ABSTRACT
CD4 T cells and antibody are required for optimal acquired immunity to Chlamydia muridarum genital tract infection, and T cell-mediated gamma interferon (IFN-γ) production is necessary to clear infection in the absence of humoral immunity. However, the role of T cell-independent immune responses during primary infection remains unclear. We investigated this question by inoculating wild-type and immune-deficient mice with C. muridarum CM001, a clonal isolate capable of enhanced extragenital replication. Genital inoculation of wild-type mice resulted in transient dissemination to the lungs and spleen that then was rapidly cleared from these organs. However, CM001 genital infection proved lethal for STAT1−/− and IFNG−/− mice, in which IFN-γ signaling was absent, and for Rag1−/− mice, which lacked T and B cells and in which innate IFN-γ signaling was retained. In contrast, B cell-deficient muMT mice, which can generate a Th1 response, and T cell-deficient mice with intact B cell and innate IFN-γ signaling survived. These data collectively indicate that IFN-γ prevents lethal CM001 dissemination in the absence of T cells and suggests a B cell corequirement. Adoptive transfer of convalescent-phase immune serum but not naive IgM to Rag1−/− mice infected with CM001 significantly increased the survival time, while transfer of naive B cells completely rescued Rag1−/− mice from CM001 lethality. Protection was associated with a significant reduction in the lung chlamydial burden of genitally infected mice. These data reveal an important cooperation between T cell-independent B cell responses and innate IFN-γ in chlamydial host defense and suggest that interactions between T cell-independent antibody and IFN-γ are essential for limiting extragenital dissemination.
KEYWORDS: B cell responses, Chlamydia, interferons
INTRODUCTION
Chlamydia trachomatis is the most prevalent sexually transmitted bacterial infection and a significant cause of female reproductive tract morbidity. The development of a vaccine remains a top global health priority (1). Preclinical C. trachomatis vaccine development includes utilization of the murine genital infection model for determination of protective immune responses against Chlamydia muridarum. Genital infection of mice with specific immune deficiencies has provided a method for determining the protective contribution afforded by humoral and cell-mediated immunity.
Previous studies revealed a central role for CD4 T cells in protection against primary intravaginal C. muridarum infection, and wild-type mice demonstrate clearance comparable to that in B cell-deficient mice (2). Multiple experiments have demonstrated a protective role for Th1 gamma interferon (IFN-γ) production in primary and acquired immunity against chlamydial infection (3–7). Furthermore, recent evidence suggests that antibody and CD4 T cell-derived IFN-γ optimally cooperate to protect against infection through neutrophil activation and subsequent chlamydial killing (8, 9). Thus, the requirement for Th1 cells and T cell-dependent antibody during protective adaptive responses is well accepted (10).
Earlier studies demonstrated that T cell-deficient athymic nude mice and severe combined immune-deficient (SCID) mice, which lack functional T and B lymphocytes because of impaired VDJ rearrangement, uniformly fail to resolve genital infection with the C. muridarum Weiss and Nigg strains, respectively (11, 12). SCID mice demonstrate high levels of dissemination, while IFN-γ-deficient mice exhibit enhanced chlamydial dissemination and a portion fail to resolve the genital tract infection (11). Infection of B cell-deficient mice results in a transient disseminated infection that is likely cleared through enhanced systemic CD4 T cell responses (13). Furthermore, T and B cell-deficient Rag1−/− mice that fail to express functional Rag1 proteins, required for somatic recombination, develop a lethal systemic infection after intravaginal infection with C. muridarum Weiss (14). Together, these data suggest an important, less characterized T cell-independent IFN-γ and B cell corequirement for protection against a primary disseminated infection.
We recently identified a clonal isolate (CM001) from a C. muridarum Nigg stock (6, 15) that was capable of enhanced extragenital replication compared to the parental stock. CM001 allowed us to explore the mechanisms of protection against dissemination during primary intravaginal infection of immune-deficient mice. Wild-type, B cell-deficient, and T cell-deficient mice survived the CM001 infection. However, mice lacking IFN-γ signaling and Rag1−/− mice died. Adoptive transfer of convalescent-phase immune serum or naive B cells protected Rag1−/− mice from CM001 lethality. B cell adoptive transfer to Rag1−/− hosts reduced the disseminated lung chlamydial burden to the levels found in T cell-deficient mice. These are the first studies to demonstrate a T cell-independent corequirement for B cells and innate IFN-γ for control of extragenital chlamydial infection.
RESULTS
Genetic characterization of C. muridarum clonal isolates.
We performed whole-genome sequencing of C. muridarum strains CM001, CM002, CM003, CM004, CM005, CM006, CM007, CM008, CM009, CM010, CM012, and CM021 with the goal of identifying unique genetic differences that might account for the dissemination properties of the variants. Single molecule real-time (SMRT) sequencing via a PacBio platform yielded high-quality draft genomes, with a mean coverage of between 27- and 114-fold per strain (see Table S5 in the supplemental material). This coverage was not always sufficient to achieve single contigs and to close genomes de novo (16), so we used the published sequence of CM972 (17), a strain derived from CM001 via plasmid curing (18), as a scaffold to facilitate comparisons of all isolates. A draft assembly of the conserved virulence plasmid was generated for each of the isolates, confirming its presence in all sequenced strains. Likely sequence errors (n = 5; Table S5) were identified in the CM972 sequence as single nucleotide calls that diverged from those detected in the parental CM001 strain and all other sequenced isolates, so these were excluded from further evaluation. Overall, we identified variations at a total of 119 loci among the 12 isolates.
Nine single nucleotide polymorphisms (SNPs) were mapped to seven coding sequences and one intragenic region (Table S5). Of these, five were unique to the isolates screened. These included a C-for-A substitution in ompA of CM001 (Fig. S2), predicted to exchange cysteine for glycine in the translated protein. We identified two genetic differences unique to CM001 in the ompA gene (TC_052, Y015_RS00285), which encodes the major outer membrane protein (MOMP), the C-for-A SNP described above, and a 6-bp insertion (AGCTTA). All the remaining isolates were predicted to express an MOMP with a double amino acid deletion and a glycine-to-cysteine substitution adjacent to the conserved portion of the VD4 region (Fig. S3) of the protein (19), changes that have been predicted to alter the MOMP conformation (20). Broadening the comparison of the MOMP sequence of CM001 and the other clonal isolates to C. muridarum sequences available in GenBank confirmed that it is an outlier with respect to the AR-Nigg population because its sequence is identical to the sequences of C. muridarum Weiss (21), C. muridarum Nigg (22), and C. muridarum Nigg3 derivatives (23), while all other strains sequenced expressed an MOMP protein identical to that of C. muridarum Nigg2 (21) and CmVar004 (24) (Fig. S3), clonal isolates recovered independently from the AR-Nigg population (21, 24).
C. muridarum clonal isolates reveal a variant with enhanced dissemination.
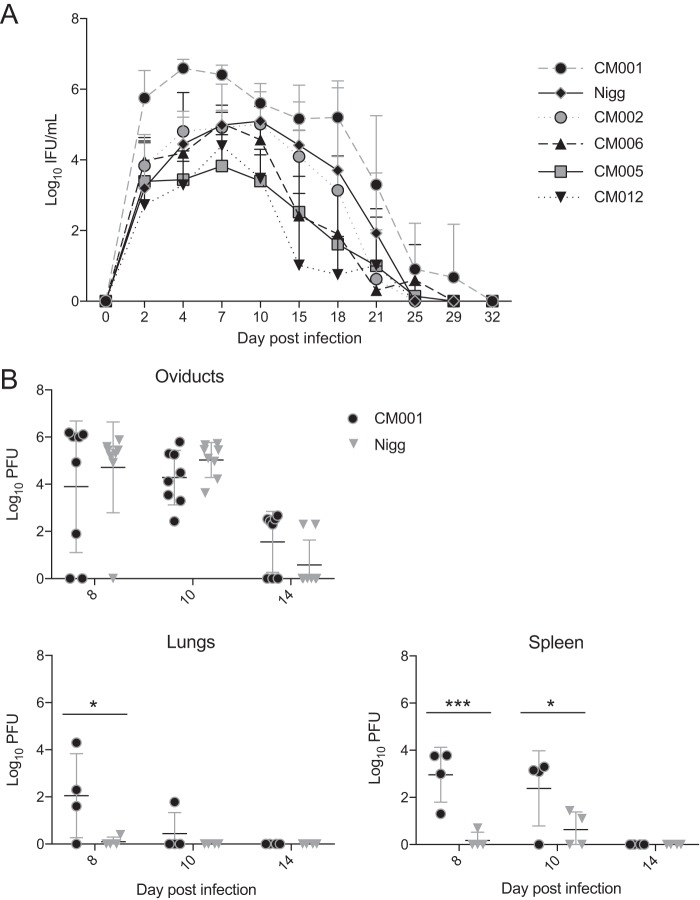
Previous studies revealed that C. muridarum Weiss and Nigg stocks contain variants with genotypic and phenotypic differences (21). We hypothesized that plaque-purified isolates (Fig. S1) derived from such stocks would yield clonal isolates that exhibited differential cervicovaginal burdens during infection. We explored this possibility by inoculating wild-type mice with a panel of plaque-purified isolates derived from a polyclonal population of C. muridarum Nigg (AR-Nigg). Groups of mice were intravaginally inoculated with Nigg or one of five different clones. Clone CM001 demonstrated a significantly increased cervicovaginal burden over the course of infection compared to that for Nigg, while infection with clone CM012 resulted in a reduced burden (Fig. 1A). However, plaque assay, a highly sensitive means to measure infectivity in cell culture (18, 25), did not reveal any differences in the efficiency of plaquing (EOP) (18) between the strains (for CM001, 3.77 × 10−1 ± 1.01 × 10−1; for Nigg, 1.77 ± 1.04 × 10−1; for CM012, 3.46 × 10−1 ± 3.76 × 10−2). We also examined the ability of these clones to ascend to the oviduct and to disseminate to the lungs and spleen. Mice intravaginally infected with CM001 demonstrated a significantly higher chlamydial burden in their lungs on day 8 and in their spleens on days 8 and 10 postinfection than mice intravaginally infected with Nigg (Fig. 1B). Both strains were undetectable in the lungs and spleen on day 14. In contrast, no significant difference in the oviduct burden between mice infected with CM001 and mice infected with Nigg was detected on any day examined. These data indicate that although both Nigg and CM001 ascend to the oviducts of wild-type mice, CM001 exhibits enhanced dissemination to the lungs and spleen during early infection.
FIG 1.
C. muridarum CM001 intravaginal infection disseminates to distal organs in wild-type mice and is rapidly cleared. (A) C57BL/6J mice were intravaginally infected with Nigg or C. muridarum clones, and the course of primary infection was monitored with lower genital tract swab specimens. Significance was determined by two-way RM ANOVA. Data represent the mean + SD for swab specimens from infected mice or swab specimens from infected and uninfected mice (n = 4 mice per group). P values for comparison of the groups over the primary infection course were as follows: P = 0.02 for Nigg versus CM001 and P = 0.05 for Nigg versus CM012. (B) CM001 and Nigg lung, spleen, and oviduct burdens were compared by determination of the numbers of PFU on the indicated days postinfection. Statistical significance, determined by two-way ANOVA, is indicated by asterisks, as follows: *, P < 0.05; ***, P < 0.001.
CM001-mediated lethality and dissemination are plasmid independent.
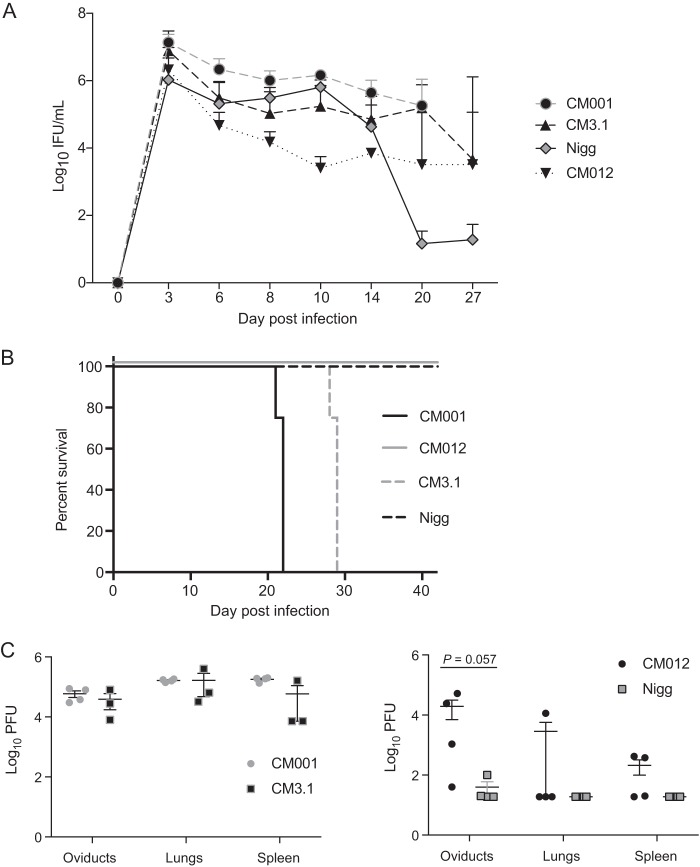
We previously observed that Rag1−/− mice intravaginally infected with plaque-purified C. muridarum (now designated CM001) succumbed to infection (15). Since C. muridarum virulence is linked to the presence of its plasmid (17, 26), we investigated if lethality and dissemination to the lungs and spleen were plasmid dependent. Rag1−/− mice infected with CM001 or plasmid-cured CM3.1 displayed a significantly increased bacterial burden compared to the Nigg-infected controls (Fig. 2A), but the infection course between the strains was not significantly different. Mice infected with CM3.1 succumbed, as did the mice infected with CM001 (Fig. 2B). Mortality was associated with high chlamydial burdens in the oviducts, lungs, and spleens of CM001- and CM3.1-infected mice (Fig. 2C). Mice infected with CM012 and Nigg demonstrated a low burden in the lungs and spleens at the time of sacrifice on day 40. Mice infected with CM018, a clonal isolate that had spontaneously lost the chlamydial virulence plasmid, also survived their infection (data not shown). These data collectively show that CM001 lethality in Rag1−/− mice is plasmid independent and is associated with a high chlamydial burden in distal organs at the time of death.
FIG 2.
C. muridarum CM001 intravaginal infection in Rag1−/− mice is associated with fatal dissemination to distal organs. (A) Rag1−/− mice were intravaginally infected with Nigg, CM001, CM012, or plasmid-deficient CM3.1, and the course of primary infection was monitored with lower genital tract swab specimens. Significance was determined by two-way RM ANOVA. Data represent the mean + SD for 4 mice per group. P values for comparison of the groups over the primary infection course were as follows: P = 0.01 for Nigg versus CM001, P < 0.001 for Nigg versus CM3.1, P = 0.01 for CM012 versus CM001, P = 0.001 for CM012 versus CM3.1, and P = not significant for the remaining group comparisons. (B) Mice were euthanized after reaching a body condition score of 2−, and an exact log-rank test was used to analyze survival differences between the Nigg- and CM001- or CM3.1-infected groups (P < 0.01). (C) Chlamydial loads in the lungs, spleen, and both oviducts were determined by plaque assay (by determination of the number of PFU) at the time of euthanasia (left) or on day 40 (right). Statistical significance was determined by the Mann-Whitney U test.
IFN-γ signaling is required for protection against CM001 lethality.
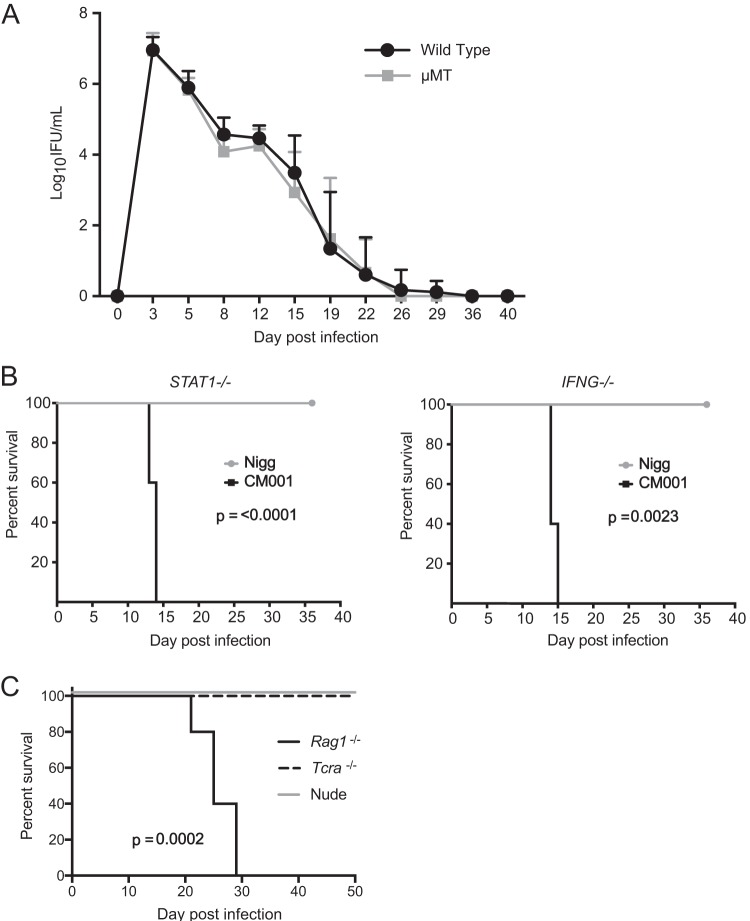
B cell-deficient mice clear disseminated infection with C. muridarum Nigg (13), but IFNG−/− mice demonstrate extragenital dissemination, and a portion of the mice succumb to infection with strain Weiss (11). We next investigated if CM001 infection of these immune-deficient mice would yield similar results. B cell-deficient mice survived infection with CM001 and demonstrated a course of infection clearance similar to that demonstrated by wild-type mice (Fig. 3A). However, CM001 infection of IFNG−/− and STAT1−/− mice, deficient in IFN-γ signaling, proved lethal, while Nigg-infected mice survived (Fig. 3B). Based on these results, we extended our analysis to determine if Th1 cells were dispensable for protection against CM001. We had previously noted that Tcra−/− mice do not succumb to CM001 infection (6) and nude mice develop chronic chlamydial genital infection with strain Nigg (12), so we were curious to discover if they would succumb to CM001 intravaginal infection like Rag1−/− mice. Nude mice survived the intravaginal CM001 infection (Fig. 3C), indicating that IFN-γ prevents lethal CM001 infection in the absence of T cells and suggests a B cell corequirement for the control of dissemination.
FIG 3.
Genetic ablation of IFN-γ signaling results in CM001-mediated lethality. (A) C57BL/6J (n = 24) and muMt (n = 14) deficient mice were intravaginally infected with CM001, and the course of primary infection was monitored with lower genital tract swab specimens. Data represent the mean + SD. Significance was determined by two-way RM ANOVA (P was not significant). (B) STAT1−/− (Nigg group, n = 10; CM001 group, n = 5) and IFNG−/− (5 mice per group) mice were intravaginally infected with Nigg or CM001. Animal welfare was monitored daily, and an exact log-rank test was used to analyze survival differences between Nigg- and CM001-infected STAT1−/− (P < 0.0001) and IFNG−/− (P = 0.0023) mice. (C) Nude, Rag1−/−, and Tcra−/− mice were infected with CM001 and monitored daily (P = 0.0002 for nude mice versus both Rag1−/− and Tcra−/− mice).
B cells cooperate with IFN-γ to protect against CM001 lethality in the absence of T cells.
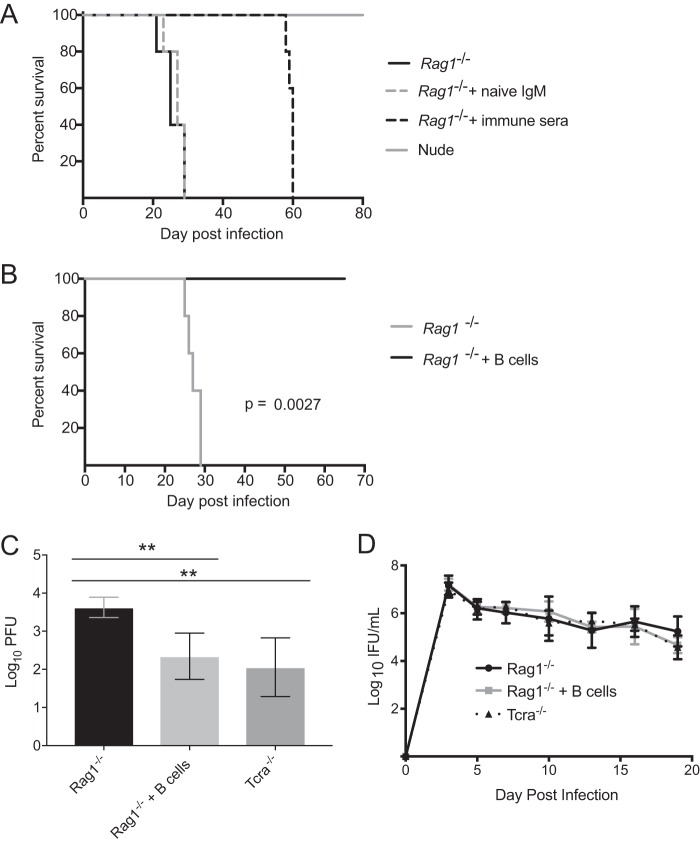
A recent study revealed that convalescent-phase immunoglobulin and IFN-γ are required for neutrophil activation and chlamydial clearance (9). Based on this finding, we hypothesized that adoptive transfer of wild-type mouse convalescent-phase immune serum, consisting of T cell-dependent antibody, to Rag1−/− mice would delay the lethal response. Adoptive transfer of immune serum significantly increased the survival time compared to that for mice receiving naive IgM or the untreated controls (Fig. 4A). We next investigated if Rag1−/− mice could be completely rescued in the absence of T cells or T cell-dependent antibody. Adoptive transfer of B cells derived from the spleens of naive T cell-deficient mice rescued Rag1−/− mice from an otherwise lethal CM001 infection (Fig. 4B). T cell-deficient mice and B cell-reconstituted Rag1−/− mice demonstrated a respective 1.2- and 1.5-log reduction in the lung chlamydial burden on day 19 postinfection compared to that in the Rag1−/− controls (Fig. 4C). The chlamydial burden in the genital tract was not significantly different between groups over the course of primary infection (Fig. 4D). These data collectively demonstrate the ability of B cells to mediate protection against extragenital chlamydiae in the absence of T cell helper and effector functions.
FIG 4.
B cells and IFN-γ cooperate to prevent CM001 lethality independently of T cells. (A) Nude mice and Rag1−/− mice receiving an intravenous transfer of naive mouse IgM, convalescent-phase immune serum, or PBS (n = 5 per group) were infected with CM001 and monitored daily. An exact log-rank test was used to analyze the survival differences between nude mice (P < 0.0001) and Rag1−/− mice (P = 0.0026) receiving immune serum. (B) Rag1−/− mice were mock treated or intravenously injected with 3 × 105 B cells (n = 5 per group) from Tcra−/− mice prior to CM001 infection and were monitored daily (P = 0.0027). (C) Tcra−/− (n = 3), Rag1−/− (n = 5), and B cell-reconstituted Rag1−/− (n = 5) mice were infected with CM001 and sacrificed on day 19 postinfection. The chlamydial load in the lungs was determined by plaque assay (determination of the number of PFU). Statistical significance, determined by one-way ANOVA, is indicated by asterisks, as follows: **, P < 0.01. (D) The course of primary infection was monitored with lower genital tract swab specimens (P was not significant). Significance was determined by two-way RM ANOVA.
DISCUSSION
This study highlights the previously underappreciated contribution of IFN-γ and B cells to the control of extragenital chlamydial dissemination, particularly during early infection, after murine intravaginal inoculation. We did not detect differences in the extent of extragenital infection between wild-type mice infected with CM001 and wild-type mice infected with the other isolates when studied after day 10, coincident with the development of a Chlamydia-specific T cell response (6, 13). Cell culture assays of replication or infectivity failed to reveal differences between CM001 and its siblings that might account for these findings. However, when we compared the consequences of infection with CM001 in immune-deficient mice to those of infection with the other isolates, we observed high chlamydial burdens in the lungs and spleens of moribund Rag1−/− mice that were euthanized after intravaginal infection. These findings reveal that immunologic control of extragenital chlamydiae after intravaginal inoculation is an important component of host defense by preventing pneumonia and weight loss, which are ultimately fatal (11, 14).
This lethal phenotype allowed us to identify determinants necessary to prevent fatal chlamydial dissemination. CM001 intravaginal infection of mice with defective IFN-γ signaling resulted in universal lethality at between 10 and 14 days postinfection. These observations are consistent with the findings of previous studies demonstrating a protective role for IFN-γ in the early control of dissemination (11, 27, 28). However, Rag1−/− mice, lacking both T and B cells, succumbed to CM001 infection after 20 days postinfection, despite the presence of IFN-γ (Table 1). This result suggests a potential cooperation between B cells and IFN-γ in prevention of chlamydial dissemination. Indeed, Rag1−/− mice were transiently rescued by the adoptive transfer of convalescent-phase immune serum from wild-type mice. This result demonstrates that antibody generated in the presence of T cell help can afford protection against disseminated chlamydial infection, in the absence of an effector Th1 response. Furthermore, transfer of naive B cells taken from T cell-deficient mice completely rescued Rag1−/− mice from lethality. This protection was associated with a decreased chlamydial burden in the lungs of genitally infected mice and suggests a protective role for T cell-independent antibody. These experiments reveal a previously undescribed T cell-independent role for B cells in concert with IFN-γ for protection against C. muridarum disseminated disease.
TABLE 1.
Outcome of CM001 infection and immune status of mouse strainsa
| Mouse genotype | Mortality | Clearance | IFN-γ signaling | Antibody | αβ T cells |
|---|---|---|---|---|---|
| Wild type | No | Yes | Yes | Yes | Yes |
| Rag1−/− | Yes | No | Yes | No | No |
| STAT1−/− | Yes | No | No | Yes | Yes |
| IFNG−/− | Yes | No | No | Yes | Yes |
| muMT | No | Yes | Yes | No | Yes |
| Tcra−/− | No | No | Yes | Yes | No |
| Rag1−/− + B cells | No | No | Yes | Yes | No |
The survivability of infection, clearance of infection, and immune profiles were compared in wild-type and gene-deficient C57BL/6J mice.
Genomic sequencing of the clonal isolates derived from the parental Nigg population revealed very little diversity among the strains. Nevertheless, we noted variations in the lengths of homopolymeric G and C tracts situated close to the start codon of three genes, TC_0436, TC_0440, and TC_0447, indicating the potential for slip-strand mispairing (29) during DNA replication to throw expression in or out of phase (see Fig. S2 in the supplemental material). Interestingly, these proteins comprise part of a conserved, eight-member, paralogous family of phosphatidylcholine-hydrolyzing phospholipase D (PLD) proteins (30, 31) containing NucT motifs (32), indicative of a PLD subfamily with nuclease activity. The extracellular, outer membrane-associated NucT nuclease of Helicobacter pylori enables it to use extracellular DNA as a purine source (33). Although the role of these proteins in C. muridarum is unknown, their high sequence homology suggests that they have a similar function and may be subject to antigenic variation, with expression altering in response to selective pressures. All of the strains investigated were predicted to express at least one of the three proteins, and we did not detect genetic variation in the remaining members of the family.
We were particularly interested in identifying genetic variation unique to CM001 because it appeared to be more virulent than the other isolates in both wild-type and immune-deficient mice. We have demonstrated the importance of the chlamydial virulence plasmid to infectivity (18, 25, 34) and in eliciting damaging innate inflammation (26), but the presence or absence of the plasmid did not influence the ability of the C. muridarum isolates to kill the immune-deficient mice examined in this study. CM3.1, a plasmid-deficient derivative of CM001, still caused lethal infection of Rag1−/− mice, while CM018, a spontaneous plasmid-deficient isolate from the Nigg population, did not. CM3.1 resembled its parent by also killing IFNG−/− and STAT1−/− mice. Our analysis revealed two sequence differences in ompA, a gene encoding MOMP, the trimeric beta-barrel porin that comprises ∼60% of the outer membrane protein component (20). The ompA sequence present in CM001 was conserved in CM972 and CM3.1 (data not shown) but not in any other isolates investigated. These changes mapped to the conserved region of VD4 (Fig. S4), a portion of MOMP that has proven difficult to resolve structurally and that differentiates it from other beta-barrel bacterial porins (20). It has been proposed that this stretch of the protein forms a loop that is important for eukaryotic cell attachment (35). Our cell culture-based infectivity assays did not reveal any differences in infectivity between CM001 and other plasmid-containing C. muridarum isolates. However, differences in infectious burden were observed, potentially illustrating the impact of the VD4 conformation on chlamydial attachment, replication, and dissemination in vivo. Our comparison of the C. muridarum sequences available in GenBank indicated that many independent laboratory stocks express MOMPs identical to the MOMP expressed by CM001, suggesting that this represents the wild-type conformation. In the case of the parental Nigg stock, prolonged cell culture (21, 36) in the absence of selective immune pressure appears to have supported the outgrowth of variants, predominantly those represented by CM002 and the remaining isolates that we characterized, but also isolates with other ompA mutations (24). Yeruva et al. recently reported their isolation of a virulent clone from the Nigg stock that they determined was virulent based on its ability to kill Rag1−/− mice (36). It would be interesting to determine if the ompA in this clone is identical to that in CM001. These observations may also help reconcile the irregular chlamydial lethality reported in immune-deficient mouse models, as these differences are likely due to genetic variation between laboratory stocks.
Our analyses were limited to delineating general requisites for protection against disseminated extragenital infection. While IFN-γ and B cells were shown to play a pivotal role in protection, the cellular source of IFN-γ and the mechanism of B cell protection were not investigated. However, previous experiments have demonstrated that NK cells are a critical source of IFN-γ in the absence of T cells (37), and B cell protection is likely dependent on their production of antibody, based on the observation that antibody-deficient (AID−/− μS−/−) B cells are unable to control dissemination from the genital tract (38). A role for antibody is further suggested by the transient protection of Rag1−/− mice from CM001 lethality by administration of convalescent-phase serum. However, we cannot rule out the possibility of antibody-independent mechanisms of protection, such as cytokine production (39). Recent investigation suggests that IgG and IFN-γ are necessary for neutrophil activation prior to chlamydial killing (9), but only T cell-dependent antibody production was investigated. Lastly, we did not determine the mechanism for dissemination; however, previous data suggest that C. muridarum may directly ascend from the genital tract into the peritoneal cavity (11). However, the possibility of dissemination through the lymphatics and blood cannot be excluded, and future investigation of this process may help further define the role of B cells and IFN-γ in host defense.
Significant T cell-independent immunity has been described for Ehrlichia muris (40), Salmonella (41), and Streptococcus pneumoniae (42). Our analyses indicate that T cell-independent B cell responses contribute to protection against lethal extragenital chlamydial infection. Interestingly, spleen-derived B cells adopt a predominately marginal zone phenotype in vivo after adoptive transfer to lymphopenic hosts (43), and neutrophil-derived cytokines can directly stimulate marginal zone B cells for T cell-independent IgG (44). T cell-independent IgG could activate neutrophils to phagocytose bacteria (42, 45), which would be enhanced in the presence of IFN-γ (9, 46). However, we cannot rule out an alternative model where antibodies limit dissemination and, independently, IFN-γ provides cell-autonomous immunity in lung epithelial cells and/or alveolar macrophages (47). Detectable, low levels of IgG have been observed in nude mice intravaginally infected with C. muridarum (12), and T cell-independent IgG can provide robust protection against viral (48) and bacterial infections (42). It is tempting to speculate that MOMP-reactive IgG is responsible for controlling chlamydial dissemination, since bacterial porins have been shown to constitute a rare group of protein antigens capable of inducing protective T cell-independent antibodies (41). If the murine model of extragenital dissemination and immunological control reflects human C. trachomatis infection, then this T cell-independent protection may help prevent the development of reactive arthritis and Fitz-Hugh-Curtis syndrome.
In conclusion, we have demonstrated that IFN-γ and B cells cooperate to provide protection against a lethal disseminated chlamydial infection independently of T cells. Examination of T cell-independent responses may further reveal detailed mechanisms of extragenital host defense against Chlamydia. Increased understanding of T cell-independent mechanisms of antichlamydial host defense may also inform methods to augment the T cell responses generated with targeted vaccines.
MATERIALS AND METHODS
Strains, cell lines, and culture conditions.
The Chlamydia muridarum Nigg stock (AR-Nigg) was obtained from Roger Rank at the University of Arkansas for Medical Sciences (21). A group of clonal variants was derived from this population via plaque assay (18, 49). Individual plaques were harvested, diluted, and repurified via plaques before being amplified to high-titer stocks for subsequent experiments. A total of 24 isolates were recovered, and a subset (strains CM002, CM005, CM006, CM007, CM008, CM009, CM010, CM012, CM018, and CM021) was selected for further analysis. Also included in the study were strains CM001, the parent of the avirulent, plasmid-cured strain CM972 (18, 26), and CM3.1, a derivative of CM972 with normal infectivity (26). Plaque-purified C. muridarum strains were propagated in Mycoplasma-free L929 cells (50) and titrated as inclusion-forming units (IFU) (51), using a fluorescently tagged antichlamydial lipopolysaccharide monoclonal antibody (Bio-Rad). Intracellular growth kinetics and infectivity in cell culture were evaluated for the isolates as previously described (18, 25, 26).
Sequencing.
Gradient-purified bacterial preparations (52) were treated with RNase/DNase to degrade the nucleic acid carried over from before isolation of genomic DNA using a MasterPure complete DNA and RNA extraction kit (Epicentre, Madison, WI). Genomic DNA was sheared and processed for single molecule real-time (SMRT) sequencing on a PacBio RSII sequencer (Pacific Biosciences, Menlo Park, CA), as previously described (53). Data were analyzed using the hierarchical genome assembly process (HGAP) method (54), and the assembled contigs were polished using the Quiver program (PacBio). Whole-genome alignments of the clonal isolates and reference sequence were performed using the progressive Mauve algorithm (55), supported in the Geneious program (version 11.0; Biomatters, Auckland, New Zealand). Where indicated, Sanger sequencing of the PCR-amplified products was performed to resolve ambiguities or to confirm predicted sequence polymorphisms.
Animals.
Six- to 8-week-old female C57BL/6J (stock no. 000664), B6.129S2-Ighmtm1Cgn/J (muMT negative; stock no. 002288), B6.129S(Cg)-Stat1tm1Dlv/J (STAT1−/−, stock no. 012606), B6.129S7-Ifngtm1Ts/J (IFNG−/−; stock no. 002287), B6.129S7-Rag1tm1Mom/J (Rag1 knockout; stock no. 002216), B6.129S2-Tcratm1Mom/J (Tcra−/−; stock no. 002116), and B6.Cg-Foxn1nu/J (C57BL/6 nude; stock no. 000819) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Mice were given food and water ad libitum in an environmentally controlled pathogen-free room with a cycle of 12 h of light and 12 h of darkness. All animal experiments were approved by the Institutional Animal Care and Use Committees at the University of Pittsburgh and University of North Carolina.
Murine Chlamydia infection and monitoring.
Female mice at least 8 weeks old were subcutaneously injected with 2.5 mg medroxyprogesterone (Depo-Provera; Upjohn) 5 to 7 days prior to infection to induce a state of anestrous (56). Mice were intravaginally inoculated with 5 × 105 inclusion-forming units (IFU) of Nigg or the C. muridarum clones diluted in 30 μl sucrose-sodium phosphate-glutamic acid buffer. The mice were monitored for cervicovaginal shedding via the collection of endocervical swab specimens (51) over the course of their infection (57). Ascending genital infection and extragenital dissemination were confirmed via quantitation of the bacterial load in homogenized oviduct tissues, spleens, and lungs from sacrificed mice (26). Animal welfare was monitored daily, and immune-deficient mice were euthanized if they achieved a body conditioning score of 2− (58).
B cell isolation and adoptive transfer.
B cells were isolated from the spleens of naive Tcra−/− mice by negative magnetic selection (Miltenyi Biotech), according to the manufacturer's protocol. A sample of isolated cells was analyzed by flow cytometry to confirm 98% CD45+ CD5− CD19+ resting B cell purity using LIVE/DEAD Fixable Yellow (Life Technologies) in combination with the following fluorochrome-labeled antibodies from BD Biosciences: anti-CD45 (clone 30-F11), anti-CD5 (clone 53-7.3), and anti-CD19 (clone 1D3). Stained cells were analyzed on a CyAN ADP analyzer (Beckman Coulter), and the resultant data were analyzed using FlowJo software. At least 3 × 106 spleen-derived B cells were injected intravenously into medroxyprogesterone-treated Rag1−/− mice 5 days prior to intravaginal infection.
C57BL/6J mice were bled at sacrifice following resolution of intravaginal C. muridarum Nigg infection to recover convalescent-phase immune serum (10). Serum was pooled and complement inactivated at 56°C for 30 min. Naive mouse-derived IgM (Rockland Immunochemicals, Inc.) was dialyzed overnight prior to intraperitoneal (i.p.) injection. Naive medroxyprogesterone-treated Rag1−/− mice received i.p. injections of phosphate-buffered saline (PBS), 0.5 ml of immune serum, or 200 μg IgM on days −1, 0, 1, 3, 7, 10, and 13 of infection.
Statistical analysis.
Differences between the means for experimental groups after infection were calculated using the Mann-Whitney U test or a two-way repeated-measures (RM) analysis of variance (ANOVA). Comparisons of animal survival were performed by an exact log rank test. Prism software (GraphPad Software) was utilized for statistical analyses, and values of P of ≤0.05 were considered significant.
Accession number(s).
The genome sequencing data for the strains sequenced in this study are deposited under NCBI BioProject accession number PRJNA435732.
Supplementary Material
ACKNOWLEDGMENTS
We thank John Alcorn for generously donating the STAT1−/− mice. We also thank Wujuan Zhong, Evan Powell, and Garth Ehrlich for technical assistance.
This work was supported by National Institutes of Health-National Institute of Allergy and Infectious Diseases grants R01 A105624 and U19 A1084024 (to T.D.).
We have no financial conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00143-18.
REFERENCES
- 1.Poston TB, Gottlieb SL, Darville T. 19 January 2017. Status of vaccine research and development of vaccines for Chlamydia trachomatis infection. Vaccine. doi: 10.1016/j.vaccine.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 2.Ramsey KH, Soderberg LS, Rank RG. 1988. Resolution of chlamydial genital infection in B-cell-deficient mice and immunity to reinfection. Infect Immun 56:1320–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perry LL, Feilzer K, Caldwell HD. 1997. Immunity to Chlamydia trachomatis is mediated by T helper 1 cells through IFN-gamma-dependent and -independent pathways. J Immunol 158:3344–3352. [PubMed] [Google Scholar]
- 4.Gondek DC, Olive AJ, Stary G, Starnbach MN. 2012. CD4+ T cells are necessary and sufficient to confer protection against Chlamydia trachomatis infection in the murine upper genital tract. J Immunol 189:2441–2449. doi: 10.4049/jimmunol.1103032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morrison SG, Su H, Caldwell HD, Morrison RP. 2000. Immunity to murine Chlamydia trachomatis genital tract reinfection involves B cells and CD4(+) T cells but not CD8(+) T cells. Infect Immun 68:6979–6987. doi: 10.1128/IAI.68.12.6979-6987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poston TB, Qu Y, Girardi J, O'Connell CM, Frazer LC, Russell AN, Wall M, Nagarajan UM, Darville T. 2017. A chlamydia-specific TCR-transgenic mouse demonstrates Th1 polyfunctionality with enhanced effector function. J Immunol 199:2845–2854. doi: 10.4049/jimmunol.1700914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W, Murthy AK, Guentzel MN, Seshu J, Forsthuber TG, Zhong G, Arulanandam BP. 2008. Antigen-specific CD4+ T cells produce sufficient IFN-gamma to mediate robust protective immunity against genital Chlamydia muridarum infection. J Immunol 180:3375–3382. doi: 10.4049/jimmunol.180.5.3375. [DOI] [PubMed] [Google Scholar]
- 8.Naglak EK, Morrison SG, Morrison RP. 2016. Gamma interferon is required for optimal antibody-mediated immunity against genital Chlamydia infection. Infect Immun 84:3232–3242. doi: 10.1128/IAI.00749-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naglak EK, Morrison SG, Morrison RP. 2017. Neutrophils are central to antibody-mediated protection against genital Chlamydia. Infect Immun 85:e00409-17. doi: 10.1128/IAI.00409-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrison SG, Morrison RP. 2005. A predominant role for antibody in acquired immunity to chlamydial genital tract reinfection. J Immunol 175:7536–7542. doi: 10.4049/jimmunol.175.11.7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotter TW, Ramsey KH, Miranpuri GS, Poulsen CE, Byrne GI. 1997. Dissemination of Chlamydia trachomatis chronic genital tract infection in gamma interferon gene knockout mice. Infect Immun 65:2145–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rank RG, Soderberg LS, Barron AL. 1985. Chronic chlamydial genital infection in congenitally athymic nude mice. Infect Immun 48:847–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li LX, McSorley SJ. 2013. B cells enhance antigen-specific CD4 T cell priming and prevent bacteria dissemination following Chlamydia muridarum genital tract infection. PLoS Pathog 9:e1003707. doi: 10.1371/journal.ppat.1003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sturdevant GL, Caldwell HD. 2014. Innate immunity is sufficient for the clearance of Chlamydia trachomatis from the female mouse genital tract. Pathog Dis 72:70–73. doi: 10.1111/2049-632X.12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frazer LC, Sullivan JE, Zurenski MA, Mintus M, Tomasak TE, Prantner D, Nagarajan UM, Darville T. 2013. CD4+ T cell expression of MyD88 is essential for normal resolution of Chlamydia muridarum genital tract infection. J Immunol 191:4269–4279. doi: 10.4049/jimmunol.1301547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koren S, Phillippy AM. 2015. One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly. Curr Opin Microbiol 23:110–120. doi: 10.1016/j.mib.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Lei L, Chen J, Hou S, Ding Y, Yang Z, Zeng H, Baseman J, Zhong G. 2014. Reduced live organism recovery and lack of hydrosalpinx in mice infected with plasmid-free Chlamydia muridarum. Infect Immun 82:983–992. doi: 10.1128/IAI.01543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Connell CM, Nicks KM. 2006. A plasmid-cured Chlamydia muridarum strain displays altered plaque morphology and reduced infectivity in cell culture. Microbiology 152:1601–1607. doi: 10.1099/mic.0.28658-0. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez-Maranon MJ, Bush RM, Peterson EM, Schirmer T, de la Maza LM. 2002. Prediction of the membrane-spanning beta-strands of the major outer membrane protein of Chlamydia. Protein Sci 11:1854–1861. doi: 10.1110/ps.3650102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feher VA, Randall A, Baldi P, Bush RM, de la Maza LM, Amaro RE. 2013. A 3-dimensional trimeric beta-barrel model for Chlamydia MOMP contains conserved and novel elements of Gram-negative bacterial porins. PLoS One 8:e68934. doi: 10.1371/journal.pone.0068934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramsey KH, Sigar IM, Schripsema JH, Denman CJ, Bowlin AK, Myers GA, Rank RG. 2009. Strain and virulence diversity in the mouse pathogen Chlamydia muridarum. Infect Immun 77:3284–3293. doi: 10.1128/IAI.00147-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg SL, Eisen J, Fraser CM. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res 28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Yang Z, Sun X, Tang L, Ding Y, Xue M, Zhou Z, Baseman J, Zhong G. 2015. Intrauterine infection with plasmid-free Chlamydia muridarum reveals a critical role of the plasmid in chlamydial ascension and establishes a model for evaluating plasmid-independent pathogenicity. Infect Immun 83:2583–2592. doi: 10.1128/IAI.00353-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeruva L, Myers GS, Spencer N, Creasy HH, Adams NE, Maurelli AT, McChesney GR, Cleves MA, Ravel J, Bowlin A, Rank RG. 2014. Early microRNA expression profile as a prognostic biomarker for the development of pelvic inflammatory disease in a mouse model of chlamydial genital infection. mBio 5:e01241-14. doi: 10.1128/mBio.01241-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell M, Darville T, Chandra-Kuntal K, Smith B, Andrews CW Jr, O'Connell CM. 2011. Infectivity acts as in vivo selection for maintenance of the chlamydial cryptic plasmid. Infect Immun 79:98–107. doi: 10.1128/IAI.01105-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Connell CM, Ingalls RR, Andrews CW Jr, Scurlock AM, Darville T. 2007. Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J Immunol 179:4027–4034. doi: 10.4049/jimmunol.179.6.4027. [DOI] [PubMed] [Google Scholar]
- 27.Williams DM, Grubbs BG, Schachter J, Magee DM. 1993. Gamma interferon levels during Chlamydia trachomatis pneumonia in mice. Infect Immun 61:3556–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rank RG, Ramsey KH, Pack EA, Williams DM. 1992. Effect of gamma interferon on resolution of murine chlamydial genital infection. Infect Immun 60:4427–4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson IR, Owen P, Nataro JP. 1999. Molecular switches—the ON and OFF of bacterial phase variation. Mol Microbiol 33:919–932. doi: 10.1046/j.1365-2958.1999.01555.x. [DOI] [PubMed] [Google Scholar]
- 30.Koonin EV. 1996. A duplicated catalytic motif in a new superfamily of phosphohydrolases and phospholipid synthases that includes poxvirus envelope proteins. Trends Biochem Sci 21:242–243. doi: 10.1016/S0968-0004(96)30024-8. [DOI] [PubMed] [Google Scholar]
- 31.Ponting CP, Kerr ID. 1996. A novel family of phospholipase D homologues that includes phospholipid synthases and putative endonucleases: identification of duplicated repeats and potential active site residues. Protein Sci 5:914–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Celma L, Corbinais C, Vercruyssen J, Veaute X, de la Sierra-Gallay IL, Guerois R, Busso D, Mathieu A, Marsin S, Quevillon-Cheruel S, Radicella JP. 2017. Structural basis for the substrate selectivity of Helicobacter pylori NucT nuclease activity. PLoS One 12:e0189049. doi: 10.1371/journal.pone.0189049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liechti GW, Goldberg JB. 2013. Helicobacter pylori salvages purines from extracellular host cell DNA utilizing the outer membrane-associated nuclease NucT. J Bacteriol 195:4387–4398. doi: 10.1128/JB.00388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Connell HA, Niu C, Gilbert ES. 2007. Enhanced high copy number plasmid maintenance and heterologous protein production in an Escherichia coli biofilm. Biotechnol Bioeng 97:439–446. doi: 10.1002/bit.21240. [DOI] [PubMed] [Google Scholar]
- 35.Baehr W, Zhang YX, Joseph T, Su H, Nano FE, Everett KD, Caldwell HD. 1988. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc Natl Acad Sci U S A 85:4000–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeruva L, Pouncey DL, Eledge MR, Bhattacharya S, Luo C, Weatherford EW, Ojcius DM, Rank RG. 2017. MicroRNAs modulate pathogenesis resulting from chlamydial infection in mice. Infect Immun 85:e00768-16. doi: 10.1128/IAI.00768-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tseng CK, Rank RG. 1998. Role of NK cells in the early host response to chlamydial genital infection. Infect Immun 66:5867–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L, McSorley S. 2014. Antibody, but not B cell-dependent antigen presentation, is required for preventing bacteria dissemination following Chlamydia muridarum genital tract infection, p 273–276. In Proceedings of the Thirteenth International Symposium on Human Chlamydial Infections International Chlamydia Symposium, San Francisco, CA. [Google Scholar]
- 39.Ganapamo F, Dennis VA, Philipp MT. 2001. CD19(+) cells produce IFN-gamma in mice infected with Borrelia burgdorferi. Eur J Immunol 31:3460–3468. doi:. [DOI] [PubMed] [Google Scholar]
- 40.Bitsaktsis C, Nandi B, Racine R, MacNamara KC, Winslow G. 2007. T-cell-independent humoral immunity is sufficient for protection against fatal intracellular ehrlichia infection. Infect Immun 75:4933–4941. doi: 10.1128/IAI.00705-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gil-Cruz C, Bobat S, Marshall JL, Kingsley RA, Ross EA, Henderson IR, Leyton DL, Coughlan RE, Khan M, Jensen KT, Buckley CD, Dougan G, MacLennan IC, Lopez-Macias C, Cunningham AF. 2009. The porin OmpD from nontyphoidal Salmonella is a key target for a protective B1b cell antibody response. Proc Natl Acad Sci U S A 106:9803–9808. doi: 10.1073/pnas.0812431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deniset JF, Surewaard BG, Lee WY, Kubes P. 2017. Splenic Ly6G(high) mature and Ly6G(int) immature neutrophils contribute to eradication of S. pneumoniae. J Exp Med 214:1333–1350. doi: 10.1084/jem.20161621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guay HM, Mishra R, Garcea RL, Welsh RM, Szomolanyi-Tsuda E. 2009. Generation of protective T cell-independent antiviral antibody responses in SCID mice reconstituted with follicular or marginal zone B cells. J Immunol 183:518–523. doi: 10.4049/jimmunol.0900068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puga I, Cols M, Barra CM, He B, Cassis L, Gentile M, Comerma L, Chorny A, Shan M, Xu W, Magri G, Knowles DM, Tam W, Chiu A, Bussel JB, Serrano S, Lorente JA, Bellosillo B, Lloreta J, Juanpere N, Alameda F, Baro T, de Heredia CD, Toran N, Catala A, Torrebadell M, Fortuny C, Cusi V, Carreras C, Diaz GA, Blander JM, Farber CM, Silvestri G, Cunningham-Rundles C, Calvillo M, Dufour C, Notarangelo LD, Lougaris V, Plebani A, Casanova JL, Ganal SC, Diefenbach A, Arostegui JI, Juan M, Yague J, Mahlaoui N, Donadieu J, Chen K, Cerutti A. 2011. B cell-helper neutrophils stimulate the diversification and production of immunoglobulin in the marginal zone of the spleen. Nat Immunol 13:170–180. doi: 10.1038/ni.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panda S, Zhang J, Tan NS, Ho B, Ding JL. 2013. Natural IgG antibodies provide innate protection against ficolin-opsonized bacteria. EMBO J 32:2905–2919. doi: 10.1038/emboj.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szomolanyi-Tsuda E, Brien JD, Dorgan JE, Garcea RL, Woodland RT, Welsh RM. 2001. Antiviral T-cell-independent type 2 antibody responses induced in vivo in the absence of T and NK cells. Virology 280:160–168. doi: 10.1006/viro.2000.0766. [DOI] [PubMed] [Google Scholar]
- 47.Finethy R, Coers J. 2016. Sensing the enemy, containing the threat: cell-autonomous immunity to Chlamydia trachomatis. FEMS Microbiol Rev 40:875–893. doi: 10.1093/femsre/fuw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szomolanyi-Tsuda E, Seedhom MO, Carroll MC, Garcea RL. 2006. T cell-independent and T cell-dependent immunoglobulin G responses to polyomavirus infection are impaired in complement receptor 2-deficient mice. Virology 352:52–60. doi: 10.1016/j.virol.2006.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banks J, Eddie B, Schachter J, Meyer KF. 1970. Plaque formation by Chlamydia in L cells. Infect Immun 1:259–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Connell CM, AbdelRahman YM, Green E, Darville HK, Saira K, Smith B, Darville T, Scurlock AM, Meyer CR, Belland RJ. 2011. Toll-like receptor 2 activation by Chlamydia trachomatis is plasmid dependent, and plasmid-responsive chromosomal loci are coordinately regulated in response to glucose limitation by C. trachomatis but not by C. muridarum. Infect Immun 79:1044–1056. doi: 10.1128/IAI.01118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kelly KA, Robinson EA, Rank RG. 1996. Initial route of antigen administration alters the T-cell cytokine profile produced in response to the mouse pneumonitis biovar of Chlamydia trachomatis following genital infection. Infect Immun 64:4976–4983. (Erratum, 65:2508, 1997.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caldwell HD, Kromhout J, Schachter J. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun 31:1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eutsey RA, Powell E, Dordel J, Salter SJ, Clark TA, Korlach J, Ehrlich GD, Hiller NL. 2015. Genetic stabilization of the drug-resistant PMEN1 Pneumococcus lineage by its distinctive DpnIII restriction-modification system. mBio 6:e00173-15. doi: 10.1128/mBio.00173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 55.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tuffrey M, Taylor-Robinson D. 1981. Progesterone as a key factor in the development of a mouse model for genital-tract infection with Chlamydia trachomatis. FEMS Microbiol Lett 12:111–115. doi: 10.1111/j.1574-6968.1981.tb07622.x. [DOI] [Google Scholar]
- 57.Darville T, Andrews CW Jr, Laffoon KK, Shymasani W, Kishen LR, Rank RG. 1997. Mouse strain-dependent variation in the course and outcome of chlamydial genital tract infection is associated with differences in host response. Infect Immun 65:3065–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burkholder T, Foltz C, Karlsson E, Linton CG, Smith JM. 2012. Health evaluation of experimental laboratory mice. Curr Protoc Mouse Biol 2:145–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.