Abstract
The structural flexibility of β-karyopherins is critical to mediate the interaction with transport substrates, nucleoporins, and the GTPase Ran. In this paper, we provide structural evidence that the molecular recognition of the transport adaptor snurportin by importin β follows the population selection mechanism. We have captured two drastically different conformations of importin β bound to the snurportin importin β binding domain trapped in the same crystallographic asymmetric unit. We propose the population selection may be a general mechanism used by β-karyopherins to recognize transport substrates.
Transport factors of the importin β-superfamily (also known as β-karyopherins) include at least 20 different known members in humans (1). β-Karyopherins can function as both import and export receptors, and their interaction with transport cargos is either direct or mediated by adaptors. At the structural level, β-karyopherins share a common helical architecture consisting of 18–21 repeated HEAT repeats (2). During transport, β-karyopherins interact simultaneously with import and export cargos and nucleoporins, in a process that is finely regulated by the small GTPase Ran.
In the past 10 years, several β-karyopherin crystal structures have been determined. Structures of human importin β1 and yeast karyopherin β2 have been determined in complex with specific import cargos (3–6) and RanGTP (7, 8). More recently, the crystal structures of the export factors CRM1 (9, 10), Cse1 (11, 12), Xpot (13), and Exportin-5 (14) were reported (2, 15), revealing the drastically distinct conformation adopted by these factors in their cytoplasmic and nuclear state (2, 15–17). Overall, β-karyopherins are flexible solenoids fluctuating between a relaxed and highly strained tertiary structure as a result of cargo binding. This structural switch can occur in at least two ways. In the case of import factor karyopherin β2 (6, 8, 18) and export factor Cse1 (11, 12), the structural rearrangements accompanying ligand binding are the result of discrete hinge movements of entire domains moving with respect to each other. This is also termed segmental conformational heterogeneity (18). In contrast, export factor Xpot in the empty cytosolic state and in the nuclear ternary complex with tRNA and Ran (13) shows differences in the conformation of the protein that are the cumulative result of small changes distributed over the entire span of the solenoid.
The highly strained conformation of karyopherins bound to their substrates raises the question of how an entropically disfavored conformational state can be stably populated in solution to allow efficient transport through the nuclear pore complex (NPC). In the recognition of the importin β binding (IBB) domain by importin β, the charge complementarity between the acidic concave surface of importin β and the highly positive IBB domain provides the energetic contribution to overcome the energetic cost of distorting the unstrained empty importin β (19), thus forcing its tertiary structure into a highly strained, cargo-bound conformation (2, 15). Thermodynamically, the strained conformation of importin β bound to the IBB domain corresponds to a tertiary structure in which backbone and side chain conformations do not fall in local energy minima. In turn, the enthalpic gain caused by surface complementarity outweighs the entropic cost of being “strained”, making the overall free energy of binding negative. Matsuura and Stewart described the strained conformation of export factor Cse1p bound to Kap60p and RanGTP (11) as a “spring-loaded” solenoid. In the case of the importin β bound to the sIBB domain, it is difficult to determine solely on the basis of the previous crystal structure if the spring-loaded conformation of importin β is the product of a protein-protein induced fit (20) or if the sIBB domain selects at equilibrium a highly strained conformer of importin β, thereby leading to a dynamic population shift, as suggested in the population selection model (21).
In this paper, we report a new crystal form of human importin β bound to the snurportin IBB domain that contains two importin β • sIBB domain (residues 25–65) complexes in the asymmetric unit in drastically different conformations. This new structure sheds light on the role of conformational selection in the recognition of IBB domains by importin β.
EXPERIMENTAL PROCEDURES
Protein Purification and Crystallization
Expression and purification of human importin β (4, 22) and αIBB (residues 11–54), sIBB (residues 1–65), and sIBB (residues 25–65) peptides were previously described (22). Crystals of importin β bound to a chemically synthesized sIBB domain (residues 25–65) were obtained using the hanging drop vapor diffusion method by mixing together 2 µL of a gel filtration-purified importin β • sIBB domain (residues 25–65) complex at 15 mg/mL with 2 µL of 18% PEG 8000 and 50 mM sodium chloride (pH 6.5) at 293 K. Crystals appeared within 3–5 days, were cryo-protected in 27.5% ethylene glycol, and were frozen in liquid nitrogen.
Data Collection, Structure Solution, and Refinement
Diffraction data were collected at MacCHESS beamline A1 on a Quantum-210 CCD detector and at beamline X6A at the Brookhaven National Synchrotron Light Source (NSLS) on a Quantum Q4 CCD detector. Data indexing, integration, and scaling were conducted with HKL2000 (23). The structure of importin β in a complex with the sIBB domain (residues 25–65) was determined by molecular replacement using Phaser (24). The best solution was obtained using two search models corresponding to the previously determined importin β • sIBB domain (residues 25–65) complex [Protein Data Bank (PDB) entry 2P8Q] (complex A) (22) and an N-terminal fragment of importin β spanning HEATs 1–11 (complex B). An initial Fo−Fc electron density difference map calculated with phases obtained from the partial phasing model allowed us to build the remaining C-terminal HEATs 12–19 of complex B. In complex A, the electron density for the entire sIBB domain (residues 25–65) is very clear and was included in the final atomic model. In contrast, in complex B, only sIBB domain residues 39–64 were modeled, as no density is seen for N-terminal residues 25–38. Model building was done using COOT (25), and refinement of atomic coordinates was conducted with phenix.refine from the PHENIX software suite (26), using iterative cycles of positional refinement and grouped B-factor refinement with individual HEAT repeats and sIBB domains defined as individual TLS domains. The final model has an Rfactor of 26.35% and an Rfree of 31.29%. The model has good geometry (see Table 1), with 82.7% (1365) of the residues in the most favored regions of the Ramachandran plot. Only 0.3% of the residues in the structure, corresponding to importin β residues Gly396, Lys710, and Pro785 in complex A and Gly396 and Asn702 in complex B, are in disallowed regions. All outliers fall in loops connecting HEAT repeat helices and have positions of α-carbon and carbonyl oxygen well-defined in the final 2Fo − Fc electron density map. Refinement and data collection statistics are listed in Table 1.
Table 1.
Crystallographic Data Collection and Refinement Statistics
| PDB entry 3LWW | |
|---|---|
| Data Collection | |
| wavelength (Å) | 0.918 |
| space group | P1211 |
| unit cell dimensions (Å) | a = 101.65, b = 101.73, c = 101.96 |
| unit cell angles (deg) | α = γ = 90, β = 110.88 |
| resolution range (Å) | 15–3.15 |
| B value from Wilson plot (Å2) | 73.90 |
| total no. of observations | 63234 |
| no. of unique observations | 32721 |
| completeness (%) | 99.2 (69.0)a |
| Rsymb (%) | 12.5 (53.5)a |
| 〈I〉/〈σ(I)〉 | 16.3 (1.2)a |
| Refinement | |
| no. of reflections (15–3.15 Å) | 59672 |
| Rwork/Rfreec (%) | 26.35/31.29 |
| no. of water molecules | 0 |
| B value of model (Å2) | 88.17 |
| rmsdd from ideal bond lengths (Å) | 0.003 |
| rmsdd from ideal bond angles (deg) | 0.716 |
| Ramachandran plot (%) (residues) | |
| core region | 82.7 (1365) |
| allowed region | 15.9 (262) |
| generously allowed region | 1.1 (18) |
| disallowed region | 0.3 (5) |
The numbers in parentheses refer to the statistics for the outer resolution shell (3.26–3.15 Å).
Rsym = Σi,h|I(i,h) − 〈I(h)〉|/Σi,h|I(i,h)|, where I(i,h) and 〈I(h)〉 are the ith and mean measurements of intensity of reflection h, respectively.
The Rfree value was calculated using 5% of the data.
Root-mean-square deviation.
Structure Analysis
Interface surface area and solvation free energy (ΔiG) calculations were performed using the PISA server (27). All figures in the paper were prepared using Pymol (28). Atomic coordinates and experimental structure factors have been deposited in the Protein Data Bank (entry 3LWW).
Circular Dichroism
Circular dichroism (CD) spectra were recorded using 20 µM αIBB or sIBB (residues 1–65) or sIBB (residues 25–65) domains in 20 mM sodium phosphate buffer and 10 mM NaCl (pH 7.4). Spectra were recorded at 20 °C between 185 and 260 nm with a Jasco J-810 spectropolarimeter using a data point increment of 0.5 nm. The signal:noise ratio was increased by measurement of three scans per spectrum; spectra were averaged, and the buffer signal was subtracted. 2,2,2-Trifluoroethanol (TFE) was added to concentrations of 30 and 50% (w/w).
RESULTS
A New Crystal Form of Importin β Bound to the Snurportin IBB Domain
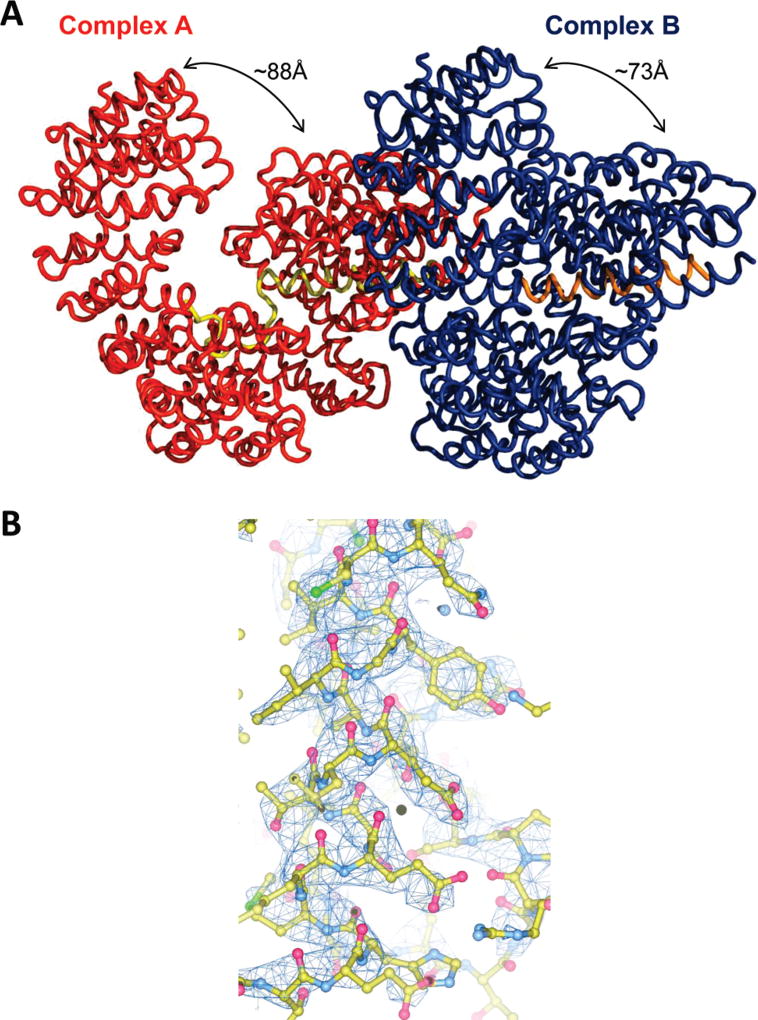
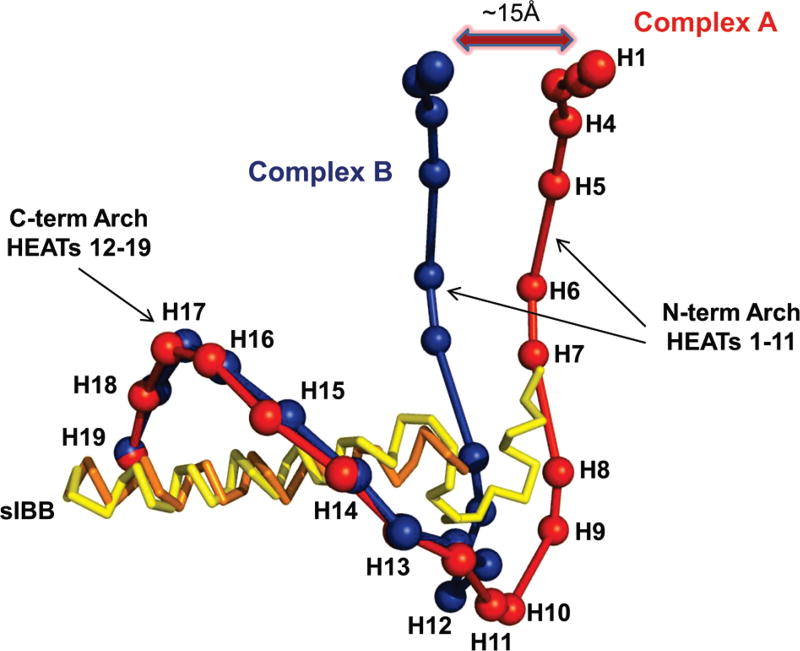
We have determined a new crystal form that contains two importin β • sIBB domain (residues 25–65) complexes in the asymmetric unit in drastically different conformations (Figure 1A). The new crystal form belongs to a primitive monoclinic space group with nearly identical unit cell axis lengths of ~101 Å (Table 1). The structure was determined by molecular replacement, and despite the limited resolution of diffraction data (~3.15 Å), the quality of the resulting electron density (Figure 1B) was sufficiently good to build a complete atomic model of both importin β • sIBB domain complexes trapped in the crystallographic asymmetric unit (Figure 1A). In the first complex (complex A), importin β is bound to an entire sIBB domain (residues 25–65) in a closed conformation that is nearly identical to the high-resolution structure previously reported (rmsd of 1.32Å) (22). Unexpectedly, the second complex in the asymmetric unit (complex B) presents an even more strained conformation of importin β with N- and C-termini interacting with each other. Superimposition of the two importin β • sIBB domain complexes in the asymmetric unit best describes the extraordinary structural plasticity of this protein (Figure 2). Importin β can be considered to be formed by two arches, spanning N- and C-terminal HEAT repeats 1–11 and 12–19, respectively. Whereas the two complexes in the asymmetric unit have superimposable C-termini, the N-terminal arches exhibit dramatic differences. In complex A, N- and C-termini are ~88Å from each other, while in complex B, this distance is only ~73Å (Figure 2). In addition, the N-terminal domain of complex B is rotated ~10° relative to the N-terminal domain of complex A. Overall, the rmsd for α-carbons between importin β • sIBB domain complexes A and B is ~2.65Å (~2.60Å if the sIBB domain is omitted). The structure of importin β in complex B represents the most strained conformation of this transport receptor visualized crystallographically to date.
Figure 1.
Conformational plasticity of importin β bound to the sIBB domain. (A) Asymmetric unit content with two importin β • sIBB domain complexes in different conformations shown as coils. Importin β is colored red and blue and the sIBB domain yellow and orange. (B) Representative 2Fo − Fc electron density computed at 3.15 Å resolution and contoured 1.5σ above background. This figure was generated with Pymol (28).
Figure 2.
Superimposed beads-on-string view of the two complexes in the asymmetric unit. The two importin β structures are colored blue and red, while the sIBB domains are colored yellow and orange. The maximum distance between importin β N-termini in complexes A and B is ~15Å.
Comparison with Previous Structures of Importin β Bound to the sIBB Domain
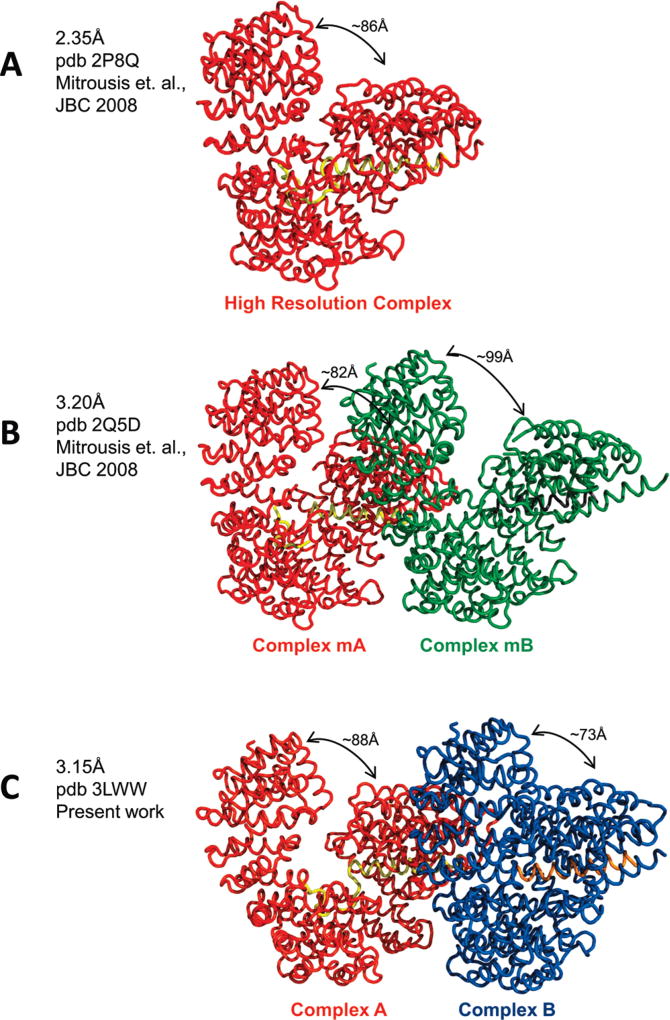
We previously reported the high-resolution structure of importin β bound to the sIBB domain (residues 25–65) determined to ~2.35 Å resolution (PDB entry 2P8Q) (Figure 3A) (22). This structure presents a strained conformation of importin β very similar to that seen in complex with the IBB domain of importin α (rmsd for α-carbons of 0.96 Å) (4). A lower-resolution structure of the importin β • sIBB domain (residues 25–65) complex was also determined (PDB entry 2Q5D) after a short (~30 min) dehydration in highly concentrated solutions of PEG 8000. This crystal form reveals two importin β complexes in the same asymmetric unit, in slightly different conformations (rmsd of ~2.14 Å) (Figure 3B) (22). In one complex (complex mA), importin β adopts a closed conformation nearly identical to the high-resolution structure of the importin β • sIBB domain complex (rmsd of ~1.2 Å) (Figure 3A,B) (22). In the second complex in the asymmetric unit (complex mB), importin β is distinctly open, with C-terminal HEAT repeats 12–19 swung up to 20 Å from the corresponding position in the closed conformation. The open tertiary structure of importin β in complex mB resembles that of Kap95p, the yeast homologue of importin β, in complex with RanGTP (7). The rmsd for α-carbons between importin β in mA and mB is ~2.05 Å.
Figure 3.
Comparison of the new crystal form with previously determined structures of the importin β • sIBB domain (residues 25–65) complex: (A) 2.35Å structure (PDB entry 2P8Q) with one importin β • sIBB domain (residues 25–65) complex in the asymmetric unit, (B) 3.2 Å structure (PDB entry 2Q5D) with two importin β • sIBB domain (residues 25–65) complexes (indicated as mA-mB) in the same asymmetric unit, and (C) 3.15 Å structure presented in this paper (PDB entry 3LWW) with complexes A and B in the same asymmetric unit. All structures in panels A–C are aligned using importin β in complex A as a reference that is colored red.
Comparison of the new crystal form with those deposited in PDB entries 2P8Q and 2Q5D is useful to fully appreciate the spectrum of conformational rearrangements in importin β triggered upon sIBB domain binding. The less strained conformation of importin β in complex A is very similar (rmsd of ~1.32Å) to the high-resolution structure of the importin β • sIBB domain complex (PDB entry 2P8Q) and to importin β in complex mA of PDB entry 2Q5D (rmsd of ~1.35Å) (Figure 3A–C) (22). In contrast, more dramatic structural differences exist between importin β in complex B and the high-resolution structure (rmsd of ~3.0 Å) or importin β in complex mB (22) (rmsd of ~2.4 Å) (Figure 3A–C). Likewise, the asymmetric unit content of the new crystal form taken as a “whole” (complexes A and B) is vastly different from complexes mA and mB previously described (rmsd of ~4.6 Å) (Figure 3B,C). Crystallographically, structure factor amplitudes for the new 3.15 Å crystal form scale to the previous 3.2Å structure (PDB entry 2Q5D) (22)with an Rfactor of >50%, strengthening the idea that these two structures are intrinsically different. Thus, the new crystal form of importin β bound to the sIBB domain (residues 25–65) is distinct from previously determined structures of the importin β • sIBB domain complex and expands the spectrum of importin β conformers observed crystallographically.
Intramolecular Contacts Stabilize the Closed Conformation of Importin β in Complex B
As the pH used to obtain this new crystal form was close to physiological (~6.5), we were curious to determine the forces stabilizing importin β in the two conformations seen in the asymmetric unit. Using PISA (27), we carefully analyzed intermolecular binding interfaces between importin β and the sIBB domain, as well as intramolecular contacts within importin β forms in the asymmetric unit. Interestingly, we found that importin β in complex B forms an extended network of intramolecular contacts between N-terminal HEATs 3–7 and C-terminal HEATs 16–19. Most notably, two intramolecular salt bridges are found between the Oδ2 atom of importin β Asp71 and the amine nitrogen (Nz) atom of Lys710 and the Oε2 atom of Glu299 and the terminal group of Arg852. These contacts, together with a hydrogen bond between the Oδ2 atom of Asp71 and the amine nitrogen (Nz) atom of Lys710, appear to greatly stabilize the closed conformation of importin β in complex B. As a result of the highly strained conformation of the importin β backbone in complex B, the N-terminal sIBB domain moiety between residues 25 and 38 fails to be stabilized by the acidic surface of the protein and is not seen in the electron density. Instead, in complex A, this region of the sIBB domain is exceptionally well-defined and makes contacts with residues exposed on the surface of importin β among HEATs 7–12 (Figure 2). Consequently, the calculated interface area between the sIBB domain and importin β is larger in complex A (1993.9Å2) than in complex B (1162.7 Å2), where one-third of the sIBB domain is not visible. This results in solvation free energies (ΔiG) upon formation of the importin β • sIBB domain complex of −10.5 and −4.1 kcal/mol for complexes A and B, respectively. Thus, on the basis of the higher negative value ofΔiG caused by intermolecular contacts, one would predict complex A to be more populated at equilibrium, as compared to the highly strained complex B. In our structure, however, the extensive network of intramolecular contacts within the N- and C-termini of importin β in complex B compensates for the reduced binding surface area with the sIBB domain, making complexes A and B sufficiently populated at equilibrium to be crystallized. We conclude that the highly strained conformation of importin β can be stabilized by the enthalpic energy of binding resulting from inter- or intramolecular contacts. Consequently, the intrinsic structural plasticity of importin β, more than the absolute number of contacts with the IBB domain, is the key to generating the electrostatic complementarity required to outweigh the energetic cost of being strained.
Structure of the sIBB Domain in Solution
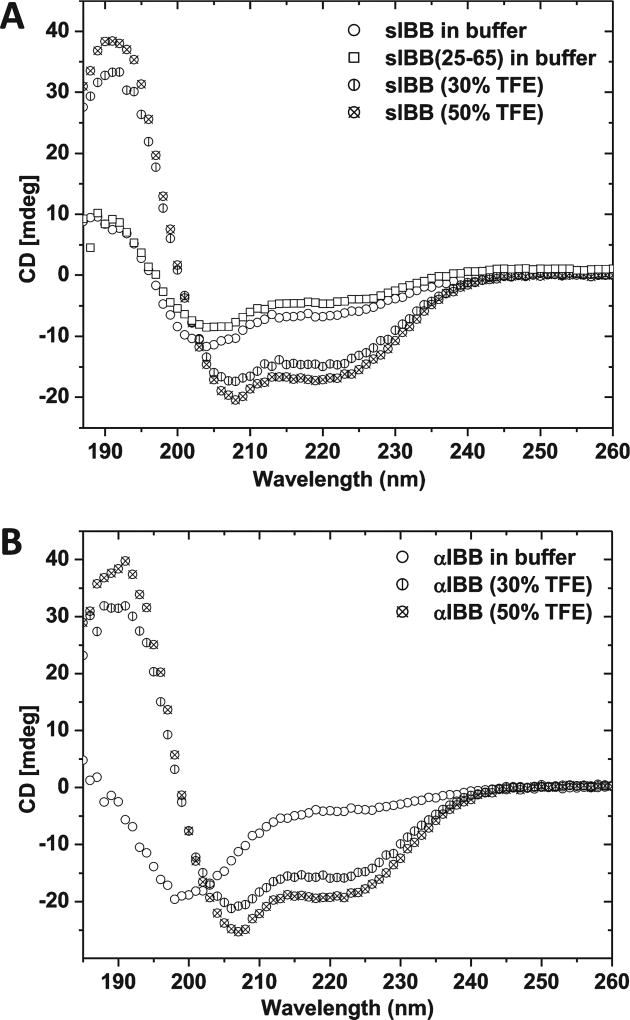
Importin β bound to the αIBB domain was also captured crystallographically in slightly different conformations (4). However, in this case, different conformers of the importin β • αIBB domain complex crystallized as individual species containing only one conformation of importin β in the crystal lattice. Likewise, the sIBB domain was recently found to modulate importin β affinity for nucleoporins, suggesting that despite the high degree of sequence similarity, αIBB and sIBB domains have distinctly different functional properties (29). To understand how the sIBB domain affects importin β plasticity, we investigated if the structure of this short domain in solution is different from that seen in the crystal structure. Circular dichroism (CD) spectra of the sIBB domain (residues 1–65) recorded at pH 7.4 displayed two double minima in the ellipticity at 222 and 208 nm, suggesting this domain is partially helical in solution (Figure 4A). Notably, adding 30 or 50% trifluoroethanol (TFE) increased the magnitude of the α-helical signal 2-fold, to yield a final helical content roughly equivalent to that of the sIBB domain structure seen in complex A. A similar propensity to adopt a helical conformation was previously observed for the αIBB domain (30), which, however, is completely unstructured in the absence of TFE (Figure 4B) and becomes folded upon binding to importin β (4). In the crystal structure of CRM1 bound to snurportin, the 11 N-terminal residues of snurportin fold into an α-helix, which functions as the nuclear export signal (NES) (9, 10). To rule out the possibility that the sIBB domain (residues 1–65) displays helical content due to the NES helix, we repeated the CD analysis using a smaller sIBB domain spanning residues 25–65, which showed helical content identical to that of the sIBB domain (residues 1–65) (Figure 4A). Thus, the helical structure of sIBB seen in solution by CD resides in its C-terminus, likely between residues 40 and 65, as shown in the structure of snurportin bound to CRM1 (10). These data further support the idea of an intrinsic structural difference between the sIBB and αIBB domains, which is directly dictated by the different primary sequence and propensity to adopt helical structure in solution.
Figure 4.
Folding of sIBB domains in solution. Circular dichroism analysis of (A) the sIBB domain (residues 1–65 or 25–65) and (B) the αIBB domain (residues 11–54) at pH 7.4. The sIBB domain is partially helical in solution, while the αIBB domain adopts a completely random coil conformation. In both panels A and B, addition of trifluoroethanol (TFE) to a final concentration of 30 or 50% rapidly induced helix formation.
DISCUSSION
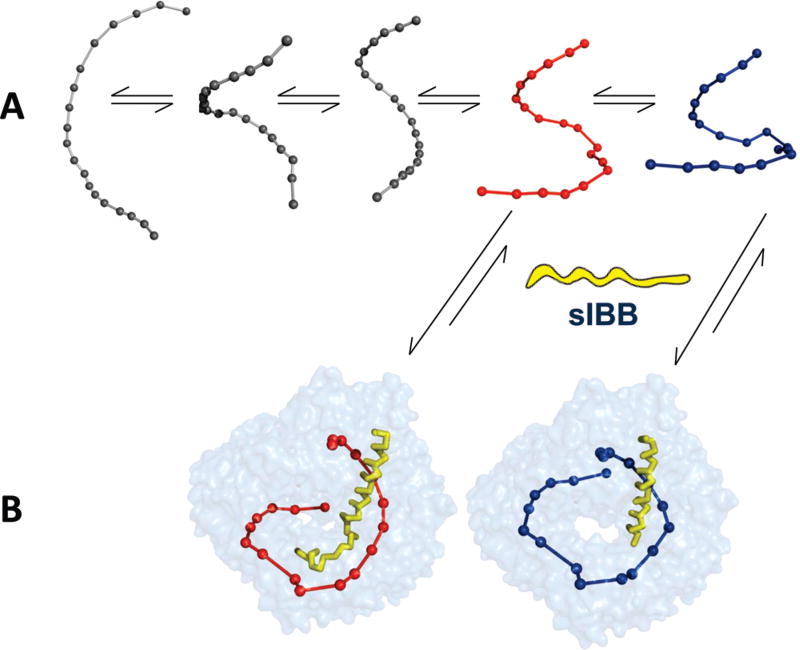
Efficient and specific recognition of transport cargos is essential to ensuring nucleocytoplasmic transport. Although importin β can also bind certain cargos directly (3), most import substrates are bound indirectly via adaptor proteins such as importin α and snurportin. In both cases, importin β recognizes only a short N-terminal region of the adaptor, known as the IBB domain. Although several structures of importin β bound to IBB domains have been determined crystallographically (4, 22), it remains unclear how the IBB domain is specifically recognized by importin β in the cytoplasm to initiate a round of nuclear import. Two models have been proposed to describe how β-karyopherins recognize their substrates (20, 21, 31) and could be used to interpret the recognition of IBB domains by importin β. According to the induced fit hypothesis, importin β changes its structure upon binding to the sIBB domain to adopt a conformation that is more complementary to the IBB domain (e.g., the strained solenoid). This in turn results in a large release of energy that locks the structure of importin β in a stably populated strained conformation. Thermodynamically, this conformation is not the native state, which is defined as the structure with the lowest energy. In contrast, according to the population selection model (20, 21, 31), importin β exists in solution as an ensemble of conformers, freely interexchanging at equilibrium, because of the low free energy of interconversion. A ligand such as the sIBB domain might interact with one or more importin β “conformers” in solution, thus shifting the equilibrium of the entire population to that specific conformer (Figure 5A). In this study, we have identified two populations of importin β bound to two slightly distinct sIBB domains, trapped in the same crystallographic asymmetric unit. If the importin β • sIBB domain complex was the final product of an “induced fit recognition” (e.g., the “lock and key”), we would expect to see only one stably populated complex in the same crystal lattice. Alternatively, we may expect different crystal forms with importin β adopting slightly distinct conformations as a result of crystal contact interference, crystallization agents, or a nonphysiological pH. Instead, the presence of two drastically different populations of importin β • sIBB domain complexes trapped in the same crystallographic asymmetric unit lends support to the population selection model. We propose that in solution importin β exists as an ensemble of different conformers (Figure 5A). Selection of a given population by the sIBB domain shifts the equilibrium to formation of a tight complex that becomes sufficiently populated in solution to be crystallized (Figure 5B).
Figure 5.
Population selection model proposed for recognition of the sIBB domain by importin β. (A) Putative conformers of importin β are shown as beads-on-string views to emphasize the different degrees of intramolecular opening. (B) Two importin β conformers with different structures are stabilized by the sIBB domain and hence trapped in the same crystal lattice.
Two molecular determinants appear to influence the selection of importin β conformers. First is the enthalpic complementarity between importin β and the sIBB domain, which provides the energy necessary to strain the importin β solenoid structure. As shown, this can be achieved either by extensive intermolecular contacts between importin β and the sIBB domain or by a reduced intermolecular component compensated by extensive intramolecular contacts between importin β N- and C-termini. Second, the IBB domain secondary structure, or simply its propensity to fold in solution, governs the mode of recognition by importin β. As demonstrated by CD spectroscopy, the sIBB domain, and not its counterpart, the αIBB domain, adopts a helical structure in solution, likely between C-terminal residues 40 and 65. We propose this folded structure functions as a template to select importin β conformers, thereby shifting the equilibrium of the importin β population to a defined number of conformers that yield high enthalpic complementarity bound to the sIBB domain. Thus far, three different crystal forms of the importin β • sIBB domain complex have been determined crystallographically, providing atomic information about three structurally distinct conformers of importin β (Figure 3): (i) the strained conformation of importin β seen in the high-resolution structure (PDB entry 2P8Q) as well as in complexes mA and A of PDB entries 2Q5D (22) and 3LWW, respectively; (ii) the unstrained conformation of importin β in complex mB of PDB entry 2Q5D (22); and (iii) the ultrastrained importin β seen in complex B of PDB entry 3LWW (this paper). This scenario is completely different from the recognition of the αIBB domain (4, 30, 32), which is completely unfolded in solution and adopts a helical structure only upon binding to importin β. Despite the high degree of similarity between the primary sequences of the α- and sIBB domains and their comparable binding affinity for importin β (29), the two peptides have different propensities to fold in solution, which results in a distinct ability to select importin β conformers in solution. Although slightly different conformations of importin β were previously observed in discrete crystal forms (4), two populations of importin β bound to the αIBB domain were never found in the same crystal form. It is interesting to speculate that the folding upon binding of αIBB is also accompanied by a concomitant selection of one importin β conformer that yields the greatest binding complementarity, in a scenario that closely resembles an induced fit.
In conclusion, the structural work presented in this paper expands the repertoire of importin β conformers determined crystallographically in complex with the sIBB domain. The plasticity seen in our new crystal form argues against the induced fit model and supports the idea of a dynamic selection of strained conformations of importin β by the sIBB domain. Future studies will have to determine whether population selection is a general feature of β-karyopherins recognizing folded substrates such as the sIBB domain while induced fit is the prevalent recognition mode for cargos that fold upon binding, such as the IBB domain of importin α.
Acknowledgments
We thank Kaylen Lott for critical reading of the manuscript and Gregory Mitrousis for help with crystallization of importin β. We are thankful to the beam line staff at macCHESS and NSLS beamline X6A for beam time and help in data collection.
Footnotes
This work was supported by National Institutes of Health Grant GM074846-01A1 to G.C.
References
- 1.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 2.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 3.Cingolani G, Bednenko J, Gillespie MT, Gerace L. Molecular basis for the recognition of a nonclassical nuclear localization signal by importin β. Mol. Cell. 2002;10:1345–1353. doi: 10.1016/s1097-2765(02)00727-x. [DOI] [PubMed] [Google Scholar]
- 4.Cingolani G, Petosa C, Weis K, Muller CW. Structure of importin-β bound to the IBB domain of importin-α. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 5.Lee SJ, Sekimoto T, Yamashita E, Nagoshi E, Nakagawa A, Imamoto N, Yoshimura M, Sakai H, Chong KT, Tsukihara T, Yoneda Y. The structure of importin-β bound to SREBP-2: Nuclear import of a transcription factor. Science. 2003;302:1571–1575. doi: 10.1126/science.1088372. [DOI] [PubMed] [Google Scholar]
- 6.Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherin β2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee SJ, Matsuura Y, Liu SM, Stewart M. Structural basis for nuclear import complex dissociation by RanGTP. Nature. 2005;435:693–696. doi: 10.1038/nature03578. [DOI] [PubMed] [Google Scholar]
- 8.Chook YM, Blobel G. Structure of the nuclear transport complex karyopherin-β2-Ran × GppNHp. Nature. 1999;399:230–237. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- 9.Dong X, Biswas A, Suel KE, Jackson LK, Martinez R, Gu H, Chook YM. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature. 2009;458:1136–1141. doi: 10.1038/nature07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monecke T, Guttler T, Neumann P, Dickmanns A, Gorlich D, Ficner R. Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science. 2009;324:1087–1091. doi: 10.1126/science.1173388. [DOI] [PubMed] [Google Scholar]
- 11.Matsuura Y, Stewart M. Structural basis for the assembly of a nuclear export complex. Nature. 2004;432:872–877. doi: 10.1038/nature03144. [DOI] [PubMed] [Google Scholar]
- 12.Cook A, Fernandez E, Lindner D, Ebert J, Schlenstedt G, Conti E. The structure of the nuclear export receptor Cse1 in its cytosolic state reveals a closed conformation incompatible with cargo binding. Mol. Cell. 2005;18:355–367. doi: 10.1016/j.molcel.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 13.Cook AG, Fukuhara N, Jinek M, Conti E. Structures of the tRNA export factor in the nuclear and cytosolic states. Nature. 2009;461:60–65. doi: 10.1038/nature08394. [DOI] [PubMed] [Google Scholar]
- 14.Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 15.Conti E, Muller CW, Stewart M. Karyopherin flexibility in nucleocytoplasmic transport. Curr. Opin. Struct. Biol. 2006;16:237–244. doi: 10.1016/j.sbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 16.Suel KE, Cansizoglu AE, Chook YM. Atomic resolution structures in nuclear transport. Methods. 2006;39:342–355. doi: 10.1016/j.ymeth.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madrid AS, Weis K. Nuclear transport is becoming crystal clear. Chromosoma. 2006;115:98–109. doi: 10.1007/s00412-005-0043-3. [DOI] [PubMed] [Google Scholar]
- 18.Cansizoglu AE, Chook YM. Conformational heterogeneity of karyopherin β2 is segmental. Structure. 2007;15:1431–1441. doi: 10.1016/j.str.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukuhara N, Fernandez E, Ebert J, Conti E, Svergun D. Conformational variability of nucleo-cytoplasmic transport factors. J. Biol. Chem. 2004;279:2176–2181. doi: 10.1074/jbc.M309112200. [DOI] [PubMed] [Google Scholar]
- 20.Zachariae U, Grubmuller H. A highly strained nuclear conformation of the exportin Cse1p revealed by molecular dynamics simulations. Structure. 2006;14:1469–1478. doi: 10.1016/j.str.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Nevo R, Stroh C, Kienberger F, Kaftan D, Brumfeld V, Elbaum M, Reich Z, Hinterdorfer P. A molecular switch between alternative conformational states in the complex of Ran and importin β1. Nat. Struct. Biol. 2003;10:553–557. doi: 10.1038/nsb940. [DOI] [PubMed] [Google Scholar]
- 22.Mitrousis G, Olia AS, Walker-Kopp N, Cingolani G. Molecular basis for the recognition of snurportin 1 by importin β. J. Biol. Chem. 2008;283:7877–7884. doi: 10.1074/jbc.M709093200. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 28.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 29.Lott K, Bhardwaj A, Mitrousis G, Pante N, Cingolani G. The importin β binding domain modulates the avidity of importin β for the nuclear pore complex. J. Biol. Chem. 2010;285:13769–13780. doi: 10.1074/jbc.M109.095760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cingolani G, Lashuel HA, Gerace L, Muller CW. Nuclear import factors importin α and importin β undergo mutually induced conformational changes upon association. FEBS Lett. 2000;484:291–298. doi: 10.1016/s0014-5793(00)02154-2. [DOI] [PubMed] [Google Scholar]
- 31.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koerner C, Guan T, Gerace L, Cingolani G. Synergy of silent and hot spot mutations in importin β reveals a dynamic mechanism for recognition of a nuclear localization signal. J. Biol. Chem. 2003;278:16216–16221. doi: 10.1074/jbc.M301137200. [DOI] [PubMed] [Google Scholar]