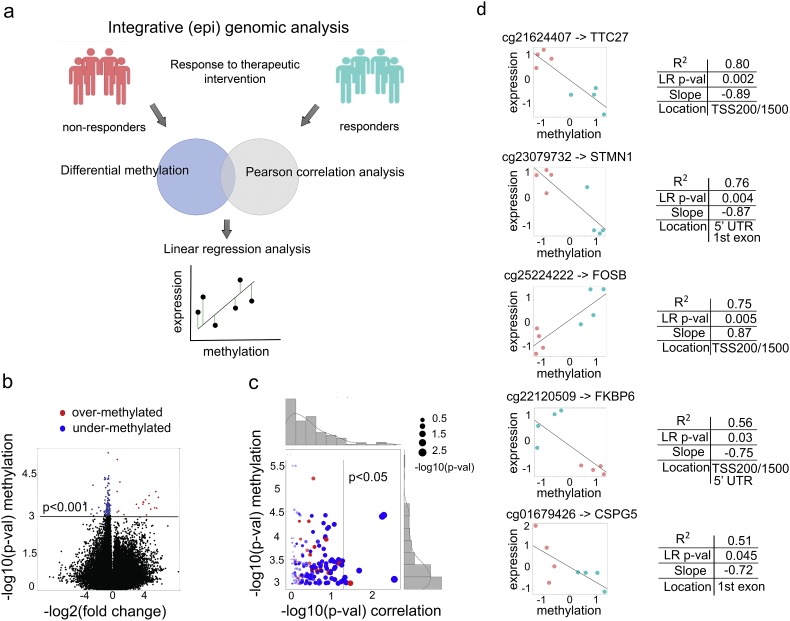
Fig. 4.
Integrative (epi) genomic analysis identifies a 5 site-gene panel. (a) Schematic representation of the integrative (epi) genomic analysis: (top) identification of differentially methylated sites; (middle) Pearson correlation analysis (i.e., pre-screening) between methylation levels of the sites and mRNA expression levels of the site-harboring genes; (bottom) linear regression analysis to identify sites that can explain expression changes of the site-harboring genes. (b) Volcano plot of the differentially methylated sites, with under-methylated (blue) and over-methylated (red) sites indicated (at p-value < 0.001, n = 144). (c) Scatter plot depicting the relationship between differential methylation (y-axis) and site-gene (i.e., methylation-expression) Pearson correlation (x-axis) for the 144 significantly methylated sites from Fig. 4b. (d) Linear regression analysis between site methylation values and mRNA expression of their site-harboring genes (linear regression at p < 0.05 shown), with non-responder (coral) and responder (aquamarine) samples indicated. The x and y axes depict z-scored methylation M-values and DESeq2 normalized expression values.