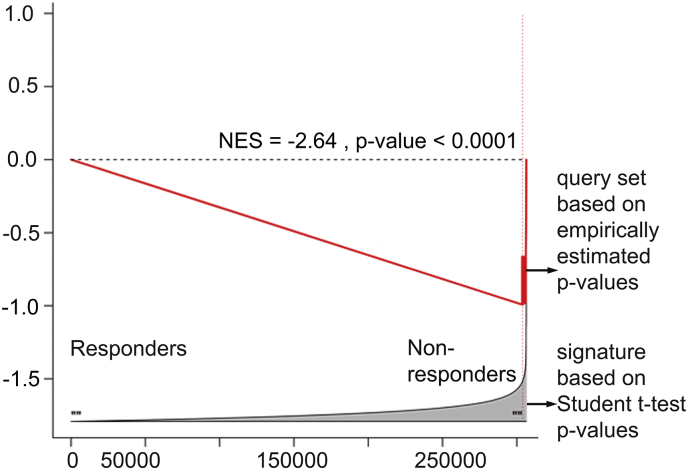
Supplementary Fig. 1.
GSEA comparison of parametric and non-parametric differential methylation signatures. GSEA comparing a parametric ADT resistance differential methylation signature, based on two-sample two-tail Student t-test p-values (used as a reference signature), and a non-parametric ADT resistance differential methylation signature, based on empirically estimated p-values (from 10,000 site permutations, sites with p < 0.001, used as a query gene set).