Abstract
Commensal microorganisms influence a variety of host functions in the gut, including immune response, glucose homeostasis, metabolic pathways and oxidative stress, among others. This study describes how Salmonella Typhi, the pathogen responsible for typhoid fever, uses similar strategies to escape immune defense responses and survive within its human host. To elucidate the early mechanisms of typhoid fever, we performed studies using healthy human intestinal tissue samples and “mini-guts,” organoids grown from intestinal tissue taken from biopsy specimens. We analyzed gene expression changes in human intestinal specimens and bacterial cells both separately and after colonization. Our results showed mechanistic strategies that S. Typhi uses to rearrange the cellular machinery of the host cytoskeleton to successfully invade the intestinal epithelium, promote polarized cytokine release and evade immune system activation by downregulating genes involved in antigen sampling and presentation during infection. This work adds novel information regarding S. Typhi infection pathogenesis in humans, by replicating work shown in traditional cell models, and providing new data that can be applied to future vaccine development strategies.
Abbreviations: STY, Salmonella Typhi; STM, Salmonella Typhimurium; M cells, Microfold cells; CFU, colony forming units; LB, Luria Burtoni broth; DMEM, Dulbecco's Modified Eagle Medium; PBS, phosphate buffered saline; DTT, dithiothreitol; EDTA, ethylenediaminetetraacetic acid; ISC, intestinal stem cell; DAPT, N-[2S-(3,5-difluorophenyl)acetyl]-L-alanyl-2-phenyl-1,1-dimethylethyl ester-glycine; qPCR, quantitative reverse transcriptase polymerase chain reaction; FITC, fluorescein isothiocyanate; TEER, trans-epithelial electrical resistance; RNA, ribonucleic acid; LDH, lactate dehydrogenase; H&E, hematoxylin and eosin; PAS, periodic acid Schiff; TEM, transmission electron microscopy; IL, interleukin
Keywords: Typhoid fever, Salmonella, Snapwell™ system, Human tissue, Terminal ileum, Immune system, Innate immunity, Immune evasion, Host-pathogen interaction, Vaccine development, Intestinal organoids, Organoid monolayer
Highlights
-
•
Salmonella Typhi targets human-specific pathways by inducing host transcriptional changes.
-
•
These pathways include cytoskeletal rearrangement, polarized cytokine release, and hampering host immune defense system
-
•
The overall outcome of Salmonella interaction with the target human host is to avoid immune response in order to efficiently infect and propagate.
Salmonella remains a severe human health threat without sufficient therapeutic options, which is mainly due to the lack of key information about the steps in early pathogenesis that lead to infection. Using human ex vivo models, we have identified key steps in S. Typhi's early development, suggesting that S. Typhi exploits human host machinery, not only to invade, but also to avoid a robust immune response. These findings identify possible targets for the development of more efficient vaccine candidates against human salmonellosis.
1. Introduction
Bacterial pathogens represent a significant global burden to human health resulting in chronic infection, significant mortality, certain cancers, and diminished quality of life [23,38]. Salmonella enterica serovar Typhi (STY) is a human-restricted, gastrointestinal pathogen whose successful infection results in Typhoid fever or chronic infection [30]. Typhoid fever is frequently fatal when untreated in pediatric or immunocompromised populations [9,31,51,79]. It affects an estimated 11.9 to 26.9 million people annually [6,49,50], with estimated financial burdens equaling roughly a third of the gross national income for patients in undeveloped areas of Southeast Asia [35]. Current treatments depend on antibiotics; however, STY is rapidly developing antibiotic resistance, thereby increasing both the risk and severity of infection.
STY is a Gram-negative, enteric pathogen of the genus Salmonellae, species enterica, subspecies enterica. The subspecies enterica represents two serovars: Typhimurium and Typhi. Genetically related, but phenotypically divergent, S. Typhimurium (STM) can cause localized inflammation of the small intestine, diarrhea and cramping. Conversely, STY infection has an incubation period of up to two weeks and results in systemic bacteremia with limited or no gastrointestinal symptoms. Comparative genomics between the two serovars reveals 480 STM-unique genes and 600-STY-unique genes [66]; notable STY genetic acquisitions include a capsule-encoding specific pathogenicity island (SPI)[78], two human-like serine-threonine kinases [73] and a typhoid toxin [18,21]. Numerous STY genes contain mutations relative to the STM homologs, with 0.6% of the STM genome encoding pseudogenes compared to 5% in the STY genome [66].
Current STY vaccination strategies include products that are expensive to produce and store. The attenuated oral vaccine Ty21a, which requires several immunizations to create a sustained immune response, confers protection in 62–96% of vaccinated individuals [41]. Alternative vaccine candidates currently in phase I and phase II studies have been generated by targeting virulence genes related to acid resistance, stress response, osmolarity and invasion. These gene targets are combined with gene deletions resulting in limited intracellular replication [24]. In most STY vaccine candidates, deleted genes attenuate infection. A notable exception is the constitutive expression of the STY-specific [82] immune-capsule suppressing regulator [78] tviA gene [24]. Despite the development of several vaccine strains against STY, no strategy confers long-term protection in a cost-effective manner.
Vaccine development against typhoid fever is hindered by significant assumptions about how S. Typhi causes infection in its human host. Specifically, no data are available regarding STY interaction with the small intestinal mucosa as the first step in the cascade of events ultimately leading to infection. Moreover, there have been no studies evaluating bacterial gene expression and consequences on host gene expression that occur during these critical early moments of infection. Despite fundamental differences in the pathology of infected humans relative to infected mice, and significant genomic differences between the two serovars, our current understanding of STY infection is mainly derived from the STM mouse model. Moreover, the two-week incubation period between STY exposure and onset of disease symptoms demonstrates a critical window in which STY infection is active prior to the onset of clinical disease. In recent years, human-derived organoid systems [19,64,67,80,84] or three-dimensional cell line models [29,60,68,69] have gained traction as a strategy to study host-pathogen interactions. These models have enabled new insight into cellular response during early STM infection to identify the role of bacterial genes n, such as the SPI-1 operon [60], or STM manipulation of signaling cascades [84]; however, few studies combining STY and human-derived organoid monolayers have been published.
Our work overcomes these shortfalls by placing STY bacteria directly onto human small intestinal tissue. In this study, we use an ex vivo, human intestinal tissue infection model and a human organoid-derived monolayer model. Infected biopsies were analyzed for transcriptional changes, cytokine profiling and electron microscopy, with specific mechanisms explored using the organoid monolayer model. This work sought to detail critical early events in Typhoid fever development to understand pathogenic mechanisms in human-derived tissue with the goal of identifying novel targets for vaccine development.
2. Materials and Methods
2.1. Bacterial Strains, Growth Conditions and Biopsy Infection
For all experiments, Salmonella enterica serovar Typhi strain Ty2 (STY, ATCC® Number: 700931) or serovar Typhimurium strain SL1344 (STM) (kind gift of Bobby Cherayil, Massachusetts General Hospital, Boston MA) were re-streaked bi-monthly on LB-agar plates. For experiments conducted under traditional laboratory conditions, an overnight culture from a single colony of STY or STM was prepared in Miller formula LB-broth (Sigma, St. Louis, MO) at 37 °C with shaking at 225 rpm. The next day, overnight cultures were diluted 1:50 into secondary subcultures and grown to log phase. To prepare the bacteria grown under pro-invasion conditions, a day culture in Miller formula LB broth from a single colony of STY or STM was started at 37 °C without shaking. After 4-8 h growth, a secondary culture diluted 1:50 in Miller LB was prepared and grown overnight at 37 °C without shaking. Bacteria were normalized to an OD600 of 0.5 prior to experimentation, pelleted by centrifugation and resuspended in warm Dulbecco's Modified Eagle Medium (DMEM) (Gibco, Grand Island, NY) for use in infection. All biopsies were infected with 1 × 108 bacteria in 250 μL DMEM for a maximum of 2 h. Biopsies were mounted using a snapwell system (Corning, Corning NY) with orientation confirmed by a dissecting microscope. Mounted biopsies acclimated for 30 m prior to removal of media and replacement with control DMEM or DMEM containing STY or SL1344. After 2 h of infection, apical and basolateral medium and biopsies were collected. For gentamicin protection assays, after 2 h infection the apical surface was washed 3× in phosphate buffered saline (PBS), and DMEM containing 5μg/mL gentamicin (Gibco) was added for 30 m at 37 °C. Afterward, biopsies were washed in PBS (Gibco) and subsequently homogenized in 0.5% Triton-X (Sigma) in PBS for serial dilution plating to determine CFU/mL recovery.
2.2. Isolation and Generation of Human Organoids; Preparation of Organoid-derived Epithelial Monolayers
The isolation and generation of human organoids was adapted from the protocol published by VanDussen et al. [76]. Briefly, human terminal ileum biopsies were processed in dithiothreitol (DTT) and ethylenediaminetetraacetic acid (EDTA) in PBS with penicillin/streptomycin (P/S, Gibco) to isolate the crypt fractions. This process was repeated 4–5 times; the fractions with the greatest number of crypts were pooled and resuspended in Matrigel (Corning, NY). Spheres were maintained in culture in 1:1 LWRN-conditioned medium and Intestinal Stem Cell (ISC) medium supplemented with Y-27632 (Sigma or Calbiochem, La Jolla, CA) and A-8301 (Sigma or Tocris, Minneapolis, MN). Spheres were fed every 2–3 days and split prior to differentiation. Undifferentiated cells were seeded onto transwell inserts and maintained in culture until monolayers formed. 48 h prior to experiment, monolayers were treated with apical N-[2S-(3,5-difluorophenyl)acetyl]-L-alanyl-2-phenyl-1,1-dimethylethyl ester-glycine (DAPT) (Calbiochem) to induce differentiation. Monolayers were assessed by quantitative reverse transcriptase polymerase chain reaction (qPCR) and confocal microscopy to identify markers for differentiated epithelial cells, as well as, Trans epithelial electrical resistance (TEER) and fluorescein isothiocyanate (FITC) dextran to determine barrier integrity.
2.3. Human Donors
All protocols for recruitment of human subjects and use of human terminal ileum biopsies were approved by Massachusetts General Hospital / Partners Healthcare IRB (Protocol 2014P002001). Prospective donors without chronic medical conditions who were scheduled to undergo a diagnostic colonoscopy were screened for good general health. Exclusion criteria included pregnancy, a known diagnosis of an autoimmune disorder or any chronic medical condition that would increase the risk from a gastrointestinal biopsy, and an inability or unwillingness to provide written informed consent. Donors signing informed consent contributed four to eight biopsies that were transported to the laboratory and used immediately to maximize tissue viability.
2.4. RNA Isolation and Transcriptomic Analysis
Following treatment (mock or infection) biopsies were immediately snap frozen on dry ice and stored at −80 until use. The control biopsy group contains four samples, the infected biopsy group contains five samples and the bacteria control group contains four samples. Biopsy ribonucleic acid (RNA) was isolated in Trizol (Ambion, Carlsbad CA), using the Direct-Zol isolation kit (Zymo, Irvine, CA) following manufacturer's instructions. RNA from purified bacterial cultures was isolated using the RNAeasy kit (Qiagen, Waltham, MA) following manufacturer's instructions. Purified RNA was submitted to GENEWIZ (Frederick, MD) for RNA-sequencing, bacterial rRNA and human rRNA depletion, RNA library preparation (multiplexing and cluster generation) and sequencing on a 1x50bp SR, HiSeq 2500; rapid run flow cells were conducted by GENEWIZ. The reads generated for each RNA sample were analyzed using an Ergatis-based RNA-Seq analysis pipeline [59]. Quality control of the sequences was performed using FastQC (version 0.10.0) [3]. Human sequencing reads were aligned to the Homo sapiens reference genome GRCh38.78 using TopHat (version 2.1.1) [34]. Bacterial reads were aligned to Salmonella enterica serovar Typhi strain Ty2 genome (NCBI Reference Sequence: NC_004631.1) using Bowtie (version 0.12.9) [40]. The number of reads that aligned to the predicted coding regions were determined using HTSeq (version 0.4.7) [5]. Differential gene expression was analyzed using DESeq (version 1.5.24) [4] Full data sets were uploaded to Ingenuity Pathway (licensed to W. Flavahan), and differentially expressed genes were used to identify the pathways changed in infected samples. To identify differentially expressed STY genes, read files were uploaded into KBASE [7] and analyzed as follows: Samples were assigned to control or experimental samples to define a sample set for downstream analysis. The reads were then aligned using Bowtie2 to create an RNAseq alignment set. Transcripts were then assembled using StringTie, and differential expression was identified using CuffDiff. Further analysis was conducted to create expression matrices, heatmaps or cluster analysis. Differentially expressed genes were uploaded into KEGG to map bacterial pathways. Data is accessible in GEO under accession GSE113333.
2.5. Cytokine Analysis
Multiplex cytokine profiling was conducted using a 13plex human pro-inflammatory profiling kit in a Mesoscale Discovery System platform (Meso Scale Diagnostics, Rockville MD). Apical and basolateral samples from infected and control biopsies were assayed per manufacturer's instructions. Results were calculated to determine total volume of cytokine released. Statistical significance was determined by one-way ANOVA with Tukey post-test and represent an n of 15 controls, 10 STM and 10 STY infected. *p ≤ 0.05, **p ≤ 0.005, ***p ≤ 0.0005.
2.6. Western Blot Analysis
Snap-frozen biopsies were thawed in RIPA buffer (Sigma) + 2× protease inhibitors (Roche, Burlington NC) + 2× phosphatase inhibitors (Roche) on ice. Biopsies were homogenized using biomasher II tubes (Kimble, Vineland NJ) and protein concentration was quantified by protein assay (Biorad, Hercules, CA). 10 μg of protein were run on a 4–20% protein gel (Biorad) and transferred onto membrane (Biorad). Membranes were blocked in 5% BSA for 30 m followed by overnight incubation in the following antibodies: mouse anti-β-tubulin (RRID:AB_2715541), rabbit anti-pP44/42 MAPK (RRID:AB_2315112), rabbit anti-p-SAPK/JNK (RRID:AB_823588), rabbit anti-pP38 (RRID:AB_2139682), rabbit anti-pP65 (RRID:AB_331284) (all Cell Signaling Technologies, Danvers, MA). Densitometry was calculated as: ((P-gene/area)/(Actin/Area)experimental)/((P-gene/area)/(Actin/Area)Media Control)). Significance was calculated by paired, two-tailed t-test, *p ≤ 0.05.
2.7. Permeability and Cell Viability
Apical supernatants were assessed for lactate dehydrogenase (LDH) release using Promega Cytox Kit (Promega, Madison, WI) per manufacturer's instructions. To assess paracellular permeability, biopsies were monitored using a TEER apparatus (World Precision Instruments, Sarasota, FL). Alternatively, passage of 1μg/mL 4kD FITC Dextran was assessed by florescence in the basolateral medium using Biotek Synergy 2 and compared against serial dilutions of stock FITC dextran to generate a standard curve. No significance was determined by paired, two-tailed t-test.
2.8. Immunostaining and Microscopy Studies
For immunostaining, hematoxylin & eosin (H&E) and Periodic Acid Schiff (PAS) analysis, biopsies were fixed in 4% paraformaldehyde at room temperature for 30 m followed by storage in 70% ethanol until paraffin embedding. Sections were stained using the antibodies against the following proteins: mouse anti-Actin (RRID:AB_11004139, Invitrogen/Thermo-Fisher), rabbit anti-Zo1 (RRID:AB_2533938, Invitrogen), mouse anti-EPCaM (RRID:AB_10981962, Thermo Fisher, Waltham, MA), rabbit anti-rab5 (RRID:AB_823625, Cell Signaling Technologies), goat anti-Muc2 (RRID:AB_2146667, Santa Cruz Biotechnology, Dallas, TX), mouse anti-Salmonella (RRID:AB_1125358, BD Biosciences, San Jose, CA), rabbit-anti-Salmonella (RRID:AB_561201, Pierce/Thermo-Fisher), rabbit-anti-Salmonella-biotin conjugated (RRID:AB_1018415, Invitrogen/Thermo-Fisher) and mouse anti-tubulin (RRID:AB_2715541, Cell Signaling Technologies). Fluorescent, conjugated, secondary monoclonal antibodies were used for detection. For samples using streptavidin-biotin detection, avidin-biotin blocking was performed prior to staining (Life Technologies/Thermo-Fisher). Samples were imaged using a Nikon A1SiR confocal microscope.
For transmission electron microscopy (TEM) analysis, samples were fixed in 2%PFA/2.5% Glut in 0.1 M Sodium Cacodylate followed by mounting on grids and imaged using a transmission electron microscope (JEOL, Peabody, MA).
3. Results
3.1. Human Intestinal Biopsies Are Susceptible to STY Infection
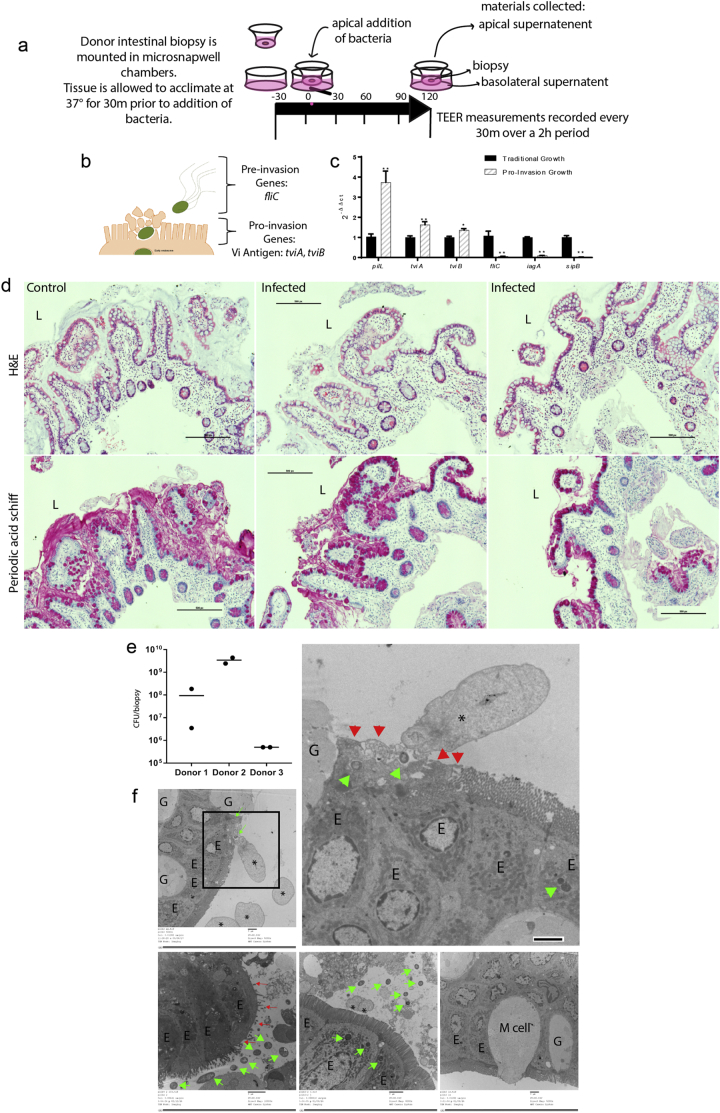
STY colonize and invade the distal ileum of humans [20]. Terminal ileum biopsies obtained during clinically indicated colonoscopies were mounted in microsnapwell devices to allow polarized exposure to microorganisms from the luminal side [8] (see schematic in Fig. 1A and donor characteristics in Table 1). The biopsies were infected with 108 CFU of STY. The bacterial inoculum was prepared by growing STY under static conditions in LB-miller to maximize the expression of genes important in epithelial cell invasion [75] (Fig. 1B,C). After 2 h, invasion and biopsy status was assessed by transmission electron microscopy (TEM), hematoxylin and eosin (H&E) staining for light microscopy, Periodic acid Schiff (PAS) staining for light microscopy and gentamicin protection assay. Intracellular bacteria were detected by TEM, H&E and gentamicin protection assay (Fig. 1D–F). Microvilli destruction and epithelial cell disorganization were observed at sites of bacterial invasion (Fig. 1F, 4B). No gross differences in H&E were observed (Fig. 1D), while minor differences in mucus staining was observed after infection by PAS staining.
Fig. 1.
Modeling S. Typhi Infection: Human intestinal biopsies are susceptible to Salmonella enterica serovar Typhi infection.
(A) A cartoon schematic detailing the microsnapwell model for infection of human intestinal biopsies. Biopsies are mounted between plastic inserts to preserve orientation by placing the basolateral surface down and the lumen-facing enterocytes in the apical opening. Biopsies are apically treated for 2 h (either media control, STY or STM).
(B) An overview of known STY gene expression prior to and during early invasion. For reference: fliC is the major subunit of flagellin used prior to invasion to facilitate epithelial cell homing and Vi antigen genes tviA and tviB are expressed during invasion.
(C) qPCR gene expression of STY bacteria in our pro-invasion inoculum relative to bacteria grown under traditional laboratory conditions.
(D) Hematoxylin & Eosin and Periodic Acid Schiff images of fixed biopsies. Scale bars represent 500 pixels, “L” indicates the lumen.
(E) Gentamicin protection assay showing the distribution of internalized STY across three donors. Invasion rates varied by donor, data presented represent counts for two separate biopsies from three distinct donors. Invasion rates for STM are 2-3× higher than STY (data not shown).
(F)Invading and intracellular STY are observed by TEM, green arrows. Cytoskeletal rearrangements are denoted by *. Microvillus destruction is marked by red arrows. M cells are marked “M cell,” enterocytes are labeled with “E,” and goblet cells are marked with “G.’ Scale bar is 2um for all TEM images.
Table 1.
Donor characteristics.
| Study arm | Total recruited | Gender | Age | Race | Ethnicity |
|---|---|---|---|---|---|
| Microsnapwell infection | 67 | 41 Males | 38 ± 31 | 2/67 Asian | 56/67 - Non-Hispanic or Latino |
| 26 Females | 2/67 AA or Black | 5/67 - Hispanic or Latino | |||
| 6/67 Other | 6/67 - Declined or unknown | ||||
| 57/67 White | |||||
| Organoid culture | 8 | 3 Male | 6 (50–70) | 8/8 White | 1/8 Hispanic or Latino |
| 5 Female | 2 (38–48) | 7/8 Non-Hispanic or Latino |
Fig. 4.
Host response to Typhi infection: barrier function and antigen trafficking.
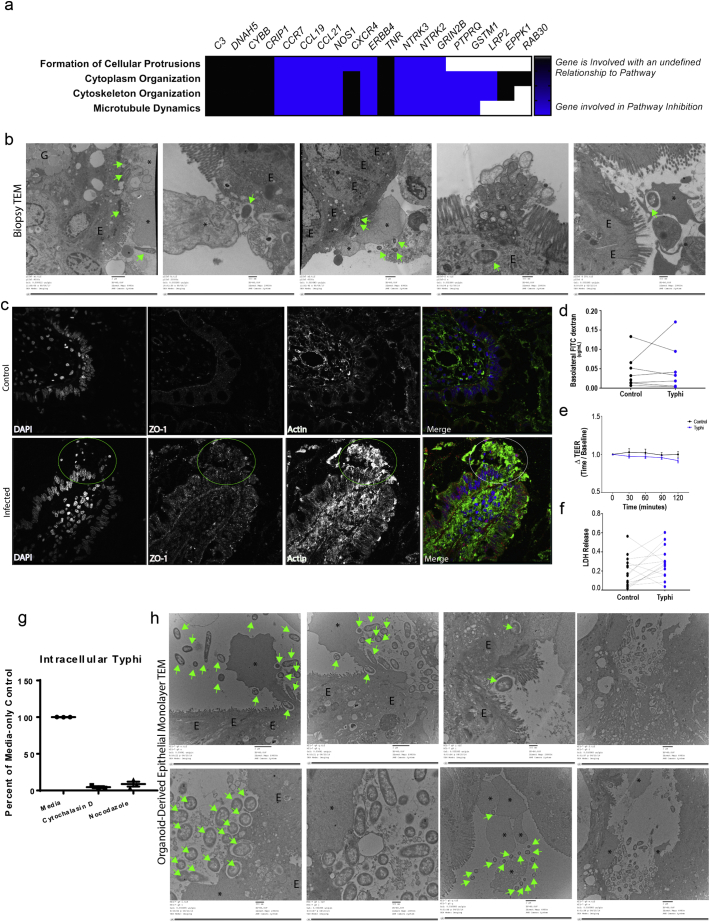
(A) Differentially expressed genes cluster in cytoskeletal modulation pathways. Genes with an unidentified relationship to the pathway are shown in black; genes involved in pathway inhibition are shown in blue.
(B, H) Reorganization of the cytoskeletal is observed upon STY association with intestinal biopsies and organoid-derived epithelial monolayers (H). Large scale bars are 2 μm, small scale bars are 500 nm.
(C) Cellular protrusions are actin-dense as observed by immunostaining (C, circled).
(D–F) No changes in paracellular permeability (D, E) are observed at early time points during infection supporting the fact that STY directly infected enterocytes of the intestinal epithelium, and that biopsy invasion is not accompanied by cell death (F).
(G) Inhibition of the cytoskeleton using the actin inhibitor cytochalasin D or microtubule inhibitor nocodazole block STY entry into organoid-derived epithelial monolayers.
(H) STY bacteria interact with the epithelium by promoting cytoskeletal rearrangement and internalization into vesicles like those observed in the infected biopsies. Invading and intracellular STY are observed by TEM, green arrows. Cytoskeletal rearrangements are denoted by * and enterocytes are labeled with “E.” Large scale bars are 2 μm, small scale bars are 500 nm.
3.2. Downregulation of Genes Involved in the Host Immune Response Is Observed Through Transcriptomic Analysis
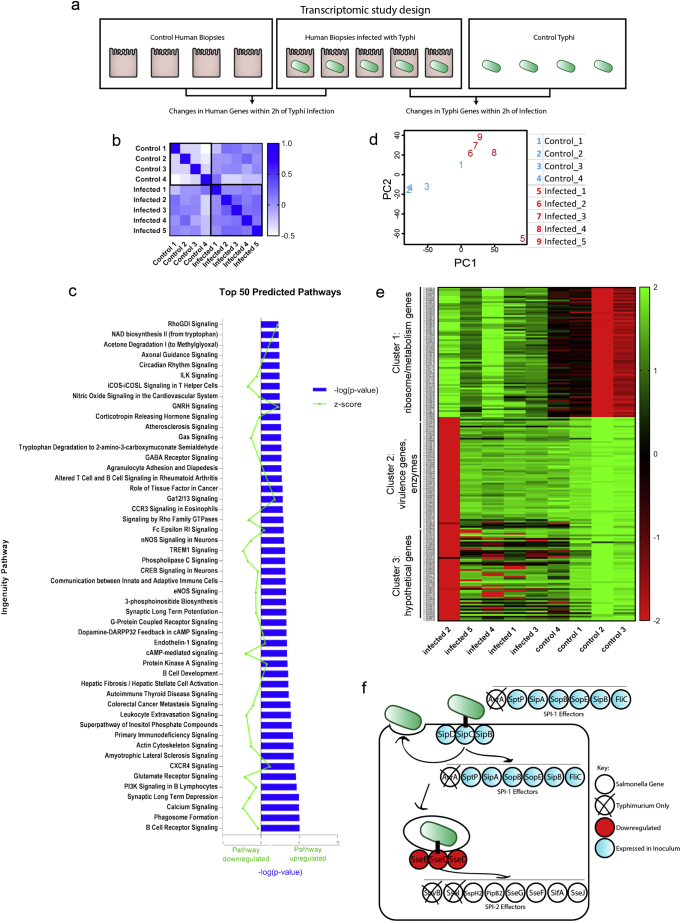
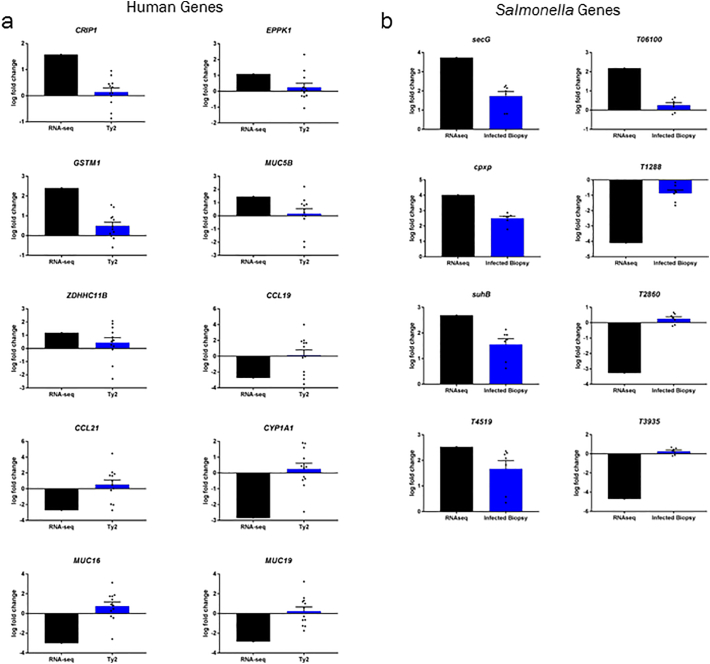
We performed RNA-sequencing to determine differentially expressed genes in both STY and host cells (Fig. 2A, qPCR validation in Supplementary Fig. 1). Gene expression profiles from non-infected control biopsies and STY-infected biopsies were compared to determine significantly upregulated (p ≤ 0.05 and >2-fold induction) and downregulated (p ≤ 0.05 and <0.5-fold reduction) genes. Similar gene expression changes were observed across samples or control groups with some anticipated differences attributed to human-to-human variability (Fig. 2B). For human-specific genes, very few genes were upregulated in response to infection (Table 2A). Regulatory RNAs, the antioxidant GSTM1, zinc-interacting protein CRIP1 and chemokine CCL25 were among the eight significantly upregulated genes in infected samples. Reducing stringency to include genes that satisfied our significance cutoff but which were expressed below our 2-fold cutoff, revealed the expression of mucin MUC5B, the cytoskeletal gene EPPK1 and several metabolism genes induced after infection. Conversely, 57 genes were significantly downregulated (Table 2B). Ultimately, the transcriptional profile of downregulated genes demonstrated significant clustering in several pathways including B-cell receptor signaling, coordination between innate and adaptive immune response, cell signaling and other pathways (Fig. 2C). This data set indicated that STY can hinder the activation of the mucosal immune response of the host during the first 2 h of invasion.
Fig. 2.
Downregulation of genes involved in the host immune response is observed through transcriptomic analysis.
(A) A schematic overview of the transcriptomic study design using human biopsies.
(B) Similarity matrix compares gene expression between samples to show that control and infected biopsies behaved like other control or infected biopsies without extensive variation.
(C) Ingenuity Pathway analysis reveals clustering of genes in the top 50 predicted pathways; −log (p-value) is shown in blue; the z-score indicating pathway upregulation or downregulation is shown in green.
(D) Principle components analysis shows that bacterial gene expression changes cluster based on infecting group or control group (C).
(E) Bacterial genes clustered in three significant groups based on transcriptional changes. Cluster one encompasses genes required for gene expression; cluster two contains virulence genes, and cluster three contains a mix of hypothetical and virulence genes.
(F) Comparison of genes utilized during STM infection overlaid with the gene expression profile of STY bacteria during invasion; it is important to note that genes expressed in our inoculum were not differentially expressed at 2 h invasion. SPI-1 effectors are important for facilitating invasion and are expressed early on; SPI-2 effectors are expressed later in infection and are important for intracellular survival. The SPI-2 genes sseB, sseC, sseD are downregulated at 2 h invasion of intestinal biopsies.
Supplementary Fig. 1.
qPCR validation of RNA-seq gene expression.
Table 2A.
Human genes upregulated after 2 h infection with S. Typhi.
| Feature.ID | p.Value | LFC infected/control | Gene name | Description (from uniprot) |
|---|---|---|---|---|
| Significant genes (P-value < 0.005 and log fold change (LFC) > 1.5) | ||||
| ENSG00000274519 | 3.88E-16 | 6.11 | mir3687–1 | Uncharacterized microRNA |
| ENSG00000231609 | 7.27E-08 | 2.96 | LOC100132215 | Uncharacterized locus |
| ENSG00000134184 | 5.53E-05 | 2.41 | GSTM1 | Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles. |
| ENSG00000185347 | 4.38E-12 | 1.91 | C14orf80 | Uncharacterized protein |
| ENSG00000237973 | 7.73E-09 | 1.86 | MTCO1P12 | Pseudogene |
| ENSG00000275783 | 8.85E-10 | 1.79 | MIR3648 | Uncharacterized microRNA |
| ENSG00000213145 | 6.81E-07 | 1.58 | CRIP1 | Seems to have a role in zinc absorption and may function as an intracellular zinc transport protein |
| ENSG00000131142 | 5.42E-09 | 1.56 | CCL25 | Potentially involved in T-cell development. Recombinant protein shows chemotactic activity on thymocytes, macrophages, THP-1 cells, and dendritic cells but is inactive on peripheral blood lymphocytes and neutrophils. |
| Significant genes (P-value > 0.005) | ||||
| ENSG00000117983 | 2.94E-08 | 1.46 | MUC5B | Gel-forming mucin that is thought to contribute to the lubricating and viscoelastic properties of whole saliva and cervical mucus. |
| ENSG00000248527 | 5.12E-07 | 1.31 | MTATP6P1 | Pseudogene |
| ENSG00000198840 | 4.32E-05 | 1.29 | ND3 | Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase |
| ENSG00000140323 | 3.12E-05 | 1.28 | DISP2 | Smoothened signaling pathway |
| ENSG00000216560 | 9.32E-06 | 1.23 | LINC00955 | Uncharacterized locus |
| ENSG00000212907 | 1.12E-05 | 1.2 | ND4L | Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase |
| ENSG00000228253 | 3.62E-06 | 1.19 | ATP8 | Mitochondrial membrane ATP synthase (F1F0 ATP synthase or Complex V) produces ATP from ADP in the presence of a proton gradient across the membrane which is generated by electron transport complexes of the respiratory chain. |
| ENSG00000206077 | 4.28E-05 | 1.19 | ZDHHC11B | Protein-cysteine S-palmitoyltransferase activity |
| ENSG00000068078 | 7.80E-05 | 1.13 | FGFR3 | Tyrosine-protein kinase that acts as cell-surface receptor for fibroblast growth factors and plays an essential role in the regulation of cell proliferation, differentiation and apoptosis. |
| ENSG00000261150 | 5.71E-05 | 1.09 | EPPK1 | Cytoskeletal linker protein that connects to intermediate filaments and controls their reorganization in response to stress |
Table 2B.
Human genes downregulated after 2 h Infection with S. Typhi.
| Feature.ID | p.Value | LFC infected/control | Gene name | Function description (from uniprot) |
|---|---|---|---|---|
| Significant genes (P-value < 0.005 and log fold change (LFC) > 1.5) | ||||
| ENSG00000242512 | 3.15E-06 | −4.03 | LINC01206 | Unknown |
| ENSG00000139304 | 5.61E-05 | −3.87 | PTPRQ | Dephosphorylates a broad range of phosphatidylinositol phosphates. |
| ENSG00000140798 | 4.56E-05 | −3.67 | ABCC12 | Probable transporter. |
| ENSG00000172995 | 1.64E-05 | −3.49 | ARPP21 | Calmodulin binding protein |
| ENSG00000280065 | 1.87E-11 | −3.31 | Unknown | |
| ENSG00000081479 | 8.79E-08 | −3.3 | LRP2 | Acts together with CUBN to mediate HDL endocytosis (By similarity).* |
| ENSG00000178568 | 1.65E-05 | −3.21 | ERBB4 | Mediates phosphorylation of SHC1 and activation of the MAP kinases MAPK1/ERK2 and MAPK3/ERK1. |
| ENSG00000163554 | 7.66E-05 | −3.2 | SPTA1 | Spectrin is the major constituent of the cytoskeletal network underlying the erythrocyte plasma membrane.* |
| ENSG00000140538 | 3.83E-10 | −3.15 | NTRK3 | NTRK3 auto-phosphorylates and activates different signaling pathways, including the phosphatidylinositol 3-kinase/AKT and the MAPK pathways, that control cell survival and differentiation. |
| ENSG00000232111 | 4.22E-05 | −3.13 | RP11-126O22.1 | Novel processed pseudogene |
| ENSG00000279003 | 2.15E-08 | −3.09 | ENSG00000279003 | Unknown |
| ENSG00000263006 | 1.26E-05 | −3.06 | ROCK1P1 | Transcribed unprocessed pseudogene |
| ENSG00000279185 | 1.51E-06 | −3 | ENSG00000279185 | Unknown |
| ENSG00000181143 | 3.26E-14 | −2.99 | MUC16 | Thought to provide a protective barrier against particles and infectious agents at mucosal surfaces |
| ENSG00000169436 | 4.07E-06 | −2.91 | COL22A1 | Acts as a cell adhesion ligand for skin epithelial cells and fibroblasts. |
| ENSG00000140465 | 5.12E-06 | −2.84 | CYP1A1 | Heme-thiolate monooxygenases.* |
| ENSG00000039139 | 2.86E-06 | −2.83 | DNAH5 | Force generating protein of respiratory cilia. Produces force toward the minus ends of microtubules. Dynein has ATPase activity; the force-producing power stroke is thought to occur on release of ADP. |
| ENSG00000205592 | 5.15E-08 | −2.83 | muc19 | May function in ocular mucus homeostasis. |
| ENSG00000148053 | 7.87E-08 | −2.82 | NTRK2 | Receptor tyrosine kinase |
| ENSG00000279184 | 8.78E-11 | −2.8 | RP3-323A16.1 | Unknown |
| ENSG00000007174 | 2.85E-07 | −2.79 | DNAH9 | Force generating protein of respiratory cilia. Produces force toward the minus ends of microtubules. |
| ENSG00000166923 | 5.46E-12 | −2.75 | GREM1 | Cytokine |
| ENSG00000186487 | 1.69E-05 | −2.74 | MYT1L | May function as a pan neural transcription factor associated with neuronal differentiation. |
| ENSG00000127241 | 5.72E-06 | −2.74 | MASP1 | Functions in the lectin pathway of complement, which performs a key role in innate immunity by recognizing pathogens through patterns of sugar moieties and neutralizing them. |
| ENSG00000172724 | 1.09E-05 | −2.73 | CCL19 | May play a role in inflammatory and immunological responses and in normal lymphocyte recirculation and homing. May play an important role in trafficking of T-cells in thymus, and T-cell and B-cell migration to secondary lymphoid organs. Binds to chemokine receptor CCR7. |
| ENSG00000137077 | 1.23E-13 | −2.7 | CCL21 | Inhibits hemopoiesis and stimulates chemotaxis. Chemotactic in vitro for thymocytes and activated T-cells, but not for B-cells, macrophages, or neutrophils. Shows preferential activity toward naive T-cells. May play a role in mediating homing of lymphocytes to secondary lymphoid organs. |
| ENSG00000175820 | 1.38E-05 | −2.69 | CCDC168 | Unknown |
| ENSG00000280156 | 8.84E-13 | −2.69 | AC006548.28 | Unknown |
| ENSG00000168702 | 5.33E-05 | −2.68 | LRP1B | Potential cell surface proteins that bind and internalize ligands in the process of receptor-mediated endocytosis. |
| ENSG00000165323 | 9.60E-07 | −2.65 | FAT3 | May play a role in the interactions between neurites derived from specific subsets of neurons during development. |
| ENSG00000101638 | 1.22E-05 | −2.64 | ST8SIA5 | This protein is involved in the pathway protein glycosylation, which is part of protein modification. |
| ENSG00000089250 | 3.31E-05 | −2.64 | NOS1 | Produces nitric oxide which is a messenger molecule with diverse functions throughout the body. |
| ENSG00000172023 | 1.92E-11 | −2.54 | REG1B | Uncharacterized protein |
| ENSG00000273079 | 2.60E-08 | −2.51 | GRIN2B | NMDA receptor subtype of glutamate-gated ion channels with high calcium permeability and voltage-dependent sensitivity to magnesium. |
For the full list of downregulated genes, please see Supplementary Table 1.
For the full list of downregulated genes, please see Supplementary Table 1.
3.3. Invading STY Showed Differential Transcriptomic Profile Compared to Control Bacteria Related to Key Transcriptional and Invasive Pathways Important for Virulence
In general, pathogenic bacteria produce effector proteins that exploit host pathways to promote their survival. Therefore, we evaluated changes in bacterial gene expression occurring concurrently with the changes in host gene expression in the same tissue explants. Similar gene expression profiles were detected in control bacteria relative to the bacteria infecting the gut mucosa- with a single notable exception (Fig. 2D) -that was further emphasized during cluster analysis (Fig. 2E). To identify patterns in gene expression, clusters of genes with similar expression patterns were generated using KBASE (Fig. 2E), with the most significant groups showing similarity in gene expression changes for cluster 1: ribosome/metabolism genes; cluster 2: virulence genes and hypothetical proteins; and cluster 3: hypothetical proteins and unknown function. Surprisingly, the gene expression profile observed during STY infection (Fig. 2E, Table 3A, Table 3B) was remarkably different from what is predicted during STM infection (Fig. 2F). STM requires SPI-1[1,32,75,85], rpoS and ompR [24] expression, which was observed in our bacterial inoculum and cultures grown in DMEM, but downregulated in bacteria invading the human gut mucosa. STY upregulated genes included numerous ribosome genes and enzymes. Upregulated enzymes (T1853, T0310, T4519 and T3866) encode eukaryotic-like serine/threonine kinases, acetyltransferases and inositol monophosphatase (Table 3A) which may have implications in the modulation of host signaling. Multiple components of the general secretory pathway (secD, secF, secG, secY) were also upregulated (Supplementary Fig. 1), suggesting that non-T3SS proteins are also utilized during infection. Several components of the SPI-2 operon including sseB, sseA, ssaG, ssaM, ssaR, ssaS and ssaO were downregulated relative to bacteria grown in DMEM alone (Table 3B, Supplementary Figs. 2, 2F), indicating that our model is capturing early events during S. Typhi pathogenesis, as expression of SPI-2 occurs during intracellular growth for STM. Interestingly, the SPI-7 operon-encoded Vi antigen is expressed in our inoculum with several genes located within the SPI-7 operon upregulated during biopsy infection (tviD, tviE), consistent with previous reports generated by using bovine epithelial mucosa [62].
Table 3A.
Select S. Typhi genes upregulated after 2 h infection.
| pGene | Gene | Log fold change | q-Value | Function |
|---|---|---|---|---|
| T_RS16275 | t3210 | 3.72 | 0.0004 | Preprotein translocase subunit SecG |
| T_RS09445 | t1853 | 3.43 | 0.0004 | Beta-hydroxydecanoyl-ACP dehydratase |
| T_RS18075 | t3559 | 3.38 | 0.0004 | Repressor of the Cpx envelope stress response pathway. |
| T_RS24905 | T_RS24905 | 3.38 | 0.0004 | Hypothetical protein |
| T_RS20765 | t4085 | 3.35 | 0.0004 | Preprotein translocase subunit SecY |
| T_RS24820 | T_RS24820 | 3.22 | 0.0004 | Hypothetical protein |
| T_RS24855 | T_RS24855 | 3.16 | 0.0004 | Hypothetical protein |
| T_RS04520 | t0882 | 3.13 | 0.0004 | Cold-shock protein CspJ |
| T_RS23945 | T_RS23945 | 3.1 | 0.0004 | Hypothetical protein |
| T_RS24535 | T_RS24535 | 3.02 | 0.0004 | Hypothetical protein |
| T_RS20790 | t4090 | 2.97 | 0.0004 | DNA-directed RNA polymerase subunit alpha |
| T_RS22875 | t4496 | 2.93 | 0.0021 | Pyrbi operon leader peptide |
| T_RS19685 | t3869 | 2.83 | 0.0004 | Hypothetical protein |
| T_RS23875 | T_RS23875 | 2.71 | 0.0004 | Hypothetical protein |
| T_RS01565 | t0310 | 2.71 | 0.0004 | Inositol monophosphatase |
| T_RS03930 | t0780 | 2.7 | 0.0004 | Glucose-1-phosphate cytidylyltransferase |
| T_RS19190 | t3772 | 2.67 | 0.0004 | Xanthine permease |
| T_RS03270 | t0646 | 2.6 | 0.0039 | Membrane protein |
| T_RS03285 | t0650 | 2.59 | 0.0004 | Phosphoenolpyruvate dependent, sugar transporting phosphotransferase system. |
| T_RS03290 | t0649 | 2.59 | 0.0004 | Phosphofructokinase |
| T_RS02365 | t0466 | 2.58 | 0.0004 | Acyltransferase |
| T_RS10080 | T_RS10080 | 2.57 | 0.0011 | Hypothetical protein |
| T_RS10085 | t1980 | 2.57 | 0.0011 | Translation initiation factor IF-1 |
| T_RS08805 | t1730 | 2.56 | 0.0004 | Hypothetical protein |
| T_RS22990 | t4521 | 2.53 | 0.0004 | Hypothetical protein |
| T_RS22995 | t4519 | 2.53 | 0.0004 | Serine/threonine protein kinase |
| T_RS23000 | t4520 | 2.53 | 0.0004 | Serine/threonine protein kinase |
| T_RS19345 | t3803 | 2.53 | 0.0004 | LPS core heptose(II) kinase RfaY |
| T_RS05655 | t1106 | 2.49 | 0.0004 | Hypothetical protein |
| T_RS02155 | T_RS02155 | 2.48 | 0.0004 | |
| T_RS08555 | t1680 | 2.46 | 0.0016 | Sodium‑potassium/proton antiporter ChaA |
| T_RS03960 | t0786 | 2.44 | 0.0004 | Glycosyl transferase |
| T_RS08780 | t1725 | 2.44 | 0.0004 | Beta-ketoacyl-ACP reductase |
| T_RS02535 | t0501 | 2.43 | 0.0004 | Colicin V production protein |
| T_RS20635 | t4059 | 2.42 | 0.0004 | Elongation factor G |
| T_RS03925 | t0779 | 2.39 | 0.0004 | CDP-6-deoxy-delta-3,4-glucoseen reductase |
| T_RS14115 | t2782 | 2.39 | 0.0004 | Hypothetical protein |
| T_RS16300 | t3215 | 2.38 | 0.0004 | RNA binding protein found associated to pre-50S subunit of the ribosome.* |
| T_RS22250 | t4371 | 2.38 | 0.0004 | Integrase |
| T_RS03965 | t0787 | 2.37 | 0.0004 | Glycosyl transferase |
| T_RS22890 | t4499 | 2.36 | 0.0004 | Ornithine carbamoyltransferase |
| T_RS12270 | t2413 | 2.36 | 0.0004 | Trigger factor |
| T_RS08595 | t1688 | 2.35 | 0.0004 | Adenylosuccinate lyase |
| T_RS21345 | t4196 | 2.34 | 0.0004 | Hypothetical protein |
| T_RS00790 | t0157 | 2.34 | 0.0004 | Pyruvate dehydrogenase complex repressor |
| T_RS17570 | t3461 | 2.31 | 0.0004 | DNA-binding protein HU-alpha |
| T_RS16745 | t3300 | 2.31 | 0.0004 | Fis family transcriptional regulator |
| T_RS12085 | t2377 | 2.31 | 0.0004 | Adenine phosphoribosyltransferase |
| T_RS00330 | t0067 | 2.3 | 0.0004 | Carbamoyl-phosphate synthase small subunit |
| T_RS00865 | t0172 | 2.29 | 0.0004 | Hypothetical protein |
| T_RS03955 | t0785 | 2.27 | 0.0004 | Transporter |
| T_RS16250 | t3205 | 2.27 | 0.0004 | Transcription termination protein NusA |
| T_RS03175 | t0627 | 2.26 | 0.0004 | Hypothetical protein |
| T_RS14120 | t2783 (iacP) | 2.23 | 0.0004 | Acyl carrier protein |
| T_RS12185 | t2397 | 2.22 | 0.0004 | Nitrogen regulatory protein P-II 2 |
| T_RS03945 | t0784 | 2.21 | 0.0004 | CDP-paratose 2-epimerase |
| T_RS03950 | t0783 | 2.21 | 0.0004 | CDP-paratose synthase |
| T_RS19320 | t3798 | 2.21 | 0.0004 | Hypothetical protein |
| T_RS19325 | t3799 | 2.21 | 0.0004 | Hypothetical protein |
| T_RS03940 | t0782 | 2.18 | 0.0004 | LPS biosynthesis protein |
| T_RS09545 | t1875 | 1.62 | 0.0067 | Hypothetical protein |
| T_RS00205 | t0042 | 1.61 | 0.0004 | Transcriptional activator NhaR |
| T_RS22130 | t4347 | 1.57 | 0.0004 | Vi polysaccharide export inner-membrane protein VexB |
| T_RS22140 | t4350 | 1.56 | 0.0037 | Vi polysaccharide biosynthesis protein TviD |
| T_RS22145 | t4349 | 1.56 | 0.0037 | Vi polysaccharide biosynthesis protein TviE |
For the full list of upregulated STY genes, please see Supplementary Table 2.
For the full list of upregulated STY genes, please see Supplementary Table 2.
Table 3B.
Select S. Typhi genes downregulated after 2 h Infection.
| Locus | Gene | Log fold change | q-Value | Function |
|---|---|---|---|---|
| T_RS05760 | t1128 | −4.6998 | 0.000303 | Heat-shock protein |
| T_RS20010 | t3935 | −4.68863 | 0.0047827 | Membrane protein |
| T_RS16050 | t3164 | −4.42922 | 0.0356647 | Serine/threonine dehydratase |
| T_RS06575, | t1288 | −4.05222 | 0.0017506 | Type III secretion system protein SsaS |
| T_RS15430 | t3045 | −3.931 | 0.0361737 | Hypothetical protein |
| T_RS14420 | t2844 | −3.85909 | 0.0022412 | Type I-E CRISPR-associated protein Cse1/CasA |
| T_RS09110 | t1790 | −3.82746 | 0.0267944 | N-acetylneuraminate epimerase |
| T_RS01550 | t0307 | −3.74357 | 0.003615 | Anaerobic sulfite reductase subunit B |
| T_RS18320 | t3606 | −3.66574 | 0.0054552 | Hypothetical protein |
| T_RS00180 | t0037 | −3.62981 | 0.0033564 | Sulfatase |
| T_RS23565 | t4633 | −3.62105 | 0.0106418 | Fimbrial chaperone protein |
| T_RS15480 | t3054 | −3.57317 | 0.0234569 | Amidohydrolase |
| T_RS07840 | t1540 | −3.5345 | 0.0046498 | Membrane protein |
| T_RS20210 | t3974 | −3.52218 | 0.0198834 | Ribokinase |
| T_RS09100 | t1788 | −3.50146 | 0.0095141 | MFS transporter |
| T_RS05770 | t1130 | −3.4989 | 0.0031178 | Hypothetical protein |
| T_RS11895 | t2339 | −3.48786 | 0.0253715 | Allantoin permease |
| T_RS06445 | t1262 | −3.42977 | 7.20E-05 | EscC/YscC/HrcC family type III secretion system outer membrane ring protein |
| T_RS06380 | t1249 | −3.42 | 0.0464325 | Hypothetical protein |
| T_RS02200 | t0435 | −3.39407 | 0.0292125 | Purine-nucleoside phosphorylase |
| T_RS06525 | t1278 | −3.383 | 2.18E-06 | Pathogenicity island protein |
| T_RS09630 | t1891 | −3.37E+00 | 0.0149459 | Hypothetical protein |
| T_RS06460 | t1265 | −3.36098 | 0.0492594 | Chaperone for SseB and SseD |
| T_RS01555 | t0308 | −3.33498 | 0.0106668 | Sulfite reductase subunit alpha |
| T_RS13295 | t2616 | −3.30635 | 0.0073327 | Chaperone protein ClpB |
| T_RS06385 | t1250 | −3.30289 | 0.0493247 | Pseudo hypothetical protein, frameshifted |
| T_RS02205 | t0436 | −3.2902 | 0.016477 | Xanthosine permease |
| T_RS06035 | t1180 | −3.2816 | 0.0254963 | Pyrimidine (deoxy)nucleoside triphosphate pyrophosphohydrolase |
| T_RS06540 | t1281 | −3.26407 | 5.89E-05 | Type III secretion system protein SsaM, |
| T_RS14505 | t2860 | −3.26E+00 | 0.0025029 | Fimbrial protein SteE |
| lpfA | t3659 | −3.25935 | 0.0271651 | Fimbrial protein |
| T_RS00190 | t0039 | −3.25E+00 | 0.0180323 | Hypothetical protein |
| T_RS23005 | t4522 | −3.09E+00 | 0.0466588 | Polarity suppression protein |
| modA,modB | t2105 | −3.06609 | 0.0034703 | Part of ModCBA molybdate transporter |
| T_RS13735 | t2702 | −3.05094 | 0.0136309 | NrdH-redoxin |
| T_RS20205 | t3973 | −3.05052 | 0.0497433 | Hypothetical protein |
| T_RS11900 | t2340 | −2.99758 | 0.03144 | MFS transporter |
| T_RS06580, | t1289 | −2.95076 | 0.0011772 | Type III secretion system protein SsaT |
| T_RS06555 | t1284 | −2.91116 | 7.20E-05 | SSaO Salmonella pathogenicity island 2 protein |
| T_RS16100 | t3175 | −2.83436 | 0.0014239 | PTS galactitol/fructose transporter subunit II |
| T_RS09515 | t1868 | −2.80141 | 7.14E-05 | Hypothetical protein |
| T_RS15520 | t3062 | −2.79169 | 0.002574 | Molybdenum ABC transporter substrate-binding protein |
| T_RS16110 | t3177 | −2.77916 | 0.0346406 | D-tagatose-bisphosphate aldolase, class II, non-catalytic subunit |
| T_RS21165 | t4162 | −2.7735 | 7.20E-05 | Hypothetical protein |
| T_RS19870 | t3906 | −2.77198 | 0.0376913 | Cyclic-guanylate-specific phosphodiesterase |
| T_RS01545 | t0306 | −2.76366 | 0.0243692 | Sulfite reductase subunit C |
| T_RS23560 | t4632 | −2.75952 | 0.0347524 | Frameshifted |
| T_RS06505 | t1274 | −2.73865 | 0.0360844 | Type III secretion system needle protein SsaG |
| T_RS19490 | t3831 | −2.71975 | 0.0499517 | Hypothetical protein |
| T_RS16450 | t3244 | −2.68263 | 0.011345 | Glutamine amidotransferase |
| T_RS02280 | t0450 | −2.67886 | 0.0466588 | Hypothetical protein |
| T_RS09750 | t1914 | −2.67E+00 | 0.0006626 | Methyltransferase |
| T_RS23210 | t4563 | −2.66886 | 0.0212746 | Membrane protein |
| T_RS23145 | t4550 | −2.6688 | 0.0477512 | SAM-dependent methyltransferase |
| T_RS08510 | t1671 | −2.64519 | 0.0003457 | Nitrate reductase molybdenum cofactor assembly chaperone |
| T_RS06895 | t1352 | −2.6331 | 0.0101017 | Tail fiber protein |
| T_RS14890 | t2935 | −2.6154 | 0.0110453 | Transcriptional regulator |
| T_RS04790 | t0937 | −2.59229 | 0.0423495 | Cell wall hydrolase |
| T_RS18330 | t3608 | −2.58732 | 0.0054552 | Hypothetical protein |
| T_RS07830 | t1538 | −2.56658 | 0.0114123 | ABC transporter ATP-binding protein |
| T_RS18345 | t3610 | −2.48E+00 | 0.000754 | Coproporphyrinogen III oxidase |
| sseA | t1265 | −2.39E+00 | 0.0404711 | Type III secretion system chaperone SseA |
| T_RS12810 | t2522 | −2.27364 | 0.0137239 | Fimbrial chaperone protein |
| T_RS06465 | t1266 | −2.27192 | 0.0169695 | SSeB necessary for the correct localization of SseC and SseD on the bacterial cell surface |
For the full list of downregulated STY genes, please see Supplementary Table 3.
For the full list of downregulated STY genes, please see Supplementary Table 3.
3.4. Cytokine Release Is Uncoupled from Transcriptional Replenishment and Mediated by Blocked MAPK and NF-kB Signaling Pathways
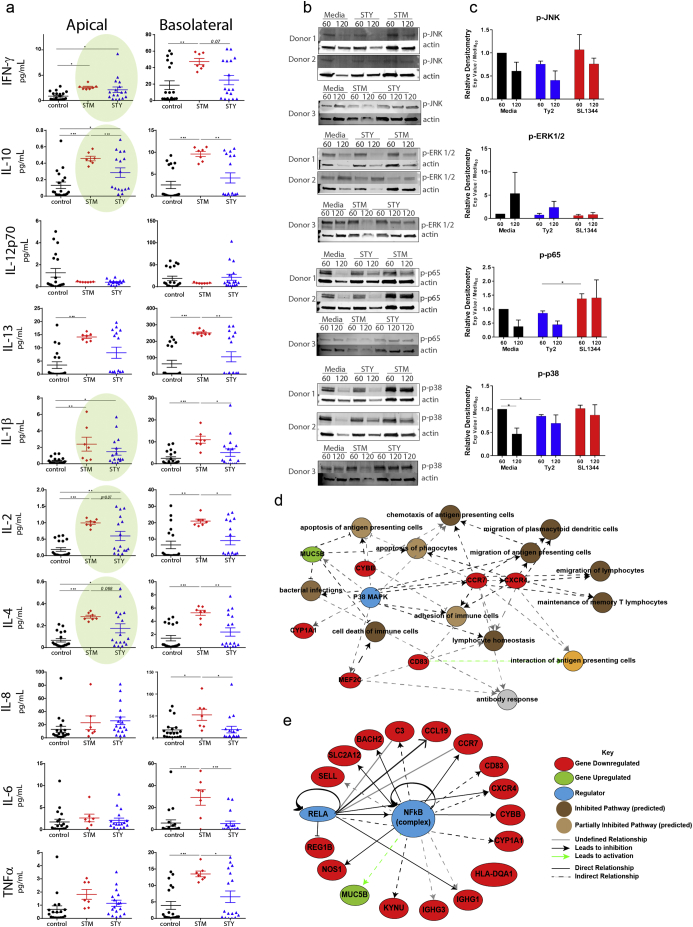
During clinical progression of typhoid fever [11,47,56,63,70], limited gastrointestinal symptoms are reported suggesting that STY invades with limited immune activation [82]. Using supernatants from infected biopsies, cytokine release was assayed using a multiplex ELISA method. Compared to control biopsies, infected biopsies demonstrated differential cytokine production both directionally (apically and basolaterally) and in response to infection with either STM or STY. The cytokine release profile was predominantly apical for STY and bidirectional for STM. For most cytokines, STM trigged a robust and uniform cytokine release, while STY infection led to mild cytokine release (e.g., IL-10, IL-2, IL-4) (Fig. 3A). Only half of the biopsies assayed secreted IL-13, IL-10, IL-4 and IL-2 in response to infection. The cytokine release profile for IFN-γ, IL-8, IL-1β and TNFα was more uniform across all donors. As much of the STY-induced cytokine release profile is skewed toward the apical pole, there was no significant increase in cytokine release on the basolateral pole. Significant basolateral release was observed with STM infection, both relative to control or STY-infected biopsies, demonstrating that the biopsies are viable and capable of responding to pathogen infection.
Fig. 3.
Tissue response to S. Typhi infection reflects differential apical cytokine release.
(A) Multiplexed ELISA analysis of pro-inflammatory and regulatory cytokines in control biopsies, STM or STY treated (black, red, blue, respectively).
(B) As cytokine release is observed but no transcriptional upregulation is detected (per RNA-seq data set), intracellular signal transduction was assessed by western blot analysis to determine the phosphorylation state of NF-kB and MAPK from samples mounted in the microsnapwell system after exposure to media, STM or STY at time points of 60 min and 120 min.
(C) Densitometry analysis of signaling blots.
(D) Pathway analysis demonstrates a central role for MAPK in regulated cellular processes and gene expression important in development of immune responses. Note: All genes shown here are downregulated and their expression patterns predict inhibition of cell recruitment.
(E) NF-kB is a central regulator of gene expression; its activation is critical to expression of genes downregulated in our RNAseq data set.
Despite significant cytokine release, no changes in cytokine transcripts were detected in the RNAseq data set (Fig. 2), suggesting that infection leads to cytokine release from stored intracellular pools and that bacterial exposure likely blocks transcription of new cytokines. The signaling cascades that lead to cytokine transcription are downstream of pattern recognition receptors' activation. Initiation of signaling cascades branching through MAPK and NF-κB were assayed by Western blot analysis of infected biopsies. After 60 min treatment in the microsnapwell chamber, phosphorylated p65, JNK, ERK and p38 was detected in all biopsies. A reduction in the phosphorylation state of p38, p65 and JNK was observed at 120 min post incubation for the control and STY infected samples, but remained phosphorylated in the STM infected biopsies (Fig. 3B). Analysis of our RNAseq data set predicted p38 (Fig. 3D) and NF-κB (Fig. 3E) to be important regulators of the gene expression changes observed during infection. The p38 MAPK regulation of numerous immune functions (blue, Fig. 3D), with genes downregulated (red) or upregulated (green) after infection, is consistent with STY inhibition of these pathways to disarm the host innate and adaptive immune responses during early infection. These pathways have been identified as targets of S. Typhi in published literature demonstrating NF-κB inhibition in monocytes [74] or MAPK inhibition in the gallbladder epithelium [67].
3.5. STY Targets the Host Cytoskeleton to Infect the Intestinal Epithelium
Bacteria often manipulate the host cytoskeleton during invasion [14,25,43,52,61,72]. Within our RNAseq data set, genes involved in cytoskeletal reorganization were downregulated. Formation of cellular protrusions, cytoplasm organization, cytoskeleton reorganization and microtubule dynamics were all identified as pathways predicted to be affected by STY infection (Fig. 4A). TEM analysis revealed STY invading the intestinal mucosa via the apical surface of the enterocyte (Fig. 4B). Sites of bacterial invasion are accompanied by microvilli destruction and long protrusions of host cytoskeleton originating from enterocytes. While M cells were observed by TEM (Fig. 1F), no association of STY with M cells was observed. Furthermore, cytoskeletal projections observed in Fig. 4B were also observed in organoid-derived epithelial monolayers (Fig. 4H), suggesting that microvilli dissolution and remodeling of the host cytoplasm creates a surface suitable for invasion reproducible in both models.
Subsequently, immunostaining of the cytoskeletal protein actin and the related tight junction protein, zonula occludens-1 (ZO-1), showed actin projections into the lumen beyond the normal cellular structure, while no changes in ZO-1 localization was detected (Fig. 4C). Despite changes in the cytoskeleton, no changes in paracellular permeability were observed during this early phase of infection as measured by mucosa-to-serosa 4kD FITC-dextran flux (Fig. 4D) and TEER (Fig. 4E). Additionally, no significant increase in LDH release was detected, indicating that the tissue biopsies were not undergoing cell death in response to bacterial infection (Fig. 4F). To further explore the role of the cytoskeleton in STY invasion, human organoid-derived epithelial monolayers were treated with medium containing the actin-inhibitor cytochalasin D or the microtubule inhibitor nocodazole prior to infection with STY. Inhibitor treatment prevented STY invasion relative to untreated control monolayers (Fig. 4G). Lastly, TEM of infected organoid-derived epithelial monolayers recapitulated phenotypes observed in the infected biopsies including cytoskeleton rearrangement, microvilli destruction and vesicle-contained intracellular bacteria (Fig. 4H).
3.6. Intracellular Trafficking, Activation of Immune Pathways and the Human-restricted Nature of STY
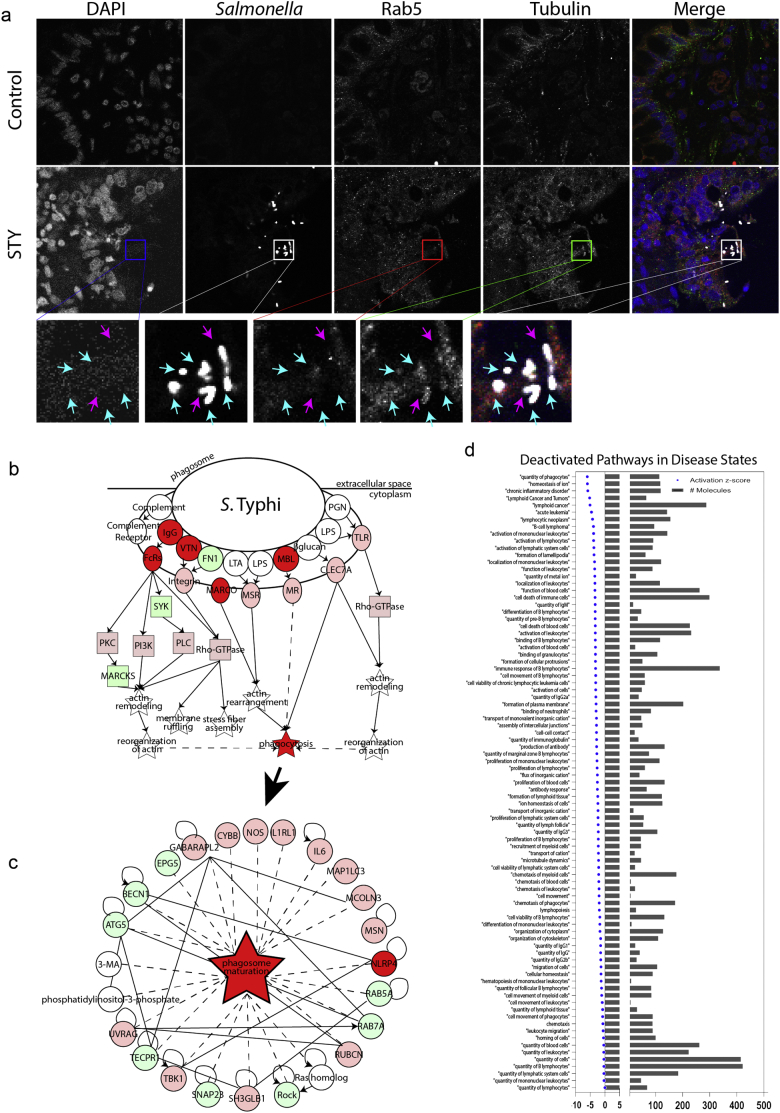
In addition to manipulating the cytoskeleton for cell entry, bacterial exploitation of the structural proteins dictates how the vesicles mature, migrate and target bacteria for clearance [71]. During early invasion, bacteria co-localized with tubulin (Fig. 5A) but they were not present in vesicles labeled with the early endosome protein Rab5. Intracellular bacteria contained within vesicles by TEM (Figs. 1F, 4B) were also observed, but the vesicle identity remains unclear. It is possible that Rab proteins are not present on these vesicles, as STM effector proteins target Rab proteins to block maturation of the autophagolysosome [13,28]. Our understanding of STY trafficking inside the epithelial cell is extremely limited. Furthermore, in our transcriptomic data set, we observe downregulation of several genes critical to bacterial clearance (including CYBB and NOS) in the mature phagosome (Fig. 5B). These data suggest that STY disarms the cellular machinery needed to kill intracellular bacteria by downregulating functional phagosome genes.
Fig. 5.
Inside the enterocyte: STY localization and survival.
(A) Immunostaining of STY-infected biopsies with cytoskeletal protein tubulin, intracellular early endosome marker Rab5, STY and genomic material (DAPI). STY colocalizes with tubulin during biopsy invasion but exists independent of Rab5. Inset images demonstrating co-localization are marked with a blue arrow; independent STY are marked with a pink arrow.
(B) Loss of intracellular early endosome marker indicates masking of cellular localization by STY. RNAseq data predicts inhibition of phagosome formation and maturation; downregulated genes are in red, upregulated genes are in green.
(C) Inhibition of phagosome maturation critically impairs antigen harvest and bacterial clearance, which are overarching themes in disease pathways predicted to be downregulated in response to STY infection of terminal ileum biopsies.
(D) Predicted deactivated disease states related to the immune response deciphered from downregulation of genes during STY infection. The negative Z-score indicates a downregulation of pathways; the number of genes refers to the number of genes downregulated during STY infection that corresponds to the pathway listed. Notable deactivated disease states include quantity of phagocytes, activation of lymphocytes, localization of mononuclear cells, activation of lymphocytes and leukocyte migration.
4. Discussion
Enteric pathogens continue to be a significant public health burden in both developed and developing countries [12,38,42,44]. STY remains an elusive vaccine target [24,41], which is partly due to critical gaps in knowledge regarding how STY causes disease. Some of these gaps are addressed for the first time in this manuscript.
4.1. STY Blocks the Immune Response during Infection
STY produces a polysaccharide capsule known as the Vi antigen, which is thought to protect the bacterial cell from detection by pattern recognition receptors [33,75,81,82]. We conclude that STY further blocks activation of the innate immune system based on depressed or uninduced gene transcription, impaired signal transduction and apical cytokine release observed during human biopsy infection. STY expression of the Vi antigen during human infection (which includes the upregulated genes vexD, vexB, tviE, and tviD) is well-documented [82], and it masks lipopolysaccharide, peptidoglycan and flagellin from the host pattern recognition receptors. Several additional expressed STY genes that may also block innate immune activation by targeting signal transduction intermediates include the γ-proteobacteria conserved suhB gene [54], which encodes an inositol monophosphatase and functions in the modulation of inositol signaling [48].
Legionella manipulates inositol signaling to enable creation of the Legionella-containing vacuole for survival [26]. Likewise, STM replicates within the Salmonella-containing vesicle (SCV). Generation of the SCV is dependent on the SPI-1 effectors SopB and SopE/E2[27], which modulate inositol signaling and Rab proteins, respectively. The suhB gene overcomes the rate-limiting step in inositol signaling to affect second messenger signaling, autophagy and cell death pathways [26]. Two additional upregulated STY specific-enzymes encode eukaryotic-like serine-threonine kinases (T4519, T4520). In data derived from the infection of immortalized macrophage cell lines, T4519 is required for both intracellular survival and IL-6 and TNF-α production, as well as for NFkB phosphorylation [73]. In line with our data, impaired activation of signal transduction pathways not only prevents gene transcription changes and overall immune response development, but also blocks vesicular trafficking. Salmonella-mediated impaired vesicle maturation prevents lysosomal or autophagolysosomal bacterial killing [28,57], thereby allowing bacterial survival and preventing bacterial antigens to stimulate the class I/II major histocompatibility complex (MHC) antigen presentation pathways for the generation of adaptive immune responses. Combined, our data suggest that STY evade innate and adaptive immune responses through multiple and complementary approaches that begin immediately upon invasion and are actively regulated through secreted factors delivered to host enterocytes or the intestinal lumen.
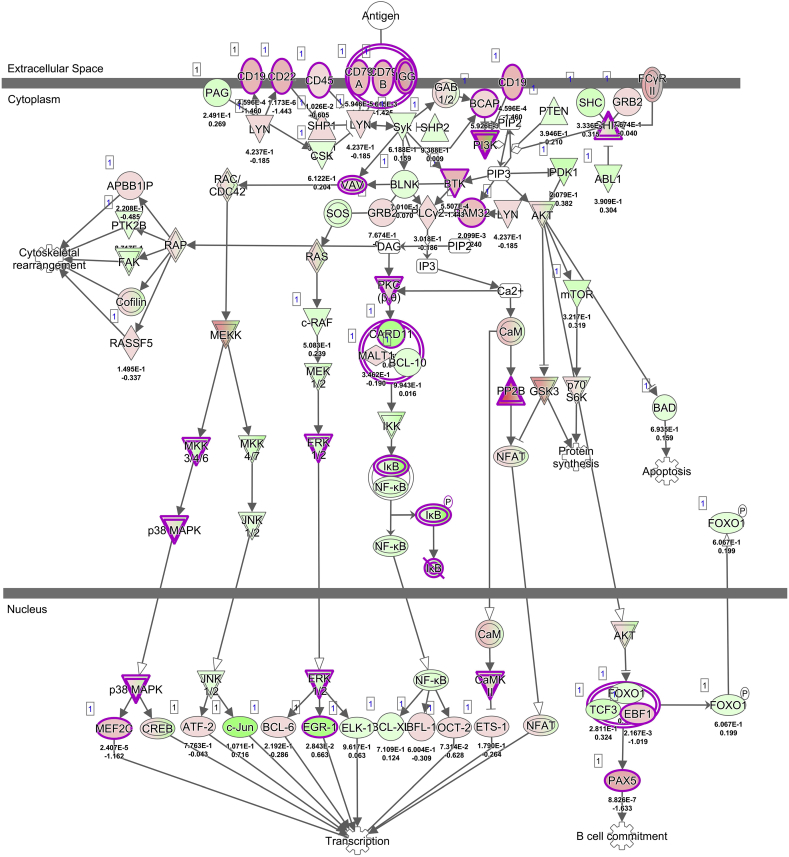
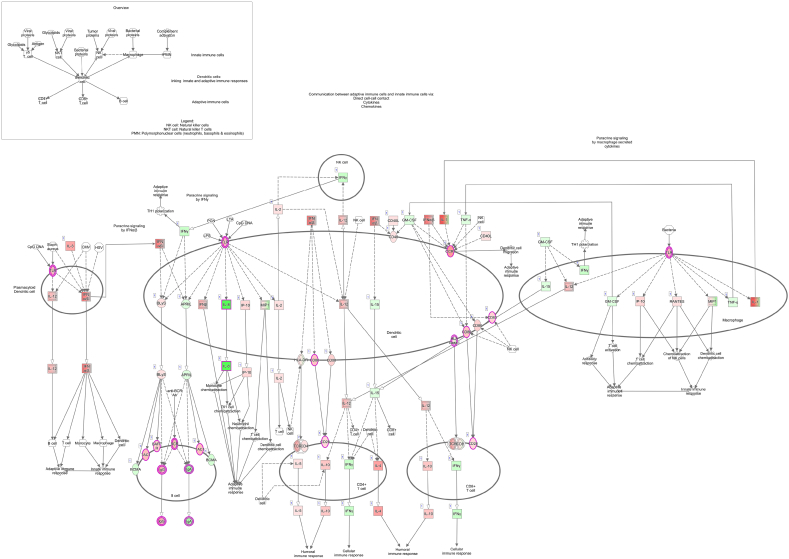
Furthermore, in our model, major MHC-II and reactive oxygen species genes (NADPH-oxidase genes and nitric oxide synthase NOS) are transcriptionally downregulated in response to infection, which may further prevent the host response to STY. Additionally, important immune system genes are downregulated, indicating critical breakpoints in B-cell and T-cell maturation, activation and overall lymphoid cell recruitment to the sites of infection (Supplementary Fig. 3, Supplementary Fig. 4). Our data, which support clinical observations that STY exposure fails to provoke an immune response, also identifies bacterial genes expressed in the modulation of the host immune response. Further studies and deletion constructs are pivotal to understand the function of these enzymes during STY infection. Our findings provide novel information on mechanisms responsible for the host cell's inability to kill STY. These mechanisms are linked to the inhibition of pathways preventing antigen recognition and, ultimately, the development of an adaptive immune response. This concept is supported by our pathway analysis showing differentially expressed genes associated with the deactivation of pathways involved in the crosstalk between innate and adaptive immune responses (Fig. 5C, Supplementary Fig. 3), B-cell receptor signaling (Supplementary Fig. 4) and the maturation of dendritic cells. Ultimately, the downregulation of cell surface genes such as CD22, CD79A and IgG, as well as intracellular signaling genes PI3K and transcription factor PAX5, underscores an inability of the STY infected host cell to activate the adaptive immune system, providing insight into how STY escape clearance by circumventing the host immune system activation.
Supplementary Fig. 3.
Crosstalk between innate and adaptive immune systems.
Supplementary Fig. 4.
B cell receptor signaling pathway.
4.2. STY Infects Enterocytes
Historically, enteric pathogens are thought to enter the host by crossing the gastrointestinal epithelium via specialized M-cells [16,17,46]. Through TEM analysis, we found numerous epithelial cells in various states of STY invasion. Although research supports a role for M-cells in STM infection [17], and to a lesser extent in STY infection [24], we observed that STY infection was predominantly via enterocytes in both tissue explants and gut organoid-derived monolayers, although previous studies have described mechanisms for apical invasion of enterocytes for STM [2]. Common themes observed in infected cells included microvilli destruction and cytoskeletal rearrangement affectingboth actin and tubulin. Furthermore, cytoskeletal projections created a site of invasion that seemed to recruit other STY bacterial cells to enter at the same location. We hypothesize that quorum sensing (QS) facilitates invasion in an energetically favorable manner, by which initial invading bacteria promote microvilli destruction and cytoskeletal rearrangement, and perhaps the release of QS molecules, resulting in cooperative invasion at the same site. STY produces QS molecules homologous to QseBC [53]; a communication system required for enterohemorrhagic E. coli pathogenesis [58]. Therefore, we hypothesize that the community of invading STY works together to suppress the immune activation in the same cell, enriching its chances of survival by creating invasion “hot spots.” The conserved QS genes in the qse and lux/lsr operon are expressed in both our inoculum and invading bacteria, but not differentially expressed during invasion. Curiously, homologs of peptide-based QS genes are differentially expressed during invasion. Peptide-initiated QS is frequently observed in Gram-positive bacteria [65,77] but only the ‘extracellular death factor’ peptide QS signaling pathway has been described in Gram-negative E. coli [36,37,39]. QS molecules are produced in response to population density and to regulate virulence factors [45,65]. It is well-documented that QS molecules induce expression of SPI-1 in STM to promote invasion [15,22,53,55]. Importantly, pathogenesis of enterohemorrhagic E. coli depends on host and commensal derived QS molecules to initiate colon invasion, whereas the invasion stimulus for STY, and the potential role for QS, remains an area warranting future investigation.
4.3. Host-Specificity of Infection: STM vs. STY
STM causes infection in a genetically susceptible, antibiotic-treated mouse [10]. Hallmarks of infection include edema, neutrophil recruitment and bacterial infiltration of the liver, lymph nodes, spleen and cecum [10]. As STM can cause systemic infection, it is used as a model for STY infection in humans. When mice are exposed to STY, the resulting infection is self-limiting within the GI tract and shares very few clinical and pathogenic traits with human disease. Conversely, human infection with STM bacteria results in local gastrointestinal infection that resolves within a few days. Genetically, the two serovars are nearly identical; however STY has gained additional virulence genes [66,83] (notably, SPI-7) and is thought to have significantly fewer protein-coding genes than STM due to insertion sequences, truncations and mutations [66]. Despite genetic similarities, we observed extensive and divergent phenotypes with human infection by STY as compared to STM. As such, we noticed that the host genes upregulated during STY infection were nearly all divergent between human and mice. For example, the cytokine CCL25 and the mitochondrial enzyme ATP8 are both found in humans but not in mice (Table 4). Other genes, like the cytoskeletal remodeling gene EPPK1, shares only 75.5% protein identity with its mouse homologue, which allows us to speculate that some of the host specificity of infection is a direct extension of the genes upregulated during infection. Other variables are important here: e.g., human MUC2 shares only 76% identity with its mouse homologue and the MUC2 product functions as a barrier protecting the intestinal epithelium from bacteria. As mice and humans have different pathogen susceptibility and crossing the mucus barrier is a significant challenge to infection, it would be interesting to study if the protein homology differences contribute to infection susceptibility. Further studies are required to evaluate the relative importance of each of the affected genes and virulence factors during the initial stages of infection.
Table 4.
Human-mouse protein homology.
| Gene name | Protein % identity |
||
|---|---|---|---|
| M. musculus | P. troglodytes | ||
| Genes upregulated upon infection | GSTM1 | 83.90% | 97.70% |
| C14orf80 | 69.40% | 99.50% | |
| CRIP1 | 97.40% | 100%a | |
| CCL25 | 54.30% | 98% | |
| MUC5B | 61.60% | 91%a | |
| ND3 | 67.00% | 94.80% | |
| DISP2 | 76.40% | 99.40% | |
| ND4L | 67.30% | 99.00% | |
| ATP8 | Human onlyb | 92.60% | |
| ZDHHC11B | Human onlyc | 95.20% | |
| FGFR3 | 93.40% | 95.80% | |
| EPPK1 | 75.50% | 98.20% | |
| Control genes | RAB5 | 98% | 100% |
| ACTIN | 100% | 100% | |
| RAB7 | 100% | 100% | |
| TUBULIN | 100% | 100% | |
| ZO1 | 91.10% | 99.90% | |
| MUC2 | 76% | 78%d | |
Please see Supplementary Fig. 5 for additional alignment information.
Please see Supplementary Fig. 5 for additional alignment information.
Predicted protein sequence.
Protein alignment covers 98% with 48% identity - alignment in Supplementary Fig. 5.
Protein alignment covers 78% with 59% identity - alignment in Supplementary Fig. 5.
Protein alignment covers 46% with 78% identity.
Current vaccine candidates target the following genes in S. Typhi: rpoS, galE, galK, ilvD, vexD, phoP, phoQ, aroC, ssaV, aroD, htrA and tviA. Based on the data presented here, only vexD is upregulated during early infection of human terminal ileum biopsies. Several genes are downregulated (phoPQ, ssaV), while others remain unchanged (galK, ilvD, aroC, aroD, htrA, tviA). These virulence, metabolism and regulatory genes have been considered important targets for initial vaccine candidates [24,41] by creating strains with limited intracellular replication capacity and attenuated virulence. Based on the results of these studies, we propose targeting additional genes specifically upregulated by STY during early infection, namely enzymes and cytoskeletal rearrangement genes. During infection, invading bacteria are efficiently shutting down the host's ability to recognize and respond to infection by manipulating the epithelial cell and blocking recruitment of immune cells to the site of infection. Deletion of genes utilized during early invasion will likely prevent STY from shutting down the host response, thereby permitting infection recognition and restoring the ability to recruit immune cells to the site of infection. Additionally, genes that target RNA degradation may also hold promise as possible targets for vaccine development. By eliminating the bacterium's ability to suppress host gene expression, perhaps we can restore the development of a strong, protective, adaptive immune response.
4.4. An Expanded Model for STY Pathogenesis
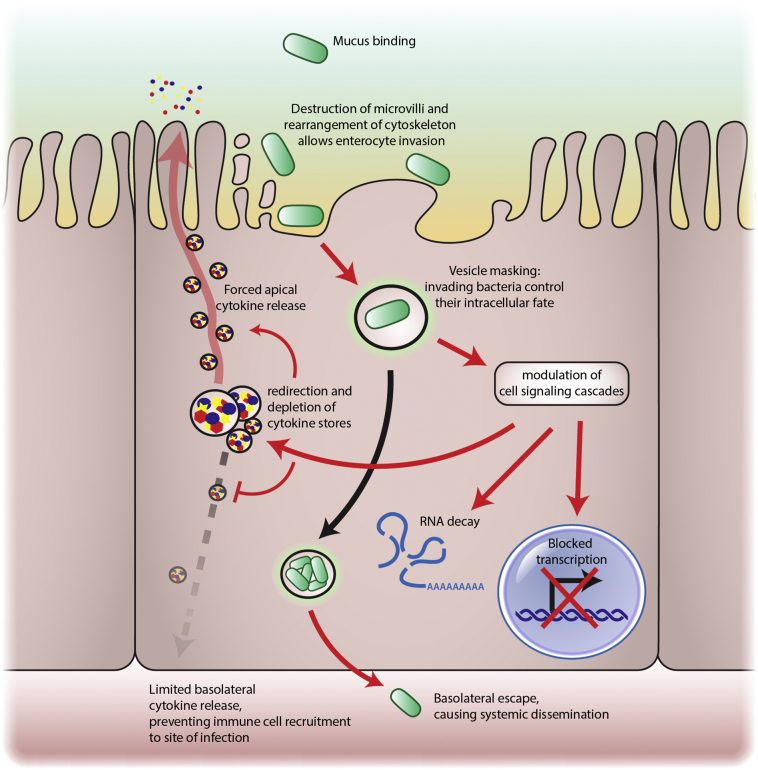
As it becomes increasingly clear that our extrapolated data from murine STM studies are insufficient to accurately recapitulate STY pathogenesis in the human host, we have developed a new model that closely resembles the human gut microenvironment to serve as a framework to study STY pathogenesis. Based on our data we propose the following model (Fig. 6): STY invades enterocytes of the terminal ileum through cytoskeletal rearrangement by targeting actin and tubulin networks. Upon entry, the bacteria target Rab proteins to conceal the vesicle from the cellular sorting pathways (a fraction of STY might even escape to the cytosol). During this time, the bacteria block host response transcription by targeting signal transduction pathways normally triggered by bacterial antigens. To prevent recruitment of immune cells to the site of infection, the release of cytokines and chemokines is re-directed via the apical surface toward the lumen of the intestine. By crippling the host recognition of invasion (i.e., by reduced or absent triggering of signal transduction and transcriptional downregulation of antibacterial or cytokine genes) STY exploits a “Trojan horse” strategy to prevent detection. STY is protected from host immune defenses and the bacteria move undetected to the basolateral pole and continue systemic dissemination in the host.
Fig. 6.
An expanded model for STY pathogenesis. Taken together, our data demonstrates STY bacteria employ a “Trojan Horse” strategy to invade enterocytes and dismantle the immune response from the intracellular environment. We propose that STY invades enterocytes after rearranging the enterocyte cytoskeleton. Invading bacteria are contained within vesicles that are modified to block vesicular trafficking. The secretion of effector proteins modulates eukaryotic cell signaling cascades and transcription. Cytokine stores are released toward the apical pole to prevent recruitment of immune cells to the site of infection. Finally, STY traverse to the basolateral pole and transition to the systemic phase of disease.
The following are the supplementary data related to this article.
Secretion systems expressed by STY during early invasion.
Supplementary Fig. 5.
Expanded human-mouse protein homology table.
Full table of downregulated human genes.
Full table of upregulated STY genes.
Full table of downregulated STY genes.
Funding
These studies were supported, in part, by NIAID, NIH, DHHS grants R01-AI036525 (to MBS), U19-AI082655 [Cooperative Center on Human Immunology] (to MBS and AF), and U19-AI109776 [Center of Excellence for Translational Research (CETR)] to MBS. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases, the National Institutes of Health, NIAID, NIH.
We thank Dana-Farber/Harvard Cancer Center in Boston, MA, for the use of the Specialized Histopathology Core, which provided embedding and sectioning service. Dana-Farber/Harvard Cancer Center is supported in part by an NCI Cancer Center Support Grant # NIH 5 P30 CA06516.
The EM core was supported by NIH/NINDS P30NS045776. Support for the Philly Dake Electron Microscope Facility was provided by NIH 1S10RR023594S10 and by funds from the Dake Family Foundation.
Conflict of Interest
The authors have no conflicts to disclose.
Author Contributions
KPN, SS, RL, LI, CSF, MBS, CF, AF, and MRF were responsible for study design, data generation and data analysis. SP and RL were responsible for donor identification, recruitment and sample collection. DKVK aided in TEM experiments. ZY, CF, KPN and WF were responsible for transcriptomic and related bioinformatics analysis. All authors contributed to manuscript preparation.
Acknowledgements
The authors would like to thank the Diane Capen for her expertise and skill in preparing the samples for TEM analysis. We would also like to thank members of the Fasano, Fiorentino and Faherty laboratories as well as the members of the University of Maryland Cooperative Center on Human Immunology (CCHI) for their thoughtful feedback and discussions during the development of the project. A special acknowledgment to Susie Flaherty for her careful editing of the manuscript.
Contributor Information
K.P. Nickerson, Email: kpnickerson@mgh.harvard.edu.
A. Fasano, Email: afasano@mgh.harvard.edu.
References
- 1.Agbor T.A., Mccormick B.A. Salmonella effectors: Important players modulating host cell function during infection. Cell Microbiol. 2011:1858–1869. doi: 10.1111/j.1462-5822.2011.01701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agbor T.A., Demma Z.C., Mumy K.L., Bien J.D., McCormick B.A. The ERM protein, Ezrin, regulates neutrophil transmigration by modulating the apical localization of MRP2 in response to the SipA effector protein during Salmonella typhimurium infection. Cell Microbiol. 2011;13(12):2007–2021. doi: 10.1111/j.1462-5822.2011.01693.x. https://doi.org/10.1111/j.1462-5822.2011.01693.x (Blackwell Publishing Ltd) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews S. Babraham bioinformatics - FastQC a quality control tool for high throughput sequence data. 2018. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (no date). Available at: (accessed 2017)
- 4.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antillón N.M., Warren J.L., Crawford F.W., Weinberger D.M., Kürüm E., Pak G.D. The burden of typhoid fever in low- and middle-income countries: A meta-regression approach. PLoS Negl Trop Dis. 2017;11(2):e0005376. doi: 10.1371/journal.pntd.0005376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arkin A.P., Stevens R.L., Cottingham R.W., Maslov S., Henry C.S., Dehal P. bioRxiv. 2016. The DOE systems biology knowledgebase (KBase)http://www.biorxiv.org/content/early/2016/12/22/096354 Available at: [Google Scholar]
- 8.Asmar R., El Panigrahi P., Bamford P., Berti I., Not T., Coppa G.V. Host-dependent zonulin secretion causes the impairment of the small intestine barrier function after bacterial exposure. Gastroenterology. 2002 doi: 10.1053/gast.2002.36578. [DOI] [PubMed] [Google Scholar]
- 9.Azmatullah A., Qamar F.N., Thaver D., Zaidi A.K., Bhutta Z. Systematic review of the global epidemiology, clinical and laboratory profile of enteric fever. J Glob Health. 2015;5(2):20407. doi: 10.7189/jogh.05.020407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barthel M., Hapfelmeier S., Quintanilla-Martínez L., Kremer M., Rohde M., Hogardt M. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun. 2003;71(5):2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basnyat B., Maskey A.P., Zimmerman M.D., Murdoch D.R. Enteric (typhoid) fever in travelers. Clin Infect Dis. 2005;41(10):1467–1472. doi: 10.1086/497136. [DOI] [PubMed] [Google Scholar]
- 12.Blackwelder W.C., Biswas K., Wu Y., Kotloff K.L., Farag T.H., Nasrin D. Statistical methods in the global enteric multicenter study (GEMS) Clin Infect Dis0320. 2012;55(Suppl. 4) doi: 10.1093/cid/cis788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birmingham C.L., Smith A.C., Bakowski M.A., Yoshimori T., Brumell J.H. Autophagy controls Salmonella infection in response to damage to the Salmonella -containing vacuole. J Biol Chem. 2006;281(16):11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 14.Campellone K.G. Cytoskeleton-modulating effectors of enteropathogenic and enterohaemorrhagic Escherichia coli: Tir, EspFU and actin pedestal assembly. FEBS J. 2010;277(11):2390–2402. doi: 10.1111/j.1742-4658.2010.07653.x. [DOI] [PubMed] [Google Scholar]
- 15.Choi J., Shin D., Kim M., Park J., Lim S., Ryu S. LsrR-mediated quorum sensing controls invasiveness of Salmonella typhimurium by regulating SPI-1 and flagella genes. PLoS One. 2012;7(5) doi: 10.1371/journal.pone.0037059. https://doi.org/10.1371/journal.pone.0037059 (Edited by N. J. Mantis) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corr S.C., Gahan C.C.G.M., Hill C. M-cells: Origin, morphology and role in mucosal immunity and microbial pathogenesis. FEMS Immunol Med Microbiol. 2008;52(1):2–12. doi: 10.1111/j.1574-695X.2007.00359.x. [DOI] [PubMed] [Google Scholar]
- 17.Clark M.A., Jepson M.A. Intestinal M cells and their role in bacterial infection. Int J Med Microbiol. 2003;293(39) doi: 10.1078/1438-4221-00242. http://www.urbanfischer.de/journals/ijmm Available at: [DOI] [PubMed] [Google Scholar]
- 18.Deng L., Song J., Gao X., Wang J., Yu H., Chen X. Host adaptation of a bacterial toxin from the human pathogen salmonella typhi. Cell. 2014;159(6):1290–1299. doi: 10.1016/j.cell.2014.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forbester J.L., Goulding D., Vallier L., Hannan N., Hale C., Pickard D. ‘Interaction of Salmonella enterica Serovar typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect Immun. 2015;83(7):2926–2934. doi: 10.1128/IAI.00161-15. https://doi.org/10.1128/IAI.00161-15 (Edited by B. A. McCormick) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francis R.S., Berk R.N. 1974. Typhoid fever; pp. 583–585. (January) [DOI] [PubMed] [Google Scholar]
- 21.Galán J.E. Typhoid toxin provides a window into typhoid fever and the biology of Salmonella Typhi. Proc Natl Acad Sci. 2016:201606335. doi: 10.1073/pnas.1606335113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gart E.V., Suchodolski J.S., Welsh T.H., Alaniz R.C., Randel R.D., Lawhon S.D. Salmonella typhimurium and multidirectional communication in the gut. Front Microbiol. 2016;7:1827. doi: 10.3389/fmicb.2016.01827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.GBD 2013 Mortality and Causes of Death Collaborators Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: A systematic analysis for the global burden of disease study 2013. Lancet. 2015;385(9963):117–171. doi: 10.1016/S0140-6736(14)61682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galen J.E., Buskirk A.D., Tennant S.M., Pasetti M.F. Live attenuated human Salmonella vaccine candidates: Tracking the pathogen in natural infection and stimulation of host immunity. EcoSal Plus. 2016;7(1):1–26. doi: 10.1128/ecosalplus.esp-0010-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glotfelty L.G., Zahs A., Hodges K., Shan K., Alto N.M., Hecht G.A. Enteropathogenic E. coli effectors EspG1/G2 disrupt microtubules, contribute to tight junction perturbation and inhibit restoration. Cell Microbiol. 2014;16(12):1767–1783. doi: 10.1111/cmi.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harding C.R., Mattheis C., Mousnier A., Oates C.V., Hartland E.L., Frankel G. LtpD is a novel Legionella pneumophila effector that binds phosphatidylinositol 3-phosphate and inositol monophosphatase IMPA1. Infect Immun. 2013;81(11):4261–4270. doi: 10.1128/IAI.01054-13. https://doi.org/10.1128/IAI.01054-13 (American Society for Microbiology) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernandez L., Hueffer K., Wenk M., Galán J.E. Salmonella modulates vesicular traffic by altering phosphoinositide metabolism. Science. 2004;20(329):946–949. doi: 10.1126/science.1098188. [DOI] [PubMed] [Google Scholar]
- 28.Homer C.R., Richmond A.L., Rebert N.A., Achkar J.J., McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in crohn's disease pathogenesis. Gastroenterology. 2010;139(5):1630–1641.e2. doi: 10.1053/j.gastro.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Höner zu Bentrup K., Ramamurthy R., Ott C.M., Emami K., Nelman-Gonzalez M., Wilson J.W. Three-dimensional organotypic models of human colonic epithelium to study the early stages of enteric salmonellosis. Microbes Infect. 2006;8(7):1813–1825. doi: 10.1016/j.micinf.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 30.Hornick R.B., Greisman S.E., Woodward T.E., DuPont H.L., Dawkins A.T., Snyder M.J. Typhoid fever: Pathogenesis and immunologic control. N Engl J Med. 1970;283(13):686–691. doi: 10.1056/NEJM197009242831306. [DOI] [PubMed] [Google Scholar]
- 31.John J., Van Aart C.J.C., Grassly N.C. The burden of typhoid and paratyphoid in India: Systematic review and meta-analysis. PLoS Negl Trop Dis. 2016;10(4):1–14. doi: 10.1371/journal.pntd.0004616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson R., Byrne A., Berger C.N., Klemm E., Crepin V.F., Dougan G. The type III secretion system effector SptP of Salmonella enterica serovar Typhi. J Bacteriol. 2017 Jan 30;199(4) doi: 10.1128/JB.00647-16. (pii: e00647-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keestra-Gounder A.M., Tsolis R.M., Bäumler A.J. Now you see me, now you don't: The interaction of Salmonella with innate immune receptors. Nat Rev Microbiol. 2015;13(4):206–216. doi: 10.1038/nrmicro3428. https://doi.org/10.1038/nrmicro3428 (Nature Publishing Group) [DOI] [PubMed] [Google Scholar]
- 34.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaljee L.M., Pach A., Garrett D., Bajracharya D., Karki K., Khan I. Social and economic burden associated with typhoid fever in Kathmandu and surrounding areas: A qualitative study. J Infect Dis. 2017 doi: 10.1093/infdis/jix122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolodkin-Gal I., Engelberg-Kulka H. The extracellular death factor: Physiological and genetic factors influencing its production and response in Escherichia coli. J Bacteriol. 2008;190(9):3169–3175. doi: 10.1128/JB.01918-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolodkin-Gal I., Hazan R., Gaathon A., Carmeli S., Engelberg-Kulka H. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science. 2007;318(5850):652–655. doi: 10.1126/science.1147248. [DOI] [PubMed] [Google Scholar]
- 38.Kotloff K.L., Nataro J.P., Blackwelder W.C., Nasrin D., Farag T.H., Panchalingam S. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the global enteric multicenter study, GEMS): A prospective, case-control study. Lancet. 2013;382(9888):209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 39.Kumar S., Kolodkin-Gal I., Vesper O., Alam N., Schueler-Furman O., Moll I. Escherichia coli quorum-sensing EDF, a peptide generated by novel multiple distinct mechanisms and regulated by trans-translation. mBio. 2016;7(1) doi: 10.1128/mBio.02034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levine M. Typhoid fever vaccines. In: Plotkin Stanley A., Orenstein Walter A., Paul A., Edwards Kathryn M., editors. Vaccines. 7th edn. Elsevier, Inc; Philadelphia, PA: 2018. pp. 1114–1144.https://www-clinicalkey-com.ezp-prod1.hul.harvard.edu/#!/content/book/3-s2.0-B9780323357616000614 Available at: [Google Scholar]
- 42.Levine M.M., Kotloff K.L., Nataro J.P., Muhsen K. The global enteric multicenter study (GEMS): Impetus, rationale, and genesis. Clin Infect Dis. 2012;55(Suppl. 4) doi: 10.1093/cid/cis761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lhocine N., Arena E.T., Bomme P., Ubelmann F., Prévost M.-C., Robine S. Apical invasion of intestinal epithelial cells by Salmonella typhimurium requires villin to remodel the brush border actin cytoskeleton. Cell Host Microbe. 2015;17(2):164–177. doi: 10.1016/j.chom.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J., Platts-Mills J.A., Juma J., Kabir F., Nkeze J., Okoi C. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: A reanalysis of the GEMS case-control study. Lancet. 2016;388(10051):1291–1301. doi: 10.1016/S0140-6736(16)31529-X. https://doi.org/10.1016/S0140-6736(16)31529-X (Elsevier Ltd) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lustri B.C., Sperandio V., Moreira C.G. Bacterial chat: Intestinal metabolites and signals in host-microbiota-pathogen interactions. Infect Immun. 2017;85(12):e00476–17. doi: 10.1128/IAI.00476-17. https://doi.org/10.1128/IAI.00476-17 (Edited by H. L. Andrews-Polymenis) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mabbott N.A., Donaldson D.S., Ohno H., Williams I.R., Mahajan A. Microfold (M) cells: Important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol. 2013;6(4):666–677. doi: 10.1038/mi.2013.30. https://doi.org/10.1038/mi.2013.30 (Nature Publishing Group) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mathura K.C., Gurubacharya D.L., Shrestha A., Pant S., Basnet P., Karki D.B. Clinical profile of typhoid patients. Kathmandu Univ. Med. J. 2003;1(2):135–137. [PubMed] [Google Scholar]
- 48.Matsuhisa A., Suzuki N., Noda T., Shiba K. Inositol monophosphatase activity from the Escherichia coli suhB gene product. J Bacteriol. 1995;177(1):200–205. doi: 10.1128/jb.177.1.200-205.1995. http://www.ncbi.nlm.nih.gov/pubmed/8002619 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meiring J.E., Gibani M., TyVAC Consortium Meeting Group, B, Bentsi-Enchill A.D., Clemens J., Darton T.C. The typhoid vaccine acceleration consortium (TyVAC): Vaccine effectiveness study designs: Accelerating the introduction of typhoid conjugate vaccines and reducing the global burden of enteric fever. Report from a meeting held on 26-27 October 2016, Oxford, UK. Vaccine. 2017;35(38):5081–5088. doi: 10.1016/j.vaccine.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 50.Mogasale, V., Maskery, B., Ochiai, R. L., Lee, J. S., Mogasale, V. V, Ramani, E., Kim, Y. E., Park, J. K., Wierzba, T. F. (2014) Burden of typhoid fever in low-income and middle-income countries: A systematic, literature-based update with risk-factor adjustment, vol. 2. www.thelancet.com/lancetgh doi: https://doi.org/10.1016/S2214-109X(14)70301-8. [DOI] [PubMed]
- 51.Mogasale V.V., Mogasale V.V., Ramani E., Lee J.S., Park J.Y., Lee K.S. ‘Revisiting typhoid fever surveillance in low and middle income countries: Lessons from systematic literature review of population-based longitudinal studies. BMC Infect Dis. 2016;16:35. doi: 10.1186/s12879-016-1351-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morampudi V., Graef F.A., Stahl M., Dalwadi U., Conlin V.S., Huang T. Tricellular tight junction protein tricellulin is targeted by the enteropathogenic Escherichia coli effector EspG1, leading to epithelial barrier disruption. Infect Immun. 2017;85(1) doi: 10.1128/IAI.00700-16. https://doi.org/10.1128/IAI.00700-16 (Edited by A. J. Bäumler) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moreira C.G., Weinshenker D., Sperandio V. QseC mediates Salmonella enterica serovar typhimurium virulence in vitro and in vivo. Infect Immun. 2010;78(3):914–926. doi: 10.1128/IAI.01038-09. https://doi.org/10.1128/IAI.01038-09 (American Society for Microbiology) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.NCBI Identical protein report: Inositol monophosphatase (Escherchia coli str. K-12 substr. MG1655) 2017. https://www.ncbi.nlm.nih.gov/protein/16130458/?report=ipg Available at:
- 55.Nesse L.L., Berg K., Vestby L.K., Olsaker I., Djønne B. Salmonella typhimurium invasion of HEp-2 epithelial cells in vitro is increased by N-acylhomoserine lactone quorum sensing signals. Acta Vet Scand. 2011;53(1):44. doi: 10.1186/1751-0147-53-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ngwu B.A., Agbo J.A. Typhoid fever: Clinical diagnosis versus laboratory confirmation. Niger Med J. 2003;12(4):187–192. [PubMed] [Google Scholar]
- 57.Nickerson K.P., Homer C.R., Kessler S.P., Dixon L.J., Kabi A., Gordon I.O. The dietary polysaccharide maltodextrin promotes Salmonella survival and mucosal colonization in mice. PLoS One. 2014;9(7) doi: 10.1371/journal.pone.0101789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Njoroge J., Sperandio V. Enterohemorrhagic escherichia coli virulence regulation by two bacterial adrenergic kinases, QseC and QseE. Infect Immun. 2012;80(2):688–703. doi: 10.1128/IAI.05921-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orvis J., Crabtree J., Galens K., Gussman A., Inman J.M., Lee E. Ergatis: A web interface and scalable software system for bioinformatics workflows. Bioinformatics (Oxford, England) 2010;26(12):1488–1492. doi: 10.1093/bioinformatics/btq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Radtke A.L., Wilson J.W., Sarker S., Nickerson C.A. Analysis of interactions of Salmonella type three secretion mutants with 3-D intestinal epithelial cells. PLoS One. 2010;5(12):e15750. doi: 10.1371/journal.pone.0015750. https://doi.org/10.1371/journal.pone.0015750 (Edited by S. Bereswill) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raines S.A., Hodgkinson M.R., Dowle A.A., Pryor P.R. The Salmonella effector SseJ disrupts microtubule dynamics when ectopically expressed in normal rat kidney cells. PLoS One. 2017;12(2) doi: 10.1371/journal.pone.0172588. https://doi.org/10.1371/journal.pone.0172588 (Edited by E. Cascales) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raffatellu M., Santos R.L., Chessa D., Wilson R.P., Winter S.E., Rossetti C.A. The capsule encoding the viaB locus reduces interleukin-17 expression and mucosal innate responses in the bovine intestinal mucosa during infection with Salmonella enterica serotype Typhi. Infect Immun. 2007;75(9):4342–4350. doi: 10.1128/IAI.01571-06. https://doi.org/10.1128/IAI.01571-06 (American Society for Microbiology) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raffatellu M., Wilson R.P., Winter S.E., Bäumler A.J. Clinical pathogenesis of typhoid fever. J Infect Dev Countries. 2008:260–266. doi: 10.3855/jidc.219. [DOI] [PubMed] [Google Scholar]
- 64.Rauch I., Deets K.A., Ji D.X., von Moltke J., Tenthorey J.L., Lee A.Y. NAIP-NLRC4 Inflammasomes coordinate intestinal epithelial cell expulsion with eicosanoid and IL-18 release via activation of Caspase-1 and -8. Immunity. 2017;46(4):649–659. doi: 10.1016/j.immuni.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rutherford S.T., Bassler B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012 Nov 1;2(11) doi: 10.1101/cshperspect.a012427. (pii: a012427) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sabbagh S.C., Forest C.G., Lepage C., Leclerc J.M., Daigle F. So similar, yet so different: Uncovering distinctive features in the genomes of Salmonella enterica serovars typhimurium and Typhi. FEMS Microbiol Lett. 2010:1–13. doi: 10.1111/j.1574-6968.2010.01904.x. [DOI] [PubMed] [Google Scholar]
- 67.Scanu T., Spaapen R.M., Bakker J.M., Pratap C.B., Wu L., Hofland I. Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma. Cell Host Microbe. 2015;17(6):763–774. doi: 10.1016/j.chom.2015.05.002. https://doi.org/10.1016/J.CHOM.2015.05.002 (Cell Press) [DOI] [PubMed] [Google Scholar]
- 68.Salerno-Goncalves R., Fasano A., Sztein M.B. Engineering of a multicellular organotypic model of the human intestinal mucosa. Gastroenterology. 2011;141(2):e18–e20. doi: 10.1053/j.gastro.2011.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Salerno-Goncalves R., Fasano A., Sztein M.B. Development of a multicellular three-dimensional Organotypic model of the human intestinal mucosa grown under microgravity. J. Vis. Exp. 2016;(113) doi: 10.3791/54148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi D., Li H., Cheng A. Radiology of infectious diseases. vol. 2. 2015. Typhoid and paratyphoid fever; pp. 295–303. [Google Scholar]
- 71.Spanò S., Liu X., Galán J.E. Proteolytic targeting of Rab29 by an effector protein distinguishes the intracellular compartments of human-adapted and broad-host Salmonella. Proc Natl Acad Sci U S A. 2011;108(45):18418–18423. doi: 10.1073/pnas.1111959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stradal T.E.B., Costa S.C.P. Current topics in microbiology and immunology. 2016. Type III secreted virulence factors manipulating signaling to actin dynamics; pp. 175–199. [DOI] [PubMed] [Google Scholar]
- 73.Theeya N., Ta A., Das S.S., Mandal R.S., Chakrabarti O., Chakrabarti S. ‘An inducible and secreted eukaryote-like serine/threonine kinase of Salmonella enterica serovar typhi promotes intracellular survival and pathogenesis. Infect Immun. 2015;83(2):522–533. doi: 10.1128/IAI.02521-14. https://doi.org/10.1128/IAI.02521-14 (Edited by B. A. McCormick) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Toapanta F.R., Bernal P.J., Fresnay S., Darton T.C., Jones C., Waddington C.S. Oral wild-type salmonella typhi challenge induces activation of circulating monocytes and dendritic cells in individuals who develop typhoid disease. PLOS Negl Trop Dis. 2015;9(6) doi: 10.1371/journal.pntd.0003837. https://doi.org/10.1371/journal.pntd.0003837 (Edited by S. Baker. Public Library of Science) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tran Q.T., Gomez G., Khare S., Lawhon S.D., Raffatellu M., Bäumler A.J. The Salmonella enterica serotype Typhi Vi capsular antigen is expressed after the bacterium enters the ileal mucosa. Infect Immun. 2010;78(1):527–535. doi: 10.1128/IAI.00972-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.VanDussen K.L., Marinshaw J.M., Shaikh N., Miyoshi H., Moon C., Tarr P.I. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut. 2015;64(6):911–920. doi: 10.1136/gutjnl-2013-306651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Verbeke F., De Craemer S., Debunne N., Janssens Y., Wynendaele E., Van de Wiele C. Peptides as quorum sensing molecules: Measurement techniques and obtained levels in vitro and in vivo. Front Neurosci. 2017;11 doi: 10.3389/fnins.2017.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Virlogeux I., Waxin H., Ecobichon C., Popoff M.Y. Role of the viaB locus in synthesis, transport and expression of Salmonella typhi Vi antigen. Microbiology. 1995;141(12):3039–3047. doi: 10.1099/13500872-141-12-3039. [DOI] [PubMed] [Google Scholar]
- 79.Watson C.H., Edmunds W.J. A review of typhoid fever transmission dynamic models and economic evaluations of vaccination. Vaccine. 2015;33(S3):C42–C54. doi: 10.1016/j.vaccine.2015.04.013. https://doi.org/10.1016/j.vaccine.2015.04.013 (Elsevier Ltd) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilson S.S., Tocchi A., Holly M.K., Parks W.C., Smith J.G. A small intestinal organoid model of non-invasive enteric pathogen–epithelial cell interactions. Mucosal Immunol. 2015;8(2):352–361. doi: 10.1038/mi.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Winter S.E., Raffatellu M., Wilson P.R., Rüssmann H., Bäumler A.J. The Salmonella enterica serotype Typhi regulator TviA reduces interleukin-8 production in intestinal epithelial cells by repressing flagellin secretion. Cell Microbiol. 2008;10(1):247–261. doi: 10.1111/j.1462-5822.2007.01037.x. [DOI] [PubMed] [Google Scholar]
- 82.Winter S.E., Winter M.G., Poon V., Keestra A.M., Sterzenbach T., Faber F. Salmonella enterica Serovar Typhi conceals the invasion-associated type three secretion system from the innate immune system by gene regulation. PLoS Pathog. 2014;10(7) doi: 10.1371/journal.ppat.1004207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yap K.-P., Gan H.M., Teh C.S.J., Chai L.C., Thong K.L. Comparative genomics of closely related Salmonella enterica serovar Typhi strains reveals genome dynamics and the acquisition of novel pathogenic elements. BMC Genomics. 2014;15(1):1007. doi: 10.1186/1471-2164-15-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Y.-G., Wu S., Xia Y., Sun J. Salmonella -infected crypt-derived intestinal organoid culture system for host-bacterial interactions. Physiol Rep. 2014;2(9) doi: 10.14814/phy2.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou D., Galán J. Salmonella entry into host cells: The work in concert of type III secreted effector proteins. Microbes Infect. 2001;3(14–15):1293–1298. doi: 10.1016/s1286-4579(01)01489-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Secretion systems expressed by STY during early invasion.
Full table of downregulated human genes.
Full table of upregulated STY genes.
Full table of downregulated STY genes.