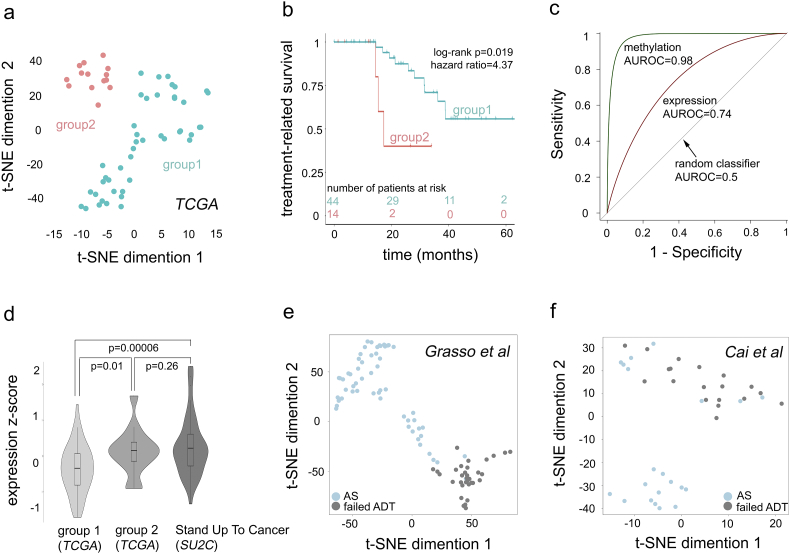
Fig. 5.
Five site-gene (epi) genomic panel predicts ADT failure in independent patient cohorts. (a) t-SNE clustering identifies two groups of patients: group 1 and group 2 (a full set of 5 dimensions considered). (b) Kaplan-Meier survival analysis identifies the significant difference in treatment-related survival (i.e., treatment response) between groups 1 and 2 from Fig. 5a. Log-rank p-value and hazard ratio are indicated. (c) ROC analysis: AUROC indicates the ability of methylated sites and expression of site harboring genes can classify patients into group 1 and group 2. (d) Violin plot for composite Stouffer integrated z-scores (see Materials and Methods) in group 1 (n = 44), group 2 (n = 14) and SU2C (n = 51) cohorts. One-tail two-sample Welch t-test p-values are indicated. (e) t-SNE clustering (all 5 dimensions considered) based on 5 site-gene panel in Grasso et al. cohort (n = 91) [46]; and (f) Cai et al. (n = 40) [45] cohort; is able to separates androgen sensitive (AS, light blue) from castration-resistant prostate cancer (failed ADT, grey) samples (sensitivity = 100% for failed ADT CRPC selection: 33/33 for Grasso et al., and 19/19 for Cai et al.).