Abstract
The fungal endophytes Neotyphodium lolii and Neotyphodium sp. Lp1 from perennial ryegrass (Lolium perenne), and related endophytes in other grasses, produce the ergopeptine toxin ergovaline, among other alkaloids, while also increasing plant fitness and resistance to biotic and abiotic stress. In the related fungus, Claviceps purpurea, the biosynthesis of ergopeptines requires the activities of two peptide synthetases, LPS1 and LPS2. A peptide synthetase gene hypothesized to be important for ergopeptine biosynthesis was identified in C. purpurea by its clustering with another ergot alkaloid biosynthetic gene, dmaW. Sequence analysis conducted independently of the research presented here indicates that this gene encodes LPS1 [Tudzynski, P., Holter, K., Correia, T., Arntz, C., Grammel, N. & Keller, U. (1999) Mol. Gen. Genet. 261, 133–141]. We have cloned a similar peptide synthetase gene from Neotyphodium lolii and inactivated it by gene knockout in Neotyphodium sp. Lp1. The resulting strain retained full compatibility with its perennial ryegrass host plant as assessed by immunoblotting of tillers and quantitative PCR. However, grass–endophyte associations containing the knockout strain did not produce detectable quantities of ergovaline as analyzed by HPLC with fluorescence detection. Disruption of this gene provides a means to manipulate the accumulation of ergovaline in endophyte-infected grasses for the purpose of determining the roles of ergovaline in endophyte-associated traits and, potentially, for ameliorating toxicoses in livestock.
Several important forage and turf grasses harbor endophytic fungi in the genus Neotyphodium that have profound effects on their grass hosts as well as on organisms that interact with infected grasses as herbivores, pathogens, or competitors (1–4). The best-studied endophyte in the U.S., Neotyphodium coenophialum, provides its host, tall fescue (Festuca arundinacea), with numerous benefits that help make tall fescue one of the most widely used plants for forage, turf, soil conservation, and land reclamation. Endophyte benefits include increases in drought tolerance, shoot growth, tillering, seed production, seed germination, phosphorus uptake, and resistance to insects and parasitic nematodes (1, 2, 4–8). The fungus is transmitted only through seed of an already infected mother plant (6, 9) and, thus, is an inherent, maternally transmitted component of a joint plant–fungus lineage (symbiotum).
Animals grazing endophyte-infected grasses may ingest significant amounts of ergovaline (10), a toxic ergopeptine produced by several Neotyphodium spp., as well as by the ergot fungus, Claviceps purpurea (11). Livestock maladies associated with ergovaline consumption (“tall fescue toxicosis”) include poor weight gain, hormonal imbalances leading to reduced fertility and lactation, and gangrene of the animals' limbs (12–15). The economic magnitude of tall fescue toxicosis in the U.S. can be gauged by an estimated $600 million loss to the beef cattle industry during 1990 (16). Considering inflation and losses to other livestock (e.g., dairy cattle, horses, and sheep), the economic cost associated with this problem may currently approach $1 billion per year.
Similar grass–endophyte relationships occur between Neotyphodium species and perennial ryegrass (Lolium perenne), a widely planted forage and turf grass. Many cultivars of perennial ryegrass are infected with Neotyphodium lolii, which produces smaller quantities of ergopeptines and larger quantities of indole-diterpene alkaloids (e.g., lolitrems), which are thought to be responsible for the mammalian toxicosis called “ryegrass staggers” (17). Other varieties of perennial ryegrass contain an endophyte derived from the hybrid N. lolii × Epichloë typhina, exemplified by isolate Lp1 (18, 19). Lp1 produces little or no lolitrems in planta but produces significant quantities of ergovaline (7, 19). Thus, in its profile of antivertebrate alkaloids, Lp1-infected perennial ryegrass more closely resembles N. coenophialum-infected tall fescue than it does typical N. lolii-infected perennial ryegrass.
Ergopeptines are nonribosomally synthesized peptides containing lysergic acid and three amino acids that vary between, and define, the molecules in the family. Ergovaline contains d-lysergic acid, l-alanine, l-valine, and l-proline, and differs from ergotamine, the most common ergopeptine produced by the ergot fungus C. purpurea, in containing l-valine in place of l-phenylalanine (11). Ergopeptines are synthesized via a thiotemplate mechanism similar to that responsible for the biosynthesis of numerous microbial peptide antibiotics and toxins (20). An enzyme system with peptide synthetase activities for lysergic acid and the three amino acids of ergotamine has been isolated from C. purpurea and designated lysergyl peptide synthetase (21, 22). Amino acid and lysergic acid activating modules reside on different polypeptides, LPS1 and LPS2, respectively.
Cloning of genes in the ergot alkaloid biosynthetic pathway and their eventual manipulation, via gene knockout or antisense strategies, would provide a means for identifying roles for ergot alkaloids in the plant–fungus associations in which they occur and, potentially, for ameliorating toxicoses with which these alkaloids are associated. A gene (dmaW) encoding dimethylallyltryptophan synthase, the first committed step in ergot alkaloid biosynthesis, has been cloned from Claviceps fusiformis American Type Culture Collection 26245 and its function has been demonstrated by expression in yeast (23). This gene also has been cloned from isolate P1 of C. purpurea (24) and, independent of the work described in this paper, was found to be closely linked in that fungus to a gene (designated cpps1) with the capacity to encode a three-module peptide synthetase containing a region of near identity with a sequenced fragment of LPS1 (24). Herein we demonstrate, through gene knockout analysis, that a similar peptide synthetase gene (which we have named lpsA) is essential for ergopeptine biosynthesis in Neotyphodium sp. Lp1 and that ergovaline can be eliminated from a Neotyphodium–grass symbiosis by genetic modification of the endophyte.
Materials and Methods
Fungal Culture and General Methodology.
Neotyphodium sp. Lp1 was provided by G. C. M. Latch and M. J. Christensen (AgResearch, Palmerston North, New Zealand). Fungal culture, routine molecular biology techniques, and detection of peptide synthetase genes by degenerate PCR with primers designed to polypeptide sequences GKPKG and YKTGDL were as described previously (25). With the exception of the degenerate PCR reactions and the quantitative PCR reactions (described separately, below), PCR reactions contained 50 mM Tris⋅HCl (pH 9.0), 15 mM (NH4)2SO4, 1.5 mM MgCl2, 0.1% Triton X-100, 200 μM of each dNTP, 1 μM each of two specific primers (sequence provided under individual applications), template DNA (as described under individual applications), and 1 unit of DyNAzyme EXT polymerase (MJ Research, Watertown, MA) in a total of 50 μl. After an initial denaturation step at 95°C for 5 min (during which DyNAzyme EXT polymerase was added, to simulate a “hot start”), reactions were incubated for 35 cycles of 94°C for 1 min, 59°C for 1 min, and 72°C for 3 min 45 s. Southern blot hybridizations were as described by Panaccione (25), with alterations in washing conditions noted where applicable. Screening of the N. lolii λ GEM12 genomic library with C. purpurea peptide synthetase gene fragment Cp605 (25) as probe was conducted with hybridizations at 42°C in 6× SSC (26)/5× Denhardt's (26)/0.1% SDS/0.1% Na4P2O7-10H2O/50 mM Tris⋅HCl, pH 7.5/50 μg/ml of herring sperm DNA/50% formamide. Filters were washed with 2× SSPE/EDTA (26) + 0.1% SDS at 50°C.
Gene Knockout Strategy and Fungal Transformation.
A 4.0-kb SalI fragment, from the insert of a clone isolated from the N. lolii λ GEM12 genomic library with peptide synthetase gene fragment Cp605 (25) as probe, was cloned into pUC18. On the basis of partial sequence data (GenBank accession no. AF368420), this SalI fragment is contained within the coding sequences of an N. lolii peptide synthetase gene (lpsA) and corresponds to nucleotides 4,441–8,423 of the 9,699-bp coding sequences of the C. purpurea cpps1 gene (24). Cleavage at a unique XhoI site in the cloned SalI fragment generates fragments of 1.4 and 2.6 kb. A selectable marker for the construct was prepared by amplifying a 2.7-kb fragment containing the hygromycin resistance gene and controlling sequences from pMOcosX (27) with primers 5′AGTCTCGAGCGATCCTTGAAGCTGTCCCTGAT3′ (anneals to pBR322 sequences beyond the 5′ end of the hygromycin resistance cassette) and 5′AGTCTCGAGCATTCACTAGGCAACCATGGTTAC3′ (anneals to the trpC transcription terminator region near the 3′ end of the cassette). This PCR product was digested with XhoI (sites occur only within the primer sequences) and ligated into the unique XhoI site of the cloned 4.0-kb lpsA fragment described above. The resulting construct was liberated from the plasmid in linear form by SalI digestion and purified by agarose gel electrophoresis before transformation.
Because Neotyphodium sp. Lp1 had been transformed previously, and transformants were maintained as endophytes on reintroduction into plants (28), it was used as recipient in transformations for gene knockout. Protoplasts were prepared as described by Murray et al. (28) but with 7 mg/ml of Driselase (InterSpex, Foster City, CA) and 1.3 mg/ml of Novozym 234 (lot 4859, InterSpex) as lysing enzymes. Protoplasts were transformed with the linear gene knockout construct (≈1 μg in 10 μl of water) as described by Rasmussen et al. (29), except spermidine and heparin were omitted. Five equal aliquots of the transformation mixture were each mixed with 4 ml of molten regeneration medium (described below) and overlaid on previously solidified regeneration medium containing hygromycin B at 300 μg/ml (CalBiochem). The regeneration medium contained, per liter: 304 g of sucrose, 1 g of NH4NO3, 1 g of KH2PO4, 1 g of NaCl, 0.25 g of anhydrous MgSO4, 0.13 g of CaCl2-2H2O, 1 g of yeast extract, 12 g of dehydrated potato dextrose broth, 1 g of peptone, 1 g of acid hydrolysate of casein, and 6 g of agarose. Transformation plates were incubated at 20°C for 4 weeks before hygromycin-resistant colonies were transferred to potato dextrose agar to induce sporulation.
Transformants were initially screened for homologous recombination of the gene knockout construct at the lpsA locus by PCR primed from oligonucleotides LPKO-F, 5′GATGCCCGCGTTGCAGGCCATGCTGTCCAG3′ (anneals to pBR322 sequences flanking hygromycin resistance cassette), and LPKO-R, 5′CAACAGAGTGCAGTGTGATCAGTACCACCAG3′ (anneals to lpsA sequences flanking the intended site of integration), and with conidia as template (30). Because only 1 of 172 transformants yielded the desired fragment in the spore-based PCR assay, the transformants were screened again in a more sensitive assay by preparing pooled cultures of five transformants each, isolating DNA with GeneClean Spin kits (Bio 101), and amplifying by PCR as described above. The presence of amplifiable DNA in the preparations was confirmed by amplification of a wild-type sized fragment from primers 5′CCAACGAAAGAAAGACAGCCGTCTAGACC3′ and 5′GTTCAAATGATGACCAAGCAACGACAGCC3′, to generate a 3.2-kb fragment from within lpsA. The knockout and selected ectopic transformants were purified to nuclear homogeneity by culturing from single uninucleate conidia (18). DNA from these strains was analyzed in Southern blot hybridizations (25) with the 4.0-kb SalI fragment (used to direct the gene knockout) as probe and with final washes with 2× SSPE/EDTA at 65°C.
Synthesis and Analysis of Grass–Endophyte Symbiota.
Endophytes were introduced into etiolated endophyte-free seedlings of perennial ryegrass cv. Rosalin according to the procedure of Latch and Christensen (31). The frequency of infection was evaluated by immunoblotting the base of the first tiller (32) with antisera raised against N. coenophialum and which detects hyphae of other Neotyphodium spp (33). Plants were propagated for 6 months in 15-cm pots in a greenhouse with supplemental lighting to give a 14 h 23°C day/10 h 20°C night, then sampled for ergovaline and quantitative PCR analyses.
Ergopeptines were extracted from 300 mg of dried clippings (primarily leaf blade) essentially as described by Hill et al. (34), except that, in the initial step, plant material was ground to a fine powder in liquid nitrogen and, in the final step, methanol eluates were concentrated to 50 μl with a Centrivap concentrator (Labconco, Kansas City, MO) and diluted 1:1 with water. To provide an internal standard for quantification, 100 ng of ergotamine-tartrate (Sigma) was added to the plant material before extraction. Ergopeptines were analyzed by HPLC on a Microsorb C18, 5-μm particle, 4.6 × 250 mm column (Varian), as described by Annis and Panaccione (35), except that fluorescence was detected on the basis of excitation at 310 nm and emission at 420 nm. Ergovaline standard was generously provided by F. T. Smith of Auburn University.
An estimate of the extent of endophyte colonization in a subset of the same plant samples analyzed by HPLC was obtained by a modification of the quantitative PCR (qPCR) procedure of Groppe and Boller (36). DNA was extracted (37) from 100 mg (dry weight) of clippings and quantified based on A260 and fluorescence on staining with ethidium bromide. Mimic DNA for qPCR was constructed by PCR primed from oligonucleotides 5′TTACCGAACTGGCGACATCCTCGACGCGTTCCCGTG3′ and 5′CATGGGCTGTCGTTGCTTGTTGCCAGAAGCCTGTCA 3′, in which the 5′-most 18 nucleotides provide primer annealing sites for lpsA-specific primers used in subsequent qPCR reactions and the remaining 18 nucleotides prime amplification of the mimic molecule corresponding to the β-tubulin gene from Neotyphodium sp. Lp1 [bases 38–535 of the published sequence (18)]. qPCR amplification reactions contained 150 ng of symbiotum DNA and 10-fold dilutions of the mimic DNA molecule (from 10 pg to 1 ag), 50 mM Tris⋅HCl (pH 9.0), 1.5 mM MgCl2, 0.1% Triton X-100, 200 μM of each dNTP, 1 μM each of lpsA specific primers (5′TTACCGAACTGGCGACAT3′ and 5′CATGGGCTGTCGTTGCTT3′), and 0.5 units of Taq DNA polymerase (Promega). Amplification conditions were an initial denaturation at 94°C for 2 min followed by 30 cycles of 94°C for 1 min, 63°C for 45 s, and 72°C for 1 min, with a final extension step at 72°C for 5 min. Amplification of the mimic template yielded a product of 536 bp, whereas amplification of the lpsA fragment yielded a product of 287 bp. Primers annealed to sequences in lpsA that were not interrupted by gene knockout. PCR products were separated by electrophoresis, photographed under UV light, and analyzed densitometrically by scion image software (Scion, Frederick, MD). Densities in images were determined to be within the linear range of quantification by comparing values to those of standards amplified from known concentrations of mimic DNA. Estimates of fungal DNA in the symbiota containing the ectopic transformant strain were adjusted to account for two additional copies of the relevant region of lpsA resulting from ectopic integration of the construct.
Results
Identification of an Ergovaline-Associated Peptide Synthetase Gene.
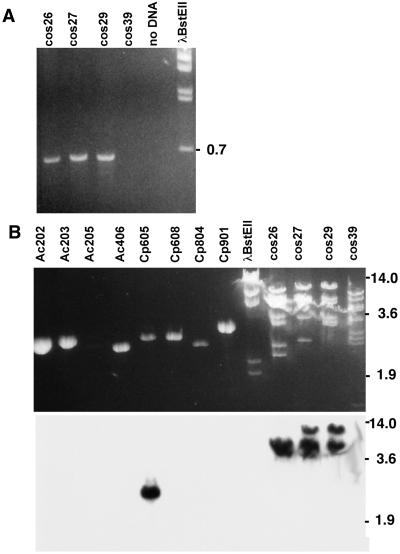
A peptide synthetase gene that was an appropriate target for gene knockout analysis was identified as a result of several observations. PCR with degenerate primers designed to anneal to conserved regions within peptide synthetase genes (25) amplified a product of the expected size from cosmids containing the ergot alkaloid biosynthetic gene, dmaW, of C. purpurea ATCC 20102 (38) (Fig. 1A). This PCR product hybridized strongly to one particular peptide synthetase gene fragment, clone Cp605 from C. purpurea ATCC 34501, among fragments of several different peptide synthetase genes previously cloned from ergopeptine-producing fungi (Fig. 1B) (25, 35). DNA sequence analysis (not shown) demonstrated 93% identity between the PCR product and clone Cp605. The close linkage of this peptide synthetase gene with the known ergot alkaloid biosynthetic gene, dmaW, suggested very strongly that it was involved in ergopeptine biosynthesis. Subsequently, Tudzynski et al. (24) published the characterization of a homologous peptide synthetase gene found by chromosome walking from dmaW (which they designated cpd1) in C. purpurea strain P1. The gene described by Tudzynski et al. (24) is capable of encoding a three-module peptide synthetase that contains a region with near identity to a sequenced peptide fragment from the purified enzyme, LPS1, and also contains a region of 93% DNA sequence identity with Cp605 from C. purpurea ATCC 34501.
Figure 1.
Identification of peptide synthetase sequences linked to C. purpurea dmaW. (A ) PCR with cosmids containing C. purpurea American Type Culture Collection (ATCC) 20102 dmaW as template and primers designed to polypeptide sequences GKPKG and YKTGDL [based on conserved regions in peptide synthetase genes (25)] results in the expected ≈700-bp product from dmaW-containing cosmids 26–7A (cos26), 27–1A (cos27), and 29–5A (cos29). [Cosmid 39–2F (cos39) contains a dmaW pseudogene; it does not appear to contain a peptide synthetase gene.] (B) The PCR product generated from cosmid 27–1A in A hybridizes with EcoRI fragments of the cosmid from which it was generated and also with Cp605, previously cloned from C. purpurea ATCC 34501 but not with any of the other peptide synthetase gene fragments previously cloned and characterized from ergopeptine-producing fungi (25, 35). Fragment sizes (in kb) indicate the relative mobility of fragments of BstEII-digested bacteriophage λ.
By low-stringency hybridization with clone Cp605, we isolated a homologue of the C. purpurea peptide synthetase gene from an available bacteriophage λ library of N. lolii isolate Lp19. Functional analysis of this N. lolii gene, which we designate lpsA [for lysergyl peptide synthetase, after the nomenclature of Riederer et al. (21)], was conducted in the closely related fungus Neotyphodium sp. Lp1 for several reasons. In contrast to N. lolii Lp19, which produces significant quantities of indole-diterpenes, Lp1 produces ergovaline as its primary antiherbivore alkaloid (19). Also, Lp1 had been transformed previously, and transformants were stable through at least one generation in planta (28). Moreover, Lp1 contains a single copy of lpsA, as compared with multiple copies detected in C. purpurea (Fig. 2). Interestingly, in the limited sample examined, lpsA has apparent homologues in related fungi that produce ergopeptines (C. purpurea and Epichloë festucae), but not in relatives that do not produce ergopeptines (C. fusiformis, Atkinsonella hypoxylon, and Cordyceps militaris) (Fig. 2).
Figure 2.
Distribution of lpsA among several members of the Clavicipitaceae. Genomic DNA (4 μg) from A. hypoxylon ATCC 64993 (lane 1), C. fusiformis ATCC 26245 (lane 2), C. purpurea ATCC 34501 (lane 3), Neotyphodium sp. Lp1 (lane 4), E. festucae ATCC 56430 (lane 5), and C. militaris ATCC 26848 (lane 6) was digested with EcoRI before electrophoresis and transfer. Probe was prepared from the 4.0-kb SalI fragment of N. lolii lpsA described in the Materials and Methods. Final washing of the blot was with 2× SSPE/EDTA at 60°C. Fragment sizes (in kb) indicate the relative mobility of HindIII-digested bacteriophage λ.
Knockout of lpsA.
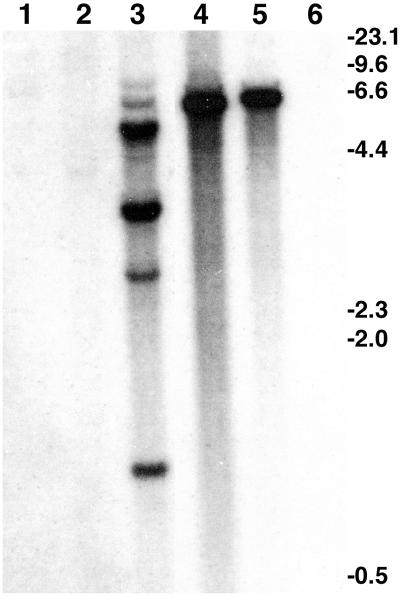
A linear construct for gene knockout was prepared in which the hygromycin resistance marker from pMOcosX (27) was flanked by internal fragments (1.4 and 2.6 kb) of lpsA (Fig. 3A). This construct was introduced into Lp1 by transformation, and a total of 172 transformants were analyzed for homologous recombination by PCR-based assays (Fig. 3A). Only one transformant produced a PCR product of the size predicted for a strain in which homologous recombination had taken place between the introduced construct and the native lpsA gene (Fig. 3B). The knockout transformant was purified to nuclear homogeneity by culturing from single conidia, and two single-conidial isolates (ko1.1 and ko1.2) were studied further. Integration of the gene knockout construct at the homologous site and lack of any duplicate copies of lpsA were confirmed by Southern blot analysis of genomic DNA digested with SalI. As predicted from maps of lpsA (Fig. 3A), the knockout eliminated the 4.0-kb SalI fragment and replaced it with a 6.7-kb fragment (Fig. 3C). No additional copies of lpsA or ectopic integration events were indicated by the hybridization patterns of ko1.1 and ko1.2. The presence of a 6.7-kb SalI fragment in the ectopic transformant (Ec, Fig. 3C), in which the wild-type fragment of 4.0 kb is still present, is likely because of integration of a concatemer of the knockout construct (which is a 6.7-kb SalI fragment) at a single ectopic site.
Figure 3.
Knockout of lpsA in Neotyphodium sp. Lp1. (A) Strategy for disrupting the internal 4.0-kb SalI fragment and for detecting the integration by PCR (symbols > and < indicate where primers LPKO-F and LPKO-R, respectively, anneal to prime amplification of a 2.7-kb fragment) and Southern blot hybridization (increased length of the disrupted SalI fragment is indicated). S = SalI; X = XhoI; E = EcoRI; Hyg = hygromycin resistance. (B) PCR product from primers LPKO-F and LPKO-R, indicating homologous recombination (lane 3 only) from spores of one of several transformants (lanes 1–4). Lane 5 = BstEII-digested bacteriophage λ DNA. (C) Southern blot probed with the 4.0-kb SalI fragment of N. lolii lpsA (used to direct the integration). Fragment sizes (in kb) are derived from BstEII digested bacteriophage λ DNA. Strain names are described in Results.
Analyses of Symbiota Containing lpsA Knockout Strains.
Symbiota were synthesized comprising perennial ryegrass systemically infected with strains ko1.1 and ko1.2 and with a single-conidial strain of a transformant containing an ectopically integrated knockout construct. These strains established symbioses with host plants with similar frequencies. The two single-conidial strains of the knockout mutant colonized 12 of 49 (24%) and 12 of 42 (29%) of the inoculated seedlings, whereas the ectopic transformant colonized 8 of 31 (26%) inoculated seedlings. The success rates observed are typical for artificial introduction of compatible endophytes into perennial ryegrass (31).
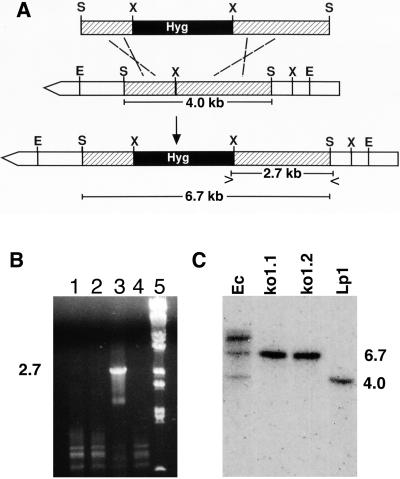
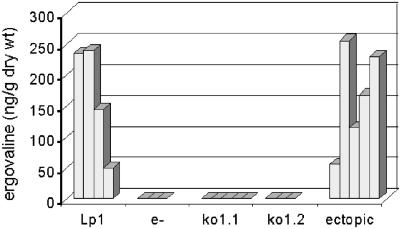
Concentrated extracts of clippings from wild-type Lp1 symbiota, five separately synthesized symbiota containing the lpsA knockout strain ko1.1, three separately synthesized symbiota containing ko1.2, and five separately synthesized symbiota containing an ectopic transformant (Ec) were analyzed for ergovaline by fluorescence HPLC (Fig. 4). None of the knockout-containing symbiota produced detectable ergovaline (detection limit 2 ng/g of tissue). The primary clippings from the ectopic transformant symbiota produced quantities of ergovaline comparable to that of established symbiota containing the Lp1 parental strain (Fig. 5). Because clipping (simulated herbivory) of leaves has been reported to increase ergovaline concentrations in regrowth of other grass–endophyte symbiota (39, 40), we analyzed five samples of regrown leaf material from symbiota containing the ectopic transformant and found an average of 220 ng of ergovaline/g of dry weight. All eight samples of regrowth from clipped knockout-containing symbiota lacked detectable ergovaline. All data were normalized to ergotamine that was added just before extraction as an internal standard. Prior extractions of samples of the various symbiota without added ergotamine demonstrated that none of the symbiota produced ergotamine. Because samples were prepared with hard layer silica gel cleanup columns described by Rottinghaus et al. (40), later-eluting, inactive ergovalinine, and ergotaminine isomers were not detected.
Figure 4.
Traces from HPLC chromatograms of extracts of the indicated symbiota. Std = standards; Ec = ectopic transformant; e− = endophyte free. Under the described conditions, ergovaline (Ev) had a retention time of 6.4 min, and ergotamine (Et) had a retention time of 10.2 min. An equal quantity of ergotamine (not produced in Neotyphodium-perennial ryegrass associations) was added to each sample before extraction as an internal standard.
Figure 5.
Ergovaline in ng/g of dry weight of extracted leaf clippings. Each bar represents data from primary clippings of an independently synthesized symbiotum containing the indicated endophyte strain or no endophyte (e−).
In addition to ergovaline, a peak with a retention time of 4.5 min that was prominent in wild-type and ectopic transformant-containing symbiota was not detected in the knockout-containing symbiota. The shoulder in this area of the chromatogram obscured whether the compound was absent or greatly reduced compared with its levels in ergovaline-producing symbiota. This compound had chromatographic and spectral properties of an ergot alkaloid and did not cochromatograph with any of the ergopeptines isolated from ergot bodies of C. purpurea (not shown).
In quantitative PCR analyses, the lpsA knockout strains appeared to colonize perennial ryegrass to the same extent (mean of 0.35% of sample DNA was of endophyte origin) as wild-type Lp1 (0.34% of DNA from endophyte) or the ectopic transformant (0.44% of DNA from endophyte) (Table 1). These data indicate that colonization of perennial ryegrass by this endophyte is unrelated to ergovaline. Furthermore, the establishment of levels of colonization by the knockout strains that were similar to those observed with control endophytes is critical to interpreting the ergovaline data presented above. Collectively, qPCR data indicate that the Lp1-derived endophytes contributed ≈0.37% of the total DNA isolated from these tissues.
Table 1.
Relative colonization of perennial ryegrass by fungal endophytes as assessed by quantitative PCR of symbiotum DNA
| Endophyte | Fungal DNA
|
|
|---|---|---|
| pg* | %† | |
| Neotyphodium sp. Lp1 | 504 | 0.34 |
| Endophyte-free | n.d.‡ | n.d. |
| Ectopic, plant 1§ | 432 | 0.29 |
| Ectopic, plant 2 | 899 | 0.60 |
| ko1.1, plant 1 | 215 | 0.14 |
| ko1.1, plant 2 | 899 | 0.60 |
| ko1.2, plant 1 | 467 | 0.31 |
| ko1.2, plant 2 | 504 | 0.34 |
Per 150 ng symbiotum DNA.
Of symbiotum DNA.
None detected.
Plant 1 or 2 = independent symbiota with same endophyte.
Discussion
Through gene knockout analysis, we have established the function of a peptide synthetase required for ergovaline production and demonstrated that ergovaline can be eliminated from a Neotyphodium grass symbiosis by genetic modification of the endophyte. The construction of ergovaline-deficient knockout symbiota gives us the tools to investigate the role of ergovaline in the various costs and benefits generally associated with endophyte infection of grasses.
Neotyphodium spp. are characteristically slow growing and recalcitrant to laboratory analyses. Our efforts to analyze gene function in Neotyphodium sp. Lp1 were hindered by a low frequency of homologous recombination. We used a one-step replacement via double crossover with the intent of increasing stability of the integrated DNA, which needs to be maintained in the genome indefinitely to assess the effects of the knockout on the grass–fungus association and the organisms that interact with this association. This type of recombination event may occur less frequently than a simpler single crossover involved in some gene disruption strategies. Nonetheless, the phenotype of the peptide synthetase knockout presented here is supported by additional observations and data. The clustering of the C. purpurea homologue of this peptide synthetase gene with dmaW (Fig. 1; ref. 24) provides a strong indication of its role in ergopeptine biosynthesis. Similarly, the distribution of apparent homologues of N. lolii lpsA among ergopeptine producers and related nonproducers correlates with their ability to produce ergopeptines (Fig. 2). Moreover, Southern blot hybridization indicated that the transformation construct in the knockout transformant integrated only at its homologous site (Fig. 3). Similarly, symbiota containing a transformant in which the same construct integrated ectopically produced ergovaline at levels comparable to symbiota containing wild-type Lp1 (Figs. 4 and 5). Finally, the strains derived from knockout and ectopic transformants maintained the expected phenotypes in several independent symbiota, which, because perennial ryegrass is an obligate outcrosser, represent several different plant genotypes.
Our data indicate that the absence of ergovaline in the knockout endophyte does not affect the balance of the symbiosis in terms of the extent of endophyte colonization and the transmission of endophyte through tillers. From one perspective, this is not surprising, because there are related endophytes of other grasses (e.g., Neotyphodium uncinatum in meadow fescue) that do not produce ergovaline and maintain stable symbioses. Nonetheless, little is known of the natural role of these compounds and, because other metabolites (with potentially redundant functions) vary among other Neotyphodium–grass associations, it could not be assumed a priori that ergovaline was unimportant in the establishment and maintenance of the plant–fungus association.
Further characterization of the alkaloid profile of the lpsA knockout, with particular attention to any changes in addition to the loss of ergovaline, will be pursued in future studies. The unidentified peak eluting at 4.5 min that was eliminated or reduced in knockout-containing symbiota (Fig. 4) has a retention time similar to a peak observed in ergovaline analyses of N. coenophialum-infected tall fescue (ref. 40, R.D.J. and D.G.P., unpublished data). In liquid chromatography-MS analyses, Shelby et al. (41) described didehydroergovaline (which is proposed to differ from ergovaline by containing a double bond in the valine or proline side chains) as the second most abundant ergopeptine from N. coenophialum-infected tall fescue. On the basis of its concentration in endophyte-infected tall fescue, didehydroergovaline would be the only other ergopeptine detected in our HPLC system. It is also the only ergopeptine described that elutes earlier than ergovaline in a reverse-phase HPLC system (41). Unfortunately, no purified didehydroergovaline is available as standard. We hypothesize that all ergopeptines would be eliminated by disruption of the peptide synthetase-encoding gene, just as all forms of the nonribosomally synthesized peptides HC- and AM-toxin are eliminated by disruption of the appropriate peptide synthetase genes in Cochliobolus carbonum (42) and Alternaria alternata (43), respectively.
The availability of ergovaline-producing and nonproducing symbiota synthesized from symbionts with otherwise similar genetic backgrounds will allow us to investigate the roles of ergovaline in negative and positive traits associated with endophyte infection of grasses. The contribution of ergovaline to animal toxicity, relative to other known and unknown endophyte metabolites, has not been directly demonstrated. Moreover, potential roles for ergovaline in other endophyte-associated traits, such as nematode resistance and anti-insect activities, remain to be investigated. A direct genetic test of the importance of ergovaline in animal toxicosis and other endophyte-conferred traits is highly desirable and now within reach.
Acknowledgments
We thank Forrest Smith for the ergovaline standard; G. C. M. Latch and M. J. Christensen for Lp1; Nichole Smith for assistance in screening transformants; Erin Zervos, Satish Bhatia, and Walter Hollin for general technical assistance; George Rottinghaus, Richard Shelby, and Linda Johnson for helpful discussions; and Jonathan Walton and Seanna Annis for commenting on an earlier version of this manuscript. This work was supported by U.S. Department of Agriculture–National Research Initiative Grant No. 98–35303-6663 and published with the approval of the Director of the West Virginia Agricultural and Forestry Experiment Station as Scientific Article no. 2789.
Abbreviation
- qPCR
quantitative PCR
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF368420).
References
- 1.Clay K. Annu Rev Ecol Syst. 1990;21:275–295. [Google Scholar]
- 2.Schardl C L, Phillips T D. Plant Dis. 1997;81:430–437. doi: 10.1094/PDIS.1997.81.5.430. [DOI] [PubMed] [Google Scholar]
- 3.Clay K, Holah J. Science. 1999;285:1742–1744. doi: 10.1126/science.285.5434.1742. [DOI] [PubMed] [Google Scholar]
- 4.Bacon C W, White J F., Jr . In: Microbial Endophytes. Bacon C W, White J F Jr, editors. New York: Dekker; 2000. pp. 237–261. [Google Scholar]
- 5.Ball D M, Pedersen J F, Lacefield G D. Am Sci. 1993;81:370–379. [Google Scholar]
- 6.Bacon C W, Richardson M D, White J F., Jr Crop Sci. 1997;37:1415–1425. [Google Scholar]
- 7.Bush L P, Wilkinson H H, Schardl C L. Plant Physiol. 1997;114:1–7. doi: 10.1104/pp.114.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malinowski D P, Belesky D P. Crop Sci. 2000;40:923–940. [Google Scholar]
- 9.Schardl C L. Annu Rev Phytopathol. 1996;34:109–130. doi: 10.1146/annurev.phyto.34.1.109. [DOI] [PubMed] [Google Scholar]
- 10.Lyons P C, Plattner R D, Bacon C W. Science. 1986;232:487–489. doi: 10.1126/science.3008328. [DOI] [PubMed] [Google Scholar]
- 11.Brunner R, Stutz P L, Tscherter H, Stadler P A. Can J Chem. 1979;57:1638–1641. [Google Scholar]
- 12.Bacon C W. J Anim Sci. 1995;73:861–870. doi: 10.2527/1995.733861x. [DOI] [PubMed] [Google Scholar]
- 13.Cross D L, Redmond L M, Strickland J R. J Anim Sci. 1995;73:899–908. doi: 10.2527/1995.733899x. [DOI] [PubMed] [Google Scholar]
- 14.Paterson J, Forcherio C, Larson B, Samford M, Kerley M. J Anim Sci. 1995;73:889–898. doi: 10.2527/1995.733889x. [DOI] [PubMed] [Google Scholar]
- 15.Porter J K. J Anim Sci. 1995;73:871–880. doi: 10.2527/1995.733871x. [DOI] [PubMed] [Google Scholar]
- 16.Hoveland C. Agr Ecosyst Environ. 1993;44:3–12. [Google Scholar]
- 17.Rowan D D. Agr Ecosyst Environ. 1993;44:103–122. [Google Scholar]
- 18.Schardl C L, Leuchtmann A, Tsai H-F, Collett M A, Watt D M, Scott D B. Genetics. 1994;136:1307–1317. doi: 10.1093/genetics/136.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen M J, Leuchtmann A, Rowan D D, Tapper B A. Mycol Res. 1993;97:1083–1092. [Google Scholar]
- 20.Marahiel M A, Stachelhaus T, Mootz H D. Chem Rev. 1997;97:2651–2673. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 21.Riederer B, Han M, Keller U. J Biol Chem. 1996;271:27524–27530. doi: 10.1074/jbc.271.44.27524. [DOI] [PubMed] [Google Scholar]
- 22.Walzel B, Riederer B, Keller U. Chem Biol. 1997;4:223–230. doi: 10.1016/s1074-5521(97)90292-1. [DOI] [PubMed] [Google Scholar]
- 23.Tsai H-F, Wang H, Gebler J C, Poulter C D, Schardl C L. Biochem Biophys Res Commun. 1995;216:119–125. doi: 10.1006/bbrc.1995.2599. [DOI] [PubMed] [Google Scholar]
- 24.Tudzynski P, Holter K, Correia T, Arntz C, Grammel N, Keller U. Mol Gen Genet. 1999;261:133–141. doi: 10.1007/s004380050950. [DOI] [PubMed] [Google Scholar]
- 25.Panaccione D G. Mycol Res. 1996;100:429–436. [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 27.Orbach M J. Gene. 1994;150:159–162. doi: 10.1016/0378-1119(94)90877-x. [DOI] [PubMed] [Google Scholar]
- 28.Murray F R, Latch G C M, Scott D B. Mol Gen Genet. 1992;233:1–9. doi: 10.1007/BF00587554. [DOI] [PubMed] [Google Scholar]
- 29.Rasmussen J B, Panaccione D G, Fang G-C, Hanau R M. Mol Gen Genet. 1992;235:74–80. doi: 10.1007/BF00286183. [DOI] [PubMed] [Google Scholar]
- 30.Xu J-R, Hamer J E. Fung Genet Newsl. 1995;42:80. [Google Scholar]
- 31.Latch G C M, Christensen M J. Ann Appl Biol. 1985;107:17–24. [Google Scholar]
- 32.Gwinn K D, Collins-Shepard M H, Reddick B B. Phytopathology. 1991;81:747–748. [Google Scholar]
- 33.An Z-q, Siegel M R, Hollin W, Tsai H-F, Schmidt D, Schardl C L. Appl Environ Microbiol. 1993;59:1540–1548. doi: 10.1128/aem.59.5.1540-1548.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hill N S, Rottinghaus G E, Agee C S, Schultz L M. Crop Sci. 1993;33:331–333. [Google Scholar]
- 35.Annis S L, Panaccione D G. Can J Microbiol. 1998;44:80–86. doi: 10.1139/w97-130. [DOI] [PubMed] [Google Scholar]
- 36.Groppe K, Boller T. Appl Environ Microbiol. 1997;63:1543–1550. doi: 10.1128/aem.63.4.1543-1550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor B, Powell A. Focus. 1982;4:4–6. [Google Scholar]
- 38.Wang J. Ph.D. thesis. Lexington: Univ. of Kentucky; 2000. [Google Scholar]
- 39.Bazely D R, Vicari M, Emmerich S, Filip L, Lin D, Inman A. J Appl Ecol. 1997;34:847–860. [Google Scholar]
- 40.Rottinghaus G E, Garner G B, Cornel C N, Ellis J L. J Agric Food Chem. 1991;39:112–115. [Google Scholar]
- 41.Shelby R A, Olsovska J, Havlicek V, Flieger M. J Agric Food Chem. 1997;45:4674–4679. [Google Scholar]
- 42.Panaccione D G, Scott-Craig J S, Pocard J A, Walton J D. Proc Natl Acad Sci USA. 1992;89:6590–6594. doi: 10.1073/pnas.89.14.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson R D, Johnson L, Itoh Y, Kodama M, Otani H, Kohmoto K. Mol Plant–Microbe Interact. 2000;13:742–753. doi: 10.1094/MPMI.2000.13.7.742. [DOI] [PubMed] [Google Scholar]