Abstract
eXamine is a Cytoscape app that displays set membership as contours on top of a node-link layout of a small graph. In addition to facilitating interpretation of enriched gene sets of small biological networks, eXamine can be used in other domains such as the visualization of communities in small social networks.
eXamine was made available on the Cytoscape App Store in March 2014, has since registered more than 7,200 downloads, and has been highly rated by more than 25 users. In this paper, we present eXamine's new automation features that enable researchers to compose reproducible analysis workflows to generate visualizations of small, set-annotated graphs.
Keywords: functional subnetwork modules, visualization, enrichment, graph drawing, network community, Euler diagram
Introduction
The Cytoscape app eXamine visualizes a small graph and a collection of node sets. The main purpose of eXamine is to aid in the interpretation of a small subnetwork module extracted from a large biological network 1. Cytoscape apps like jActiveModules 2 or external tools like Heinz 3 extract subnetwork modules from a protein-protein interaction network given gene expression data. To interpret the identified subnetwork module, a frequent follow-up analysis is to compute enrichment of the nodes of the identified module in terms of known annotations such as from the Gene Ontology (GO) 4, 5 or from the Kyoto Encyclopedia of Genes and Genomes (KEGG) 6. These annotations are a collection of node sets. eXamine provides a visual analysis approach that facilitates interpretation of the subnetwork module and the identified node sets by biologists. Given a small collection of node sets, eXamine generates a visualization of a small graph as a node-link layout together with contours for the selected sets ( Figure 1). More specifically, the layout is computed using a variation of the algorithm described in 7. This algorithm preserves topological distances along inter-module links as much as possible, while making sure that none of the nodes overlap. In addition, spanning graphs are derived for those sets that have been selected by the user. These graphs are included in the computation of topological distances between nodes, pulling the nodes closer together. The spanning graphs are also used to draw the set contours, by dilating and eroding the associated links to form the rounded shapes that visually encompass and connect nodes.
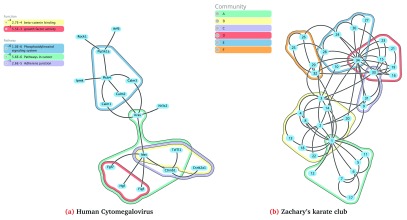
Figure 1. eXamine displays set membership as contours on top of a node-link layout.
The graphs generated in the two use cases using eXamine’s automation features.
As an alternative to eXamine, the existing group layout of Cytoscape can be used to show node partitions by visualizing disjoint sets in separate circles. The Venn and Euler diagram app 8 for Cytoscape visualizes overlapping sets. However, in both cases network and group analysis are visually separate. The RBVI Cytoscape plugins 9 provide a group viewer that aggregates groups into meta-nodes, without making group overlaps explicit.
Here, we present a new version of eXamine that uses Cytoscape’s recently introduced automation features. We demonstrate eXamine’s new automation features using two use cases. The first use case replicates the case study provided in the original eXamine publication 1. The second use case, the analysis of a social network, demonstrates that eXamine is applicable to other domains beyond computational biology. eXamine’s new automation features enable the composition of reproducible analysis workflows that generate appealing visualizations of small, set-annotated graphs.
Methods
Implementation
eXamine is implemented in Java and is available as an app for Cytoscape 3. We used WebCola algorithms http://ialab.it.monash.edu/webcola/ to simultaneously lay out nodes, links, and set contours. We refer to the original publication 1 for additional implementation details regarding the used visualization techniques. Since the typical analysis workflows of eXamine consist of relatively simple commands that do not require streaming of complex data, we implemented the automation features through the ‘Command’ interface rather than the ‘Function’ interface. As a result, eXamine’s commands can be either executed from the command line or through Cytoscape’s REST (CyREST) interface from a Jupyter notebook.
Operation
As eXamine requires Cytoscape 3.6 to run, eXamine has the same system requirements as Cytoscape 3.6. eXamine can be operated through the Cytoscape graphical user interface (GUI) or through the new ‘Command’ interface of Cytoscape. We refer to the original publication 1 for GUI instructions. In the following, we will describe and use the new ‘Command’ interface of eXamine.
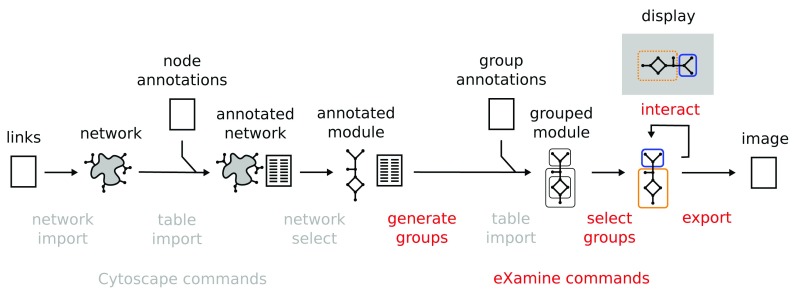
Table 1 provides a summary of the API of the eXamine commands and their parameters. To enable workflow authors to use eXamine’s automation most effectively, we also generated Swagger-based documentation that describes the commands and arguments. This documentation can be accessed in the Cytoscape menu: Help → Automation → CyREST Command API. Figure 2 shows a typical workflow of eXamine analysis.
Table 1. Application Programming Interface (API) of Cytoscape commands provided by eXamine.
Figure 2 shows an example workflow using the below commands.
| Command | Argument (type) | Description |
|---|---|---|
|
examine generate
groups |
Generates eXamine groups from a given set of columns of the
node table |
|
| ○ network (CyNetwork)
○ selectedGroupColumns (List) ○ useAllNodes (Boolean) |
Network for which to generate the groups
Columns of the node table from which the groups will be created Indicates whether to use all nodes for group generation, or only those nodes currently selected in Cytoscape |
|
|
examine select
groups |
Select groups to be visualized by eXamine | |
| ○ selectedGroup (List) | Group identifiers to select | |
|
examine remove
groups |
Removes all groups generated by eXamine | |
| ○ network (CyNetwork) | Network from which to remove the groups | |
|
examine update
settings |
Updates visualization settings | |
| ○ network (CyNetwork)
○ labelColumn (String) ○ urlColumn (String) ○ scoreColumn (String) ○ showScore (Boolean) ○ selectedGroupColumns (List) |
Network to visualize
Column of the node table that contains the node labels Column of the node table that contains URLs with additional information about the nodes Column that contains annotation enrichment scores Indicates whether to show annotation enrichment scores Group columns of the node table to show in the visualization |
|
| examine interact | Shows the selected network and groups in an interactive
visualization window |
|
| examine export | Generates an SVG file with a visualization of the selected
network and groups |
|
| ○ path (String) | Filename |
Figure 2. Example workflow that is supported by the eXamine automation commands.
First, we import a network, followed by importing node annotations that associate each node with of set of groups. Next, we optionally select a smaller subnetwork to visualize. We generate an internal representation of the groups, and import additional group annotations. After selecting the groups to visualize, we export an image of the visualization. Alternatively, we can launch a window that allows the user to select different groups.
Use cases
To illustrate the new ‘Command’ interface of eXamine, we present two use cases. In the first use case, we describe a workflow to study a small subnetwork module extracted from the KEGG mouse network 6. In the second use case, we describe a workflow to study Zachary’s karate club, a well-known social network. Both workflows are available as Jupyter notebooks. The workflows require Cytoscape v3.6 and a recent Python version (Python v2.7 or Python v3.6).
Use case 1: Dysregulated signaling in Human Cytomegalovirus
The Human Cytomegalovirus (HCMV) is a highly-contagious herpes virus. Previously 1, we used eXamine to interpret a small subnetwork module (17 nodes and 18 edges) extracted from the KEGG mouse network using Heinz 3 given gene expression data of an HCMV-infected mouse cell line. Node sets of this subnetwork module were annotated using enriched pathways from KEGG and enriched terms from GO. Below, we provide Python code that uses Cytoscape’s and eXamine’s automation feautures to generate Figure 1.
-
1.
To begin, we define a helper function that creates HTTP GET requests.
import requests PORT_NUMBER = 1234 BASE = ’http://localhost:’ + str (PORT_NUMBER) + ’/v1/’ BASE_URL = "https://raw.githubusercontent.com/ls–cwi/eXamine/master/data/" def executeRestCommand(namespace="", command="", args={}): postString = BASE + "commands/" + namespace + "/" + command firstarg = True for arg in args: postString += ("?" if firstarg else "&") + arg + "=" + args[arg] if (firstarg): firstarg = False res = requests.get(postString) return res
-
2.
We import the full KEGG mouse network using the ‘network import url‘ command provided by Cytoscape.
executeRestCommand("network", "import url", {"indexColumnSourceInteraction" : "2", "indexColumnTargetInteraction" : "2", "url" : BASE_URL + "edges.txt"}) -
3.
We import node annotation, containing set membership information, using the ‘table import url‘ command provided by Cytoscape.
executeRestCommand("table", "import url", {"firstRowAsColumnNames" : "true", "keyColumnIndex" : "1", "startLoadRow" : "1", "dataTypeList" : "s,s,f,f,f,s,s,s,sl,sl,sl,sl", "url" : BASE_URL + "nodes_induced.txt"}) -
4.
Using the ‘network select’ command provided by Cytoscape, we select the nodes that comprise the subnetwork module identified by Heinz 3.
executeRestCommand("network", "select", {"nodeList":"Module:small"}) -
5.
Using the ‘examine generate groups’ command, we generate group nodes for each member of the specified sets that occur in the module (the nodes we previously selected).
executeRestCommand("examine", "generate groups", {"selectedGroupColumns" : "Function,Pathway"}) -
6.
We import additional annotations that describe the generated groups using the ‘import table url’ command of Cytoscape.
executeRestCommand("table", "importurl", {"firstRowAsColumnNames" : "true", "keyColumnIndex" : "1", "startLoadRow" : "1", "url" : BASE_URL + "sets_induced.txt"}) -
7.
As described in 1, we consider groups from the sets Pathway and Function using the ‘examine update settings’ command. In addition, we specify the columns that contain node labels and URLs as well as group scores and annotations.
executeRestCommand("examine", "update settings", {"labelColumn" : "Symbol", "urlColumn" : "URL", "scoreColumn" : "Score", "showScore" : "true", "selectedGroupColumns" : "Component, Function"}) -
8.
Using the ‘examine select groups’ command, we select the same groups as described in 1.
executeRestCommand("examine", "select groups", {"selectedGroups" : "GO:0008013,GO:0008083,mmu04070,mmu05200,mmu04520"}) -
9.
Finally, we export the visualization to a scalable vector graphics (SVG) file using the ‘examine export’ command. Figure 1a shows the resulting graph.
executeRestCommand("examine", "export", {"path": "fig1a.svg"}) -
Alternatively, using the ‘examine interact’ command, we can launch an interactive visualization window that allows us to select different groups.
executeRestCommand("examine","interact",{})
Use case 2: Zachary’s karate club
We consider the graph ‘Zachary’s karate club’, which is an undirected social network of friendships between 34 members of a karate club at a US university in the 1970s 10. Here, we use eXamine to visualize the six overlapping communities of this network identified in 11.
-
1.
We use the same helper function as defined in the previous use case.
-
2.
We import the network using the ‘network import url‘ command provided by Cytoscape.
executeRestCommand("network", "import url", {"indexColumnSourceInteraction" : "2", "indexColumnTargetInteraction" : "2", "url" : BASE_URL + "edges_karate.gml"}) -
3.
We import node annotation, containing set membership information, using the ‘table import url‘ command provided by Cytoscape.
executeRestCommand("table", "import url", {"firstRowAsColumnNames" : "true", "keyColumnIndex" : "1", "startLoadRow" : "1", "dataTypeList" : "s,sl", "url" : BASE_URL + "nodes_karate_induced.txt"}) -
4.
Using the ‘network select’ command provided by Cytoscape, we select all nodes of the network.
executeRestCommand("network" , "select", {"nodeList":"all"}) -
5.
Using the ‘examine generate groups’ command, we generate group nodes for each member of the specified sets that annotate the nodes of the network.
executeRestCommand("examine", "generate groups", {"selectedGroupColumns" : "Community"}) -
6.
We consider groups from the set Community using the ‘examine update settings’ command.
executeRestCommand("examine", "update settings", {"labelColumn" : "label", "urlColumn" : "label", "showScore" : "false", "selectedGroupColumns" : "Community"}) -
7.
We select all six groups using the ‘examine select groups’ command.
executeRestCommand("examine", "select groups", {"selectedGroups" : "A,B,C,D,E,F"}) -
8.
Finally, we export the visualization to an SVG file using the ‘examine export’ command. Figure 1b shows the resulting graph.
executeRestCommand("examine", "export", {"path": "fig1b.svg"})
Discussion
eXamine is limited to visualizing small, relatively sparse networks. It is not possible to use eXamine to construct a comprehensive layout if the network consists of hundreds of nodes or if there are dozens of annotation sets to visualize at the same time. This is a natural limitation of any visualization approach based on node-link diagrams and set contours.
eXamine currently uses Cytoscape’s CyREST API to import networks and their annotations. If apps that provide gene enrichment analysis functionality, such as BiNGO 12, would expose this functionality through the CyREST API, we envision updating the workflow to include this type of upstream analysis.
Summary
eXamine is a Cytoscape app for a set-oriented visual analysis approach for small annotated graphs that displays set membership as contours on top of a node-link layout ( Figure 1). In this paper, we presented new automation features for eXamine that are accessible through Cytoscape’s REST API ( Table 1). As such, researchers can embed eXamine in reproducible and well-documented workflows that generate appealing visualizations of small, set-annotated graphs ( Figure 2). We demonstrated two such workflows in the context of computational network biology and social network analysis.
Data and software availability
1. Cytoscape app store: http://apps.cytoscape.org/apps/examine
2. Source code: https://github.com/ls-cwi/eXamine
3. Latest commit as of release: https://github.com/ls-cwi/eXamine/commit/adeea322cdd6a787baca98adfe70c4165c4d892c
4. Archived source code as at time of publication: https://doi.org/10.5281/zenodo.1218569 13
5. License: GPL v2.0
6. Jupyter notebooks: https://github.com/ls-cwi/eXamine/blob/master/doc/tutorial/eXamineNotebook/eXamineTutorial.ipynb and https://github.com/ls-cwi/eXamine/blob/master/doc/tutorial/eXamineNotebook/eXamineTutorial2.ipynb.
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
[version 1; referees: 1 approved
References
- 1. Dinkla K, El-Kebir M, Bucur CI, et al. : eXamine: exploring annotated modules in networks. BMC Bioinformatics. 2014;15:201. 10.1186/1471-2105-15-201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ideker T, Ozier O, Schwikowski B, et al. : Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18(Suppl 1):S233–S240. 10.1093/bioinformatics/18.suppl_1.S233 [DOI] [PubMed] [Google Scholar]
- 3. Dittrich MT, Klau GW, Rosenwald A, et al. : Identifying functional modules in protein-protein interaction networks: an integrated exact approach. Bioinformatics. 2008;24(13):i223–31. 10.1093/bioinformatics/btn161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ashburner M, Ball CA, Blake JA, et al. : Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. The Gene Ontology Consortium: Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017;45(D1):D331–D338. 10.1093/nar/gkw1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kanehisa M, Furumichi M, Tanabe M, et al. : KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D36. 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dwyer T, Koren Y, Marriott K: IPSEP-COLA: an incremental procedure for separation constraint layout of graphs. IEEE Trans Vis Comput Graph. 2006;12(5):821–828. 10.1109/TVCG.2006.156 [DOI] [PubMed] [Google Scholar]
- 8. Venn and Euler Diagrams: Venn and Euler Diagrams. Reference Source [Google Scholar]
- 9. RBVI Cytoscape Plugins: RBVI Cytoscape Plugins - Cytoscape Group Support. Reference Source [Google Scholar]
- 10. Zachary WW: An information flow model for conflict and fission in small groups. J Anthropol Res. 1977;33(4):452–473. 10.1086/jar.33.4.3629752 [DOI] [Google Scholar]
- 11. Chen W, Liu Z, Sun X, et al. : A game-theoretic framework to identify overlapping communities in social networks. Data Min Knowl Discov. 2010;21(2):224–240. 10.1007/s10618-010-0186-6 [DOI] [Google Scholar]
- 12. Maere S, Heymans K, Kuiper M: BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–3449. 10.1093/bioinformatics/bti551 [DOI] [PubMed] [Google Scholar]
- 13. Dinkla K, Spohr P, El-Kebir M, et al. : ls-cwi/eXamine: Automation Submission Version (Version v2.1.1). Zenodo. 2018. Data Source [Google Scholar]