Abstract
Therapeutic resistance is a central problem in clinical oncology. We have developed a systematic genome-wide computational methodology to allow prioritization of patients with favorable and poor therapeutic response. Our method, which integrates DNA methylation and mRNA expression data, uncovered a panel of 5 differentially methylated sites, which explain expression changes in their site-harboring genes, and demonstrated their ability to predict primary resistance to androgen-deprivation therapy (ADT) in the TCGA prostate cancer patient cohort (hazard ratio = 4.37). Furthermore, this panel was able to accurately predict response to ADT across independent prostate cancer cohorts and demonstrated that it was not affected by Gleason, age, or therapy subtypes. We propose that this panel could be utilized to prioritize patients who would benefit from ADT and patients at risk of resistance that should be offered an alternative regimen. Such approach holds a long-term objective to build an adaptable accurate platform for precision therapeutics.
Keywords: Therapeutic resistance, DNA methylation, mRNA expression, Epigenomics, Androgen-deprivation, Prostate cancer
Highlights
-
•
Integrative DNA methylation and mRNA expression analysis discovers a panel of markers of treatment resistance.
-
•
This panel can predict patients with predisposition to resistance and those who would benefit from the therapy.
-
•
Our approach is applicable to a wide range of therapeutic regimens.
Therapeutic resistance is an emerging clinical problem, with detrimental implications in oncology. Here, we propose a computational approach that integrates genomic and epigenomic data to prioritize patients at risk of treatment resistance. We have integrated DNA methylation and mRNA expression patient profiles, which defined a comprehensive panel of markers of therapeutic response. We have demonstrated that this panel predicts patients with predisposition to resistance and those who would benefit from the therapy. Even though driven by a critical need to investigate resistance to androgen-deprivation therapy in prostate cancer, our approch is applicable to a wide range of therapeutic regimens.
1. Introduction
Prostate cancer is the most common malignancy and one of the leading causes of death in American men [1, 2, 3]. Since prostate cancer initiation and progression depend on androgens [4, 5, 6], androgen-deprivation has been the mainstay of treatment for patients with advanced disease. Even though majority of patients initially respond to androgen-deprivation therapy (ADT), remission lasts 2–3 years on average, with eventual relapse and progression to castration-resistant disease, which is nearly always metastatic and lethal [7,8]. Resistance to ADT and the paucity of the therapeutic options for patients with castration-resistant disease are among major clinical challenges in prostate cancer management [9, 10, 11].
While multifaceted and heterogeneous, prostate cancer is characterized by the scarcity of genomic mutations [12] and absence of well-defined subtypes [13, 14, 15], thus making therapeutic management challenging and suggesting that more complex mechanisms (e.g., interplay of epigenomic and genomic mechanisms) might play a role in treatment response. In the last decade, epigenomics has been at the center of scientific interest, including recognition of its role in cancer initiation and progression [16, 17, 18, 19, 20]. In recent years, one of the most commonly observed epigenomic means, chromatin accessibility (i.e., DNA methylation), has received significant attention due to its role in cell development [21], genomic imprinting (i.e., biological process through which a gene carries information about its ancestor) [22], aging [23] and carcinogenesis [24,25]. DNA methylation (Fig. 1) is defined by an addition of methyl group to the fifth position of cytosine (converting it to 5-methylcytosine). In mammals, methylation of cytosine often occurs in regions where cytosine is followed by guanine (connected through phosphate molecule), named a CpG site [26,27]. A DNA region with frequent occurrences of CpG sites is commonly known as a CpG island or CGI [27,28]. Interestingly, 70% of gene promoter regions are associated with the CGIs, which can alter gene regulation [26]. In fact, if CGI within the promoter region is methylated, it becomes occupied by the Methylated DNA Binding Protein (MDBP) [29], which competes with transcription factor binding. MDBP can act as a transcription repressor or enhancer [30,31], depending on the transcription process it interferes with. In cancer, importance of the CGIs was initially observed among retinoblastoma patients, where CGI hyper-methylation led to silencing of Rb gene [32]; since then numerous groups have demonstrated the significant role of DNA methylation in oncogenesis [25,33, 34, 35].
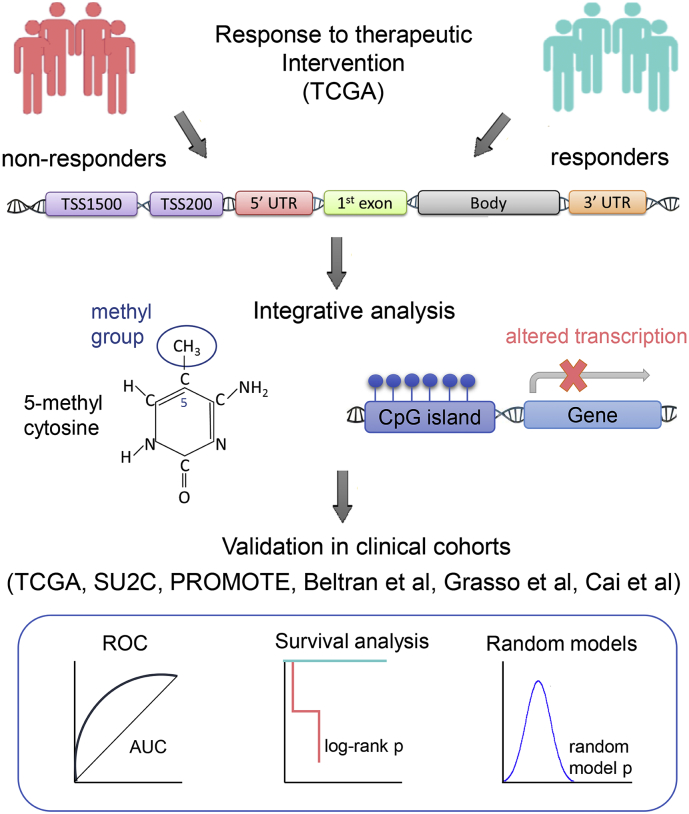
Fig. 1.
Schematic representation of the systematic integrative approach. (Top) Non-responder and responder groups are compared for differentially methylated events/sites. (Middle) Differential methylation is integrated with expression of site-harboring genes. (Bottom) Candidate site-gene panel is evaluated for clinical significance.
In recent years, studies started to link aberrant level of DNA methylation to cellular transformation and clonal expansion [36,37], often implicated in therapeutic response and resistance. For instance, hyper-methylation of MLH1 has been shown to be associated with increased resistance to cisplatin in ovarian cancer [38]; hyper-methylation of HOXC10 has been found to influence resistance to anti-estrogen therapy in ER+ breast cancer [39]; hypo-methylation of ABCB1 had been associated to paclitaxel-resistant ovarian cancer [40], etc. Further, recent studies have demonstrated that integrative analysis is crucial for in-depth understanding of molecular mechanisms involved in therapeutic response, for example (i) correlation between DNA methylation and mRNA expression of FHIT has been suggested as a marker for risk management in non-small cell lung and breast cancer [41]; (ii) aberrant frequencies of genes correlated between DNA methylation (as well as copy number variation) and expression levels could identify molecular subtypes in hepatocellular carcinoma patients [42]; (iii) correlation between DNA methylation and gene expression defined transcriptional patterns in molecular subtypes of breast cancer [43], etc. Thus, a systematic investigation of the effect of DNA methylation on therapeutic response and analysis of its functional effect on the expression of the harboring genes might enhance our understanding of the mechanisms implicated in resistance and provide valuable predictive markers of predisposition to therapeutic failure.
In this study, we have developed a systematic genome-wide integrative approach to analyze DNA methylation and its causal effect on mRNA gene expression to predict response to therapeutic intervention in cancer patients (see schematics in Fig. 1). We have named this approach Epi2GenR - Epigenomic and Genomic mechanisms of treatment Resistance. We have compared (epi) genomic profiles from primary tumors of prostate cancer patients with poor (i.e., non-responders) and favorable (i.e., responders) response to androgen-deprivation therapy and identified a panel of 5 differentially methylated sites, whose methylation changes explain expression variation in their site-harboring genes. We further tested the ability of this panel to predict therapeutic response in non-overlapping independent patient cohorts. In fact, the 5 site-gene panel was able to differentiate patients with predisposition to ADT failure from patients with favorable treatment response in TCGA-PRAD [13] (log-rank p = 0.0191, hazard ratio = 4.37) and other [44, 45, 46, 47, 48]patient cohorts (sensitivity = 100%, AUROC = 0.83, AUROC = 0.98). We have confirmed significant non-random predictive ability of the identified 5 site-gene panel and its robustness to increased false positive (FP) and false negative (FN) rates through random modeling and robustness analysis, respectively. Furthermore, we have demonstrated that the ability of this panel to predict therapeutic response does not depend on commonly used prognostic variables, such as pathological and clinical T-stage, Gleason score (i.e., pathology-based grading system of prostate tissues), age, and therapy subtype. We propose that this panel can potentially be used to pre-screen patients to prioritize those who would benefit from ADT and patients at risk of developing resistance. Our method holds a long-term potential to improve therapeutic management of cancer patients and builds a foundation for personalized therapeutic advice for patients with advanced malignancies.
2. Materials and Methods
2.1. DNA Methylation and mRNA Expression Resources
Prostate cancer patient cohorts utilized in this study come from the publicly available data sources, including The Cancer Genome Atlas - Prostate Adenocarcinoma (TCGA-PRAD), Stand up to Cancer (SU2C), Grasso et al. (GSE35988), Cai et al. (GSE32269), Sboner et al. (GSE16560), Beltran et al., and Prostate Cancer Medically Optimized Genome-Enhanced Therapy (PROMOTE) datasets (Table 1): (i) TCGA-PRAD [13] cohort was downloaded from Genomics Data Commons (GDC, https://gdc.nci.nih.gov/) on November 15, 2016. Information about type and time of treatment was obtained and synthesized from the clinical, follow-up, and the treatment data files, obtained from the TCGA GDC legacy archive (https://portal.gdc.cancer.gov/legacy-archive). For the purpose of this study we selected patients with primary tumors (obtained after radical prostatectomy), which were treated with adjuvant androgen deprivation therapy (ADT) and further monitored for disease progression (n = 66), which were suited to study primary ADT resistance. TCGA-PRAD DNA methylation was profiled on Illumina Infinium Human Methylation (HM450) array and RNA-seq was profiled on Illumina HiSeq 2000; (ii) Stand up to Cancer (SU2C) [48] contained tumors from metastatic castration-resistant prostate cancer (CRPC, n = 51, raw sequencing data for 51 patients were available for download from dbGaP phs000915.v1.p1) obtained from bone or soft tissue biopsies, profiled on Illumina HiSeq 2500 platform; (iii) Grasso et al. [46] dataset was obtained from Gene Expression Omnibus (GEO) GSE35988 and contained prostatectomy samples of primary tumors from patients with hormone-naïve prostate cancer (n = 58) and metastatic CRPC samples at rapid autopsy (n = 33), profiled on Agilent-014850 Whole Human Genome Microarray 4x44K G4112F; (iv) Cai et al. [45] dataset was obtained from GEO GSE32269 and contained primary tumors from patients with hormone-naïve prostate cancer isolated by laser capture microdissection (LCM) from frozen biopsies (n = 21) and CRPC bone metastasis obtained through CT guided bone marrow biopsies (n = 19), profiled on Affymetrix Human Genome U133A array; (v) Beltran et al. [44]: data were downloaded from dbGaP (phs000909.v1.p1) and contained tumors from metastatic castration-resistant prostate cancer (CRPC, neuroendocrine samples excluded, n = 34) obtained from lung, soft tissue and spinal cord biopsies, profiled on Illumina Genome Analyzer II; (vi) Prostate Cancer Medically Optimized Genome-Enhanced Therapy (PROMOTE) [47]: data were downloaded from dbGaP (phs001141.v1.p1) and contained tumors from metastatic CRPC that received 12 weeks of Abiraterone acetate and failed this treatment (n = 29), obtained from tissue biopsies and profiled on Illumina HiSeq 2500 platform; and as a negative control, we utilized (vii) Sboner et al. [49] dataset obtained from GEO GSE16560, which consisted of not treated primary tumors obtained from transurethral resection of the prostate (TURP) (n = 281) and profiled on 6 k cDNA-mediated annealing, selection, ligation, and extension (DASL) microarray platform.
Table 1.
Prostate cancer cohorts utilized for discovery and validation.
| Dataset | Description | N | Platform | Source |
|---|---|---|---|---|
|
TCGA-PRAD, The Cancer Genome Atlas – Prostate Adenocarcinoma [13] |
Primary tumors obtained after radical prostatectomy and treated with adjuvant androgen deprivation therapy (ADT). | 66 | Illumina Infinium Human Methylation (HM450) array Illumina HiSeq 2000 |
Genomics Data Commons (GDC) (https://gdc.nci.nih.gov/) |
|
SU2C, Stand Up To Cancer [48] |
Metastatic CRPC samples obtained from bone or soft tissue biopsies. | 51 | Illumina HiSeq 2500 | phs000915.v1.p1 |
| Grasso et al. [46] | Androgen-sensitive: localized androgen-naïve primary tumors after prostatectomy. CRPC: CRPC metastatic samples obtained at rapid autopsy. |
58 33 |
Agilent-014850 Whole Human Genome Microarray 4x44K G4112F | GSE35988 |
| Cai et al. [45] | Androgen-sensitive: androgen-naïve primary tumors isolated by laser capture microdissection (LCM) from frozen biopsies. CRPC: CRPC bone metastasis obtained from CT guided bone marrow biopsies. |
21 19 |
Affymetrix Human Genome U133A array | GSE32269 |
| Sboner et al. [49] | Primary tumors not subjected to treatments and obtained from transurethral resection of the prostate (TURP). | 281 | Human 6 k Transcriptionally Informative Gene Panel for DASL | GSE16560 |
| Beltran et al. [44] | Metastatic CRPC samples obtained from lung, soft tissue and spinal cord biopsies. | 34 | Illumina Genome Analyzer II | phs000909.v1.p1 |
|
PROMOTE, Prostate Cancer Medically Optimized Genome-Enhanced Therapy [47] |
Metastatic CRPC samples, obtained from tissue biopsies, treated with Abiraterone acetate for 12 weeks (with subsequent treatment failure). | 29 | Illumina HiSeq 2500 | phs001141.v1.p1 |
N = number of patients.
2.2. DNA Methylation and mRNA Expression Data Analysis and Integration
Our study introduces a framework (Fig. 1) to effectively integrate DNA methylation with mRNA expression patient profiles to identify markers of primary treatment resistance. DNA methylation profiles on Illumina Infinium Human Methylation depict methylation levels of CpG sites, located across TSS 200/1500 (TSS 1500 and TSS 200), 5′ UTR, 1st exon, gene body, and 3′ UTR regions and are reported as β (Beta) values. We adopted suggestion in Du et al. to convert β to M-values, better suited for statistical analysis [50], where . All statistical computing in this manuscript was performed using R studio version 1.0.143 (R version 3.4.3). Differential methylation signature was defined as a list of methylation sites ranked by their differential methylation between patients with poor and favorable ADT response, using two-tail two-sample Student t-test (t.test function in R). We utilized DESeq2 R package [51] for RNAseq data normalization and processing.
Initial essential step in our analysis was to evaluate if sites from a particular region (i.e., TSS 200/1500, 5′ UTR, 1st exon, gene body, 3′ UTR) were enriched in the differential methylation signature. For this purpose, we used Fisher Exact Test (FET) [52] and Gene Set Enrichment Analysis (GSEA) [53]. For FET, 500 top most differentially methylated sites were used to evaluate if a specific region is over-represented in the top 500 sites compared to the rest of the signature and was conducted using fisher.test function in R. In GSEA, differential methylation signature was used as a reference and sites associated to specific regions were utilized as query sets. Normalized Enrichment Score (NES) and p-value for significance were estimated using 1000 site permutations. GSEA was implemented in R studio, v 3.3.2.
To identify differentially methylated sites that have functional effect on the site-harboring genes, we measured their association through Pearson correlation [54] (also confirmed with Spearman correlation [54]), estimated between differentially methylated sites (M-values) and their site-harboring genes (DESeq2 normalized counts), using cor.test function in R. To further evaluate a potential cause-effect relationship, we employed the linear least squares regression analysis [55] where each methylation M-value was considered as predictor (i.e., independent) variable and an mRNA expression value of the site-harboring gene was considered as response (i.e., dependent) variable, estimated using lm function in R. Our analysis identified a panel of 5 site-gene pairs, which are differentially methylated between patients with poor and favorable treatment response and can explain expression variation in their site-harboring genes, which increases the probability of identifying (epi) genomic markers with functional role in therapeutic resistance.
2.3. Evaluation of Clinical Significance of our Findings
To evaluate ability of the identified 5 site-gene panel to predict therapeutic response in independent datasets, we subjected prostate cancer patient cohorts to t-distributed stochastic neighbor embedding clustering (t-SNE), a widely-used dimensionality reduction technique, well suited for visualization of high-dimensional datasets in a two (or three) dimensional space [56]. In particular, t-SNE considers all pairs of high-dimensional (i.e., 5-dimentional in our case) points and converts their high-dimensional similarity into conditional probabilities in such a way that similar points (i.e., patient profiles) are assigned a high conditional probability, and dissimilar points are assigned a low conditional probability (i.e., defining Probability Distribution H in a high-dimensional space). Further, t-SNE defines Probability Distribution L over the same pairs of points (i.e., patient profiles) in the low-dimensional (i.e., 2-dimentional) space, and it aims to minimize the Kullback–Leibler divergence [57] between the Probability Distribution H and Probability Distribution L with respect to the locations of the points (i.e., patient profiles) in the space. Therefore, patients with similar 5 site-gene patterns will be projected as nearby points and patients with dissimilar 5-site gene patterns will be projected as distant points in the two-dimensional space. Differences in therapeutic response in the identified patient groups were evaluated through Kaplan-Meier treatment-related survival analysis [58] and Cox proportional hazard model [59] using survival and survminer packages in R [60,61]. Log-rank and Wald tests were used to estimate statistical significance of the Kaplan-Meier survival analysis and Cox proportional hazard model, respectively, using R coxph function from survival package [62].
To compare the ability of the DNA methylation and mRNA expression of the 5 site-gene panel to effectively classify patient groups, we utilized receiver operating characteristics (ROC) analysis [63] on multiple (i.e., multivariable) logistic regression model, where identified patient groups were used as a response variable and M-value/RNA-seq distributions of 5 site-gene panel were used as inputs. ROC curves were evaluated using area under the curve (AUROC) [64], with AUROC = 0.5 being a random classifier. The logistic regression analysis was implemented using glm [65] function and ROC analysis using roc function from pROC package in R [66].
To evaluate if expression of the 5 site-gene panel in primary tumors was comparable to CRPC metastasis and to demonstrate that molecular profiles of patients that received ADT and developed Biochemical Recurrence are similar to patients that failed ADT with metastatic disease, we compared TCGA-PRAD and SU2C datasets, for which we combined their raw RNA-seq counts with subsequent normalization using DESeq2 R package [51]. To compare expression levels of the 5 site-gene panel across these datasets, we first scaled each gene (i.e., each gene was z-scored with respect to the mean expression of this gene across the combined TCGA-SU2C datasets) and defined a composite expression z-score for the 5 site-gene panel for each patient. In particular, to define the composite expression z-score for each patient, we combined z-scores of the 5 genes from the 5 site-gene panel using Stouffer integration [67] (stouffer function from vulcan package in R [68]). Distributions for such composite z-scores were then compared between TCGA-PRAD and SU2C patient cohorts using one-tail two-sample Welch t-test.
Finally, to confirm ability of our 5 site-gene panel to identify and distinguish samples with CRPC ADT failure, we utilized independent patient cohorts for (i) t-SNE clustering; and (ii) multiple (i.e., multivariable) logistic regression followed by ROC analysis. In particular, to demonstrate that the 5 site-gene panel allows the effective identification of the CRPC ADT failure samples, we subjected patient cohorts in Grasso et al. [46] and Cai et al. [45] to t-SNE clustering [56], with all 5 dimensions considered. Furthermore, to show the ability of the 5-site-gene panel to effectively distinguish between CRPC ADT failure samples and TCGA-PRAD samples with favorable response (i.e., group 1), similarly to SU2C cohort, we combined raw counts for patients cohorts in Beltran et al. [44] and PROMOTE [47] with the TCGA-PRAD cohort and subjected them to multiple logistic regression (where samples with CRPC ADT failure and samples with favorable ADT response were used as a response variable and gene expression distributions of 5 site-gene panel were used as inputs) followed by ROC analysis.
2.4. Multimodal Performance Evaluation of our Model
For multimodal performance assessment of our model, we have evaluated (i) advantages of our model over other commonly used methods, such as expression, methylation, and correlation data alone; (ii) non-randomness of its predictive ability through comparison to 5 site-gene pairs selected at random; (iii) robustness of our findings through evaluation of how well our model can classify patients at varying levels of noise; and demonstrated that (iv) predictive ability of our panel is not affected by the commonly used prognostic clinical variables, such as pathological and clinical T-stage, Gleason score, age, and therapy subtypes.
Comparative analysis to expression, methylation, and correlation data alone was done using Kaplan-Meier survival analysis, hazard ratio, and concordance index (i.e., c-index), which measures how well our model can predict therapeutic response (with 1.0 being the highest predictive ability, equivalent to AUROC = 1 or 100%). C-index was estimated using concordance.index function from survcomp package in R [69].
To evaluate non-randomness of our predictions, we utilized random model built through selection of 5 site-gene pairs at random 10,000 times. Nominal p-value for the model was estimated as the number of times Kaplan-Meier log-rank p-values for random 5 site-gene pairs reached or outperformed log-rank p-value for the original 5 site-gene panel.
To evaluate the robustness of our model, we tested its predictive ability with the increase of False Negative (FN) and False Positive (FP) rates. Let us define an iteration i = 1… 58. For False Negative estimation, at each iteration i, exactly i patients were selected at random and removed from the validation cohort using sample function from R. Each iteration i was repeated 10,000 times except when i = 1 and 2: for i = 1, it was run 58 times as total number of ways 1 patient can be chosen from 58 patients is 58; for i = 2, it was run 1000 times as the total number of ways 2 patients can be chosen from 58 patients is ((58 choose 2) = 1653). For False Positive estimation, at each iteration i, exactly i patients were added to the validation cohort: random patients were generated as follows: (1) probability of an event was generated at random based on the actual data from the original un-altered validation set; (2) patient group was chosen at random, based on the probability of choosing a patient from the original un-altered validation set; (3) time to event or follow-up were chosen through random number generator using sample function in R. As FN and FP rates were increased, we clustered the newly formed noise-enriched patient set and subjected it to Kaplan-Meier survival analysis, reporting the median log-rank p-values for each i, sampled from 10,000 iterations.
To confirm that fluctuations in the signature threshold levels do not affect predictive power of our model, we evaluated performance of our model while varying (i) methylation signature threshold (originally p < 0.001) between 0.0001 and 0.005; and (ii) correlation threshold (originally p < 0.05) between 0.02 and 0.05. For each threshold point, we used multiple logistic regression, where TCGA-PRAD patient groups with poor and favorable treatment response were used as a response variable and M-values of site harboring genes below the corresponding threshold were used as inputs, followed by ROC analysis.
Finally, to assess if the predictive ability of the 5 site-gene panel is independent of commonly used prognostic variables such as pathological and clinical T-stage, Gleason score, patient age and therapy subtypes, we performed univariable and multivariable Cox proportional hazard model analysis using coxph function from survival package in R [62].
3. Results
3.1. Overview
To identify molecular mechanisms that differentiate favorable and poor treatment response, we have compared DNA methylation profiles of patients that failed ADT (i.e., non-responders) with profiles of patients with favorable response to ADT (i.e., responders) (see schematics Fig. 1), which defined a therapeutic failure signature. Region-based analysis of methylation sites identified TSS 1500, TSS 200, 5′ UTR, and 1st exon regions with significant contribution (i.e., enrichment) in the therapeutic failure signature. We followed this discovery with the integrative analysis of DNA methylation and gene expression profiles, which identified methylation sites that are significantly associated (i.e., through Pearson correlation) and can explain expression variation (i.e., through linear regression analysis) of their site-harboring genes. Identified candidates were then subjected to validation (i.e., their ability to predict treatment response) in independent non-overlapping clinical patient cohorts, using Kaplan-Meier survival analysis [58] (log-rank p = 0.0191, hazard ratio = 4.37), t-distributed stochastic neighbor embedding clustering [56] (sensitivity =100%), and ROC analysis [63] (AUROC = 0.83, AUROC = 0.98). We have compared performance of our model to methylation, expression, and correlation alone and demonstrated that our model outperforms these data types in correctly classifying patients at risk of ADT resistance. Furthermore, we evaluated statistical significance of these predictions through random modeling (random model 1 p = 0.010, random model 2 p = 0.011) and robustness analysis (FN threshold = 31%, 18/58; FP threshold = 9%, 5/58) to demonstrate non-random robust classification of patients into ADT response groups. Finally, to demonstrate that our model is not affected by commonly used prognostic clinical variables, such as pathological and clinical T-stage, Gleason score, age, and therapy subtypes, we utilized multivariable Cox proportional hazard model [59], demonstrating that none of these variables were predictive of ADT response and they did not affect predictive ability of the 5 site-gene panel.
3.2. Time-Course Analysis of Treatment Response Identifies Signature of Therapeutic Failure
To evaluate treatment response to ADT in prostate cancer patients, we analyzed HumanMethylation450 [70] DNA methylation profiles of primary prostate tumors from The Cancer Genome Atlas (TCGA-PRAD) [13] patient cohort (Table 1). While relatively recent, TCGA-PRAD represents the most comprehensive cohort of ADT treatment administration with clinical follow-up to date. Samples in TCGA-PRAD study represent localized prostate cancer samples that had been obtained through radical prostatectomy from patients that did not receive any neoadjuvant (i.e., prior to prostatectomy) treatment. Following prostatectomy, patients were monitored for adjuvant (i.e., post-operative) ADT administration and disease progression, such as Biochemical Recurrence (BCR, defined by a rapid rise of prostate-specific antigen, PSA, in patient blood), local or distant metastases, and prostate cancer-related lethality (i.e., death due to prostate cancer). For this study, we focused on patients that received adjuvant ADT and had available follow-up clinical data (n = 66), suited to study primary ADT resistance.
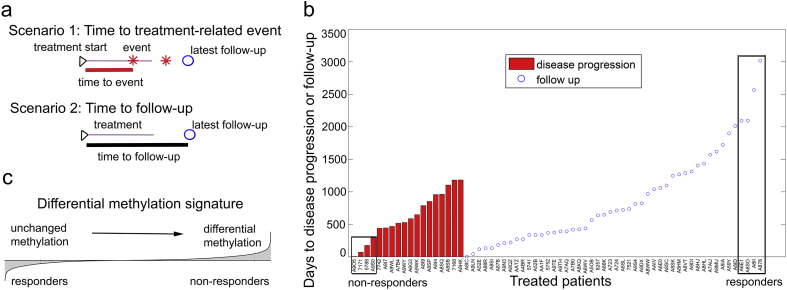
Each patient was evaluated with respect to the start of his androgen-deprivation regimen and time to disease progression (BCR, local or distant metastasis, or lethality) or time to follow-up (if no event occurred, such patients were considered censored). If a patient had an event during the course of the treatment or within 1.5 years after the end of the treatment (Fig. 2a, Scenario 1), time to treatment failure was defined as time between the treatment start and such an event (Fig. 2b, red bars). At the same time, if a patient did not experience a treatment-related event, time for his treatment-related disease-free survival was estimated as time between the treatment start and time to his latest follow-up (Fig. 2a, Scenario 2, Fig. 2b, blue circles).
Fig. 2.
Analysis of therapeutic response defines signature of ADT resistance. (a) Schematic representation of a treatment time-course. Scenario 1: Time to treatment-related event: event occurred during the course of the treatment or within 1.5 years after the treatment end. Time between treatment start and a treatment-related event indicated. Scenario 2: Time to follow-up: time between treatment-start and latest follow-up indicated. (b) ADT response in the TCGA-PRAD cohort. Red vertical bars correspond to time between treatment start and event. Blue circles define censored patients (without events), indicating time between treatment start and latest follow-up. Non-responder (n = 4) and responder (n = 4) (Table S1) patients are indicated. (c) Schematic depiction of the differential methylation signature between non-responder and responder patients, sorted from sites whose methylation did not change (left tail) to sites with significant differential methylation (right tail). Signature was defined as a list of sites ordered by –log10 (p-value) from the two-sample two-tail t-test comparing non-responder and responder patient groups.
To define epigenomic changes that differentiate therapeutic failure and success, we compared DNA methylation profiles of patients with most rapid treatment failure to patients with longest treatment-related disease-free survival. For this, we ranked patients based on their treatment-related disease-free survival time (Fig. 2b) and compared those that fall into the most left and right distribution tails (roughly, patients outside of a 90% confidence interval), which defined (i) non-responders as patients that experienced treatment failure within 1 year of treatment (n = 4) and (ii) responders as patients with treatment-related disease-free survival over 5.5 years (n = 4) (Fig. 2b).
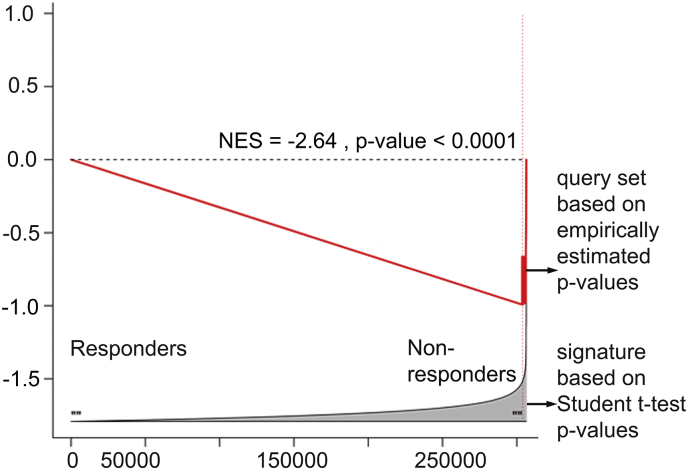
We compared age, Gleason score, and tumor aggressiveness at diagnosis (which includes pathological and clinical T- stage) between the two groups (Table S1) and did not observe significant difference in clinical aggressiveness in non-responder compared to responder groups (average age in non-responders group = 60.5 and in responders group = 60.0; average Gleason score in non-responders group was 8.25 and in responders group = 9.0). We then compared M-value transformed DNA methylation profiles of non-responders and responders using two-sample two-tail t-test (see Materials and Methods, Dataset S1), followed by ranking of the methylation sites based on the t-test p-values, from the least differentially methylated (Fig. 2c, left tail) to the most differentially methylated (Fig. 2c, right tail), which defined a “differential methylation signature” of ADT resistance (i.e., treatment failure). We paralleled this analysis with a non-parametric signature reconstruction, where an empirical p-value for each site was estimated after 10,000 random site permutations (followed by an FDR correction) and obtained virtually identical results (NES = −2.64, p < 0.001), confirming robustness of our signature reconstruction (Fig. S1).
3.3. TSS 200/1500, 5′ UTR, and 1st Exon Regions Are Enriched in Treatment Resistance Signature
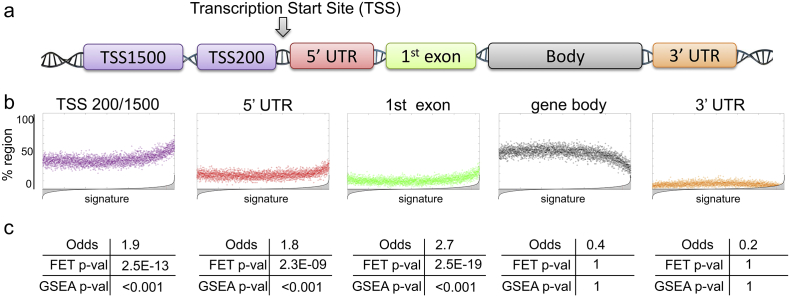
Following reconstruction of the differential methylation signature, we sought to evaluate the contribution of each region (profiled on HumanMethylation450 array, Fig. 3a) to resistance to ADT. Regions profiled on the HumanMethylation450 include TSS 200 (i.e., −200 base pairs upstream of the Transcription Start Site, TSS) or TSS 1500 (i.e., between -200 and -1500 base pairs upstream of the TSS), 5′UTR (5′ untranslated region), 1st exon, gene body, and 3′UTR (3′ untranslated region) (Fig. 3a). Not to overlook proximal and distal regulatory elements, we considered TSS 200 and TSS 1500 together for subsequent analysis (i.e., referred to as TSS 200/1500). It is noteworthy to mention that a single site can be associated to several regions due to possible multiple splice variants of a site-bearing gene.
Fig. 3.
Methylation sites from TSS 200/1500, 5′UTR, and 1st exon regions are enriched in methylation signature of ADT resistance. (a) Schematic diagram showing methylation regions profiled on the HumanMethylation450 array. (b) Scatter plots depicting enrichment of each region (i.e., site location) in the differential methylation signature. A reference methylation signature (as in Fig. 2c, ranked from the least differentially methylated (left) to the most differentially methylated (right)) is divided into 100 site-long steps and contribution of each region is calculated as % region inside each step. (c) Odds, Fisher Exact Test (FET) and Gene Set Enrichment Analysis (GSEA) demonstrate statistical significance of the region enrichment from Fig. 3b. For odds and FET, 500 most differentially methylated sites were considered for significance testing. For GSEA, differential methylation signature (Fig. 2c) was utilized as a reference and sites from each region were utilized as query sets. P-value was estimated with 1000 site permutations.
To evaluate contribution of each region, we tested its enrichment in the differential methylation signature. First to visualize enrichment of each region, we divided the differential methylation signature into 100 site-long steps. Each step was evaluated with respect to the percentage (i.e., fraction) of sites that fall into TSS 200/1500, 5′UTR, 1st exon, body, or 3′UTR regions (Fig. 3b). Right-side upward- pointing “horn” indicates over-representation (i.e., enrichment) of sites from a specific region in the most differentially methylated part of the signature (i.e., right tail), as is evident from analysis for TSS 200/1500, 5′ UTR, and 1st exon regions (Fig. 3b). We evaluated statistical significance of such enrichments using Fisher Exact Test (FET) [52] and Gene Set Enrichment Analysis (GSEA) [53] (see Materials and Methods), which confirmed statistical significance of enrichment for TSS 200/1500 (FET p = 2.5E-13, GSEA p < 0.001), 5′UTR (FET p = 2.3E-09, GSEA p < 0.001), and 1st exon (FET p = 2.5E-19, GSEA p < 0.001) regions in the differential methylation signature, while body and 3′ UTR regions did not show any enrichment (Fig. 3c). Given these results, we considered TSS 200/1500, 5′ UTR and 1st exon regions for our subsequent therapeutic failure analysis.
3.4. Integrative (epi) Genomic Analysis Identifies a 5 Site-Gene Panel of ADT Resistance
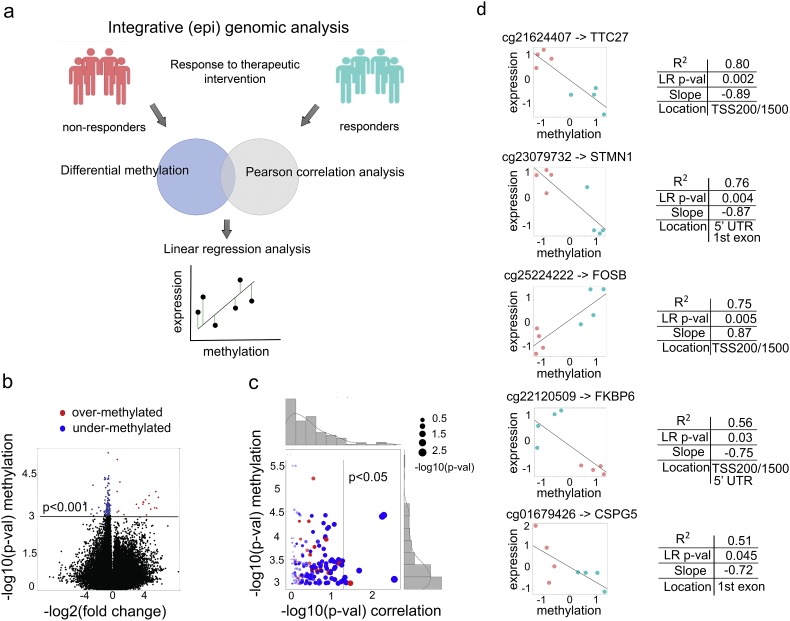
To elucidate markers of ADT resistance, we sought to categorize differentially methylated sites and identify those that have a significant association and can explain variation in the expression of the site-harboring genes (general strategy in Fig. 4a). For this, we focused on sites from TSS 200/1500, 5′ UTR, and 1st exon regions with significant differential methylation (t-test p < 0.001, n = 144, Dataset S1) between non-responders and responders (Fig. 4b). Our ultimate goal was to identify a potential “cause-effect” relationship, where differentially methylated sites (n = 144) would have a potential functional “causal” effect on the expression changes in the site-harboring genes. As a pre-screen for such relationship, we utilized Pearson correlation analysis [54] between methylation M-values (see Materials and Methods) for each site and mRNA expression levels (i.e., DESeq2 [51] normalized counts) for each corresponding site-harboring gene. Such analysis was done for each site-gene pair and identified differentially methylated sites with significant positive (or negative) association to their corresponding genes (i.e., Pearson correlation p < 0.05, n = 8) (Fig. 4c).
Fig. 4.
Integrative (epi) genomic analysis identifies a 5 site-gene panel. (a) Schematic representation of the integrative (epi) genomic analysis: (top) identification of differentially methylated sites; (middle) Pearson correlation analysis (i.e., pre-screening) between methylation levels of the sites and mRNA expression levels of the site-harboring genes; (bottom) linear regression analysis to identify sites that can explain expression changes of the site-harboring genes. (b) Volcano plot of the differentially methylated sites, with under-methylated (blue) and over-methylated (red) sites indicated (at p-value < 0.001, n = 144). (c) Scatter plot depicting the relationship between differential methylation (y-axis) and site-gene (i.e., methylation-expression) Pearson correlation (x-axis) for the 144 significantly methylated sites from Fig. 4b. (d) Linear regression analysis between site methylation values and mRNA expression of their site-harboring genes (linear regression at p < 0.05 shown), with non-responder (coral) and responder (aquamarine) samples indicated. The x and y axes depict z-scored methylation M-values and DESeq2 normalized expression values.
Our next step was to test these site-gene pairs to determine the extent to which methylation values can explain variation in the expression changes of their site-harboring genes. For this, we utilized linear least squares regression analysis [55], where a methylation M-value was considered as
predictor (i.e., independent) variable and an mRNA expression value was considered as response (i.e., dependent) variable. Linear regression analysis identified a panel of 5 site-gene pairs (Fig. 4d), where differentially methylated sites could explain from 51% to 80% variation (i.e., as defined by the coefficient of determination, R2) of the site-harboring genes: TTC27 (R2 = 0.80, p = 0.002), STMN1 (R2 = 0.76, p = 0.004), FOSB (R2 = 0.75, p = 0.005), FKBP6 (R2 = 0.56, p = 0.03), and CSPG5 (R2 = 0.51, p = 0.045). Interestingly, the differentially methylated site harbored by FOSB showed a positive relationship (i.e., positive slope) with FOSB mRNA expression while sites harbored by FKBP6, TTC27, CSPG5 and STMN1 showed a negative relationship (i.e., negative slope) (Fig. 4d), which indicates that changes in methylation levels might interfere with transcriptional regulation by a repressor or an activator, respectively.
3.5. Validation in Independent Patient Cohorts
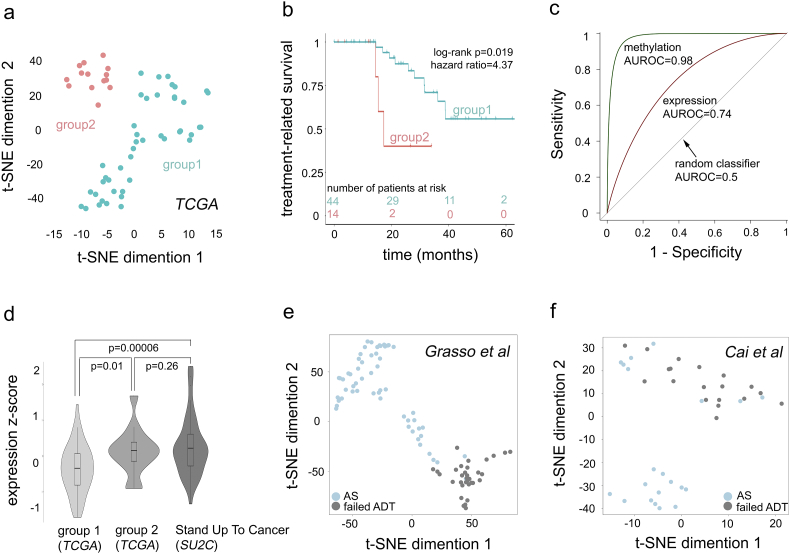
We next evaluated the ability of the 5 site-gene panel to predict therapeutic response to ADT in independent non-overlapping patient cohorts. Our validation sets have been chosen to demonstrate several points, including that (i) our 5 site-gene panel is capable of distinguishing between primary tumors with poor and favorable treatment response; (ii) molecular profiles of patients that were administered ADT and developed Biochemical Recurrence are similar to profiles of patients that genuinely failed the ADT with metastases and developed CRPC; (iii) our 5 site-gene panel can effectively identify CRPC samples; and (iv) our 5 site-gene panel can accurately distinguish between the primary prostate cancer samples with favorable treatment response and CRPC samples that failed ADT treatment. For this, we started with a TCGA-PRAD cohort (n = 58, TCGA-PRAD validation set) of ADT treated patients (Table 1), excluding non-responders (n = 4) and responders (n = 4) to avoid over-fitting. T-distributed stochastic neighbor embedding clustering (t-SNE), a widely-used dimensionality reduction technique [56], done on the methylation levels of the identified 5 site-gene panel, classified patients from the validation set into two groups (i.e., group 1 and group 2) (see STAR Methods, Fig. 5a).
Fig. 5.
Five site-gene (epi) genomic panel predicts ADT failure in independent patient cohorts. (a) t-SNE clustering identifies two groups of patients: group 1 and group 2 (a full set of 5 dimensions considered). (b) Kaplan-Meier survival analysis identifies the significant difference in treatment-related survival (i.e., treatment response) between groups 1 and 2 from Fig. 5a. Log-rank p-value and hazard ratio are indicated. (c) ROC analysis: AUROC indicates the ability of methylated sites and expression of site harboring genes can classify patients into group 1 and group 2. (d) Violin plot for composite Stouffer integrated z-scores (see Materials and Methods) in group 1 (n = 44), group 2 (n = 14) and SU2C (n = 51) cohorts. One-tail two-sample Welch t-test p-values are indicated. (e) t-SNE clustering (all 5 dimensions considered) based on 5 site-gene panel in Grasso et al. cohort (n = 91) [46]; and (f) Cai et al. (n = 40) [45] cohort; is able to separates androgen sensitive (AS, light blue) from castration-resistant prostate cancer (failed ADT, grey) samples (sensitivity = 100% for failed ADT CRPC selection: 33/33 for Grasso et al., and 19/19 for Cai et al.).
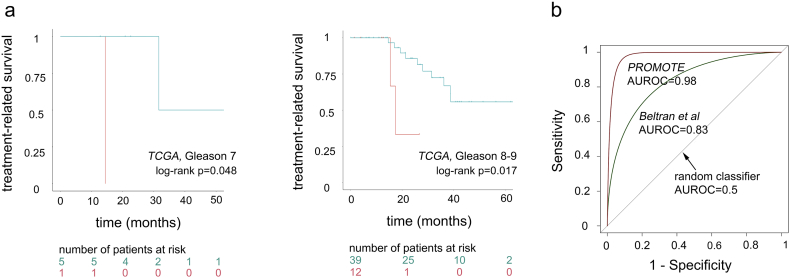
The next essential step in our predictive analysis was to evaluate if these patient groups significantly differed in their response to androgen-deprivation treatment. For this, we compared treatment-related disease-free survivals (as defined previously, Fig. 2a-b) between the groups using Kaplan-Meier survival analysis [58], which demonstrated significant difference in treatment response between the groups (see STAR Methods, Fig. 5b) (log-rank p = 0.0191). Patients in groups 1 (aquamarine) experienced treatment-related disease progression events at a slower rate, while events related to androgen-deprivation therapy in group 2 (coral) occurred at a much faster rate (hazard ratio = 4.37, p = 0.031). We further evaluated if patient separation was effected by Gleason score, a commonly used clinical prognostic variable. For this, we evaluated patients with Gleason score 7 and Gleason score 8 + 9 separately and demonstrated that they did not affect treatment differences between the group 1 and group 2 (Fig. S2a, Gleason score 7 log-rank p = 0.048; Gleason score 8 + 9 log-rank p = 0.017) patient classification.
Given potential cause-effect relationship in the 5 site-gene panel, we further confirmed the effect of the expression of site-harboring genes on the separation between the two groups through Receiver Operating Characteristic (ROC) analysis [63], whose performance was evaluated using area under the ROC, AUROC [64] (see STAR Methods, Fig. 5c), where AUROC = 0.5 indicates a random classifier and AUROC = 1 indicates a complete separation of the patient groups. ROC analysis demonstrated that both methylation levels of 5 sites (AUROC = 0.98) as well as expression levels of site-harboring genes (AUROC = 0.74) significantly contributed to the group separation and thus can be utilized for patient classification.
In addition to validation in the TCGA-PRAD cohort, we further evaluated if the identified 5 site-gene panel can determine failed ADT response in (i) Stand Up To Cancer (SU2C) [48] patient cohort with castration-resistant prostate cancer (CRPC) metastatic samples (n = 51); (ii) Grasso et al. [46] patient cohort with androgen-naive primary tumors (n = 58) and CRPC metastatic samples (n = 33); (iii) Cai et al. [45] patient cohort with androgen-naïve primary tumors (n = 21) and CRPC bone metastasis (n = 19) (Fig. 5d-f, Table 1); (iv) Beltran et al. [44] patient cohort with CRPC metastatic samples (n = 34); and (v) Prostate Cancer Medically Optimized Genome-Enhanced Therapy (PROMOTE) [47] patient cohort with CRPC metastatic samples that were treated with Abiraterone acetate for 12 weeks with subsequent treatment failure (n = 29) (Fig. S2b, Table 1). First, to evaluate if expression levels of the 5 site-gene panel in primary tumors with poor ADT response (i.e. group 2) are comparable to metastatic CRPC samples (i.e., metastatic samples with ADT failure) and demonstrate that profiles of patients that received ADT after surgery with subsequent BCR are similar to the profiles of patients who have failed ADT with metastatic disease, we compared expression levels from the 5 site-gene panel in the TCGA-PRAD patient cohort (group 1 and group 2, primary tumors) and SU2C (CRPC metastatic samples) (see Materials and Methods, Fig. 5d), which demonstrated that genes from the 5 site-gene panel (i) substantially differ between patient with favorable ADT response (i.e., group 1) and poor ADT response (i.e., group 2) (p = 0.01) as well as between patients with favorable ADT response (i.e., group 1) and CRPC metastasis (p = 0.00006); and (ii) have similar expression patterns in patients with poor ADT response (i.e., group 2) and CRPC metastatic samples (p = 0.26) (Fig. 5d), demonstrating that 5-site gene panel has comparable expression levels in primary tumors with poor ADT response and metastatic CRPC samples. Subsequently, to further confirm ability of our 5 site-gene panel to identify samples with CRPC ADT failure, we subjected expression profiles from patient cohorts in Grasso et al. [46] and Cai et al. [45] to t-SNE clustering (see Materials and Methods), which demonstrated the ability of our panel to separate CRPC (grey) from androgen sensitive (AS) (light blue) samples (sensitivity to cluster CRPC into one group was 100% in both datasets) (Fig. 5e-f). Finally, to confirm the ability of the 5-site-gene panel to effectively distinguish between CRPC ADT failure samples and TCGA-PRAD samples with favorable ADT response, we compared patient profiles in Beltran et al. [44] and PROMOTE [47] to the patients from the TCGA-PRAD with favorable treatment response (i.e., group 1) through multiple logistic regression followed by ROC analysis (see Materials and Methods), which demonstrated that our 5 site-gene panel can effectively distinguish between TCGA-PRAD with favorable ADT response and Beltran et al. [44] (AUROC = 0.83) and PROMOTE [47] (AUROC = 0.98) (Fig. S2b). Taken together our findings indicate a significant ability of our 5-site gene panel to predict ADT failure in diverse prostate cancer cohorts.
3.6. Multimodal Comparative Analysis Demonstrates Statistical Significance of the Predictive Model
For multimodal performance assessment of our model, we have evaluated (i) advantages of our model over methylation, expression, and correlation data alone; (ii) non-randomness of its predictive ability through comparison to 5 site-gene pairs selected at random; (iii) robustness of our findings through evaluation of how well our model can classify patients at varying levels of noise; and demonstrated that (iv) predictive ability of our panel is not affected by the commonly used prognostic clinical variables, such as pathological and clinical T-stage, Gleason score, age, and therapy subtypes.
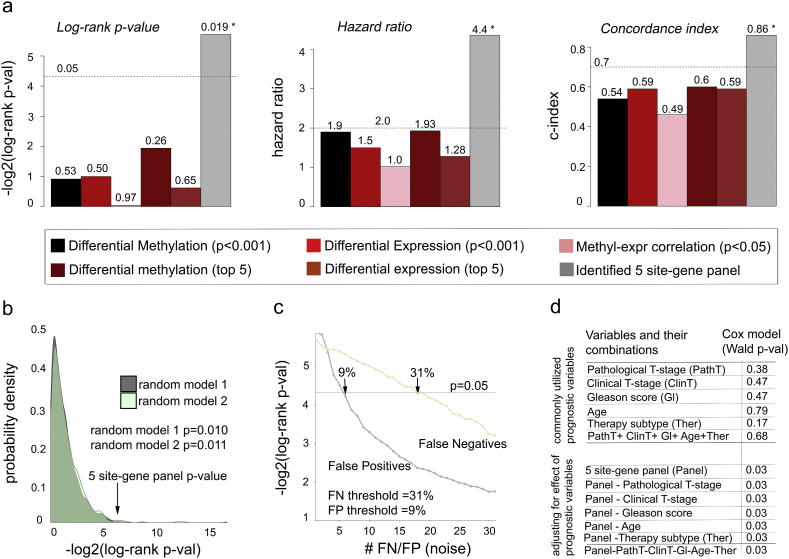
To assess advantages of our model over other commonly used methods, we have compared the ability of the 5 site-gene panel to predict ADT failure to (i) differentially methylated sites alone (two-tail two-sample Student t-test p < 0.001); (ii) differentially expressed genes alone (two-tail two-sample Student t-test p < 0.001); and (iii) site-gene pairs identified from the correlation analysis (Pearson correlation p < 0.05); (iv) top 5 differentially expressed genes; and (v) top 5 differentially methylated genes which have also been utilized by [71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85] and achieved significant results in predicting disease progression. We have compared the ability of our model to predict ADT response to the therapeutic predictive ability of methylation, expression and correlation alone through Kaplan-Meier survival analysis (results reported through log-rank p-value and hazard ratio) and concordance index (i.e., c-index) (see Materials and Methods) in the TCGA-PRAD validation set and demonstrated that our 5 site-gene panel outperforms these data types in correctly classifying patients at risk of ADT resistance (Fig. 6a).
Fig. 6.
Multimodal comparative analysis demonstrates significance of the 5 site-gene panel. (a) Comparing 5 site-gene panel (grey bars) to commonly used methods, including differential methylation (black bars), differential expression (red bars),Pearson correlation between methylation and expression (pink bars), top 5 differentially methylated sites (dark red), and top 5 differentially expressed genes (brown) through log-rank p-value, hazard ratio and concordance index. * indicates statistically significant changes (log-rank p = 0.019; HR p = 0.03; c-index p = 0.0001) (b) Random models to evaluate the ability of the 5 site-gene pairs chosen at random to separate patients into groups with different treatment response. Distributions of log-rank p-values from the random models indicate the significance of the predictive ability of our identified 5 site-gene panel. (c) Robustness analysis measuring predictive ability of the identified 5 site-gene panel across increasing FP and FN rates. (d) Multivariable Cox proportional hazard model demonstrates that commonly used prognostic clinical variables do not predict ADT response and do not affect predictive ability of the identified panel (Wald test Cox p-values indicated).
Furthermore, we evaluated non-randomness of the predictive ability of our 5 site-gene panel through comparison to 5 site-gene pairs selected at random (see Materials and Methods). For this, we defined two random models, where 5 site-gene pairs were selected at random from the pool of (i) all sites from the HumanMethylation450 platform (Fig. 6b, random model 1, dark grey); and (ii) sites from TSS 200/1500, 5′ UTR, and 1st exon regions (Fig. 6b, random model 2, light green). Five random sites were sampled 10,000 times in the TCGA-PRAD validation set and then evaluated for their ability to predict therapeutic response through Kaplan-Meier survival analysis. Empirical p-value for each random model was estimated as a number of times log-rank p-values for the randomly selected sites reached or outperformed the log-rank p-value for our original 5 site-gene panel, which demonstrated non-randomness of the 5 site-gene panel predictive ability (random model 1 p = 0.010; random model 2 p = 0.011) (Fig. 6b).
To test robustness of the predictive ability for the 5 site-gene panel, we introduced noise into our TCGA-PRAD validation set (excluding samples used for signature reconstruction to avoid overfitting, n = 58) and measured how much noise can be “tolerated” so that our 5 site-gene panel can still accurately predict treatment response. For this, we either randomly removed ADT treated patients (i.e., introduced False Negatives, FN) or randomly added ADT treated patients (i.e., introduced False Positives, FP) from or to the validation set (see Materials and Methods, Fig. 6c). Let us denote the number of patients removed or added at each iteration as i (i = 1 … 58). At each iteration, we evaluated ability of the 5 site-gene panel to classify patients and predict therapeutic response using Kaplan-Meier survival analysis. Each ith iteration was run 10,000 times and median log-rank p-values across 10,000 runs were reported (Fig. 6c). Our analysis demonstrated that the 5 site-gene panel could successfully predict therapeutic response even at 31% FN (18/58) and at 9% FP (5/58) rates (Fig. 6c), which demonstrates the robustness of its predictive ability even at high noise levels.
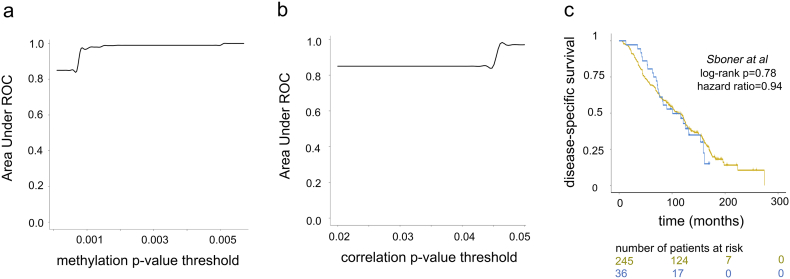
To confirm that fluctuations in the signature threshold levels do not affect power of our model to identify distinct treatment response groups, we evaluated predictive ability of our model while varying (i) methylation signature threshold (p < 0.001); and (ii) correlation threshold (p < 0.05) through multiple logistic regression at each threshold level followed by ROC analysis (see Materials and Methods), which demonstrated that our model kept its predictive power at varying methylation signature (AUROC between 0.85 and 0.99) and correlation (AUROC between 0.85 and 0.98) thresholds (Fig. S3a-b).
Finally, to confirm that the 5 site-gene panel is an indicator of primary resistance and not overall disease aggressiveness, as a negative control, we tested if the 5 site-gene panel can classify patients based on disease aggressiveness in Sboner et al. dataset [49], also known as a Swedish Watchful Waiting cohort with patients up to 30 years of clinical follow-up not subjected to treatments (n = 281, localized prostate tumors) (Table 1). The Kaplan-Meier survival analysis confirmed that predictive ability of our panel is independent of disease aggressiveness (log-rank p = 0.78, hazard ratio = 0.94, prostate cancer-related death was used as a clinical end-point) (Fig. S3c). Furthermore, to confirm this finding, we evaluated if the predictive ability of the 5 site-gene panel is independent of commonly used prognostic clinical variables [86,87], such as pathological and clinical T-stage, Gleason score, patient age and therapy subtypes, which include luteinizing hormone releasing hormone (LHRH) agonists (i.e., bind to pituitary LHRH receptor to stimulate the production of luteinizing hormone thus interfering with the yield of testosterone), LHRH antagonists (i.e., block the pituitary LHRH receptor thus completely shutting down the production of testosterone), CYP17 inhibitors (i.e., block CYP17 enzyme essential for androgen synthesis) and anti-androgens (i.e., bind to androgen receptor blocking androgen binding). For this, we performed multivariable Cox proportional hazard model analysis [59] in the TCGA-PRAD validation set, which confirmed that (i) none of these variables were predictive of ADT response and (ii) they did not affect predictive ability of the 5 site-gene panel (Fig. 6d). Given that our model has a highly accurate independent ability to predict ADT response, we anticipate that this panel can be ultimately utilized to classify patients at risk of developing resistance to ADT, prioritize patients for ADT intervention, and be incorporated into personalized and precision therapeutic platforms.
4. Discussion
In this work, we have developed a systematic genome-wide method that integrates DNA methylation and mRNA gene expression profiles to extrapolate therapeutic resistance in cancer patients. Several features of our method distinguish it from previously utilized methods used for data analysis in oncology. Firstly, it introduces a systematic (epi) genomic data driven method for predictive analysis of therapeutic resistance and is an original method of its kind to the best of our knowledge. Second distinguishing feature of our approach is in its ability to identify potential functional “cause-effect” relationships between DNA methylation sites and mRNA expression of the site-harboring genes, which outperformed genomic approaches that rely on single data type (e.g., expression or methylation data alone) or their correlation alone, and significantly increases the probability of identifying (epi) genomic markers with functional role in therapeutic resistance. Thirdly, our approach introduces a highly non-random robust technique to classify patients at risk of resistance and those who would benefit from the specific therapeutic intervention. Finally, while motivated by the emerging cases of resistance to androgen-deprivation in prostate cancer, our approach can be potentially applicable to other treatment regimens and diseases.
Our systematic integrative analysis of the ADT resistance in prostate cancer has identified a panel of 5 differentially methylated sites harbored by FKBP6, TTC27, CSPG5, FOSB, and STMN1 genes. Several of these genes have been known to play a role in carcinogenesis and treatment response in other cancer types. For instance, (i) hyper-methylation of FKBP6 has been shown to decrease cell viability and enhance progression in cervical cancer [88]; (ii) FOSB is a known regulator of differentiation during tumorigenesis in breast cancer [89]; (iii) FOSB increases tumor growth and metastases in ovarian cancer [90]; (iv) FOSB is implicated in TP4 response in triple negative breast cancer [91] (v) STMN1 affects cell-cycle progression and cell mobility in non-small cell lung cancer [92]; (vi) STMN1 has been shown to have prognostic significance in breast cancer [93]; and (vii) STMN1 enhances sensitivity to treatment with paclitaxel in esophageal cancer [94]. The identified 5 site-gene panel thus constitutes valuable candidates for further therapeutic studies.
Interestingly, FOSB is a member of FOS gene family AP1 complexes, which bind to the promoter or enhancer regions of target genes [90,95] and regulate cell survival, proliferation, angiogenesis, invasion, and metastasis [90,96, 97, 98, 99]. Several studies have indicated that FOSB contributes towards increased concentration of IL-8 (interleukin-8) which influence angiogenesis, affecting cellular proliferation and metastases in ovarian cancer [90]. Recently, it has also been observed that IL-8 is associated with transcriptional activity of androgen receptor (AR), which indicates that FOSB might play an important role in ADT resistance and thus constitutes a valuable candidate for further functional validation.
In recent years, clinical oncology has witnessed the emergence of a so-called neuroendocrine prostate cancer phenotype, with strong ties to failed response to androgen-deprivation treatment [100, 101, 102]. In fact, several studies have shown that a substantial number of patients treated with enzalutamide or abiraterone relapse and develop neuroendocrine features [44,103]. Interestingly, one of the genes we identified, FOSB, has been shown to contribute to disease progression in small bowel neuroendocrine tumors [104]. Thus, it would be a crucial subsequent step to evaluate the clinical relevance of our gene panel in neuroendocrine phenotype.
In summary, we have introduced a systematic genome-wide integrative approach that identified an (epi) genomic panel of 5 site-genes which are predictive of response to ADT. We propose that this panel can be utilized to pre-screen patients and identify those (i) who are at higher risk of developing resistance to ADT and who should potentially be advised an alternative therapeutic regimen (such as chemotherapy, radiation therapy etc.), thereby avoiding ADT side effects and improving disease-course; and (ii) who would benefit from ADT, making it their priority therapy choice. Furthermore, this panel could be utilized to prioritize patients for prospective clinical trials with a long-term objective to extend this effort to build an adaptable accurate platform for precision therapeutics.
The following are the supplementary data related to this article.
Supplementary Fig. 1.
GSEA comparison of parametric and non-parametric differential methylation signatures. GSEA comparing a parametric ADT resistance differential methylation signature, based on two-sample two-tail Student t-test p-values (used as a reference signature), and a non-parametric ADT resistance differential methylation signature, based on empirically estimated p-values (from 10,000 site permutations, sites with p < 0.001, used as a query gene set).
Supplementary Fig. 2.
Treatment-related survival analysis of candidate 5 site-gene panel in TCGA, Beltran et al, and PROMOTE patient cohorts. (a) Treatment-related Kaplan-Meier survival analysis of candidate 5 site-gene panel in TCGA Gleason 7 and Gleason 8-9, demonstrating that therapeutic predictive ability of the identified 5 site-gene panel is independent of Gleason score. Log-rank p-values are indicated. (b) ROC analysis comparing TCGA-PRAD (group1) with Beltran et al (green) and PROMOTE (brown) patient cohorts. AUROC is indicated.
Supplementary Fig. 3.
Multimodal analysis of signature thresholds and negative control dataset. (a) Area Under ROC distribution (y-axis) as a function of varying methylation signature threshold (x-axis). (b) Area Under ROC distribution (y-axis) as a function of varying correlation threshold (x-axis). (c) Kaplan-Meier survival analysis on Sboner et al dataset demonstrates that therapeutic predictive ability of the identified 5 site-gene panel is independent of disease progression. Log-rank p-value and hazard ratio are indicated.
Clinical characteristics of non-responder and responder patients
Differential methylation signature of ADT resistance
Acknowledgments
Acknowledgements
We would like to thank Regev Schweiger (Tel Aviv University) and Sarra Rahem (Rutgers University) for useful discussions. We would like to acknowledge the support and computational resources from the Office of Advanced Research Computing at Rutgers Biomedical and Health Sciences.
Funding Sources
Antonina Mitrofanova is supported by the Prostate Cancer Foundation Young Investigator Award, Rutgers School of Health Professions Dean's Research grant, and Rutgers start-up funds. Sheida Hayati is supported by the New Jersey Commission on Cancer Research Pre-Doctoral Fellowship (DHFS17PPC013).
Conflicts of Interest
The authors declare no competing financial interests.
Author Contributions
S.P. performed computational and bioinformatics data analysis, S.H. performed analysis on genomic locations of the identified sites, N.J.E. performed pathway analysis, J.S.P. advised on statistical methods utilized in the manuscript, S.P. and A.M. designed the overall study and prepared figures, A.M. supervised the data analysis, and S.P. and A.M. wrote the manuscript. All authors provided discussion and comments on the manuscript.
References
- 1.Bray F., Ren J.S., Masuyer E., Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133–1145. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 2.Haas G.P., Delongchamps N., Brawley O.W., Wang C.Y., De La Roza G. The worldwide epidemiology of prostate Cancer: perspectives from autopsy studies. Can J Urol. 2008;15:3866–3871. [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Huggins C., Hodges C.V. Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J Clin. 1972;22:232–240. doi: 10.3322/canjclin.22.4.232. [DOI] [PubMed] [Google Scholar]
- 5.Lonergan P.E., Tindall D.J. Androgen receptor signaling in prostate cancer development and progression. J Carcinogenesis. 2011;10:20. doi: 10.4103/1477-3163.83937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scher H.I., Sawyers C.L. Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol. 2005;23:8253–8261. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 7.Karantanos T., Corn P.G., Thompson T.C. Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene. 2013;32:5501–5511. doi: 10.1038/onc.2013.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lallous N., Volik S.V., Awrey S., Leblanc E., Tse R., Murillo J. Functional analysis of androgen receptor mutations that confer anti-androgen resistance identified in circulating cell-free DNA from prostate cancer patients. Genome Biol. 2016;17:10. doi: 10.1186/s13059-015-0864-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandrasekar T., Yang J.C., Gao A.C., Evans C.P. Mechanisms of resistance in castration-resistant prostate cancer (CRPC) Translational Andrology and Urology. 2015;4:365–380. doi: 10.3978/j.issn.2223-4683.2015.05.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoyanova T., Riedinger M., Lin S., Faltermeier C.M., Smith B.A., Zhang K.X. Activation of Notch1 synergizes with multiple pathways in promoting castration-resistant prostate cancer. Proc Natl Acad Sci. 2016;113:E6457–E6466. doi: 10.1073/pnas.1614529113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wallace T.J., Torre T., Grob M., Yu J., Avital I., Brücher B. Current approaches, challenges and future directions for monitoring treatment response in prostate Cancer. J Cancer. 2014;5:3–24. doi: 10.7150/jca.7709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marzese D.M., Hoon D.S.B. Emerging technologies for studying DNA methylation for the molecular diagnosis of cancer. Expert Rev Mol Diagn. 2015;15:647–664. doi: 10.1586/14737159.2015.1027194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abeshouse A., Ahn J., Akbani R., Ally A., Amin S., Andry Christopher D. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoenborn J.R., Nelson P., Fang M. Genomic profiling defines subtypes of prostate cancer with the potential for therapeutic stratification. Clin Cancer Res. 2013;19:4058–4066. doi: 10.1158/1078-0432.CCR-12-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen M.M., Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baxter E., Windloch K., Gannon F., Lee J.S. Epigenetic regulation in cancer progression. Cell Biosci. 2014;4:45. doi: 10.1186/2045-3701-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhingra P., Martinez-Fundichely A., Berger A., Huang F.W., Forbes A.N., Liu E.M. Identification of novel prostate cancer drivers using RegNetDriver: a framework for integration of genetic and epigenetic alterations with tissue-specific regulatory network. Genome Biol. 2017;18:141. doi: 10.1186/s13059-017-1266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma S., Kelly T.K., Jones P.A. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urbanucci A., Barfeld S.J., Kytölä V., Itkonen H.M., Coleman I.M., Vodák D. Androgen receptor deregulation drives Bromodomain-mediated chromatin alterations in prostate Cancer. Cell Rep. 2017;19:2045–2059. doi: 10.1016/j.celrep.2017.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao L., Shen H., Laird P.W., Farnham P.J., Berman B.P. Inferring regulatory element landscapes and transcription factor networks from cancer methylomes. Genome Biol. 2015;16:105. doi: 10.1186/s13059-015-0668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith Z.D., Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 22.Butler M.G. Genomic imprinting disorders in humans: a mini-review. J Assist Reprod Genet. 2009;26:477–486. doi: 10.1007/s10815-009-9353-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson A.A., Akman K., Calimport S.R.G., Wuttke D., Stolzing A., De Magalhães J.P. The role of DNA methylation in aging, rejuvenation, and age-related disease. Rejuvenation Res. 2012;15:483–494. doi: 10.1089/rej.2012.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luczak M.W., Jagodzinski P.P. The role of DNA methylation in cancer development. Folia Histochem Cytobiol. 2006;44:143–154. [PubMed] [Google Scholar]
- 25.Wajed S.A., Laird P.W., Demeester T.R. DNA methylation: an alternative pathway to Cancer. Ann Surg. 2001;234:10–20. doi: 10.1097/00000658-200107000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deaton A.M., Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Illingworth R.S., Bird A.P. CpG islands – ‘a rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 28.Gardiner-Garden M., Frommer M. CpG Islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X.Y., Ehrlich K.C., Wang R.Y., Ehrlich M. Effect of site-specific DNA methylation and mutagenesis on recognition by methylated DNA-binding protein from human placenta. Nucleic Acids Res. 1986;14:8387–8397. doi: 10.1093/nar/14.21.8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sengupta P.K., Ehrlich M., Smith B.D. A methylation-responsive MDBP/RFX site is in the first exon of the collagen alpha2(I) promoter. J Biol Chem. 1999;274:36649–36655. doi: 10.1074/jbc.274.51.36649. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X.-Y., Asiedu C.K., Supakar P.C., Khan R., Ehrlich K.C., Ehrlich M. Binding sites in mammalian genes and viral gene regulatory regions recognized by methylated DNA-binding protein. Nucleic Acids Res. 1990;18:6253–6260. doi: 10.1093/nar/18.21.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greger V., Passarge E., Hopping W., Messmer E., Horsthemke B. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum Genet. 1989;83:155–158. doi: 10.1007/BF00286709. [DOI] [PubMed] [Google Scholar]
- 33.Conerly M., Grady W.M. Insights into the role of DNA methylation in disease through the use of mouse models. Disease Models & Mechanisms. 2010;3:290–297. doi: 10.1242/dmm.004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 35.Laird P.W., Jaenisch R. The role of DNA methylation in cancer genetic and epigenetics. Annu Rev Genet. 1996;30:441–464. doi: 10.1146/annurev.genet.30.1.441. [DOI] [PubMed] [Google Scholar]
- 36.Chen C.-C., Lee K.-D., Pai M.-Y., Chu P.-Y., Hsu C.-C., Chiu C.-C. Changes in DNA methylation are associated with the development of drug resistance in cervical cancer cells. Cancer Cell Int. 2015;15:98. doi: 10.1186/s12935-015-0248-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones P.A., Baylin S.B. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 38.Gifford G., Paul J., Vasey P.A., Kaye S.B., Brown R. The acquisition of hMLH1 methylation in plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clin Cancer Res. 2004;10:4420–4426. doi: 10.1158/1078-0432.CCR-03-0732. [DOI] [PubMed] [Google Scholar]
- 39.Pathiraja T.N., Nayak S., Xi Y., Jiang S., Garee J.P., Edwards D.P. Epigenetic reprogramming of HOXC10 in endocrine-resistant breast cancer. Sci Transl Med. 2014;6:229ra41. doi: 10.1126/scitranslmed.3008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eyre R., Harvey I., Stemke-Hale K., Lennard T.W., Tyson-Capper A., Meeson A.P. Reversing paclitaxel resistance in ovarian cancer cells via inhibition of the ABCB1 expressing side population. Tumour Biol. 2014;35:9879–9892. doi: 10.1007/s13277-014-2277-2. [DOI] [PubMed] [Google Scholar]
- 41.Zöchbauer-Müller S., Fong K.M., Maitra A., Lam S., Geradts J., Ashfaq R. 5′ CpG island methylation of the <em>FHIT</em> gene is correlated with loss of gene expression in lung and breast cancer. Cancer Res. 2001;61:3581–3585. [PubMed] [Google Scholar]
- 42.Woo H.G., Choi J.-H., Yoon S., Jee B.A., Cho E.J., Lee J.-H. Integrative analysis of genomic and epigenomic regulation of the transcriptome in liver cancer. Nat Commun. 2017;8:839. doi: 10.1038/s41467-017-00991-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhee J.-K., Kim K., Chae H., Evans J., Yan P., Zhang B.-T. Integrated analysis of genome-wide DNA methylation and gene expression profiles in molecular subtypes of breast cancer. Nucleic Acids Res. 2013;41:8464–8474. doi: 10.1093/nar/gkt643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beltran H., Rickman D.S., Park K., Chae S.S., Sboner A., Macdonald T.Y. Molecular characterization of neuroendocrine prostate Cancer and identification of new drug targets. Cancer Discov. 2011;1:487–495. doi: 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai C., Wang H., He H.H., Chen S., He L., Ma F. ERG induces androgen receptor-mediated regulation of SOX9 in prostate cancer. J Clin Invest. 2013;123:1109–1122. doi: 10.1172/JCI66666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grasso C.S., Wu Y.M., Robinson D.R., Cao X., Dhanasekaran S.M., Khan A.P. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kohli M., Wang L., Xie F., Sicotte H., Yin P., Dehm S.M. Mutational landscapes of sequential prostate metastases and matched patient derived xenografts during enzalutamide therapy. PLoS One. 2015;10 doi: 10.1371/journal.pone.0145176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson D., Van Allen E.M., Wu Y.M., Schultz N., Lonigro R.J., Mosquera J.M. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sboner A., Demichelis F., Calza S., Pawitan Y., Setlur S.R., Hoshida Y. Molecular sampling of prostate cancer: a dilemma for predicting disease progression. BMC Med Genet. 2010;3:8. doi: 10.1186/1755-8794-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du P., Zhang X., Huang C.-C., Jafari N., Kibbe W.A., Hou L. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fisher R.A. On the interpretation of χ 2 from contingency tables, and the calculation of P. J R I State Dent Soc. 1922;85:87–94. [Google Scholar]
- 53.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mukaka M.M. A guide to appropriate use of correlation coefficient in medical research. Malawi Med J. 2012;24:69–71. [PMC free article] [PubMed] [Google Scholar]
- 55.Chatterjee S., Hadi A.S. Regression analysis by example. Fourth Edition. 2006. Simple linear regression; pp. 21–51. [Google Scholar]
- 56.Maaten L.V.D., Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 57.Hershey J.R., Olsen P.A. Acoustics, speech and signal processing, 2007. ICASSP 2007. IEEE international conference on, 2007. IEEE; 2007. Approximating the Kullback Leibler divergence between Gaussian mixture models. (IV-317-IV-320) [Google Scholar]
- 58.Goel M.K., Khanna P., Kishore J. Understanding survival analysis: Kaplan-Meier estimate. Int J Ayurveda Res. 2010;1:274–278. doi: 10.4103/0974-7788.76794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bender R., Augustin T., Blettner M. Generating survival times to simulate Cox proportional hazards models. Stat Med. 2005;24:1713–1723. doi: 10.1002/sim.2059. [DOI] [PubMed] [Google Scholar]
- 60.Kassambara A., Kosinski M., Biecek P. R package version 0.3. 2017. survminer: drawing survival curves using ggplot2; p. 1. [Google Scholar]
- 61.Therneau T. A package for survival analysis in S. R package version 2.38. 2015. http://CRAN.R-project.org/package=survival Retrived from.
- 62.Therneau T., Grambsch P. Springer; 2010. Modeling survival data: extending the cox model (statistics for biology and health) [Google Scholar]
- 63.Hajian-Tilaki K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J Int Med. 2013;4:627–635. [PMC free article] [PubMed] [Google Scholar]
- 64.Hanley J.A., Mcneil B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 65.Zeileis A., Kleiber C., Jackman S. Regression models for count data in R. J Stat Softw. 2008;27:1–25. [Google Scholar]
- 66.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.-C. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stouffer S.A., Suchman E.A., Devinney L.C., Star S.A., Williams Jr R.M. vol. 1. 1949. The American soldier: adjustment during army life.(studies in social psychology in World War II) [Google Scholar]
- 68.Alvarez M.J., Shen Y., Giorgi F.M., Lachmann A., Ding B.B., Ye B.H. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat Genet. 2016;48:838. doi: 10.1038/ng.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schröder M.S., Culhane A.C., Quackenbush J., Haibe-Kains B. Survcomp: an R/Bioconductor package for performance assessment and comparison of survival models. Bioinformatics. 2011;27:3206–3208. doi: 10.1093/bioinformatics/btr511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morris T.J., Beck S. Analysis pipelines and packages for Infinium HumanMethylation450 BeadChip (450k) data. Methods (San Diego, Calif) 2015;72:3–8. doi: 10.1016/j.ymeth.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Amaro A., Esposito A.I., Gallina A., Nees M., Angelini G., Albini A. Validation of proposed prostate cancer biomarkers with gene expression data: a long road to travel. Cancer Metastasis Rev. 2014;33:657–671. doi: 10.1007/s10555-013-9470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barfeld, S. J., Urbanucci, A., Itkonen, H. M., Fazli, L., Hicks, J. L., Thiede, B., Rennie, P. S., Yegnasubramanian, S., Demarzo, A. M. & Mills, I. G. c-Myc Antagonises the transcriptional activity of the androgen receptor in prostate Cancer affecting key gene networks. EBioMedicine, 18, 83–93. [DOI] [PMC free article] [PubMed]
- 73.Benzon B., Zhao S.G., Haffner M.C., Takhar M., Erho N., Yousefi K. Correlation of B7-H3 with androgen receptor, immune pathways and poor outcome in prostate cancer: an expression-based analysis. Prostate Cancer Prostatic Dis. 2017;20:28–35. doi: 10.1038/pcan.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bibikova M., Chudin E., Arsanjani A., Zhou L., Garcia E.W., Modder J. Expression signatures that correlated with Gleason score and relapse in prostate cancer. Genomics. 2007;89:666–672. doi: 10.1016/j.ygeno.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 75.Cai, J., Li, B., Zhu, Y., Fang, X., Zhu, M., Wang, M., Liu, S., Jiang, X., Zheng, J., Zhang, X., et al. Prognostic biomarker identification through integrating the gene signatures of hepatocellular carcinoma properties. EBioMedicine, 19, 18–30. [DOI] [PMC free article] [PubMed]
- 76.Geybels M.S., Wright J.L., Bibikova M., Klotzle B., Fan J.-B., Zhao S. Epigenetic signature of Gleason score and prostate cancer recurrence after radical prostatectomy. Clin Epigenetics. 2016;8:97. doi: 10.1186/s13148-016-0260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haldrup C., Mundbjerg K., Vestergaard E.M., Lamy P., Wild P., Schulz W.A. DNA methylation signatures for prediction of biochemical recurrence after radical prostatectomy of clinically localized prostate Cancer. J Clin Oncol. 2013;31:3250–3258. doi: 10.1200/JCO.2012.47.1847. [DOI] [PubMed] [Google Scholar]
- 78.Litovkin, K., VAN Eynde, A., Joniau, S., Lerut, E., Laenen, A., Gevaert, T., Gevaert, O., Spahn, M., Kneitz, B. & Gramme, P. 2015. DNA methylation-guided prediction of clinical failure in high-risk prostate cancer. PLoS One, 10, e0130651. [DOI] [PMC free article] [PubMed]
- 79.Massie C.E., Mills I.G., Lynch A.G. The importance of DNA methylation in prostate cancer development. J Steroid Biochem Mol Biol. 2017;166:1–15. doi: 10.1016/j.jsbmb.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 80.Mitrofanova, A., Aytes, A., Zou, M., Shen, Michael M., Abate-Shen, C. & Califano, A. Predicting drug response in human Prostate cancer from preclinical analysis of In vivo mouse models. Cell Rep, 12, 2060-2071. [DOI] [PMC free article] [PubMed]
- 81.Risk M.C., Knudsen B.S., Coleman I., Dumpit R.F., Kristal A.R., Lemeur N. Differential gene expression in benign prostate epithelium of men with and without prostate cancer: evidence for a prostate cancer field effect. Clin Cancer Res. 2010;16:5414–5423. doi: 10.1158/1078-0432.CCR-10-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ryl, T., Kuchen, E. E., Bell, E., Shao, C., Flórez, A. F., Mönke, G., Gogolin, S., Friedrich, M., Lamprecht, F., Westermann, F., et al. Cell-cycle position of single MYC-driven Cancer cells dictates their susceptibility to a chemotherapeutic drug. Cell Systems, (237-250.e8). [DOI] [PubMed]
- 83.Wang L., Gong Y., Chippada-Venkata U., Heck M.M., Retz M., Nawroth R. A robust blood gene expression-based prognostic model for castration-resistant prostate cancer. BMC Med. 2015;13:201. doi: 10.1186/s12916-015-0442-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu Y., Davison J., Qu X., Morrissey C., Storer B., Brown L. Methylation profiling identified novel differentially methylated markers including OPCML and FLRT2 in prostate cancer. Epigenetics. 2016;11:247–258. doi: 10.1080/15592294.2016.1148867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu, K.-H., Berry, G. J., Rubin, D. L., Ré, C., Altman, R. B. & Snyder, M. Association of Omics Features with histopathology patterns in lung adenocarcinoma. Cell Systems [DOI] [PMC free article] [PubMed]
- 86.Guinney, J., Wang, T., Laajala, T. D., Winner, K. K., Bare, J. C., Neto, E. C., Khan, S. A., Peddinti, G., Airola, A., Pahikkala, T., et al. Prediction of overall survival for patients with metastatic castration-resistant prostate cancer: development of a prognostic model through a crowdsourced challenge with open clinical trial data. Lancet Oncol, 18, 132–142. [DOI] [PMC free article] [PubMed]
- 87.Halabi S., Lin C.-Y., Small E.J., Armstrong A.J., Kaplan E.B., Petrylak D. Prognostic model predicting metastatic castration-resistant prostate Cancer survival in men treated with second-line chemotherapy. JNCI J Cancer Institute. 2013;105:1729–1737. doi: 10.1093/jnci/djt280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ili C.G., Viscarra T., Araya J.C., Lopez J., Mora B., Retamal J. Abstract B28: FKBP6 gene is involved in progression of cervical cancer. Mol Cancer Res. 2016;14:B28. [Google Scholar]
- 89.Milde-Langosch K., Kappes H., Riethdorf S., Loning T., Bamberger A.M. FosB is highly expressed in normal mammary epithelia, but down-regulated in poorly differentiated breast carcinomas. Breast Cancer Res Treat. 2003;77:265–275. doi: 10.1023/a:1021887100216. [DOI] [PubMed] [Google Scholar]
- 90.Shahzad M.M., Arevalo J.M., Armaiz-Pena G.N., Lu C., Stone R.L., Moreno-Smith M. Stress effects on FosB- and interleukin-8 (IL8)-driven ovarian cancer growth and metastasis. J Biol Chem. 2010;285:35462–35470. doi: 10.1074/jbc.M110.109579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ting C.-H., Chen Y.-C., Wu C.-J., Chen J.-Y. Targeting FOSB with a cationic antimicrobial peptide, TP4, for treatment of triple-negative breast cancer. Oncotarget. 2016;7:40329–40347. doi: 10.18632/oncotarget.9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nie W., Xu M.D., Gan L., Huang H., Xiu Q., Li B. Overexpression of stathmin 1 is a poor prognostic biomarker in non-small cell lung cancer. Lab Invest. 2015;95:56–64. doi: 10.1038/labinvest.2014.124. [DOI] [PubMed] [Google Scholar]
- 93.Saal L.H., Johansson P., Holm K., Gruvberger-Saal S.K., She Q.-B., Maurer M. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A. 2007;104:7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu H.W., Jiang D., Xie Z.Y., Zhou M.H., Sun D.Y., Zhao Y.G. Effects of stathmin 1 silencing by siRNA on sensitivity of esophageal cancer cells Eca-109 to paclitaxel. Genet Mol Res. 2015;14:18695–18702. doi: 10.4238/2015.December.28.18. [DOI] [PubMed] [Google Scholar]
- 95.Lamph W.W., Wamsley P., Sassone-Corsi P., Verma I.M. Induction of proto-oncogene JUN/AP-1 by serum and TPA. Nature. 1988;334:629–631. doi: 10.1038/334629a0. [DOI] [PubMed] [Google Scholar]
- 96.Shaulian E., Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 97.Shaulian E., Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–6. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 98.Tulchinsky E. Fos family members: regulation, structure and role in oncogenic transformation. Histol Histopathol. 2000;15:921–928. doi: 10.14670/HH-15.921. [DOI] [PubMed] [Google Scholar]
- 99.Van Dam H., Castellazzi M. Distinct roles of Jun: Fos and Jun: ATF dimers in oncogenesis. Oncogene. 2001;20:2453–2464. doi: 10.1038/sj.onc.1204239. [DOI] [PubMed] [Google Scholar]
- 100.Akamatsu, S., Wyatt, Alexander W., Lin, D., Lysakowski, S., Zhang, F., Kim, S., Tse, C., Wang, K., Mo, F., Haegert, A., et al. The placental gene PEG10 promotes progression of neuroendocrine prostate cancer. Cell Rep, 12, 922–936. [DOI] [PubMed]
- 101.Beltran H., Prandi D., Mosquera J.M., Benelli M., Puca L., Cyrta J. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee, J. K., Phillips, J. W., Smith, B. A., Park, J. W., Stoyanova, T., Mccaffrey, E. F., Baertsch, R., Sokolov, A., Meyerowitz, J. G., Mathis, C., et al. N-Myc drives neuroendocrine prostate Cancer initiated from human prostate epithelial cells. Cancer Cell, 29, 536–547. [DOI] [PMC free article] [PubMed]
- 103.Epstein J.I., Amin M.B., Beltran H., Lotan T.L., Mosquera J.M., Reuter V.E. Proposed morphologic classification of prostate cancer with neuroendocrine differentiation. Am J Surg Pathol. 2014;38:756–767. doi: 10.1097/PAS.0000000000000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Miller H.C., Frampton A.E., Malczewska A., Ottaviani S., Stronach E.A., Flora R. MicroRNAs associated with small bowel neuroendocrine tumours and their metastases. Endocr Relat Cancer. 2016;23:711–726. doi: 10.1530/ERC-16-0044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical characteristics of non-responder and responder patients
Differential methylation signature of ADT resistance