were analyzed, and those that satisfied three criteria, (i) a relative abundance of >3%, (ii) a relative abundance higher than two standard deviations above the mean for all samples, and (iii) a relative abundance higher than the upper fence of a box plot, were considered outliers. A range of OTUs classified within genera known to be associated with disease, including Treponema, Proteus, Capnocytophaga, Arcobacter, Campylobacter, Haemophilus, Aeromonas, and Pseudomonas, were identified as outliers (Fig. 6A). Relatively unknown genera, including Cloacibacillus, Alloscardovia, Brevibacillus, Providencia, and Pyramidobacter, were also identified (Fig. 6A). The majority of the genera were classified as Firmicutes, followed by Proteobacteria (Fig. 6B). However, OTUs of an unclassified taxonomy (<60% similarity) at the phylum level were also identified to be outliers in a number of patients (Fig. 6A and B).
FIG 6.
Analysis to identify outlier bacterial taxa within individual patients. Abundances of OTUs within individual samples were analyzed, and those that satisfied three criteria, (i) a relative abundance of >3%, (ii) a relative abundance higher than two standard deviations above the mean for all samples, and (iii) a relative abundance higher than the upper fence of a box plot, were considered outliers. Only the first 900 OTUs were included in this analysis. (A) Frequency of OTUs identified as outliers according to classification at the genus level. uncl, unclassified. (B) Total number of genera that contain outlier OTUs per phylum.
OTUs within Sutterella (OTU0026, OTU0064, and OTU0485) were identified as outliers 32 times across the patient samples (Fig. 6A). Furthermore, OTUs within Clostridium cluster XI were identified to be outliers in a range of samples (Fig. 6A), consistent with the diagnostic results from this cohort of patients showing that C. difficile alone was responsible for 29 cases. Of interest, OTU0244, classified as Parvimonas, a genus related to Clostridium XI, was identified as an outlier in one sample identified as C. difficile. A decrease in the threshold from a 3% to a 0.5% relative abundance identified Parvimonas OTU0244 as an outlier in an additional seven patients.
DISCUSSION
The burden of acute gastroenteritis can be divided into two types, an acute burden that is experienced more frequently in low-income countries and a chronic burden that appears to affect a subset of gastroenteritis patients worldwide. It is well accepted that infection and subsequent gastroenteritis lead to shifts in the gut microbiome (4). However, few studies have examined the nature of these changes and how they may relate to the development of chronic sequelae.
The concept of enterotypes within the gut microbiome was first introduced by Arumugam et al., who showed the presence of at least three enterotypes dominated by Bacteroides, Faecalibacterium (classified as Ruminococcus at the time), or Prevotella (5). Here, two similar enterotypes dominated by Bacteroides or Faecalibacterium were found in this cohort. Taxa within the Bacteroidetes and Firmicutes showed strong correlations in network analyses, reflecting their coevolution within the human gastrointestinal tract. The presence of these two common enterotypes suggested that a large proportion of patients may not have experienced large-scale shifts in their microbial composition. In contrast, for a subset of patients, we observed the emergence of a potential Escherichia-Shigella-dominated enterotype, and this was absent in four distinct cohorts of healthy individuals. We also observed a relationship among Escherichia-Shigella, Veillonella, and Streptococcus in the network analysis, which is in line with reports of the coaggregation of these bacterial groups in previous studies (6). Predictive metagenomics comparing the Escherichia-Shigella enterotype with other patients showed a striking enrichment of proinflammatory pathways, such as bacterial invasion of epithelial cells, lipopolysaccharide biosynthesis proteins, and the RIG-I-like receptor signaling pathway, as well as a decrease in beneficial bacterial functions, such as flavone and flavonol biosynthesis. The RIG-I signaling pathway is not known to be present in prokaryotes; thus, it is not currently clear which genomes contribute to the this pathway's counts.
Stool color and consistency appeared to be strongly associated with the composition of the gut microbiota, and deviations from the norm (brown and solid) were accompanied by increases in the abundance of Escherichia-Shigella. Notably, consistent increases in the abundance of Veillonella were found in white stool. White stool results from a lack of bile, and Veillonella species are known to hydrolyze conjugated bile salts (7). The relationship between Veillonella species and the absence of bile in stool is a novel finding and should be investigated further.
The possible origins of the Escherichia-Shigella enterotype were investigated. This investigation showed that the increased abundance of this bacterial taxon within this enterotype was unlikely to be due to the causative agent itself, given that isolates from confirmed cases of viral infection and non-Gammaproteobacteria bacterial infection were also classified within this enterotype. This is supported by a recent study that reported a significant increase in the richness of Enterobacteriaceae in more-complicated cases of acute viral gastroenteritis compared to that in mild-to-moderate cases (8). We also observed that a more stringent analysis of the 16S rRNA gene sequences allowed the further differentiation of Escherichia-Shigella. In particular, one sequence with higher similarity to Salmonella clustered away from the more-abundant Escherichia-Shigella sequence. The presence of the Salmonella-classified sequence in 3/6 confirmed Salmonella cases further supports this conclusion. These results suggest that more-stringent analyses of the 16S rRNA gene sequences can be used to subdivide this taxon and that longer 16S rRNA or 23S rRNA gene sequences may hold more promise for future studies.
Differences between the microbial compositions of patients with confirmed bacterial infections and those of patients with confirmed viral infections were found in this cohort. We confirmed differences between C. difficile-colonized individuals and those infected with either rotavirus group A or norovirus genotype II. The enrichment of a range of Bacteroides OTUs in patients with viral infections suggested that their bacterial profiles may not have been altered by infection as strongly as in those with confirmed bacterial infection. This may relate to the increased presence of mucus and/or blood in the stool of bacterial gastroenteritis patients compared to that in viral gastroenteritis patients.
We designed a strict analysis to identify bacterial taxa that behaved as outliers in individual patients. The analysis identified OTUs belonging to several genera, including Treponema, Proteus, Capnocytophaga, Campylobacter, Haemophilus, Aeromonas, and Pseudomonas, some of which were previously associated with gastroenteritis (2, 9). Of interest, through this analysis, we identified one case of an enrichment of Arcobacter, a genus related to Campylobacter and previously associated with gastroenteritis (10), and at least one case related to Parvimonas, a bacterium associated with colorectal cancer and an array of extragastrointestinal diseases (11–14). A number of OTUs classified into other relatively unknown genera, including Cloacibacillus, Alloscardovia, Brevibacillus, Providencia, and Pyramidobacter, were designated outliers in some patient samples. Notably, sequences that were classified as being of an unknown bacterial origin were also detected as enriched outliers in some samples. However, it is worth noting that the identification of outliers in the data is correlative in nature, and future mechanistic studies would need to differentiate between bacterial blooms due to disease and a role in etiology.
The outlier analysis also identified a large number of occurrences of Sutterella OTUs in this cohort. Interestingly, dogs with acute diarrhea have increased levels of Sutterella within their microbiome compared to healthy dogs (15). More recently, increased abundances of Sutterella in the microbiomes of ulcerative colitis patients undergoing fecal microbiota transplantation with an increased likelihood of therapy failure were reported (16). In contrast, other studies have reported no difference in the prevalences (but not abundances) of Sutterella between inflammatory bowel disease patients and controls, suggesting that this bacterium may be a common commensal (17, 18). The importance of the increased levels of Sutterella in the gastrointestinal tracts of a subset of patients remains to be elucidated.
A strong association between the age of patients and their overall microbiota composition was observed in our patient cohort. The effect of subject age on the gut microbiome was reported previously (19); however, we found that this association was driven partly by a decrease in the relative abundance of Faecalibacterium in our older patients. Given that this bacterial taxon has been shown to possess a range of anti-inflammatory properties (20), its possible role in age-associated inflammation should be examined.
This study is not without its limitations. We observed lower levels of confirmed causative agents in our samples than in previous studies, which limited our ability to identify microbiota changes associated with specific infectious agents. Stool colors were determined through subjective methods, and a more quantitative method, such as imaging, would improve these classifications. This study examined the contribution of bacteria to disease but not the contribution of viruses, which play a significant role in etiology. We did not have information regarding the severity of the gastroenteritis episode, which would likely have a confounding effect. Furthermore, longitudinal analysis of the microbiome of these patients would establish the importance of our findings.
Our data represent the first in-depth analysis of the gut microbiota in patients with acute gastroenteritis and across different stool patterns. We identified the expansion of Escherichia-Shigella as a potential dysbiotic signature of interest in a subset of patients. Furthermore, the finding that phenotypic changes in stool color and consistency appear to be associated with specific changes in the microbiota is novel. For the first time, microbiota analysis was leveraged to identify enriched bacterial taxa in cases where no causative agent was identified by using routine diagnostic tests. These data highlight the importance of examining the microbiome to study the acute and chronic burdens of acute gastroenteritis.
MATERIALS AND METHODS
Subjects.
Fecal samples were collected from 475 adult and pediatric patients (aged 1 month to 98 years) presenting with clinical gastroenteritis (loose stools for >24 h with or without concomitant signs of fever, weight loss, and upper gastrointestinal symptoms) at the Prince of Wales Hospital (Australia). This cohort represents a subset of patients reported previously (21). Routine diagnostic testing was undertaken at the NSW Health Pathology Randwick Microbiology Laboratory to detect common gastrointestinal pathogens (21). Briefly, testing to detect Salmonella spp., Shigella spp., Campylobacter jejuni-Campylobacter coli, toxigenic Clostridium difficile, and Yersinia enterocolitica was performed by using bacterial multiplex PCR (22). Samples positive for C. jejuni-C. coli by PCR were cultivated on agar to identify the Campylobacter species present. Samples where isolation was unsuccessful were reported as being C. jejuni-C. coli positive. Detection of rotavirus group A, norovirus genotype II, adenovirus serotype 40/41, and astrovirus was performed by using viral multiplex PCR. Enzyme immunoassays were performed for the detection of Cryptosporidium and Giardia lamblia, and any positive results were confirmed by microscopy (22). Fecal samples were also classified according to their color (brown, yellow, green, red, black, white, and colorless) and consistency (solid and liquid) by two independent researchers prior to DNA extraction. Any classification discrepancies between researchers were labeled unknown.
Ethics approval and consent to participate.
This study was approved by the UNSW Biomedical Human Research Ethics Advisory Panel (UNSW reference number HC14235), and experiments were performed in accordance with relevant guidelines and regulations.
DNA extraction and 16S rRNA amplicon sequencing.
DNA was extracted from 300 mg feces by using a phenol-chloroform-isoamyl alcohol DNA extraction method (21). The 16S rRNA gene was amplified by using Kapa HiFi HotStart ReadyMix (95°C for 3 min and 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, followed by a final step at 72°C for 5 min) and earth microbiome primers (515F-806R). Indices and Illumina sequencing adapters were attached by using the Nextera XT index kit according to the manufacturer's instructions. Amplicon sequencing was performed with Illumina MiSeq 2x250-bp chemistry at the Ramaciotti Centre for Genomics. A total of 498 samples were sequenced, representing 475 patient samples and 23 random replicates from the same cohort.
Sequencing and statistical analyses.
Raw reads were analyzed by using the MiSeq operating procedures within Mothur v1.39.1 (RRID: SCR_011947) (23), which resulted in 36,427 ± 1,091 total clean reads/sample. α-Diversity was examined by using both subsampled reads (n = 2,000) and nonsubsampled read counts, and reported patterns were consistent across both data sets. Multivariate analyses, including nMDS, PERMANOVA, and PERMDISP (homogeneity of dispersions), were performed on a Bray-Curtis resemblance matrix of square-root-transformed relative abundances by using Primer-E v7. The distribution of square-root-transformed relative abundances is shown in Fig. S7A in the supplemental material.
Distance-based linear modeling (DistLM) analyses between age and the microbiota were performed on either on a Bray-Curtis resemblance matrix of square-root-transformed relative abundances or Euclidean distances across patient ages. Hierarchal clustering analysis was performed by using Calypso (24). LEfSe and PICRUSt analyses were also performed (25, 26). Phylogenetic analyses were performed in Mothur by using the Clear-cut command (23). Classification of samples into enterotypes using Dirichlet multinomial mixture models was performed in Mothur by using the get.communitytype command. A replicate analysis of the 23 patients confirmed the reproducibility of our microbiome data (Fig. S7B).
Data availability.
Sequences were submitted to the European Nucleotide Archive under accession number PRJEB23690. Data from healthy adult individuals were obtained from the European Nucleotide Archive or were reported previously (16, 27–29).
Supplementary Material
ACKNOWLEDGMENTS
N.O.K. is supported by a Cancer Institute NSW career development fellowship (15/CDF/1-11). N.C.-R. is supported by a National Health and Medical Research Council, Australia, early career fellowship (APP1111461) and a Cancer Australia priority-driven collaborative cancer research grant (1129488). H.M.M. acknowledges funding from the Ministry of Higher Education, Malaysia (UM.C/625/HIR/MOHE/CHAN/13/1).
We declare that we have no competing interests.
W.D.R., J.M., and A.P.U. were involved in study subject recruitment and sample collection. H.M.M., S.M.R., N.O.K., and W.D.R. contributed reagents and funds. N.C.-R. and A.P.U. performed the experimental work. N.O.K. performed the microbiota and statistical analyses. N.O.K. drafted the manuscript. N.O.K., N.C.-R., H.M.M., W.D.R., A.P.U., S.M.R., and J.M. read, corrected, and approved the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00060-18.
REFERENCES
- 1.World Health Organization. 2017. Diarrhoeal disease. World Health Organization, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs330/en/ Accessed 28 October 2017. [Google Scholar]
- 2.Kaakoush NO, Castaño-Rodríguez N, Mitchell HM, Man SM. 2015. Global epidemiology of Campylobacter infection. Clin Microbiol Rev 28:687–720. doi: 10.1128/CMR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Kamdar K, Khakpour S, Chen J, Leone V, Brulc J, Mangatu T, Antonopoulos DA, Chang EB, Kahn SA, Kirschner BS, Young G, DePaolo RW. 2016. Genetic and metabolic signals during acute enteric bacterial infection alter the microbiota and drive progression to chronic inflammatory disease. Cell Host Microbe 19:21–31. doi: 10.1016/j.chom.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, MetaHIT Consortium, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, et al. 2011. Enterotypes of the human gut microbiome. Nature 473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheithauer BK, Wos-Oxley ML, Ferslev B, Jablonowski H, Pieper DH. 2009. Characterization of the complex bacterial communities colonizing biliary stents reveals a host-dependent diversity. ISME J 3:797–807. doi: 10.1038/ismej.2009.36. [DOI] [PubMed] [Google Scholar]
- 7.Wei X, Yan X, Zou D, Yang Z, Wang X, Liu W, Wang S, Li X, Han J, Huang L, Yuan J. 2013. Abnormal fecal microbiota community and functions in patients with hepatitis B liver cirrhosis as revealed by a metagenomic approach. BMC Gastroenterol 13:175. doi: 10.1186/1471-230X-13-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen SY, Tsai CN, Lee YS, Lin CY, Huang KY, Chao HC, Lai MW, Chiu CH. 2017. Intestinal microbiome in children with severe and complicated acute viral gastroenteritis. Sci Rep 7:46130. doi: 10.1038/srep46130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qamar FN, Nisar MI, Quadri F, Shakoor S, Sow SO, Nasrin D, Blackwelder WC, Wu Y, Farag T, Panchalingham S, Sur D, Qureshi S, Faruque AS, Saha D, Alonso PL, Breiman RF, Bassat Q, Tamboura B, Ramamurthy T, Kanungo S, Ahmed S, Hossain A, Das SK, Antonio M, Hossain MJ, Mandomando I, Tennant SM, Kotloff KL, Levine MM, Zaidi AK. 2016. Aeromonas-associated diarrhea in children under 5 years: the GEMS experience. Am J Trop Med Hyg 95:774–780. doi: 10.4269/ajtmh.16-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kayman T, Abay S, Hizlisoy H, Atabay HI, Diker KS, Aydin F. 2012. Emerging pathogen Arcobacter spp. in acute gastroenteritis: molecular identification, antibiotic susceptibilities and genotyping of the isolated arcobacters. J Med Microbiol 61:1439–1444. doi: 10.1099/jmm.0.044594-0. [DOI] [PubMed] [Google Scholar]
- 11.Baghban A, Gupta S. 2016. Parvimonas micra: a rare cause of native joint septic arthritis. Anaerobe 39:26–27. doi: 10.1016/j.anaerobe.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Flemer B, Warren RD, Barrett MP, Cisek K, Das A, Jeffery IB, Hurley E, O'Riordain M, Shanahan F, O'Toole PW. 7 October 2017. The oral microbiota in colorectal cancer is distinctive and predictive. Gut doi: 10.1136/gutjnl-2017-314814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shtaya A, Schuster H, Riley P, Harris K, Hettige S. 2017. Oesophageal pleural fistula presenting with Parvimonas micra infection causing cervical and brain abscesses. Anaerobe 47:233–237. doi: 10.1016/j.anaerobe.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Uemura H, Hayakawa K, Shimada K, Tojo M, Nagamatsu M, Miyoshi-Akiyama T, Tamura S, Mesaki K, Yamamoto K, Yanagawa Y, Sugihara J, Kutsuna S, Takeshita N, Shoda N, Hagiwara A, Kirikae T, Ohmagari N. 2014. Parvimonas micra as a causative organism of spondylodiscitis: a report of two cases and a literature review. Int J Infect Dis 23:53–55. doi: 10.1016/j.ijid.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Suchodolski JS, Markel ME, Garcia-Mazcorro JF, Unterer S, Heilmann RM, Dowd SE, Kachroo P, Ivanov I, Minamoto Y, Dillman EM, Steiner JM, Cook AK, Toresson L. 2012. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS One 7:e51907. doi: 10.1371/journal.pone.0051907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paramsothy S, Kamm MA, Kaakoush NO, Walsh AJ, van den Bogaerde J, Samuel D, Leong RWL, Connor S, Ng W, Paramsothy R, Xuan W, Lin E, Mitchell HM, Borody TJ. 2017. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet 389:1218–1228. doi: 10.1016/S0140-6736(17)30182-4. [DOI] [PubMed] [Google Scholar]
- 17.Hansen R, Berry SH, Mukhopadhya I, Thomson JM, Saunders KA, Nicholl CE, Bisset WM, Loganathan S, Mahdi G, Kastner-Cole D, Barclay AR, Bishop J, Flynn DM, McGrogan P, Russell RK, El-Omar EM, Hold GL. 2013. The microaerophilic microbiota of de-novo paediatric inflammatory bowel disease: the BISCUIT study. PLoS One 8:e58825. doi: 10.1371/journal.pone.0058825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukhopadhya I, Hansen R, Nicholl CE, Alhaidan YA, Thomson JM, Berry SH, Pattinson C, Stead DA, Russell RK, El-Omar EM, Hold GL. 2011. A comprehensive evaluation of colonic mucosal isolates of Sutterella wadsworthensis from inflammatory bowel disease. PLoS One 6:e27076. doi: 10.1371/journal.pone.0027076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buford TW. 2017. (Dis)Trust your gut: the gut microbiome in age-related inflammation, health, and disease. Microbiome 5:80. doi: 10.1186/s40168-017-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottière HM, Doré J, Marteau P, Seksik P, Langella P. 2008. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Underwood AP, Kaakoush NO, Sodhi N, Merif J, Seah Lee W, Riordan SM, Rawlinson WD, Mitchell HM. 2016. Campylobacter concisus pathotypes are present at significant levels in patients with gastroenteritis. J Med Microbiol 65:219–226. doi: 10.1099/jmm.0.000216. [DOI] [PubMed] [Google Scholar]
- 22.Siah SP, Merif J, Kaur K, Nair J, Huntington PG, Karagiannis T, Stark D, Rawlinson W, Olma T, Thomas L, Melki JR, Millar DS. 2014. Improved detection of gastrointestinal pathogens using generalised sample processing and amplification panels. Pathology 46:53–59. doi: 10.1097/PAT.0000000000000022. [DOI] [PubMed] [Google Scholar]
- 23.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zakrzewski M, Proietti C, Ellis JJ, Hasan S, Brion MJ, Berger B, Krause L. 2017. Calypso: a user-friendly Web-server for mining and visualizing microbiome-environment interactions. Bioinformatics 33:782–783. doi: 10.1093/bioinformatics/btw725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tap J, Furet JP, Bensaada M, Philippe C, Roth H, Rabot S, Lakhdari O, Lombard V, Henrissat B, Corthier G, Fontaine E, Doré J, Leclerc M. 2015. Gut microbiota richness promotes its stability upon increased dietary fibre intake in healthy adults. Environ Microbiol 17:4954–4964. doi: 10.1111/1462-2920.13006. [DOI] [PubMed] [Google Scholar]
- 28.Oki K, Toyama M, Banno T, Chonan O, Benno Y, Watanabe K. 2016. Comprehensive analysis of the fecal microbiota of healthy Japanese adults reveals a new bacterial lineage associated with a phenotype characterized by a high frequency of bowel movements and a lean body type. BMC Microbiol 16:284. doi: 10.1186/s12866-016-0898-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujio-Vejar S, Vasquez Y, Morales P, Magne F, Vera-Wolf P, Ugalde JA, Navarrete P, Gotteland M. 2017. The gut microbiota of healthy Chilean subjects reveals a high abundance of the phylum Verrucomicrobia. Front Microbiol 8:1221. doi: 10.3389/fmicb.2017.01221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences were submitted to the European Nucleotide Archive under accession number PRJEB23690. Data from healthy adult individuals were obtained from the European Nucleotide Archive or were reported previously (16, 27–29).