ABSTRACT
Streptococcus agalactiae (group B Streptococcus [GBS]) is often a commensal bacterium that colonizes healthy adults asymptomatically and is a frequent inhabitant of the vaginal tract in women. However, in immunocompromised individuals, particularly the newborn, GBS may transition to an invasive pathogen and cause serious disease. Despite the use of the currently recommended intrapartum antibiotic prophylaxis for GBS-positive mothers, GBS remains a leading cause of neonatal septicemia and meningitis. To adapt to the various host environments encountered during its disease cycle, GBS possesses multiple two-component regulatory systems (TCSs). Here we investigated the contribution of a transcriptional regulator containing a LytTR domain, LtdR, to GBS pathogenesis. Disruption of the ltdR gene in the GBS chromosome resulted in a significant increase in bacterial invasion into human cerebral microvascular endothelial cells (hCMEC) in vitro as well as the greater penetration of the blood-brain barrier (BBB) and the development of meningitis in vivo. Correspondingly, infection of hCMEC with the ΔltdR mutant resulted in increased secretion of the proinflammatory cytokines interleukin-8 (IL-8), CXCL-1, and IL-6. Further, using a mouse model of GBS vaginal colonization, we observed that the ΔltdR mutant was cleared more readily from the vaginal tract and also that infection with the ΔltdR mutant resulted in increased cytokine production from human vaginal epithelial cells. RNA sequencing revealed global transcriptional differences between the ΔltdR mutant and the parental wild-type GBS strain. These results suggest that LtdR regulates many bacterial processes that can influence GBS-host interactions to promote both bacterial persistence and disease progression.
KEYWORDS: RNA sequencing, blood-brain barrier, cytokines, group B Streptococcus, meningitis, two-component regulatory systems, vaginal colonization
INTRODUCTION
Streptococcus agalactiae (group B Streptococcus [GBS]) is a Gram-positive, beta-hemolytic bacterium normally found in the human gastrointestinal and urogenital tracts of asymptomatic individuals. However, GBS possesses an array of virulence factors that renders it capable of causing invasive disease in susceptible hosts, including the newborn. Despite widespread intrapartum antibiotic administration to colonized mothers to prevent vertical transmission, GBS remains a leading cause of pneumonia, sepsis, and meningitis in neonates (1, 2). Over the course of its disease cycle, GBS encounters very different host microenvironments and must adapt to successfully colonize and survive in different host niches. In bacteria, the ability to efficiently adjust to different environments is commonly mediated by signaling through two-component regulatory systems (TCSs) (3, 4).
TCSs consist of a membrane-bound histidine kinase sensor that, upon activation by an external signal, phosphorylates a cytoplasmic transcriptional regulator that affects expression of downstream gene targets. Of the 21 TCSs that have been described in GBS, only a few have been well characterized (5–7). The TCS response regulator CiaR is known to play a role in promoting GBS survival within brain endothelial cells as well as host phagocytic cells, such as neutrophils and macrophages (8). The LiaR response regulator promotes GBS virulence by regulating cell wall synthesis in response to environmental stresses, such as elevated temperature and the presence of antimicrobial peptides (9). The best-studied TCS in GBS is CovS/R (control of virulence sensor/regulator), which regulates the expression of a variety of virulence factors that are important for GBS pathogenesis, such as pili, which promote GBS attachment to host surfaces, and cylE, which is involved in the expression of beta-hemolysin/cytolysin (β-H/C) (10–12). We have shown previously that GBS strains lacking the covR gene are hyperinflammatory and result in increased sepsis and meningitis in murine infection models due to increased expression of β-H/C (11). Interestingly, infection with GBS wild-type (WT), ΔcovR, or ΔcylE strains revealed that the β-H/C toxin is a key mediator in provoking an acute inflammatory response in the vaginal epithelium and that functional CovR regulation dampens cytokine production and promotes bacterial persistence in the mouse vaginal tract (13). These observations demonstrate the importance of TCS transcriptional regulators in regulating virulence factors in order to promote niche establishment.
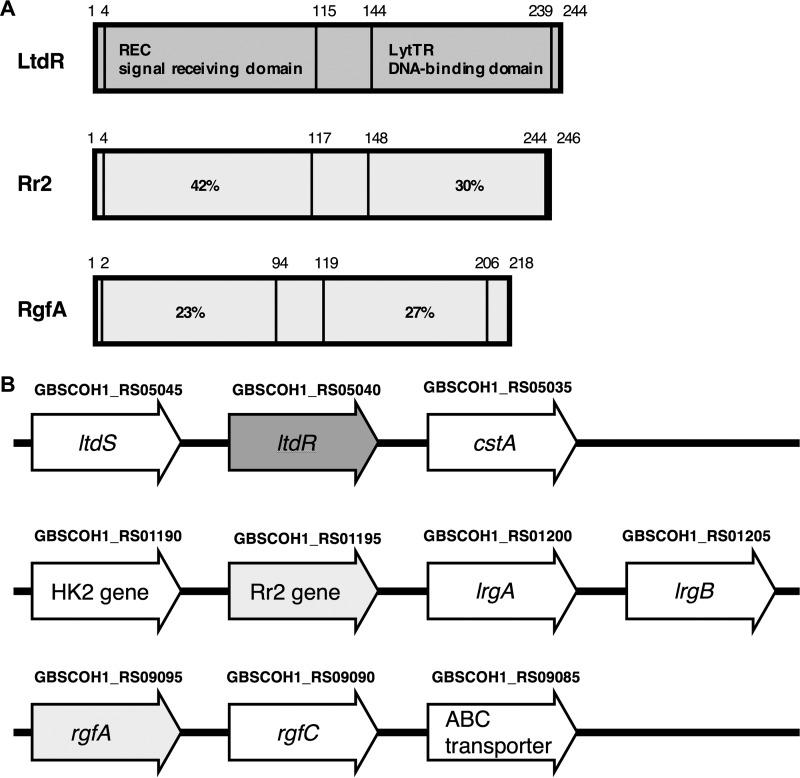
While the majority of TCS transcriptional regulators possess a helix-turn-helix (HTH) DNA-binding domain, a small percentage of transcriptional regulators contain a LytTR non-HTH domain (14–16). LytTR-containing proteins account for just ∼2.7% of prokaryotic response regulators and predominantly regulate the production of virulence factors in pathogenic bacteria (17). GBS possesses three TCS transcriptional regulators that include the LytTR DNA-binding domain (Fig. 1A). Of these, the RgfA regulator (GBSCOH1_RS09095) has been previously characterized and may contribute to virulence by mediating GBS binding to host fibrinogen (18). Another of these LytTR-containing response regulators, encoded by GBSCOH1_RS01195, has also been studied and is referred to as Rr2 (5). Rr2 is upregulated in GBS during stationary growth phase and is likely activated in response to stress conditions, such as a lack of nutrients, low pH, and the accumulation of toxic metabolites (5). The GBS Rr2 gene is contained within a locus that is homologous to loci found in the pathogens Staphylococcus aureus and Streptococcus mutans, where it has been shown to regulate the expression of murein hydrolases encoded by the adjacent lrgAB operon (19–22). In those pathogens, the Rr2 regulator is known as LytR and has been shown to be important for cell wall turnover, cell autolysis, and biofilm formation (23–25).
FIG 1.
(A) Schematic diagram of LytTR-containing two-component system transcriptional regulator proteins. Numbers above the diagram correspond to the amino acid positions. The percent identity of the primary amino acid sequences of the Rr2 and RgfA REC and LytTR domains to LtdR is indicated. (B) Schematic of the ltdR-, Rr2-, and rgfA-containing gene loci. Gene locus tags published in the NCBI reference sequence with GenBank accession number NZ_HG939456.1 are indicated above the gene annotations.
Here we investigate the third LytTR domain-containing TCS regulator (GBSCOH1_RS05040), which we designate LtdR, and its role in GBS colonization and virulence. Using targeted mutagenesis, we show, using mouse models of GBS colonization and invasive disease, that LtdR-mediated regulation affects both the development of meningitis and vaginal persistence. Additionally, we observed that the loss of LtdR results in increased GBS invasion into host cells and increased inflammatory signaling by infected cells in vitro. Finally, we performed RNA sequencing (RNA-seq) to identify LtdR-regulated genes and observed global transcriptional changes in the ΔltdR mutant strain.
RESULTS
Construction and characterization of the ltdR deletion strain.
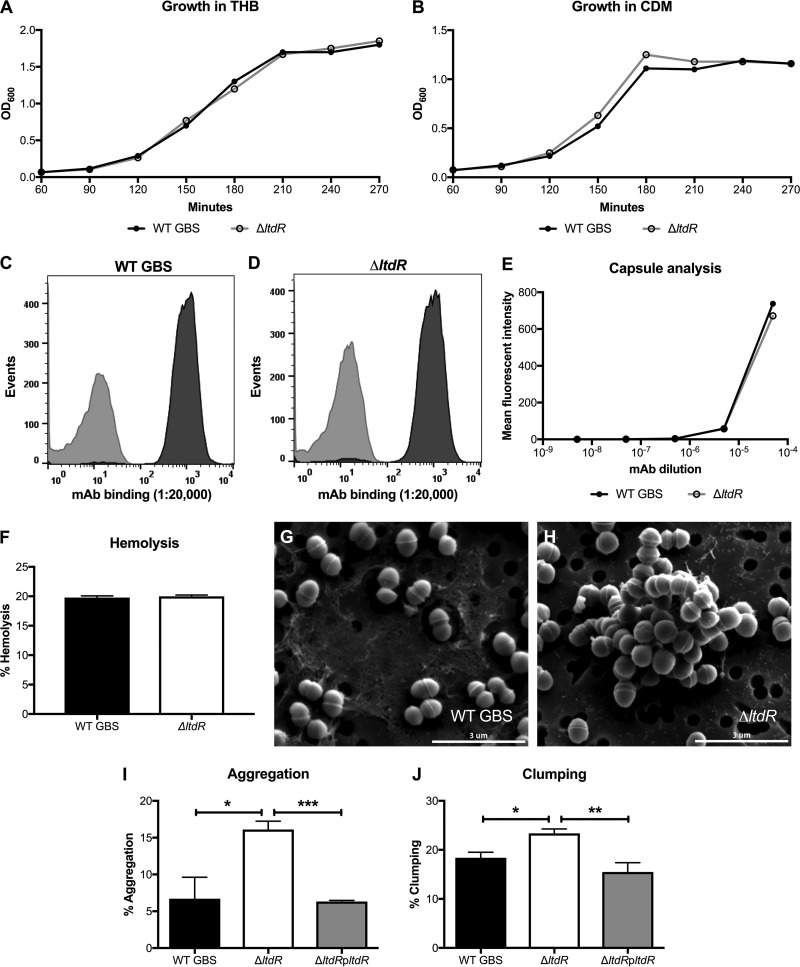
The LtdR transcriptional regulator possesses the REC signal receiver domain and the LytTR DNA-binding domain (Fig. 1A). The ltdR gene locus is distinct from the loci of the other two LytTR-containing two-component system transcriptional regulators. The ltdR gene is directly downstream of the ltdS histidine kinase sensor gene, and this operon is upstream of a gene encoding a putative carbon starvation gene. The gene encoding Rr2 is also downstream of the gene encoding its histidine kinase and upstream of the lrg murein hydrolase genes. The gene encoding the RgfA regulator appears upstream of the gene for its cognate kinase, RgfC (Fig. 1B). We performed precise insertional mutagenesis to generate the ΔltdR mutant GBS strain in the highly encapsulated, hypervirulent strain COH1 background (sequence type 17 [ST-17], serotype III) (26, 27). Additionally, the ltdR gene was cloned into the pDCerm vector to complement the ΔltdR mutant strain. Using NCBI BLAST program, we determined that the ltdR gene in COH1 is 99% homologous to ltdR genes in the GBS strains 2603V/R (ST-110, serotype V), CJB111 (ST-1, serotype V), NEM316 (ST-23, serotype III), A909 (ST-7, serotype Ia), and H36B (ST-6, serotype Ib). Analysis of growth demonstrated that the ΔltdR mutant COH1 strain grew similarly to the parental WT strain in rich medium (Todd-Hewitt broth [THB]) and chemically defined medium (CDM) (Fig. 2A and B). Bacterial numbers were also assessed by plating and determination of the number of CFU at various growth phases, and the CFU counts were similar between the WT and ΔltdR mutant strains (data not shown). Additionally, there was no difference in capsule abundance and hemolytic ability between the WT and the ΔltdR mutant strains (Fig. 2C to F). However, scanning electron microscopy revealed that the ΔltdR mutant appears to aggregate or cluster more readily than the WT strain, which could suggest a difference in cell division or surface architecture (Fig. 2G and H). Quantitative assays further demonstrated that the ΔltdR mutant exhibited increased aggregation and clumping phenotypes compared to the WT and complemented strains (Fig. 2I and J).
FIG 2.
(A and B) Growth curves for WT GBS and the ΔltdR mutant in THB (A) and CDM (B) at 37°C. (C to E) Flow cytometry using serial dilutions of a monoclonal antibody (MAb) to the serotype III capsule to determine the presence of capsule in WT GBS (C) and the ΔltdR mutant (D). A monoclonal antibody to the serotype Ia capsule was used as the isotype control. (F) Hemolysis assay comparing the hemolysis of sheep blood cells by WT GBS and the ΔltdR mutant. Representative data from 1 of at least 2 independent experiments are shown. (G and H) Scanning electron microscopy images of WT GBS (G) and ΔltdR mutant (H) strains. (I) Aggregation assay comparing aggregation of WT GBS, the ΔltdR mutant, and the complemented strain in THB. (J) Clumping assay comparing clumping of the WT GBS, the ΔltdR mutant, and the complemented strain in THB containing 0.1% fibrinogen. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
Loss of ltdR results in increased pathogenesis of meningitis in vivo.
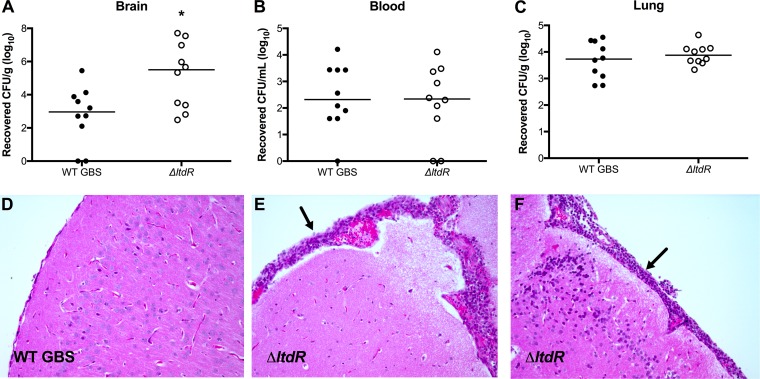
To determine whether LtdR contributes to GBS virulence in vivo, we utilized a murine model of GBS hematogenous meningitis (28–30). Mice were challenged with either WT GBS or the isogenic ΔltdR mutant as described in Materials and Methods. At the experimental endpoint, mice were euthanized and brain tissue, blood, and lung tissue were collected to determine the bacterial loads. We recovered similar numbers of ΔltdR mutant and WT GBS from mouse blood and lung; however, we observed a significant increase in the amounts of ΔltdR mutant GBS recovered from the brain tissue (Fig. 3A to C). Histopathological examination of fixed tissue revealed meningeal thickening as well as the presence of inflammatory infiltrates in the brains of mice infected with the ΔltdR mutant (Fig. 3D to F).
FIG 3.
Mouse model of GBS meningitis. (A to C) At 72 h after infection, mice were euthanized and bacterial loads in the brain (A), blood (B), and lung (C) were assessed. (D to F) Representative images of hematoxylin-eosin-stained brain sections from mice inoculated with the WT (D) or the ΔltdR mutant (E and F) GBS strain. Arrows, areas of neutrophil infiltration and meningeal thickening. Representative data from 1 of 3 independent experiments are shown. *, P < 0.05.
Loss of ltdR promotes bacterial invasion into endothelial cells and stimulates cytokine secretion in vitro.
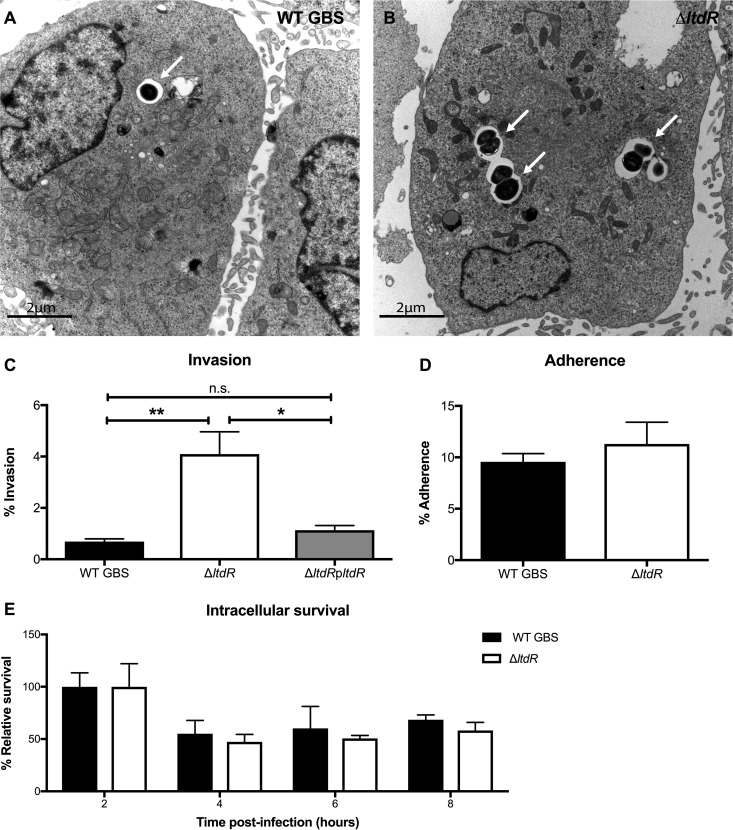
Because we observed increased bacterial loads in the brains of mice challenged with the ΔltdR mutant, we next characterized the ability of the ΔltdR mutant to invade human cerebral microvascular endothelial cells (hCMEC), the cells that constitute the blood-brain barrier (BBB). Using transmission electron microscopy, we first visualized more intracellular bacteria in brain endothelial cells following infection with the ΔltdR mutant than following infection with the WT strain (Fig. 4A and B). We then performed a quantitative cell invasion assay as described previously (28, 31) and in Materials and Methods. Monolayers of hCMEC were infected with WT, the ΔltdR mutant, or the complemented strain with an inoculum of 105 CFU/well (multiplicity of infection [MOI], 1); data are expressed as the percentage of intracellular GBS recovered after 2 h of incubation plus 2 h of treatment with antibiotics to kill extracellular bacteria. As shown in Fig. 4C, the ΔltdR mutant strain exhibited a significant increase in hCMEC invasion (P < 0.0005) compared to the WT strain. Furthermore, complementation of the ΔltdR mutant lowered the invasion ability to a level close to that of the WT (Fig. 4C). To determine whether the increased invasion observed with the ΔltdR mutant could be explained by increased attachment to host cells, we examined the amount of total cell-associated (surface-adherent plus intracellular) GBS to hCMEC; however, we observed no significant difference between the WT and the ΔltdR mutant strains (Fig. 4D). Further, to assess whether the observed increase in recovered intracellular bacteria for the ΔltdR mutant was due to enhanced survival or bacterial replication of the ΔltdR mutant within hCMEC, we performed additional experiments to measure intracellular survival over time. We recovered intracellular bacteria up to 8 h postinfection and determined that the ΔltdR mutant was able to survive inside hCMEC at levels comparable to those of WT strain COH1 (Fig. 4E). These data suggest that LtdR contributes primarily to bacterial invasion into brain endothelial cells.
FIG 4.
LtdR regulation influences GBS invasion into the brain endothelium. (A and B) Transmission electron micrographs of hBMEC infected with WT (A) or ΔltdR mutant (B) GBS. (C) Invasion of WT GBS, the ΔltdR mutant, and the complemented strain into hCMEC was quantified after a 2-h infection. (D) Adherence of WT GBS and the ΔltdR mutant strain to hCMEC was assessed after a 30-min incubation. (E) The intracellular survival of WT GBS and the ΔltdR mutant strain relative to that of the WT and the ΔltdR mutant at 2 h postinfection was determined up to 8 h postinfection. Experiments were performed at least 3 times in triplicate, and error bars represent SDs; the results of a representative experiment are shown. *, P < 0.05; **, P < 0.005; n.s., not significant.
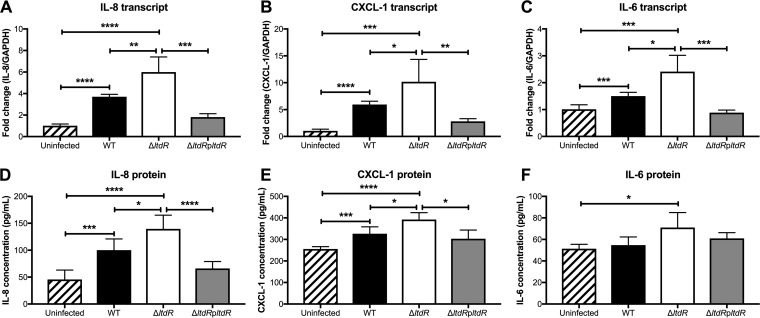
Since we observed increased inflammatory infiltrate, predominately neutrophils, in tissues surrounding the brains of mice challenged with ΔltdR mutant compared to those of mice challenged with the WT strain, we hypothesized that LtdR-mediated regulation may impact the host inflammatory response. We have shown previously that GBS infection elicits cytokine and chemokine signaling, which promotes neutrophil influx into the central nervous system (CNS) (32). To examine if cytokine/chemokine expression was altered during infection with the ΔltdR mutant, we infected hCMEC with the WT, the ΔltdR mutant, or the complemented mutant strain and measured the transcript abundance and protein production of major neutrophil chemoattractants, interleukin-8 (IL-8) and CXCL-1, as well as the proinflammatory cytokine IL-6. Cells infected with the ΔltdR mutant GBS had significantly higher levels of transcript and secreted protein than uninfected cells or cells infected with WT GBS. Complementation of the ltdR mutation lowered the level of cytokine expression by infected hCMEC to levels similar to those for WT-infected cells (Fig. 5A to F). Taken together, these data suggest that the loss of LtdR enhances GBS invasion into the brain endothelium and that this may result in the stimulation of cytokine release from host cells.
FIG 5.
LtdR impacts cytokine expression by infected hCMEC. (A to C) hCMEC were infected with GBS for 5 h, and then the cells were collected and the transcript levels of IL-8 (A), CXCL-1 (B), and IL-6 (C) were quantified by RT-qPCR. (D to F) The hCMEC supernatant was collected for detection of IL-8 (D), CXCL-1 (E), and IL-6 (F) protein secretion during GBS infection. Experiments were performed at least 3 times in triplicate, and error bars represent SDs; the results of a representative experiment are shown. *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.00005.
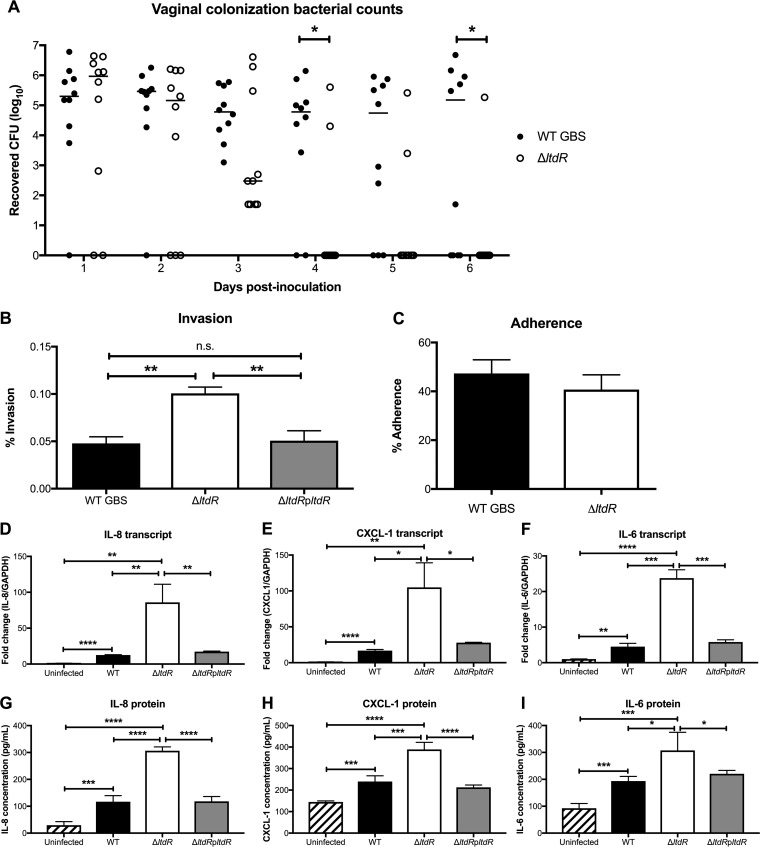
LtdR contributes to vaginal colonization.
To examine the function of LtdR in a different host niche, we utilized a murine model of GBS vaginal colonization (33) to characterize the role of LtdR regulation in colonization. Mice (8 weeks old) were treated with 17β-estradiol 1 day prior to bacterial inoculation. The GBS WT or the ΔltdR mutant (1 × 107) was inoculated directly into the vagina, and on successive days, the vaginal lumen was swabbed and the recovered bacteria were quantified on agar plates to determine bacterial persistence and changes in the bacterial load over time. Similar numbers of both strains were recovered from the mouse vagina on day 1 postinoculation. However, the animals that were challenged with ΔltdR mutant strain cleared the GBS more rapidly than the mice that were inoculated with the WT strain (Fig. 6A). This result could mean that the ΔltdR mutant was less able to attach to vaginal epithelium; however, we observed that, similar to our observations with brain endothelium, the ΔltdR mutant was more invasive than WT GBS into human vaginal epithelial cells (hVEC) but was not significantly more adherent (Fig. 6B and C). We have previously shown that GBS infection of hVEC results in activation of numerous immune pathways, including those involving cytokines and chemokines involved in leukocyte recruitment and activation (13). To determine whether LtdR plays a role in inflammatory signaling, we infected hVEC with the WT, the ΔltdR mutant, or the complemented strain and detected the highest levels of IL-8, CXCL-1, and IL-6 transcripts and secreted protein from the cells infected with the ΔltdR mutant strain (Fig. 6D to I). Thus, as we observed in brain endothelium, LtdR regulation also impacts the host inflammatory response in vaginal epithelial cells.
FIG 6.
LtdR plays a role in GBS persistence and inflammation in the vaginal tract. (A) Murine vaginal colonization model. Mice were inoculated with either the WT or the ΔltdR mutant GBS strain, and the bacterial load was monitored daily. (B) Invasion of WT GBS, the ΔltdR mutant, and the complemented strain into hVEC was quantified after a 2-h infection. (C) The adherence of WT GBS and the ΔltdR mutant was assessed after a 30-min incubation. (D to F) hVEC were infected with GBS for 5 h, and then the transcript levels of IL-8 (D), CXCL-1 (E), and IL-6 (F) were assessed by RT-qPCR. (G to I) An ELISA to quantify the IL-8 (G), CXCL-1 (H), and IL-6 (I) secreted by hVEC was performed following a 5-h infection with GBS strains. Experiments were performed at least 3 times in triplicate, and error bars represent SDs; the results of a representative experiment are shown. *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.00005; n.s., not significant.
LtdR regulation impacts many bacterial cell processes.
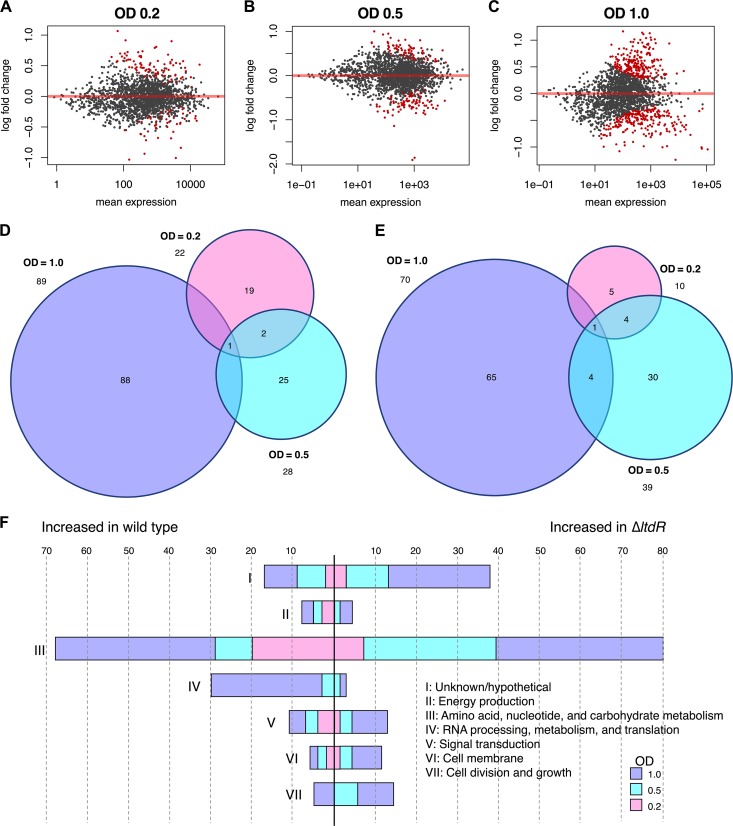
Thus far, we have observed that LtdR contributes to both colonization and invasive disease. Because GBS colonization and meningitis occur in different host niches, we hypothesized that LtdR may regulate multiple bacterial pathways to affect GBS interactions within different host cell environments. To characterize the impact of LtdR regulation on global gene transcription, we performed RNA sequencing analysis of the ΔltdR mutant as well as the parental WT strain grown to an optical density (OD) of 0.2, 0.5, and 1.0. We observed significant global changes at all growth phases, with the highest number of significantly differentially expressed genes being seen at early stationary phase (OD = 1.0); in total, 135 genes were downregulated and 109 genes were upregulated in the ΔltdR mutant compared to their levels of regulation in the WT strain (Fig. 7A to E; see also Table S1 in the supplemental material) We classified these genes based on the clusters of orthologous groups (COG) of genes (34, 35). The majority of genes differentially expressed during all three growth phases are involved in metabolic pathways. There were also increases in the expression of genes involved in RNA processing in the WT strain. Interestingly, the expression of a number of transcriptional regulators (all classified under the signal transduction COG) was higher in the ΔltdR mutant strain during early stationary phase (Fig. 7F). In our examination of the differentially expressed transcripts, we saw little overlap of genes affected between the three growth phases (Fig. 7A to E). These results suggest that the effects of LtdR regulation are variable depending on the growth phase. GBSCOH1_RS09570, which encodes a bifunctional homocysteine S-methyltransferase/methylenetetrahydrofolate reductase, was the transcript that was more abundant in the WT GBS than in the ΔltdR mutant at every time point. This enzyme functions in methionine metabolism pathways. The gene that was more abundant in the ΔltdR mutant than in the WT at all time points was GBSCOH1_RS05780, which encodes the LysR family transcriptional regulator MtaR. MtaR has previously been shown to regulate the expression of the methionine transport genes metQ1 (GBSCOH1_RS07790), pdsM (GBSCOH1_RS07785), metN (GBSCOH1_RS07780), and metP (GBSCOH1_RS07775) (36, 37). In our RNA-seq analysis, metQ1, pdsM, and metN transcript levels were significantly higher in the ΔltdR mutant at the earliest time point, while just the metQ1 transcript level was increased in the ΔltdR mutant in mid-log phase, and none of the genes in the methionine metabolism operon were significantly differentially expressed at the latest time point. Reverse transcription-quantitative PCR (RT-qPCR) was used to validate some of these and other select transcripts that were expressed at significantly higher or lower levels in the ΔltdR mutant than in the WT, as determined by RNA sequencing. We observed that the differences in transcript abundance between WT GBS and ΔltdR mutant cultures seen by RT-qPCR were similar to those seen by RNA sequencing (Table S2). We further observed that the complemented strain had significantly lower levels of expression of select transcripts that were highly upregulated in the ΔltdR mutant compared to their levels of expression in the WT strain (Fig. S1). The results of our RNA-seq and RT-qPCR analyses of the WT, ΔltdR mutant, and complemented strains indicate that LtdR may regulate many cellular pathways, including those involving methionine uptake and metabolism, as well as expression of potential virulence factors.
FIG 7.
RNA sequencing to identify LtdR-regulated processes. (A to C) MA plots highlighting genes differentially expressed between WT GBS and the ΔltdR mutant at different growth phases. Significantly differentially expressed genes (adjusted P < 0.1) are indicated in red. An OD600 of 0.2 (A), an OD600 of 0.5 (B), and an OD600 of 1.0 (C) correspond to late lag phase, exponential phase, and early stationary phase, respectively. (D and E) Venn diagrams of genes expressed at significantly (adjusted P < 0.05) higher levels in WT GBS than in the ΔltdR mutant (D) and transcripts that were present at significantly larger amounts in the ΔltdR mutant strain than in the WT (E). (F) Significantly differentially expressed genes (adjusted P < 0.05) were classified according to the cluster of orthologous groups (COG) of genes.
DISCUSSION
Bacteria use TCSs to monitor and respond to environmental signals. GBS possesses 21 TCSs (6), about twice as many as other closely related pathogens, such as Streptococcus pyogenes, Streptococcus pneumoniae, and Streptococcus mutans, all of which encode just 13 TCSs in their genomes (38–40). Three of the GBS TCS transcriptional regulators contain the rare LytTR DNA-binding domain (5). Because LytTR-containing response regulators predominantly influence virulence in Gram-positive pathogens (15, 17), we explored the role of the previously uncharacterized LtdR regulator in GBS meningitis and vaginal persistence. In this study, we show that LtdR regulation can impact GBS infection and colonization by influencing host inflammatory responses.
It is likely that GBS invasion into BBB endothelial cells is the critical first step for the development of meningitis (2). Our data suggest that LtdR plays a role in promoting GBS invasion into host cells. We found that the ΔltdR mutant was significantly more invasive than the parental WT GBS strain, without any difference in attachment to host cells or survival within cells for up to 8 h. Additionally, we were able to recover more ΔltdR mutant GBS than WT GBS from the brain tissue of infected mice, while we observed similar bacterial loads in other tissues. These observations suggest that LtdR regulates factors that specifically promote the uptake of GBS into brain endothelial cells without affecting GBS attachment to host cells. Subsequent to bacterial BBB invasion, the stimulation of host immune pathways is essential to the progression of meningitis. The release of inflammatory factors by brain endothelial cells, microglia, astrocytes, and infiltrating immune cells can exacerbate neuronal injury (41). An analysis of the global transcription profile in BBB endothelium showed that the expression of many chemokines and cytokines is increased in response to GBS infection, with IL-8, CXCL-1, and IL-6 being among the most highly induced (32). In the current study, we found that disrupting LtdR regulation results in increased expression of those three cytokines. IL-8 and CXCL-1 are major neutrophil chemoattractants known to orchestrate neutrophil activation and recruitment during GBS infection (30, 42), while IL-6, another proinflammatory cytokine that is elevated in the cerebrospinal fluid of meningitis patients, has also been shown to be secreted by brain endothelial cells during GBS infection (32). Although an inflammatory response may be beneficial as a host defense against bacterial infections, excess inflammation can exacerbate the symptoms of bacterial meningitis and can be detrimental during an acute infection (43, 44). We observed that mice infected with the ΔltdR mutant GBS strain exhibited higher brain bacterial loads and increased meningeal inflammation compared to animals challenged with WT GBS. Thus, LtdR regulation impacts bacterial BBB penetration and disease progression during GBS meningitis.
While increased inflammation can be associated with a worse prognosis in invasive bacterial infections such as meningitis, we have observed that stimulation of the host immune system is associated with bacterial clearance during vaginal colonization (13, 45). We have previously investigated the ability of different GBS strains to successfully colonize the murine vaginal tract and found that the less inflammatory strains persisted the longest (46). Here, we observed that the ΔltdR mutant GBS strain promoted increased levels of proinflammatory cytokines/chemokines from vaginal epithelial cells compared to the levels promoted by WT GBS and did not persist in the mouse vagina. These results are consistent with those of previous studies showing that the upregulation of proinflammatory signaling pathways is associated with GBS clearance from the vaginal tract (13, 47). Further, our data suggest that the LtdR/S system is necessary for limiting the expression of GBS virulence factors during colonization, thereby reducing host innate immune responses to promote stable vaginal persistence.
We have identified the GBS LtdR response regulator to be a key player in inducing inflammation in both brain endothelial cells and vaginal epithelial cells; however, the exact mechanism(s) by which GBS LtdR controls gene expression to promote proinflammatory signaling remains unclear. To further understand how the LtdS/R system contributes to colonization and disease pathogenesis, we performed RNA sequencing to compare the transcriptional profiles of the GBS WT and ΔltdR mutant strains. Our analysis revealed dramatic transcriptional differences between GBS WT and ΔltdR mutant strains. We detected the highest number of transcripts significantly differentially expressed between the two strains during the later growth phase, suggesting that LtdR regulation may be most active during stationary phase. Overall, there were 135 genes that were downregulated and 109 genes that were induced in the ΔltdR mutant compared to their expression in the WT strain; thus, LtdR appears to act as both a positive and a negative regulator. Genes that appear to be negatively regulated by LtdR include those involved in signal transduction, cell division and growth, and the cell membrane, as well as almost 40 genes that have a hypothetical or unknown function. Interestingly, in a previous study, the ltdR gene was deleted in a different GBS background, specifically, in the serotype V CJB111 strain (in a mutant designated the Δrr11 mutant) (5). While the impact of Rr11 on pathogenesis was not characterized, it was reported that Rr11 similarly regulates genes involved in metabolism, as determined using microarray analysis (5).
Many TCS sensor kinases are stimulated by stress conditions that are present during stationary phase, such as a lack of nutrients, low pH, and the accumulation of toxic metabolites, and our observations suggest that the LtdS/R TCS may also play a role in adaptation to different growth environments. Future studies to determine the signal(s) being sensed by the LtdS sensor kinase will be of interest. We saw a number of genes associated with methionine metabolism expressed at higher levels in the ΔltdR mutant than in WT GBS. GBS requires methionine for growth but cannot synthesize the amino acid, so it must scavenge methionine from its environment. Previous work has shown that a GBS strain lacking the MtaR methionine transport regulator exhibits growth defects in media with physiological methionine concentrations and is attenuated in an in vivo rat sepsis model (36, 37). While we observed no differences in growth between the WT and the ΔltdR mutant GBS strains in THB and CDM, further characterization of the role of LtdR regulation in GBS growth and survival in blood, in serum, and under other low-methionine conditions is warranted.
We also observed that the gene encoding the pore-forming toxin hemolysin III and the gene directly upstream encoding a MerR domain-containing transcriptional regulator, which likely regulates hemolysin III expression, were upregulated in the ΔltdR mutant compared to their expression in the WT. Hemolysin III from Bacillus cereus has been shown to lyse erythrocytes, and in Vibrio vulnificus the toxin is known to impact virulence in a mouse model of infection (48–50). However, nothing is known about the role of hemolysin III in GBS colonization and invasive disease. Our results suggest that LtdR regulation might affect hemolysin III expression. As the GBS β-H/C toxin is known to promote neutrophilic chemokine signaling in the BBB endothelium (28), further studies on hemolysin III and its contribution to proinflammatory responses during GBS colonization and disease progression will be of interest.
This study demonstrates the importance of LtdR regulation to the progression of GBS meningitis as well as GBS persistence within the vaginal tract during colonization. Our results are consistent with previous reports that suggest that GBS must tightly regulate expression of virulence and inflammatory factors in order to promote a colonizing state and that changes in the expression levels of these factors contribute to disease progression. Future studies will be aimed at determining the specific LtdR-regulated genes that contribute to bacterial invasion and host proinflammatory signaling, which may provide important targets for preventing GBS colonization and subsequent invasive disease.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
GBS clinical isolate COH1 (serotype III) (51) and its isogenic ΔltdR mutant were used for the experiments. The GBS strains were grown in THB (Hardy Diagnostics) at 37°C, and growth was monitored by measuring the optical density at 600 nm (OD600). Escherichia coli strain TOP10 electrocompetent cells and strain MC1061 electrocompetent cells (Invitrogen), used for plasmid propagation, were grown in LB medium. For antibiotic selection, 2 μg/ml chloramphenicol (Sigma) and 5 μg/ml erythromycin (Sigma) were incorporated into the growth medium for GBS and 300 μg/ml erythromycin was incorporated into the growth medium for E. coli. Chemically defined medium (CDM) (36) and THB were used for comparison of the growth of the GBS WT and the ΔltdR mutant. To evaluate growth in CDM or THB, GBS was initially grown to log phase (OD600, 0.3) in THB. The cells were harvested by centrifugation, washed three times in an equivalent volume of phosphate-buffered saline (PBS; Gibco), and diluted 1 to 50 into the growth medium. Growth was monitored spectrophotometrically at a wavelength of 600 nm.
Construction of the ΔltdR deletion and complemented strains.
Deletion of the ltdR gene (GBSCOH1_RS05040) was performed in the genome of COH1. The following upstream and downstream primers were used to generate the forward fusion fragment: 5′-GGGACTGACAACAGAATCTTG and 5′-CACAATGTAAGAGC(G/T)CTGCTATAACGAGG. The reverse fusion fragment was generated with upstream primer 5′-CCTCGTTATAGCAG(A/C)GCTCTTACATTGTG and downstream primer 5′-CCCAAGTGCTTTATCAATAGG. The forward and reverse amplicons included GBSCOH1_RS05035 (forward fragment) and GBSCOH1_RS05045 (reverse fragment) sequences to be used as recombination homologous arms for allelic exchange. A 1,999-bp fusion deletion product was generated with previously generated forward and reverse PCR products mixed in equal amounts in a PCR mixture with the following primers: 5′-GGGACTGACAACAGAATCTTG and 5′-CCCAAGTGCTTTATCAATAGG. High-fidelity enzyme (Roche) was used for all PCRs. The PCR product was digested and gel purified prior to cloning into the TOPO Blunt vector (Invitrogen). The resulting plasmid, TOPO/GBSCOH1_RS05040AD, was used to transform E. coli TOP10 cells (Invitrogen). Selection of clones was performed according to the manufacturer's directions. Positive clones were screened by SphI restriction endonuclease digestion and plasmid sequencing (Arizona State University). The TOPO/GBSCOH1_RS05040AD plasmid was digested with BamHI and EcoRI and ligated into a similarly digested pHY304, yielding pHY304/GBSCOH1_RS05040AD, which was transformed directly into E. coli MC1061. The plasmid was propagated and isolated from E. coli for transformation of electrocompetent GBS strain COH1. The WT GBSCOH1_RS05040 chromosomal allele was replaced with the allele encoding the deletion fragment. The resulting strain was cultured at 30°C in the presence of erythromycin. The cells were shifted to 37°C in the presence of erythromycin to select for the chromosomal integration of pHY304/GBSCOH1_RS05040AD. Integration was confirmed by the isolation of chromosomal DNA and subsequent PCR with the following primers: 5′-GGGCATTTAACGACGAAACTG and 5′-CGTCGTTTTACAACGTCGTGA. Isolates with the integrated plasmid were chosen and passaged five times in the absence of erythromycin at 30°C. The resulting strain was then cured of the plasmid by passage at 37°C and screened for sensitivity to erythromycin. The chromosome of the GBS ΔltdR mutant was verified by PCR with primers that flanked the cloned region and that were specific for the deleted sequence. For complementation studies, the full-length ltdR gene was amplified using the following primers: 5′-GGTCTAGAAAAGGTATTGTATGAAG, incorporating a XdaI restriction site, and 5′-GGCATATGGAATAACTTTTCATTAGTTA, incorporating an NdeI restriction site, and cloned into the pDCerm plasmid. The ΔltdR deletion strain was transformed with the recombinant plasmid to generate the complemented strain. The ΔltdR pltdR complemented strain was grown in the presence of erythromycin.
Hemolysis assay.
GBS strains were grown to an OD600 of 0.4 and then harvested by centrifugation and resuspended in PBS. A total of 1 × 109 CFU was added to fresh sheep blood (VWR) in V-bottom 96-well plates (Corning). The plates were sealed and incubated at 37°C with agitation for 1 h. The plates were centrifuged at 200 × g for 10 min, and 100 μl of the supernatant was transferred to a flat-bottom 96-well plate. The absorbance at 541 nm (A541) was read, and the percent hemolysis was calculated by comparing the A541 values for the GBS-treated wells to the A541 values for the wells with blood incubated with water.
Flow cytometry.
Flow cytometry to determine capsule expression was performed as described in reference 52. Briefly, bacterial stocks were washed in sterile PBS–0.5% bovine serum albumin (BSA) and then incubated with 10-fold serial dilutions (range, 1:2,000 to 1:200,000,000 final) of a monoclonal anti-serotype III antibody or the monoclonal anti-serotype Ia isotype control, washed via centrifugation, and labeled with a donkey anti-mouse IgM conjugated to Alexa Fluor 647 (Invitrogen) at a 1:2,000 dilution. All incubations were performed at 4°C with shaking. Samples were washed again and then resuspended, read on a FACSCalibur flow cytometer (BD Biosciences), and analyzed using FlowJo (v10) software. The monoclonal antibodies, which were provided by John Kearney at the University of Alabama at Birmingham, were purified. For the histograms shown in Fig. 2C and D, both the anti-serotype III and anti-serotype Ia isotype control antibodies were used at a final dilution of 1:20,000.
Aggregation and clumping assays.
GBS strains were grown to an OD600 of 0.4 and then harvested by centrifugation and resuspended in PBS (for aggregation assay) or PBS with 0.1% fibrinogen (for clumping assay) in 1.75-ml Eppendorf tubes (E & K Scientific). A total of 0.1 ml was pipetted from the top of the cell suspension after 150 min, and the turbidity was measured at 600 nm.
Mouse models of hematogenous GBS meningitis and vaginal colonization.
The animal experiments were approved by the Committee on the Use and Care of Animals at San Diego State University (SDSU; protocol number 16-10-021D) and performed using accepted veterinary standards. We utilized a mouse GBS infection model as described previously (28–30). Briefly, 8-week-old male CD-1 mice (Charles River) were injected intravenously with 1 × 108 CFU of wild-type GBS or the isogenic ΔltdR mutant. After 72 h, mice were euthanized and blood and brain tissue were collected. One half of each brain was fixed in 4% paraformaldehyde (Ricca Chemical Company) for histopathological analyses. Formalin-fixed, paraffin-embedded brains were sectioned onto glass slides. The slides were stained using hematoxylin and eosin (Sigma), and images were taken using a Zeiss upright microscope with an attached AxioCam Icc3 camera. Adobe Photoshop and Illustrator software was used to process the images. The remaining tissue was homogenized, and the brain tissue homogenates as well as blood and lung homogenates were plated on THB agar for enumeration of the bacterial CFU.
The mouse model of GBS vaginal colonization was performed as described previously (33). Ten-week-old female CD-1 mice (Charles River) were injected intraperitoneally with 0.5 mg 17β-estradiol (Sigma) that had been suspended in 100 μl sesame oil 1 day prior to inoculation with GBS. Mice were vaginally inoculated with 1 × 107 CFU of wild-type GBS or the isogenic ΔltdR mutant, and on subsequent days, the vaginal lumen was swabbed with a sterile ultrafine swab. The recovered GBS were enumerated on CHROMagar StrepB agar (DRG International).
Cell lines and infection assays.
Cells of the well-characterized human cerebral microvascular endothelial cell line (hCMEC/D3) (53–56), referred to here as hCMEC, were obtained from Millipore and were maintained in an EndoGRO-MV complete medium kit supplemented with 1 ng/ml fibroblast growth factor 2 (FGF-2; Millipore) at 37°C with 5% CO2. Human brain microvascular endothelial cells (hBMEC) were cultured as described previously (32) in RPMI 1640 (Corning Cellgro) containing 10% fetal bovine serum (Atlanta Biologicals), 10% Nu-Serum growth medium supplement (BD Biosciences), and 1% nonessential amino acids (Gibco) at 37°C with 5% CO2. Immortalized human vaginal epithelial cells (hVEC) were obtained from the American Type Culture Collection (VK2/E6E7, ATCC CRL-2616) and were maintained in keratinocyte serum-free medium (KSFM; Gibco) with 0.1 ng/ml human recombinant epidermal growth factor (EGF; Gibco) and 0.05 mg/ml bovine pituitary extract (Gibco) at 37°C with 5% CO2 as described previously (46).
Assays to determine the total number of cell surface-adherent or intracellular bacteria were performed as described previously (29). Briefly, bacteria were grown to mid-log phase to infect cell monolayers (1 × 105 CFU at a multiplicity of infection [MOI] of 1). Total cell-associated GBS were recovered following a 30-min incubation, while intracellular GBS were recovered after 2 h of infection and 2 h of incubation with 100 μg gentamicin (Sigma) and 5 μg penicillin (Sigma) to kill all extracellular bacteria. To assess intracellular survival, cells were treated with antibiotics for up to 8 h after a 2-h infection with GBS. Cells were detached with 0.1 ml of 0.25% trypsin–EDTA solution and lysed with addition of 0.4 ml of 0.025% Triton X-100 by vigorous pipetting. The lysates were then serially diluted and plated on THB agar to enumerate the bacterial CFU.
Electron microscopy.
For scanning electron microscopy, bacteria were grown to log phase in THB containing human fibrinogen (100 μg/ml). They were then fixed for 10 min using a one-step method with 2.5% glutaraldehyde, 1% osmium tetroxide, 0.1 M sodium cacodylate. Bacteria were collected on 0.4-μm-pore-size polycarbonate filters by passing the solution through a Swinnex device outfitted on a 10-ml syringe. The filters were dehydrated through a series of increasing ethanol concentrations and then dried in a Tousimis Samri-790 critical point drying machine. The dried filters were mounted on scanning electron microscopy sample stubs with double-sided carbon tape, coated with 6-nm platinum using a Quorom Q150ts high-resolution coater, and imaged with an FEI FEG450 scanning electron microscope. For transmission electron microscopy, hBMEC were grown in 24-well plates to confluence, and GBS was subsequently added to the cell monolayers as described above. Following 2 h of incubation, the monolayers were washed with PBS and collected by centrifugation. The pellets were fixed in 2% glutaraldehyde, 4% formaldehyde in 0.1 M cacodylate buffer at pH 7.4 for 2 h at room temperature. The pellets were then fixed again in 1% osmium tetroxide in 0.1 M cacodylate buffer for 1 h. After rinsing with water, samples were dehydrated with a series of increasing concentrations of ethanol and left on a rotator overnight in a 50% solution of Spurr resin and acetone. Samples were placed in 100% Spurr resin the next day and placed on a rotator for several hours before they were transferred to fresh 100% Spurr resin and polymerized in an oven for 24 h at 60°C. The sample blocks were sectioned at 50 nm on a Leica ultramicrotome and picked up on Formvar-coated copper grids. Grids with sections were stained with uranyl acetate and lead citrate, viewed using a Tecnai-12 (FEI) transmission electron microscope, and photographed using a Zeiss 215 side-mount digital camera. Images were generated using AMT image capture software. Adobe Photoshop and Illustrator software was used to process the images.
RT-qPCR and ELISA.
GBS were grown to mid-log phase, and 1 × 106 CFU (MOI, 10) was added to hCMEC or hVEC monolayers and incubated at 37°C with 5% CO2 for 6 h. The cells were then lysed, and total RNA was extracted (Macherey-Nagel) and cDNA was synthesized (Quanta Biosciences) according to the manufacturers' instructions. Primers and primer efficiencies for IL-8, CXCL-1, IL-6, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were utilized as previously described (57). IL-8, CXCL-1, and IL-6 proteins from hCMEC and hVEC supernatants were detected by enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's instructions (R&D systems). For bacterial RT-qPCR, triplicate cultures of WT GBS, the ΔltdR mutant, and the complemented strain were grown at 37°C in THB. The growth of the GBS strains was monitored by measuring the optical density, and samples were collected at an OD600 of 0.2, 0.5, and 1. Bacteria were collected and RNA was isolated as described above, and cDNA was synthesized (Quanta Biosciences). The primers for qPCR are listed in Table S3 in the supplemental material.
Generation of RNA-seq data.
Triplicate cultures of WT GBS and the ΔltdR mutant were grown at 37°C in THB. The growth of GBS strains was monitored by measuring the optical density, and samples were collected at an OD600 of 0.2, 0.5, and 1.0 to correspond with late lag, exponential, and stationary growth phases, respectively. Bacteria were isolated from the medium by centrifugation and resuspended in TRIzol reagent (Thermo Fisher). Zirconia/silica beads (diameter; 0.1 mm; BioSpec Products) were added to the bacterial suspensions, and the bacteria were lysed by beating for 2 min at maximum speed on a bead beater (BioSpec Products). RNA was isolated by following the manufacturer's protocol using a Direct-Zol RNA MiniPrep Plus kit (Zymo Research). Illumina cDNA libraries were generated using a modified version of the RNAtag-seq protocol (58). Briefly, 500 ng to 1 μg of total RNA was fragmented, depleted of genomic DNA, dephosphorylated, and ligated to DNA adapters carrying 5′-AN8-3′ barcodes of known sequence with a 5′ phosphate and a 3′ blocking group. Barcoded RNAs were pooled and depleted of rRNA using a RiboZero rRNA depletion kit (Epicentre). Pools of barcoded RNAs were converted to Illumina cDNA libraries in 2 main steps: (i) reverse transcription of the RNA using a primer designed to be specific to the constant region of the barcoded adapter with addition of an adapter to the 3′ end of the cDNA by template switching using SMARTScribe reverse transcriptase (Clontech) as described previously (59) and (ii) PCR amplification using primers whose 5′ ends target the constant regions of the 3′ or 5′ adapters and whose 3′ ends contain the full Illumina P5 or P7 sequences. cDNA libraries were sequenced on an Illumina NextSeq 500 platform to generate paired end reads.
Analysis of RNA-seq data.
Sequencing reads from each sample in a pool were demultiplexed on the basis of their associated barcode sequences using custom scripts. Up to 1 mismatch in the barcode was allowed, provided that it did not make assignment of the read to a different barcode possible. Barcode sequences were removed from the first read, as were terminal G residues from the second read that may have been added by the SMARTScribe reverse transcriptase during template switching. Reads were aligned to the NCBI reference sequence with GenBank accession number NZ_HG939456.1 using the Burrows-Wheeler aligner (60), and reads counts were assigned to genes and other genomic features using custom scripts. Differential expression analysis was conducted with the DESeq2 package (61). Visualization of raw sequencing data and coverage plots in the context of genome sequences and gene annotations was conducted using the GenomeView browser (62).
Data analysis.
GraphPad Prism (version 7.0) software was used for statistical analysis, and statistical significance was accepted at P values of <0.05. Statistical analysis for the hematogenous meningitis experiments, bacterial adherence and invasion assays, cytokine RT-qPCR, and ELISA was performed using the t test. Statistical analysis for the vaginal colonization experiments comparing the numbers of CFU of WT GBS and the ΔltdR mutant recovered from the mouse vaginal lumen on each day was performed using the Mann-Whitney test. For RNA sequencing, genes were considered significantly differentially expressed if there was a 1.5-fold difference in the level of expression and if the P value was below 0.05. Venn diagrams were calculated using the area-proportional Venn diagram tool (BioInfoRx).
Supplementary Material
ACKNOWLEDGMENTS
We thank Kwang Sik Kim and Monique Stins (Johns Hopkins) for providing hBMEC, John Kearney (University of Alabama at Birmingham) for providing anti-serotype III and anti-serotype Ia GBS IgM antibodies, and Jonathan Livny (Broad Institute of MIT and Harvard) for helpful discussions.
RNA-seq libraries were constructed and sequenced at the Broad Institute of MIT and Harvard by Jessica Alexander at the Microbial ‘Omics Core and by the Genomics Platform, respectively. The Microbial ‘Omics Core at the Broad Institute of MIT and Harvard also provided guidance on the experimental design and conducted preliminary analysis for all RNA-seq data.
This study was supported by the Rees Stealy Research Foundation/SDSU Heart Institute and San Diego Chapter ARCS scholarships to L.D. and NIH/NINDS R01-NS051247 to K.S.D.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00822-17.
REFERENCES
- 1.Thigpen MC, Whitney CG, Messonnier NE, Zell ER, Lynfield R, Hadler JL, Harrison LH, Farley MM, Reingold A, Bennett NM, Craig AS, Schaffner W, Thomas A, Lewis MM, Scallan E, Schuchat A, Emerging Infections Programs Network. 2011. Bacterial meningitis in the United States, 1998-2007. N Engl J Med 364:2016–2025. doi: 10.1056/NEJMoa1005384. [DOI] [PubMed] [Google Scholar]
- 2.Doran KS, Nizet V. 2004. Molecular pathogenesis of neonatal group B streptococcal infection: no longer in its infancy. Mol Microbiol 54:23–31. doi: 10.1111/j.1365-2958.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- 3.Beier D, Gross R. 2006. Regulation of bacterial virulence by two-component systems. Curr Opin Microbiol 9:143–152. doi: 10.1016/j.mib.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Bassler BL, Losick R. 2006. Bacterially speaking. Cell 125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Faralla C, Metruccio MM, De Chiara M, Mu R, Patras KA, Muzzi A, Grandi G, Margarit I, Doran KS, Janulczyk R. 2014. Analysis of two-component systems in group B Streptococcus shows that RgfAC and the novel FspSR modulate virulence and bacterial fitness. mBio 5:e00870-14. doi: 10.1128/mBio.00870-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glaser P, Rusniok C, Buchrieser C, Chevalier F, Frangeul L, Msadek T, Zouine M, Couve E, Lalioui L, Poyart C, Trieu-Cuot P, Kunst F. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol Microbiol 45:1499–1513. doi: 10.1046/j.1365-2958.2002.03126.x. [DOI] [PubMed] [Google Scholar]
- 7.Tettelin H, Masignani V, Cieslewicz MJ, Eisen JA, Peterson S, Wessels MR, Paulsen IT, Nelson KE, Margarit I, Read TD, Madoff LC, Wolf AM, Beanan MJ, Brinkac LM, Daugherty SC, DeBoy RT, Durkin AS, Kolonay JF, Madupu R, Lewis MR, Radune D, Fedorova NB, Scanlan D, Khouri H, Mulligan S, Carty HA, Cline RT, Van Aken SE, Gill J, Scarselli M, Mora M, Iacobini ET, Brettoni C, Galli G, Mariani M, Vegni F, Maione D, Rinaudo D, Rappuoli R, Telford JL, Kasper DL, Grandi G, Fraser CM. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc Natl Acad Sci U S A 99:12391–12396. doi: 10.1073/pnas.182380799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quach D, van Sorge NM, Kristian SA, Bryan JD, Shelver DW, Doran KS. 2009. The CiaR response regulator in group B Streptococcus promotes intracellular survival and resistance to innate immune defenses. J Bacteriol 191:2023–2032. doi: 10.1128/JB.01216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klinzing DC, Ishmael N, Dunning Hotopp JC, Tettelin H, Shields KR, Madoff LC, Puopolo KM. 2013. The two-component response regulator LiaR regulates cell wall stress responses, pili expression and virulence in group B Streptococcus. Microbiology 159:1521–1534. doi: 10.1099/mic.0.064444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landwehr-Kenzel S, Henneke P. 2014. Interaction of Streptococcus agalactiae and cellular innate immunity in colonization and disease. Front Immunol 5:519. doi: 10.3389/fimmu.2014.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lembo A, Gurney MA, Burnside K, Banerjee A, de los Reyes M, Connelly JE, Lin WJ, Jewell KA, Vo A, Renken CW, Doran KS, Rajagopal L. 2010. Regulation of CovR expression in group B Streptococcus impacts blood-brain barrier penetration. Mol Microbiol 77:431–443. doi: 10.1111/j.1365-2958.2010.07215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang SM, Cieslewicz MJ, Kasper DL, Wessels MR. 2005. Regulation of virulence by a two-component system in group B streptococcus. J Bacteriol 187:1105–1113. doi: 10.1128/JB.187.3.1105-1113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patras KA, Wang NY, Fletcher EM, Cavaco CK, Jimenez A, Garg M, Fierer J, Sheen TR, Rajagopal L, Doran KS. 2013. Group B Streptococcus CovR regulation modulates host immune signalling pathways to promote vaginal colonization. Cell Microbiol 15:1154–1167. doi: 10.1111/cmi.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev 29:231–262. doi: 10.1016/j.fmrre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Galperin MY. 2008. Telling bacteria: do not LytTR. Structure 16:657–659. doi: 10.1016/j.str.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behr S, Heermann R, Jung K. 2016. Insights into the DNA-binding mechanism of a LytTR-type transcription regulator. Biosci Rep 36:e00326. doi: 10.1042/BSR20160069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikolskaya AN, Galperin MY. 2002. A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res 30:2453–2459. doi: 10.1093/nar/30.11.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al Safadi R, Mereghetti L, Salloum M, Lartigue MF, Virlogeux-Payant I, Quentin R, Rosenau A. 2011. Two-component system RgfA/C activates the fbsB gene encoding major fibrinogen-binding protein in highly virulent CC17 clone group B Streptococcus. PLoS One 6:e14658. doi: 10.1371/journal.pone.0014658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahn SJ, Rice KC, Oleas J, Bayles KW, Burne RA. 2010. The Streptococcus mutans Cid and Lrg systems modulate virulence traits in response to multiple environmental signals. Microbiology 156:3136–3147. doi: 10.1099/mic.0.039586-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahn SJ, Qu MD, Roberts E, Burne RA, Rice KC. 2012. Identification of the Streptococcus mutans LytST two-component regulon reveals its contribution to oxidative stress tolerance. BMC Microbiol 12:187. doi: 10.1186/1471-2180-12-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunskill EW, Bayles KW. 1996. Identification of LytSR-regulated genes from Staphylococcus aureus. J Bacteriol 178:5810–5812. doi: 10.1128/jb.178.19.5810-5812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brunskill EW, Bayles KW. 1996. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J Bacteriol 178:611–618. doi: 10.1128/jb.178.3.611-618.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brunskill EW, de Jonge BL, Bayles KW. 1997. The Staphylococcus aureus scdA gene: a novel locus that affects cell division and morphogenesis. Microbiology 143(Pt 9):2877–2882. doi: 10.1099/00221287-143-9-2877. [DOI] [PubMed] [Google Scholar]
- 24.Sharma-Kuinkel BK, Mann EE, Ahn JS, Kuechenmeister LJ, Dunman PM, Bayles KW. 2009. The Staphylococcus aureus LytSR two-component regulatory system affects biofilm formation. J Bacteriol 191:4767–4775. doi: 10.1128/JB.00348-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang SJ, Xiong YQ, Yeaman MR, Bayles KW, Abdelhady W, Bayer AS. 2013. Role of the LytSR two-component regulatory system in adaptation to cationic antimicrobial peptides in Staphylococcus aureus. Antimicrob Agents Chemother 57:3875–3882. doi: 10.1128/AAC.00412-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies HD, Jones N, Whittam TS, Elsayed S, Bisharat N, Baker CJ. 2004. Multilocus sequence typing of serotype III group B streptococcus and correlation with pathogenic potential. J Infect Dis 189:1097–1102. doi: 10.1086/382087. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi S, Adderson EE, Nagano Y, Nagano N, Briesacher MR, Bohnsack JF. 1998. Identification of a highly encapsulated, genetically related group of invasive type III group B streptococci. J Infect Dis 177:1116–1119. doi: 10.1086/517408. [DOI] [PubMed] [Google Scholar]
- 28.Doran KS, Chang JC, Benoit VM, Eckmann L, Nizet V. 2002. Group B streptococcal beta-hemolysin/cytolysin promotes invasion of human lung epithelial cells and the release of interleukin-8. J Infect Dis 185:196–203. doi: 10.1086/338475. [DOI] [PubMed] [Google Scholar]
- 29.Doran KS, Engelson EJ, Khosravi A, Maisey HC, Fedtke I, Equils O, Michelsen KS, Arditi M, Peschel A, Nizet V. 2005. Blood-brain barrier invasion by group B Streptococcus depends upon proper cell-surface anchoring of lipoteichoic acid. J Clin Invest 115:2499–2507. doi: 10.1172/JCI23829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banerjee A, Kim BJ, Carmona EM, Cutting AS, Gurney MA, Carlos C, Feuer R, Prasadarao NV, Doran KS. 2011. Bacterial pili exploit integrin machinery to promote immune activation and efficient blood-brain barrier penetration. Nat Commun 2:462. doi: 10.1038/ncomms1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chi F, Wang L, Zheng X, Wu CH, Jong A, Sheard MA, Shi W, Huang SH. 2011. Meningitic Escherichia coli K1 penetration and neutrophil transmigration across the blood-brain barrier are modulated by alpha7 nicotinic receptor. PLoS One 6:e25016. doi: 10.1371/journal.pone.0025016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doran KS, Liu GY, Nizet V. 2003. Group B streptococcal beta-hemolysin/cytolysin activates neutrophil signaling pathways in brain endothelium and contributes to development of meningitis. J Clin Invest 112:736–744. doi: 10.1172/JCI200317335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patras KA, Doran KS. 16 November 2016. A murine model of group B streptococcus vaginal colonization. J Vis Exp. doi: 10.3791/54708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Natale DA, Galperin MY, Tatusov RL, Koonin EV. 2000. Using the COG database to improve gene recognition in complete genomes. Genetica 108:9–17. doi: 10.1023/A:1004031323748. [DOI] [PubMed] [Google Scholar]
- 35.Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shelver D, Rajagopal L, Harris TO, Rubens CE. 2003. MtaR, a regulator of methionine transport, is critical for survival of group B streptococcus in vivo. J Bacteriol 185:6592–6599. doi: 10.1128/JB.185.22.6592-6599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bryan JD, Liles R, Cvek U, Trutschl M, Shelver D. 2008. Global transcriptional profiling reveals Streptococcus agalactiae genes controlled by the MtaR transcription factor. BMC Genomics 9:607. doi: 10.1186/1471-2164-9-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graham MR, Smoot LM, Migliaccio CA, Virtaneva K, Sturdevant DE, Porcella SF, Federle MJ, Adams GJ, Scott JR, Musser JM. 2002. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc Natl Acad Sci U S A 99:13855–13860. doi: 10.1073/pnas.202353699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Throup JP, Koretke KK, Bryant AP, Ingraham KA, Chalker AF, Ge Y, Marra A, Wallis NG, Brown JR, Holmes DJ, Rosenberg M, Burnham MK. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol Microbiol 35:566–576. doi: 10.1046/j.1365-2958.2000.01725.x. [DOI] [PubMed] [Google Scholar]
- 40.Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A 99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim KS. 2003. Pathogenesis of bacterial meningitis: from bacteraemia to neuronal injury. Nat Rev Neurosci 4:376–385. doi: 10.1038/nrn1103. [DOI] [PubMed] [Google Scholar]
- 42.Banerjee A, Van Sorge NM, Sheen TR, Uchiyama S, Mitchell TJ, Doran KS. 2010. Activation of brain endothelium by pneumococcal neuraminidase NanA promotes bacterial internalization. Cell Microbiol 12:1576–1588. doi: 10.1111/j.1462-5822.2010.01490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brouwer MC, McIntyre P, Prasad K, van de Beek D. 2015. Corticosteroids for acute bacterial meningitis. Cochrane Database Syst Rev 2015:CD004405 doi: 10.1002/14651858.CD004405.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang YC, Olson J, Beasley FC, Tung C, Zhang J, Crocker PR, Varki A, Nizet V. 2014. Group B Streptococcus engages an inhibitory Siglec through sialic acid mimicry to blunt innate immune and inflammatory responses in vivo. PLoS Pathog 10:e1003846. doi: 10.1371/journal.ppat.1003846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheen TR, Jimenez A, Wang NY, Banerjee A, van Sorge NM, Doran KS. 2011. Serine-rich repeat proteins and pili promote Streptococcus agalactiae colonization of the vaginal tract. J Bacteriol 193:6834–6842. doi: 10.1128/JB.00094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patras KA, Rosler B, Thoman ML, Doran KS. 2015. Characterization of host immunity during persistent vaginal colonization by group B Streptococcus. Mucosal Immunol 8:1339–1348. doi: 10.1038/mi.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carey AJ, Tan CK, Mirza S, Irving-Rodgers H, Webb RI, Lam A, Ulett GC. 2014. Infection and cellular defense dynamics in a novel 17beta-estradiol murine model of chronic human group B streptococcus genital tract colonization reveal a role for hemolysin in persistence and neutrophil accumulation. J Immunol 192:1718–1731. doi: 10.4049/jimmunol.1202811. [DOI] [PubMed] [Google Scholar]
- 48.Baida GE, Kuzmin NP. 1996. Mechanism of action of hemolysin III from Bacillus cereus. Biochim Biophys Acta 1284:122–124. doi: 10.1016/S0005-2736(96)00168-X. [DOI] [PubMed] [Google Scholar]
- 49.Chen YC, Chang MC, Chuang YC, Jeang CL. 2004. Characterization and virulence of hemolysin III from Vibrio vulnificus. Curr Microbiol 49:175–179. doi: 10.1007/s00284-004-4288-5. [DOI] [PubMed] [Google Scholar]
- 50.Pei J, Millay DP, Olson EN, Grishin NV. 2011. CREST—a large and diverse superfamily of putative transmembrane hydrolases. Biol Direct 6:37. doi: 10.1186/1745-6150-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chaffin DO, Beres SB, Yim HH, Rubens CE. 2000. The serotype of type Ia and III group B streptococci is determined by the polymerase gene within the polycistronic capsule operon. J Bacteriol 182:4466–4477. doi: 10.1128/JB.182.16.4466-4477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park IH, Geno KA, Yu J, Oliver MB, Kim KH, Nahm MH. 2015. Genetic, biochemical, and serological characterization of a new pneumococcal serotype, 6H, and generation of a pneumococcal strain producing three different capsular repeat units. Clin Vaccine Immunol 22:313–318. doi: 10.1128/CVI.00647-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weksler B, Romero IA, Couraud PO. 2013. The hCMEC/D3 cell line as a model of the human blood brain barrier. Fluids Barriers CNS 10:16. doi: 10.1186/2045-8118-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weksler BB, Subileau EA, Perriere N, Charneau P, Holloway K, Leveque M, Tricoire-Leignel H, Nicotra A, Bourdoulous S, Turowski P, Male DK, Roux F, Greenwood J, Romero IA, Couraud PO. 2005. Blood-brain barrier-specific properties of a human adult brain endothelial cell line. FASEB J 19:1872–1874. doi: 10.1096/fj.04-3458fje. [DOI] [PubMed] [Google Scholar]
- 55.Llombart V, Garcia-Berrocoso T, Bech-Serra JJ, Simats A, Bustamante A, Giralt D, Reverter-Branchat G, Canals F, Hernandez-Guillamon M, Montaner J. 2016. Characterization of secretomes from a human blood brain barrier endothelial cells in-vitro model after ischemia by stable isotope labeling with aminoacids in cell culture (SILAC). J Proteomics 133:100–112. doi: 10.1016/j.jprot.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 56.Ohtsuki S, Ikeda C, Uchida Y, Sakamoto Y, Miller F, Glacial F, Decleves X, Scherrmann JM, Couraud PO, Kubo Y, Tachikawa M, Terasaki T. 2013. Quantitative targeted absolute proteomic analysis of transporters, receptors and junction proteins for validation of human cerebral microvascular endothelial cell line hCMEC/D3 as a human blood-brain barrier model. Mol Pharm 10:289–296. doi: 10.1021/mp3004308. [DOI] [PubMed] [Google Scholar]
- 57.van Sorge NM, Ebrahimi CM, McGillivray SM, Quach D, Sabet M, Guiney DG, Doran KS. 2008. Anthrax toxins inhibit neutrophil signaling pathways in brain endothelium and contribute to the pathogenesis of meningitis. PLoS One 3:e2964. doi: 10.1371/journal.pone.0002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shishkin AA, Giannoukos G, Kucukural A, Ciulla D, Busby M, Surka C, Chen J, Bhattacharyya RP, Rudy RF, Patel MM, Novod N, Hung DT, Gnirke A, Garber M, Guttman M, Livny J. 2015. Simultaneous generation of many RNA-seq libraries in a single reaction. Nat Methods 12:323–325. doi: 10.1038/nmeth.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu YY, Machleder EM, Chenchik A, Li R, Siebert PD. 2001. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques 30:892–897. [DOI] [PubMed] [Google Scholar]
- 60.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abeel T, Van Parys T, Saeys Y, Galagan J, Van de Peer Y. 2012. GenomeView: a next-generation genome browser. Nucleic Acids Res 40:e12. doi: 10.1093/nar/gkr995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.