Abstract
Background
Tuberculosis (TB) is the leading infectious cause of death worldwide. A major barrier to control of the pandemic is a lack of clinical biomarkers with the ability to distinguish active TB from healthy and sick controls and potential for development into point-of-care diagnostics.
Methods
We conducted a prospective case control study to identify candidate urine-based diagnostic biomarkers of active pulmonary TB (discovery cohort) and obtained a separate blinded “validation” cohort of confirmed cases of active pulmonary TB and controls with non-tuberculous pulmonary disease for validation. Clean-catch urine samples were collected and analyzed using high performance liquid chromatography-coupled time-of-flight mass spectrometry.
Results
We discovered ten molecules from the discovery cohort with receiver-operator characteristic (ROC) area-under-the-curve (AUC) values >85%. These 10 molecules also significantly decreased after 60 days of treatment in a subset of 20 participants followed over time. Of these, a specific combination of diacetylspermine, neopterin, sialic acid, and N-acetylhexosamine exhibited ROC AUCs >80% in a blinded validation cohort of participants with active TB and non-tuberculous pulmonary disease.
Conclusion
Urinary levels of diacetylspermine, neopterin, sialic acid, and N-acetylhexosamine distinguished patients with tuberculosis from healthy controls and patients with non-tuberculous pulmonary diseases, providing a potential noninvasive biosignature of active TB.
Funding
This study was funded by Weill Cornell Medicine, the National Institute of Allergy and Infectious Diseases, the Clinical and Translational Science Center at Weill Cornell, the NIH Fogarty International Center grants, and the NIH Tuberculosis Research Unit (Tri-I TBRU).
Keywords: Tuberculosis, Biomarker, Metabolomics, Urine
Highlights
-
•
Urinary levels of small metabolites appear capable of distinguishing cases of active pulmonary tuberculosis from sick and healthy controls.
-
•
Levels of these biomarkers decrease after 60 days of treatment in a longitudinal cohort of 20 participants.
-
•
- Many of the identified biomarkers are known inflammatory intermediates that may reflect a specific immune response to tuberculosis.
Urine tests are commonly used to enable non-invasive, rapid and point-of-care diagnosis of various infectious diseases. We identified diacetylspermine, neopterin, sialic acid and N-acetylhexosamine as potential urine-based biomarkers for tuberculosis from two independent patient cohorts. These metabolites are known inflammatory intermediates and appear to decrease with anti-tuberculosis therapy in a subset of participants followed over 2 months. If validated, these metabolites have potential to both improve our understanding of the immune reaction to active tuberculosis and facilitate the development of a much-needed clinical biomarker for tuberculosis.
1. Introduction
Tuberculosis (TB) has surpassed HIV/AIDS as the leading cause of deaths due to an infectious disease [1]. Spread simply by sharing air with an infected person, TB poses a public health threat of singular proportion due to its facile spread and rising rates of multi-drug resistant strains of Mycobacterium tuberculosis (Mtb). Early diagnosis is thus emerging as an essential, but incompletely met, need of TB control efforts [2].
A significant limitation of current TB diagnostics is their dependence on sputum-based assays. Conventional sputum microscopy and microbiologic culture remain the gold standard for diagnosis. Sputum microscopy relies on manual visualization of the bacteria, has diagnostic sensitivities that vary widely according to collection and processing techniques [3], and is not helpful in cases where TB has disseminated out of the lungs. In addition, cultures require 3–6 weeks of incubation. Nucleic acid amplification-based methods, such as GeneXpert, have greatly increased sensitivity and have a more rapid turnaround time but remain prohibitively expensive in many resource-limited settings where TB disease burden is highest. Additionally, they require sputum that is often difficult to obtain, and exhibit limited usefulness in cases of extrapulmonary disease [4,5].
Urine tests are an increasingly common diagnostic modality used to enable non-invasive, rapid, and point-of-care diagnosis of various infectious diseases, including legionella, pneumococcal pneumonia, histoplasmosis, and TB [6]. Several studies have looked at urine as a source of clinically relevant biomarkers for TB. Lipoarabinomannan (LAM) is a component of the mycobacterial cell wall that is shed into urine and capable of being detected in the urine of patients with active pulmonary TB. With an overall estimated sensitivity of 46% and specificity of 89%, programmatic implementation of urine LAM assays in HIV-infected subjects has been associated with improvements in early TB diagnosis and a 17% decrease in all-cause mortality [7]. A point-of-care urine assay with improved diagnostic performance may result in even larger reductions in mortality. Recently, IP-10, an interferon inducible protein was also shown to be increased in the urine of people with active TB and may track with treatment response further demonstrating the potential for urine as an optimal biological specimen for biomarker discovery [[8], [9], [10]].
Metabolomics is an emerging systems level technology with the potential to expand the current repertoire of diagnostic biomarkers due to its ability to enable the unbiased, multiplexed profiling and comparison of metabolites in a biological sample. Accordingly, metabolomics has been successfully used to identify novel biomarkers for different disease states [11]. Specific interest in TB has recently revealed distinct metabolite signatures present in blood, breath, and urine that may be associated with Mtb infection or treatment response [[12], [13], [14]]. Based on such studies, we hypothesized that TB might also be associated with specific metabolites that could be detected in the urine and used to both diagnose and monitor the response to treatment of patients with active pulmonary TB.
Here, we report the unbiased discovery of a urinary small molecule bio-signature that could differentiate cases of active pulmonary tuberculosis from healthy controls and/or pulmonary disease from other causes in two unrelated patient cohorts. Identified components of this signature included known products of the immune response to Mtb. If validated, this metabolic signature could facilitate the future development of a point-of-care urine test for TB.
2. Materials and Methods
2.1. Study Design
We recruited participants with active pulmonary TB and matched healthy controls from the GHESKIO Center in Haiti. Significant urinary biomarkers were then validated using banked randomized urine samples collected by the Foundation for Innovative Diagnostics (FIND) in collaboration with the WHO/TDR TB Specimen Bank on people with active TB and controls with non-tuberculous pulmonary disease.
2.2. Discovery Cohort
Participants were enrolled at the GHESKIO Center in Port-au-Prince, Haiti from September 2011 to March 2012. Participants with suspected pulmonary TB were screened for active TB by medical history, physical exam, chest radiography, sputum smear for acid-fast bacilli (AFB) and Mtb culture. Participants with one or more clinical symptoms of TB (defined as cough, dyspnea, fever, night sweats, weight loss, lymphadenopathy, and hemoptysis) and a positive sputum smear for AFB or positive sputum culture were included in the study. Cases of MDR TB were excluded. Controls were recruited at the GHESKIO Center and screened by medical exam and chest x-ray. Controls were matched to cases by age +/− 5 years, sex, and HIV status. Controls were excluded if they met any of the following exclusion criteria: symptoms of active tuberculosis at the time of study enrollment, history of tuberculosis or treatment with anti-tuberculosis medications, HIV with CD4 count <250 or AIDS defining illness, history of chronic immunosuppressive therapy, or history of chronic diseases (heart disease, lung disease, auto-immune disease, malignancy). Clean-catch urine samples were collected in sterile cups and immediately refrigerated at −4 degrees for 1–7 h. Urine was then aliquoted on ice and stored at −80 °C until shipment to NYC. All samples were collected from participants at the time of enrollment, prior to initiation of anti-tuberculosis therapy. No fasting restrictions or specific time of urine sample collection was defined.
2.3. Longitudinal Cohort
A subset of 20 participants with active TB had urine samples collected at the time of diagnosis, prior to starting anti-tuberculosis therapy and at day 60 after initiation of treatment. These participants were followed for 6 months to ensure sustained treatment response. All participants diagnosed with tuberculosis at GHESKIO were started on anti-mycobacterial treatment on-site, following World Health Organization Guidelines [15]. Tuberculosis treatment consisted of isoniazid, rifampin, pyrazinamide and ethambutol in all cases.
2.4. Validation Cohort
We obtained blinded urine samples from 50 participants with confirmed pulmonary TB and 50 participants with cough who were being evaluated for active tuberculosis but proven negative from collaborators at the Foundation for Innovative New Diagnostics (FIND)/WHO TDR TB Specimen Bank. Participants were recruited from a single center in Vietnam from June 2007 to October 2010. Participants with cough were screened for active TB with chest x-ray, sputum smear microscopy for AFB, and sputum culture. Participants were defined as having active pulmonary TB if they had positive sputum smear microscopy and Mtb culture. Control participants were included if they had negative sputum smear AFB, Mtb culture, and an alternative diagnosis. Controls were followed for an additional 3 months from the time of study enrollment to document clinical improvement off anti-mycobacterial therapy. Urine samples were collected on site, at the time of the initial evaluation, and stored at a FIND storage facility at −80 °C until shipped on dry ice to New York Presbyterian/Weill Cornell Medicine in New York for testing.
2.5. Sample Preparation
The urine samples were stored at −80 °C at Weill Cornell Medicine until processing. Samples were thawed and filtered for large particles using PALL nanosep centrifuge devices (centrifuged for 10 min at 10,000 rpm). The osmolality of each sample was measured using an Advanced Instruments model 3250 micro osmometer. Filtered substrate was then diluted with MiliQ filtered water to a standard 150 mOsm/kg H2O in order to normalize the salt concentration within each sample. Samples were then mixed with LCMS grade 0.2% formic acid in methanol at a 1:1 ratio.
2.6. HPLC-MS Analysis
Samples were separated by aqueous normal phase chromatography using a Cogent Diamond Hydride column. Samples were then analyzed using an Agilent Technologies 6230 high resolution, accurate mass TOF LC/MS. Detected ions were characterized by chromatographic retention time and ion mass. Data analysis was performed using Profinder B08 and Qualitative analysis (Agilent Technologies). Molecules were characterized by mass, time, and abundance (as reported by ion intensity) [16].
Experiments were repeated to ensure reproducibility using unfrozen urine aliquots from the discovery cohort (technical replicates). Repeat experiments were performed using 4 independent randomized subsets of cases and controls (n = 5–7 cases and 5–7 controls per experiment). Molecule abundance remained significantly different in repeat experiments and ROC curve analysis of repeated sample sets confirmed an area under the ROC curve of 87–100 for each molecule when comparing molecules within each experiment independently. Validation cohort samples were randomized and blinded prior to sample prep and HPLC analysis. Longitudinal samples were randomized prior to HPLC analysis. Sample prep and HPLC analysis of longitudinal samples was repeated three times to ensure reproducibility. Creatinine normalization did not significantly alter molecule ion counts, significance calculations or ROC curves.
2.7. Creatinine Normalization
A random subset of urine samples from the discover and validation cohort were selected and urine molecule abundance was compared prior to and after creatinine normalization. Urine creatinine concentration was measured using a creatinine colorimetric assay kit (Sigma Aldrich Catalog number MAK080). Absorbance was measured using a Spectramax M2 microplate reader at 570 nm (fluorometric (λex = 535/λem = 587 nm). If the creatinine concentration was out of the measurable range, the sample was excluded from the analysis. Creatinine measurement was done in triplicate for each urine sample. Statistical analysis was performed as described in statistical methods section. A random sample of 66 participants from the discovery (Haitian) cohort and 88 from the validation (Vietnam) cohort was analyzed before and after creatinine normalization. There was no difference in overall metabolite abundance or ROC characteristics when comparing samples that were normalized to osmolality alone vs. those that were normalized to both osmolality and creatinine. (Supplementary Table 1).
2.8. Molecule Identification
Molecule identification was done using the average exact mass for each molecule. Samples were prepared as described above and analyzed using the MS/MS feature of the Agilent Technologies model 6520 Q-TOF LC/MS. MS/MS spectral analysis was performed using Agilent Technologies Qualitative analysis program and confirmed by Agilent Molecular Structure Correlator software and Metlin and HMDB databases. Molecule identification was further confirmed by comparing MS/MS spectra of each molecule with purchased pure chemicals as standards: 3-ureidopropionic acid (product # 94295-1G, Sigma-Aldrich), 3-hydroxy-DL-kynurenine (product # H1771, Sigma-Aldrich), D-neopterin (product # N3386 Sigma-Aldrich), N1,N12-diacetylspermine (item # 17918, Cayman Chem), 7-methylguanine (product # 67073, Sigma-Aldrich), 3’-N-acetylneuraminyl-N-acetyllactosamine (product # A6936, Sigma-Aldrich), N-acetylneuraminic acid (product # 19023, Sigma-Aldrich), N-acetyl-β-D-mannosamine (product # sc-295642A, Santa Cruz Biotechnology), and N-acetylglucosamine (product # A4106, Sigma-Aldrich). Standards were analyzed alone, in a 1:1 solution of 0.2% formic acid in methanol and MilliQ H2O at a final concentration of 50 nM and in urine samples spiked with each respective standard to ensure accurate retention time and m/z value.
2.9. Statistical Methods
2.9.1. Discovery Cohort
The statistical analysis to identify molecules relied on a mixed model and factor analysis methodology (RRmix) that corrects for batch effects and other unwanted variation, and that detects a small number of molecules that are significantly different between two study groups [17]. Specifically, the model is defined as follows:
where yg is an n × 1 column vector of the gth metabolite's log-transformed, mass-to-charge ratio values from n-samples, μ is a vector of intercept parameters, βg is a 2 × 1 parameter vector, X = [1n, x] is an n × 2 matrix where the second column is an indicator vector for TB/control group status. Fg is the gth column of q × G factor matrix, Λ is the n × q loading matrix, and Wg is the n × 1 residual error vector. RRmix is fit for all metabolites simultaneously, estimates the metabolite-specific group effect while adjusting for unwanted variation such as batch effect reflected by the term, ΛFg, in the model.
This class of latent factor linear models can provide a means to handle unwanted variability without prior knowledge of its source of measurements [18] or internal controls [19]. In this approach, which has been shown to be superior to other methods [17], instead of attempting to parameterize known effects [19] or using a two-step approach [20], RRmix uses a factor analysis structure to model and adjust for latent batch effect factors. Data were separated into training and test groups, using randomly selected small subsets of cases versus controls in multiple iterations. Modeling molecule-specific mean effects as a random effect with a prior distribution that is a zero-point mass element mixture model, significantly different molecules were declared if their posterior non-null probability was >0.9 [17] or if the p-value was less than or equal to 0.0001 after adjustment for false discovery rate. Nonparametric ROC curves were then constructed using molecule abundances or ion counts as classifiers to known patient TB status.
Technical replicates were analyzed for significance using Wilcoxon rank sum test and significance was defined as p-value < 0.05. Nonparametric ROC curve analysis was performed as above.
2.9.2. Validation Cohort
Molecule abundances in the validation cohort were compared using a nonparametric test of medians and Wilcoxon rank sum test with significance defined as p-value < 0.05. Nonparametric ROC curves were then constructed using molecule abundances or ion counts as classifiers to known patient TB status.
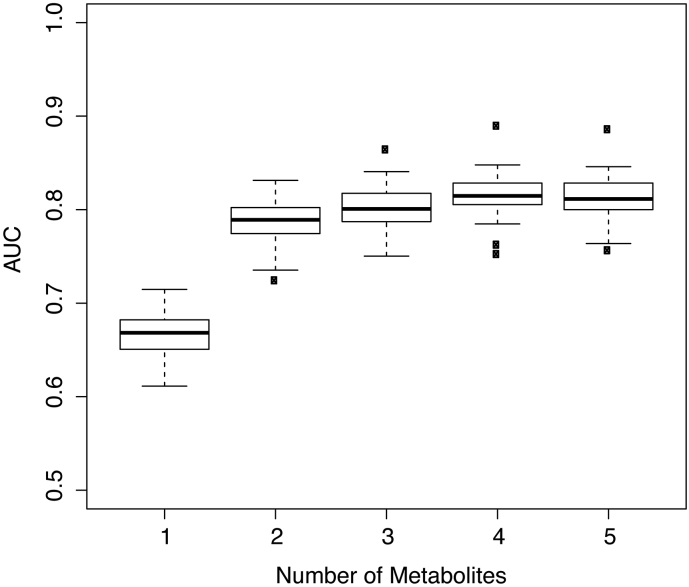
Using random forest modeling, we selected 5 molecules with a ROC AUC of at least 75% in both discovery and validation cohorts and built RF classifiers to assess relative importance of these molecules as predictors. First, we used the discovery cohort to rank relative importance of molecules by randomly selecting 80% of data and building RF and Gini importance indices [21]. After 50 iterations, biomarkers were ranked based on average of variable importance from largest to smallest. We then tested this model on the validation set 50 times. In each iteration, models with the top-ranked biomarkers were blindly tested on a randomly selected subset of 80% of the validation set. Fig. 3 shows the AUC statistics from 50 repetitions.
Fig. 3.
Performance of Molecule Combinations. Box plots showing area under the ROC curve for the top 5 molecules in combination. Using random forest modeling on the discovery set we ranked the top 5 molecules by importance and tested these molecules and molecule combinations on the validation set in 50 iterations. Molecules are shown in the following order, for which each successive molecule is added to the model: N-acetylhexosamine, sialic acid 3, neopterin, diacetylspermine, sialic acid 1. The lower quartile and upper quartile represent the 25th and 75th percentile respectively.
2.9.3. Longitudinal Cohort
Urine samples from each participant in the longitudinal cohort, at time zero and at 2 months, were prepped, randomized and analyzed in the same experiment as described previously. Mean ion counts were compared using paired t-test. Effect size was calculated using Cohen's d calculation.
Statistical analysis was performed using STATA 14 and R version 3.4.0.
2.9.4. Ethics
The study protocol and consent forms for samples collected at GHESKIO were approved by the GHESKIO Center and Weill Cornell Medicine Institutional Review Boards. All participants provided written informed consent. IRB approval was obtained from Weill Cornell Medicine for testing of unlinked and anonymous urine samples obtained from the Foundation for Innovative New Diagnostics (FIND).
3. Results
3.1. Discovery Cohort
Screening 260 adult participants at the GHESKIO Center from September 2011 to March 2012, we identified 107 subjects meeting inclusion criteria for pulmonary tuberculosis, defined as having 1 or more symptoms of active pulmonary TB and positive sputum smear for acid-fast bacilli (AFB) and/or Mtb culture. Of these, we matched 102 to asymptomatic controls enrolled at the same center during the same period. Cases were matched to controls by age +/− 5 years, gender, and HIV status (Supplementary Fig. 1). Pertinent demographic and clinical characteristics are listed in Table 1. The most common clinical symptoms among TB cases were cough (94.1%), fever (70.6%), and weight loss (67.6%). Participants with TB had a significantly lower body weight than controls (109.7 lbs. vs 136.1 lbs. p < 0.05) (Table 1). Of the control subjects, 62.7% had a positive Tuberculin Skin Test (TST), defined as over 10 mm induration at the time of enrollment.
Table 1.
Clinical characteristics of participants from the discovery (Haiti) and validation (Vietnam) cohorts.
| Discovery (Haiti) |
Validation (Vietnam) |
|||
|---|---|---|---|---|
| Control (n = 102) | Cases (n = 102) | Control (n = 50) | Cases (n = 50) | |
| Age, years (range) | 33.8 (20–71) | 33.7 (18–68) | 47.2 (19–83) | 36.6 (19–79) |
| Sex (% male) | M 53 (51.9%) | M 53 (51.9%) | 33 (66%) | 38 (76%) |
| Average Weight (range) | 136.1 lbs. (85–285) | 109.7 lbs. (78–172) | N/A | N/A |
| HIV+ | 15 (14.7%) | 16 (15.7%) | 0 (0%) | 0 (0%) |
| Average CD4 | 536.3 (390–790) | 433 (71–974) | N/A | N/A |
| On ARVs prior to enrollment | 0 | 3 | N/A | N/A |
| Symptoms | ||||
| Fever | 0 | 72 (70.6%) | 16 (32%) | 29 (58%) |
| Night sweats | 0 | 56 (54.9%) | 3 (6%) | 8 (16%) |
| Cough | 0 | 96 (94.1%) | 50 (100%) | 50 (100%) |
| Dyspnea | 0 | 19 (18.6%) | 26 (52%) | 8 (16%) |
| Weight loss | 0 | 69 (67.6%) | 4 (8%) | 18 (36%) |
| Abnormal CXR | 0 | 96 (94.1%) | 50 (100%) | 50 (100%) |
Using clean catch urine samples collected at the time of study enrollment, prior to initiation of anti-tuberculous therapy, we next compared the metabolic composition of samples from cases and matched controls. To do so, we normalized all urine samples to a standard 150 mOsm/kg H2O and analyzed them in randomized batches, each containing 30–35 cases and 30–35 controls, by untargeted liquid chromatography-coupled high resolution mass spectrometry [22]. Using a statistical method capable of simultaneously correcting for unrecognized potential batch effects and an absence of internal controls (RR mix) [17], we identified 154 molecules whose levels were significantly different in the urine of cases of TB compared to asymptomatic controls. After adjustment for multiple testing (false discovery rate p-value < 0.0001 and a Bayesian posterior non-null probability of 0.9), 49 of these 154 molecules were found to be significantly different between the two groups (Supplementary Table 2).
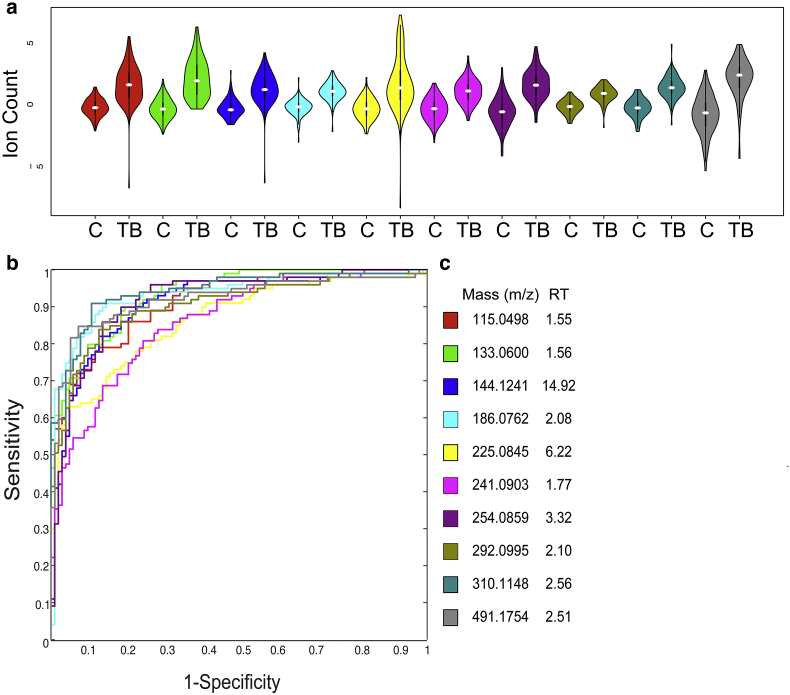
Of these 49, 10 exhibited area under the Receiver Operator Characteristic (ROC) curve values >85% (Fig. 1, Table 2). These molecules remained significantly different, with p-values of <0.0001 irrespective of adjustments for age, gender, weight, HIV status, and clinical symptoms (e.g. fever, night sweats, dyspnea, lymphadenopathy, and hemoptysis). The mean abundance of a molecule subsequently identified as neopterin (m/z 254.0859) was found to be higher in HIV infected participants than in HIV uninfected participants but remained significantly higher in TB cases irrespective of HIV status. (Supplementary Fig. 2) Subgroup analysis of samples from healthy controls failed to reveal any significant differences in the levels of these biomarkers in TST-positive vs. -negative healthy controls.
Fig. 1.
Significant Molecules in Discovery Cohort. (a) Violin plots showing overall abundance of significant urinary molecules in TB cases (TB) (n = 102) verses matched controls (C) (n = 102). Each color represents a distinct molecule shown in table. Darker black line represents 25th and 75th percentile. White line represents median value. Values are scaled to mean and log2 transformed. Values are shown in log2 scale. All molecules with p-value < 0.0001 (b) Receiver operator characteristic (ROC) curve of 10 significant molecules in discovery cohort of 102 TB cases and 102 matched controls. Each color represents a distinct molecule shown in table. (c) Table of molecules. Masses listed as mass to charge ratio (m/z) in positive mode. Retention time (RT) is listed in minutes.
Table 2.
Significant Molecules with Preliminary Identification and Area Under ROC (AUC).
| Mass (m/z) | Retention time (min) | Predicted formula | Preliminary identification | Discovery cohort |
Validation cohort |
|
|---|---|---|---|---|---|---|
| AUC (95% CI) | AUC (95% CI) | |||||
 |
115.0498 | 1.55 | C4H6N2O2 | Unknown 1 | 91.25% (87.4%–95.1%) | 58.29% (46.22%–70.36%) |
 |
133.0600 | 1.56 | C4H8N2O3 | Ureidopropionic acid | 93.47% (90.4%–96.5%) | 65.01% (53.46%–76.56%) |
 |
144.1241 | 14.92 | C14H30N4O2 | Diacetylspermine | 90.95% (86.8%–95.1%) | 80.26% (70.84%–89.67%) |
 |
186.0762 | 2.08 | C8H15NO6 | N-Acetylhexosamine | 93.24% (89.5%–97.0%) | 79.74% (70.25%–89.22%) |
 |
225.0845 | 6.22 | C10H12N2O4 | Hydroxykynurenine | 86.50% (81.5%–91.5%) | 58.97% (46.97%–70.95%) |
 |
241.0903 | 1.77 | C9H12N4O4 | Unknown 2 | 85.96% (80.1%–90.1%) | 67.39% (55.85%–78.93%) |
 |
254.0859 | 3.32 | C9H11N5O4 | Neopterin | 91.75% (87.8%–95.7%) | 78.50% (69.10%–87.90%) |
 |
292.0995 | 2.10 | C11H17NO8 | Sialic acid 1 | 90.49% (86.2%–94.8%) | 74.47% (64.15%–84.78%) |
 |
310.1148 | 2.56 | C11H19NO9 | Sialic acid 2 | 94.16% (90.9%–97.4%) | 70.85% (59.87%–81.86%) |
 |
491.1754 | 2.51 | C17H26N6O11 | Sialic acid 3 | 92.08% (88.0%–96.2%) | 75.40% (65.16%–85.63%) |
3.2. Molecule Identification
Of the 10 molecules with areas under the ROC curve of over 85%, we identified 4 by collision-induced dissociation MS/MS analysis, and comparison to reference MS/MS spectra of pure chemical standards and metabolomics databases (Metlin, HMDB). These metabolites were ureidopropionic acid, N1N12 diacetylspermine, hydroxykynurenine, and neopterin. Of the remaining 6 metabolites, we were able to determine the chemical class of 4 by MS/MS spectral analysis. These included 3 sialic acid species (numbered 1, 2 and, 3), and a N-acetylhexosamine. The remaining 2 molecules have not yet been identified. (Supplementary Figs. 3 and 4).
3.3. Validation Cohort
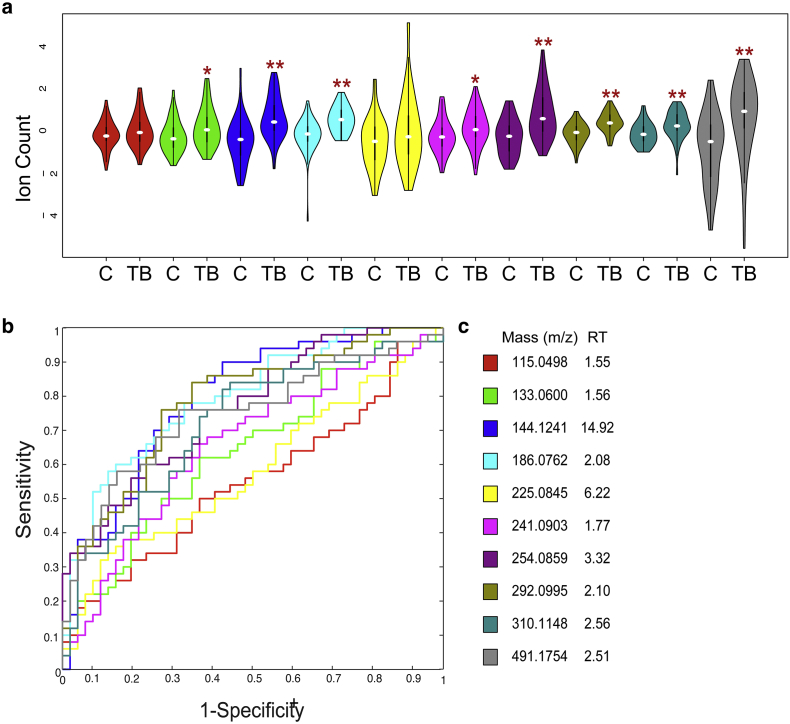
To more robustly investigate the discriminatory potential of these candidate biomarkers, we next analyzed a blinded “validation” set of urine samples from an unrelated cohort of 50 adults with active pulmonary TB and 50 controls with other respiratory tract diseases (obtained from the Foundation for Innovative New Diagnostics (FIND)). Subjects were recruited from a single center in Vietnam from June 2007 to October 2010 and were enrolled in the study if they had symptoms consistent with pulmonary TB, a chest radiograph, AFB sputum smear microscopy, and Mtb culture. TB cases were defined by positive sputum smear AFB and Mtb culture. Control participants were included if they had both negative sputum AFB and Mtb culture and an alternative diagnosis as follows: 27/50 (54%) with pneumonia, 9/50 (18%) with prior TB, 4/50 (8%) with COPD, and 10/50 (20%) with an unclear diagnosis. Urine samples were collected at the time of enrollment. Controls were followed for an additional 3 months off Mtb treatment to ensure clinical improvement. We received samples blinded and randomized. Associated clinical information was provided after metabolomic analysis was complete. Samples were normalized to 150 mOsm/kg H2O and analyzed in random batches of 25 as before. Of the 10 previously described molecules, 8 remained significantly different in this cohort. Moreover, this analysis independently identified levels of diacetylspermine, ureidopropionic acid, neopterin, sialic acids (1, 2 & 3), N-acetylhexosamine, and an unknown molecule with a molecular mass of 240 Da (m/z 241.0903) as significantly different in TB cases compared to participants with other respiratory tract disease with a p-value of <0.05. (Wilcoxon Rank Sum, Fig. 2). These molecules remained significantly different after adjustment for age, gender, and smoking status. After normalization to creatinine concentration and adjustment for age, gender, smoking status, and clinical symptoms (e.g. fever, dyspnea, night sweats, weight loss, and hemoptysis) diacetylspermine, neopterin, sialic acids (1, 2 & 3), N-acetylhexosamine, and hydroxykynurenine remained significantly different when comparing cases of pulmonary TB to controls with other non-tuberculous pulmonary disease. (p-value <0·05), suggesting that these markers may have some specificity for TB disease. As shown in Fig. 2 and Table 2, ROC curve analyses show good individual overall sensitivity and specificity with AUC values over 75%.
Fig. 2.
Significant Molecules in Validation Cohort. (a) Violin plots showing overall abundance of significant urinary molecules in TB cases (TB) (n = 50) verses controls with pulmonary disease from other causes (C) (n = 50). Each color represents a distinct molecule shown in table. Darker black line represents 25th and 75th percentile. White line represents median value. Values are scaled to mean and log2 transformed. Values are shown in log2 scale. Wilcoxon rank sum test *p-value < 0.05, **p value < 0.001. (b) Receiver operator characteristic (ROC) curve of 10 significant molecules in validation cohort of 50 TB cases and 50 controls with pulmonary disease from other causes. Each color represents a distinct molecule shown in table. (c) Table of molecules. Masses listed as mass to charge ratio (m/z) in positive mode. Retention time (RT) is listed in minutes.
We found that 5 biomarkers show good predictive performance with ROC AUC values >75% for both discovery and validation data (diacetylspermine, N-acetylhexosamine, neopterin, sialic acid 1, and sialic acid 3). To further optimize the discriminatory power of these molecules and identify potentially improved metabolite combinations, we applied Random Forest (RF) classification modeling. We initially tested the relative importance of each molecule as a predictor of TB using a randomly selected subset of 80% of the discovery data in 50 iterations. The 5 molecules were then ranked by their average Gini importance index [21]. After the biomarkers were ranked, RF classifiers built from the discovery set were blindly tested on a randomly selected subset of 80% of the validation data 50 times (Fig. 3). This model identified the specific combination of N-acetylhexosamine, sialic acid 3, neopterin, and diacetylspermine as exhibiting a ROC AUC of 82.0% in the validation cohort.
3.4. Longitudinal Cohort
Of the 102 cases in the original discovery cohort, a subset of 20 participants were followed longitudinally and had urine samples collected at the time of enrollment and at 2 months' post-initiation of anti-tuberculosis therapy. These participants were followed for an additional 4 months to ensure sustained treatment responses. Tuberculosis treatment consisted of isoniazid, rifampin, pyrazinamide, and ethambutol in all cases. Participant characteristics are shown in Supplementary Table 3. The mean age was 39 years. The most common presenting symptoms included cough (95%), weight loss (85%), fever (60%), and night sweats (60%). All participants had positive sputum AFB on enrollment, 14 (70%) had documented negative sputum AFB at 2 months. All had documented negative sputum AFB at 4 months. There were no documented relapses at 6 months of follow up. With an n of 20, we calculate over 80% power to detect a difference in mean metabolite abundance of 20% with an alpha of 0.01.
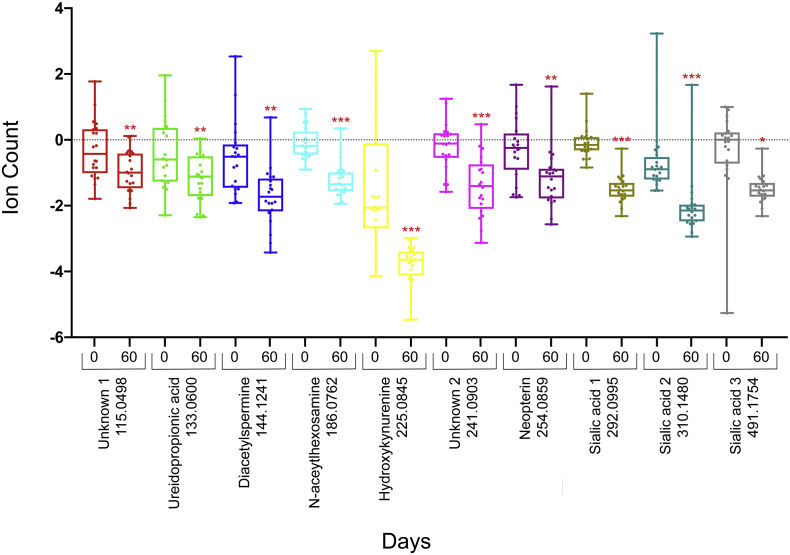
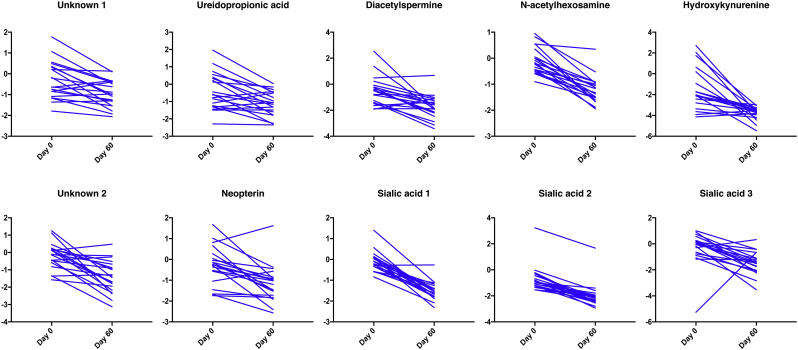
Thawed urine aliquots from each participant, prior to initiation of anti-tuberculosis therapy and after two months of treatment, were prepped and analyzed as described above. Comparing the average abundance of each of the ten identified molecules at diagnosis and after 2 months of anti-tuberculosis treatment we noted statistically significant decreases in the overall abundance of all 10 molecules after 60 days of treatment (paired t-test, Fig. 4) mean effect size 1.46 (range 0.84–3.09, Cohen's d). These trends were also observed on the individual patient level when comparing levels at the time of enrollment, prior to initiation of therapy and at 2 months (Fig. 5).
Fig. 4.
Molecule Abundance Before and After 60 Days of Treatment. Box plot showing overall abundance of urinary molecules from 20 participants before and sixty days after initiation of anti-tuberculosis therapy. The lower quartile and upper quartile represent the 25th and 75th percentile respectively. Values are scaled to mean and log2 transformed. Values are shown in log2 scale. N = 20 Paired t-test *p value < 0.01, **p value < 0.001 ***p value < 0.0001.
Fig. 5.
Line Graph Showing Overall Abundance of Urinary Molecules Before and Sixty Days after Initiation of Anti-tuberculosis Therapy. Each line represents an individual participant. Values are scaled to mean and Log2 transformed. Values are shown in log2 scale.
4. Discussion
Identification of clinically relevant, non-sputum based biomarkers capable of facilitating TB diagnosis remains a major unmet need in efforts to achieve control of the pandemic [23,24]. Such biomarkers are especially needed in characteristically difficult to diagnose populations, such as HIV-infected, children, and individuals with extrapulmonary TB, where disease prevalence and mortality are especially high. To date, few studies have looked for urine metabolite biomarkers for tuberculosis and have mostly focused on volatile organic compounds from breath or serum metabolites [[25], [26], [27]]. One recent study by Luier et al., showed changes in tryptophan, phenylalanine, and tyrosine as significant changes that occur in the urine in cases of active pulmonary tuberculosis [28]. This study, however, was limited by a small cohort of participants with TB and did not include a validation cohort.
Our study identified N-acetylhexosamine, neopterin, diacetylspermine, and sialic acids as potential discriminatory urinary biomarkers of active TB. We found that these potential biomarkers discriminated active pulmonary tuberculosis from healthy controls with an overall sensitivity and specificity of over 95% in a cohort of 204 participants from Haiti, and from sick controls in an independent blinded cohort of patients with active pulmonary tuberculosis and other non-tuberculous causes of pulmonary disease from Vietnam with an overall sensitivity and specificity of over 82%. These biomarkers remain significantly different after adjustment for age, gender, and symptomatology. Interestingly, neopterin, kynurenine, and sialic acids have all been individually reported to be increased in blood, urine or pleural fluids of patients with TB in previous studies [[29], [30], [31], [32], [33], [34]]. Neopterin has also been studied as a potential biomarker for HIV infection [35,36]. In our study, levels of neopterin were increased in HIV infected participants when compared to HIV uninfected but remained significantly higher in participants with active TB regardless of HIV status. N1,N12-diacetylspermine was similarly identified by Mahapatra et al. as a potential biomarker of TB treatment response, as well as a urinary biomarker for certain malignancies [13,37,38].
Many of the biomarkers identified in this study are well known bioactive products of activated immune cells. Neopterin and kynurenine are downstream products of macrophage INF gamma signaling [39,40]. Spermine is thought to be a negative regulator of macrophage activation [41]. N-acetylated sugars and sialic acids are thought to be involved in immune signaling through glycosaminoglycan (GAG) signaling and sialic-acid-binding immunoglobulin like receptors (Siglecs) [[42], [43], [44], [45]]. Interestingly, increasing evidence indicates that immune regulation and metabolic regulation are highly integrated and interdependent [46]. Moreover, several studies have identified specific transcriptional signatures of TB [[47], [48], [49]] while others have reported specific T cell responses [50,51]. These findings thus support the biological plausibility of a corresponding immunometabolic signature of TB, the specificity of which remains to be elucidated, but is supported by the apparent increased ROC AUC associated with the specific combination of diacetylspermine, neopterin, sialic acid 3, and N-acetylhexosamine. While the majority of identified discriminatory metabolites correspond to putatively host-derived molecules, the precise chemical identities of others remain to be determined and leave open the possibility of microbe-derived products, such as in the case of lipoarabinomannan [52].
In a subset of patients that were followed longitudinally, we found that all ten molecules decreased with anti-tuberculosis treatment. Albeit a small cohort of participants, these findings raise the possibility of an additional prognostic role for urine metabolites as potential biomarkers of treatment response. Few studies have looked at metabolite biomarkers to follow treatment response to TB drug therapy. Mahapatra et al. looked at urine metabolite changes while participants were being treated for tuberculosis. Although this study did not specifically look at metabolites that could be used for diagnosis, it also identified N1,N12-diacetylspermine as one of 12 urine metabolites detected in participants with active pulmonary TB that significantly decreased after treatment.
Taken together, this study is distinguished by the use of 2 distinct clinical cohorts of patients from different areas of the world, that included sick controls, standardized experimental and analytical techniques, and orthogonal statistical methods to decrease the chance of overfitting. Interestingly, many of the metabolites identified correspond to known products of activated immune cells, some of which have, in fact, been previously reported in isolation. Their rediscovery in this study and improved diagnostic power when combined raise the intriguing possibility that elucidation of a TB specific immune response can be aided through metabolomic assessment of urinary products. We recognize as a limitation the need for multiple reiterative studies in diverse populations to finalize biomarkers for a new diagnostic test. Our study was a first step, demonstrating the potential of using LC-MS technology to discover urinary biomarkers for TB diagnosis and identifying five candidates for future studies. The development of a urine-based point-of-care diagnostic for TB will ultimately require further elucidation of the biological origin of these metabolites as well as their performance in endemic clinical settings and additional populations, including those with latent or extrapulmonary TB. However, the discovery of a potential urinary immunometabolic signature of TB helps lay the foundation for the future development of a novel biological class of point-of-care diagnostic.
Acknowledgments
Acknowledgments
We thank the volunteers who participated in this study and the staff at both GHESKIO and FIND. We thank Livio Avril for helping to enroll and consent participants at the GHESKIO Center. We gratefully acknowledge Carl Nathan, Sabine Ehrt, Dirk Schnappinger, Selin Somerson, Kathryn Dupnik, Kohta Saito, Matthew Wipperman, Thomas Baker, the TB researchers at Weill Cornell, and the Tri-I TBRU for critical input and guidance and Robert Jansen for help with metabolite identification.
Funding Sources
This work was supported by the following funding sources: NIH T 32 training grant (T32-2T32AI007613-16) at Weill Cornell Medicine, the Clinical and Translational Science Center at Weill Cornell KL2 grant (NIH/NCATS KL2TR000458), the NIH Fogarty International Center grants R24 TW007988 and TW010062, and the Tuberculosis Research Units Networks (TBRU-N, AI111143).
Conflicts of Interest
All authors approve of the final submitted manuscript. None of the authors have a financial or other conflict of interest. Dr. Sean Collins took a position at Gilead after the completion of the study but did not have any conflicts during the time of the study.
Author Contributions
FI and LS conducted the experiments. SC, DD, ND, PJ enrolled participants and collected participant data and urine samples at GHESKIO. MHL, SS, MW and FI conducted statistical analysis. SF advised on MS and MS/MS data analysis and experimental design. JB provided violin plots. FI, DF and KR wrote the manuscript. JP, WJ, DF and KR supervised and coordinated the work. All authors reviewed the manuscript, agreed with the results and provided insight.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.04.014.
Appendix A. Supplementary data
Supplementary material
References
- 1.Organization, W. H. 2017. Global Tuberculosis Report 2017.
- 2.WHO Policy on TB Infection Control in Health-Care Facilities . 2009. Congregate Settings and Households. Geneva. [PubMed] [Google Scholar]
- 3.Steingart K.R., Henry M., Ng V., Hopewell P.C., Ramsay A., Cunningham J. Fluorescence versus conventional sputum smear microscopy for tuberculosis: a systematic review. Lancet Infect. Dis. 2006;6:570–581. doi: 10.1016/S1473-3099(06)70578-3. [DOI] [PubMed] [Google Scholar]
- 4.Denkinger C.M., Schumacher S.G., Boehme C.C., Dendukuri N., Pai M., Steingart K.R. Xpert MTB/RIF assay for the diagnosis of extrapulmonary tuberculosis: a systematic review and meta-analysis. Eur. Respir. J. 2014;44:435–446. doi: 10.1183/09031936.00007814. [DOI] [PubMed] [Google Scholar]
- 5.Dowdy D.W., Cattamanchi A., Steingart K.R., Pai M. Is scale-up worth it? Challenges in economic analysis of diagnostic tests for tuberculosis. PLoS Med. 2011;8 doi: 10.1371/journal.pmed.1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Couturier M.R., Graf E.H., Griffin A.T. Urine antigen tests for the diagnosis of respiratory infections: legionellosis, histoplasmosis, pneumococcal pneumonia. Clin. Lab. Med. 2014;34:219–236. doi: 10.1016/j.cll.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Peter J.G., Zijenah L.S., Chanda D., Clowes P., Lesosky M., Gina P. Effect on mortality of point-of-care, urine-based lipoarabinomannan testing to guide tuberculosis treatment initiation in HIV-positive hospital inpatients: a pragmatic, parallel-group, multicountry, open-label, randomised controlled trial. Lancet. 2016;387:1187–1197. doi: 10.1016/S0140-6736(15)01092-2. [DOI] [PubMed] [Google Scholar]
- 8.Cannas A., Calvo L., Chiacchio T., Cuzzi G., Vanini V., Lauria F.N. IP-10 detection in urine is associated with lung diseases. BMC Infect. Dis. 2010;10:333. doi: 10.1186/1471-2334-10-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petrone L., Cannas A., Aloi F., Nsubuga M., Sserumkuma J., Nazziwa R.A. Blood or urine IP-10 cannot discriminate between active tuberculosis and respiratory diseases different from tuberculosis in children. Biomed. Res. Int. 2015;2015:589471. doi: 10.1155/2015/589471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrone L., Cannas A., Vanini V., Cuzzi G., Aloi F., Nsubuga M. Blood and urine inducible protein 10 as potential markers of disease activity. Int. J. Tuberc. Lung Dis. 2016;20:1554–1561. doi: 10.5588/ijtld.16.0342. [DOI] [PubMed] [Google Scholar]
- 11.Nagana Gowda G.A., Raftery D. Biomarker discovery and translation in metabolomics. Curr. Metabol. 2013;1:227–240. doi: 10.2174/2213235X113019990005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frediani J.K., Jones D.P., Tukvadze N., Uppal K., Sanikidze E., Kipiani M. Plasma metabolomics in human pulmonary tuberculosis disease: a pilot study. PLoS One. 2014;9 doi: 10.1371/journal.pone.0108854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahapatra S., Hess A.M., Johnson J.L., Eisenach K.D., Degroote M.A., Gitta P. A metabolic biosignature of early response to anti-tuberculosis treatment. BMC Infect. Dis. 2014;14:53. doi: 10.1186/1471-2334-14-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phillips M., Basa-Dalay V., Bothamley G., Cataneo R.N., Lam P.K., Natividad M.P. Breath biomarkers of active pulmonary tuberculosis. Tuberculosis (Edinb) 2010;90:145–151. doi: 10.1016/j.tube.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Treatment of Tuberculosis: Guidelines. 4th ed. 2010. (Geneva) [PubMed] [Google Scholar]
- 16.Nandakumar M., Nathan C., Rhee K.Y. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat. Commun. 2014;5:4306. doi: 10.1038/ncomms5306. [DOI] [PubMed] [Google Scholar]
- 17.Salerno S., Jr., Mehrmohamadi M., Liberti M.V., Wan M., Wells M.T., Booth J.G. RRmix: a method for simultaneous batch effect correction and analysis of metabolomics data in the absence of internal standards. PLoS One. 2017;e0179530:12. doi: 10.1371/journal.pone.0179530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Causeur D., Friguet C., Houee-Bigot M., Kloareg M. Factor analysis for multiple testing (FAMT): an R package for large-scale significance testing under dependence. J. Stat. Softw. 2011;40 [Google Scholar]
- 19.de Livera A.M., Sysi-Aho M., Jacob L., Gagnon-Bartsch J.A., Castillo S., Simpson J.A. Statistical methods for handling unwanted variation in metabolomics data. Anal. Chem. 2015;87:3606–3615. doi: 10.1021/ac502439y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leek J.T., Storey J.D. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breiman L. Technical Report 670. University of California at Berkeley; 2004. Consistency for a simple model of random forests. [Google Scholar]
- 22.Patti G.J., Yanes O., Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012;13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallis R.S., Kim P., Cole S., Hanna D., Andrade B.B., Maeurer M. Tuberculosis biomarkers discovery: developments, needs, and challenges. Lancet Infect. Dis. 2013;13:362–372. doi: 10.1016/S1473-3099(13)70034-3. [DOI] [PubMed] [Google Scholar]
- 24.Wallis R.S., Wang C., Doherty T.M., Onyebujoh P., Vahedi M., Laang H. Biomarkers for tuberculosis disease activity, cure, and relapse. Lancet Infect. Dis. 2010;10:68–69. doi: 10.1016/S1473-3099(10)70003-7. [DOI] [PubMed] [Google Scholar]
- 25.Dang N.A., Janssen H.G., Kolk A.H. Rapid diagnosis of TB using GC-MS and chemometrics. Bioanalysis. 2013;5:3079–3097. doi: 10.4155/bio.13.288. [DOI] [PubMed] [Google Scholar]
- 26.Phillips M., Basa-Dalay V., Blais J., Bothamley G., Chaturvedi A., Modi K.D. Point-of-care breath test for biomarkers of active pulmonary tuberculosis. Tuberculosis (Edinb) 2012;92:314–320. doi: 10.1016/j.tube.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 27.Xu D.D., Deng D.F., Li X., Wei L.L., Li Y.Y., Yang X.Y. Discovery and identification of serum potential biomarkers for pulmonary tuberculosis using iTRAQ-coupled two-dimensional LC-MS/MS. Proteomics. 2014;14:322–331. doi: 10.1002/pmic.201300383. [DOI] [PubMed] [Google Scholar]
- 28.Luier L., Loots D.T. Tuberculosis metabolomics reveals adaptations of man and microbe in order to outcompete and survive. Metabolomics. 2016;12 [Google Scholar]
- 29.Ahmad P., Kumar A., Salahuddin A. Cerebrospinal fluid sialic acid in tuberculous meningitis. Indian Pediatr. 1985;22:191–194. [PubMed] [Google Scholar]
- 30.Cok G., Parildar Z., Basol G., Kabaroglu C., Bayindir U., Habif S. Pleural fluid neopterin levels in tuberculous pleurisy. Clin. Biochem. 2007;40:876–880. doi: 10.1016/j.clinbiochem.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 31.Imecik O., Ozer F. Diagnostic value of sialic acid in malignant pleural effusions. Chest. 1992;102:1819–1822. doi: 10.1378/chest.102.6.1819. [DOI] [PubMed] [Google Scholar]
- 32.Immanuel C., Victor L., Chelvi K.S., Padmapriyadarsini C., Rehman F., Iliayas S. Serum neopterin levels in HIV infected patients with & without tuberculosis. Indian J. Med. Res. 2005;121:220–225. [PubMed] [Google Scholar]
- 33.Suzuki Y., Suda T., Asada K., Miwa S., Suzuki M., Fujie M. Serum indoleamine 2,3-dioxygenase activity predicts prognosis of pulmonary tuberculosis. Clin. Vaccine Immunol. 2012;19:436–442. doi: 10.1128/CVI.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuksekol I., Ozkan M., Akgul O., Tozkoparan E., Al-Rashed M., Balkan A. Urinary neopterin measurement as a non-invasive diagnostic method in pulmonary tuberculosis. Int. J. Tuberc. Lung Dis. 2003;7:771–776. [PubMed] [Google Scholar]
- 35.Mildvan D., Spritzler J., Grossberg S.E., Fahey J.L., Johnston D.M., Schock B.R. Serum neopterin, an immune activation marker, independently predicts disease progression in advanced HIV-1 infection. Clin. Infect. Dis. 2005;40:853–858. doi: 10.1086/427877. [DOI] [PubMed] [Google Scholar]
- 36.Wirleitner B., Schroecksnadel K., Winkler C., Fuchs D. Neopterin in HIV-1 infection. Mol. Immunol. 2005;42:183–194. doi: 10.1016/j.molimm.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 37.Kuwata G., Hiramatsu K., Samejima K., Iwasaki K., Takahashi K., Koizumi K. Increase of N1, N12-diacetylspermine in tissues from colorectal cancer and its liver metastasis. J. Cancer Res. Clin. Oncol. 2013;139:925–932. doi: 10.1007/s00432-013-1405-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi Y., Sakaguchi K., Horio H., Hiramatsu K., Moriya S., Takahashi K. Urinary N1, N12-diacetylspermine is a non-invasive marker for the diagnosis and prognosis of non-small-cell lung cancer. Br. J. Cancer. 2015;113:1493–1501. doi: 10.1038/bjc.2015.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huber C., Batchelor J.R., Fuchs D., Hausen A., Lang A., Niederwieser D. Immune response-associated production of neopterin. Release from macrophages primarily under control of interferon-gamma. J. Exp. Med. 1984;160:310–316. doi: 10.1084/jem.160.1.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor M.W., Feng G.S. Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J. 1991;5:2516–2522. [PubMed] [Google Scholar]
- 41.Zhang M., Wang H., Tracey K.J. Regulation of macrophage activation and inflammation by spermine: a new chapter in an old story. Crit. Care Med. 2000;28:N60–6. doi: 10.1097/00003246-200004001-00007. [DOI] [PubMed] [Google Scholar]
- 42.Bordon Y. Inflammation: live long and prosper with Siglecs. Nat. Rev. Immunol. 2015;15:266–267. doi: 10.1038/nri3851. [DOI] [PubMed] [Google Scholar]
- 43.Langford-Smith A., Day A.J., Bishop P.N., Clark S.J. Complementing the sugar code: role of GAGs and sialic acid in complement regulation. Front. Immunol. 2015;6:25. doi: 10.3389/fimmu.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macauley M.S., Crocker P.R., Paulson J.C. Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 2014;14:653–666. doi: 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mulloy B., Rider C.C. Cytokines and proteoglycans: an introductory overview. Biochem. Soc. Trans. 2006;34:409–413. doi: 10.1042/BST0340409. [DOI] [PubMed] [Google Scholar]
- 46.Hotamisligil G.S. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 47.Anderson S.T., Kaforou M., Brent A.J., Wright V.J., Banwell C.M., Chagaluka G. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N. Engl. J. Med. 2014;370:1712–1723. doi: 10.1056/NEJMoa1303657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berry M.P., Graham C.M., Mcnab F.W., Xu Z., Bloch S.A., Oni T. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bloom C.I., Graham C.M., Berry M.P., Rozakeas F., Redford P.S., Wang Y. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS One. 2013;e70630:8. doi: 10.1371/journal.pone.0070630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adekambi T., Ibegbu C.C., Cagle S., Kalokhe A.S., Wang Y.F., Hu Y. Biomarkers on patient T cells diagnose active tuberculosis and monitor treatment response. J. Clin. Invest. 2015;125:1827–1838. doi: 10.1172/JCI77990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rozot V., Patrizia A., Vigano S., Mazza-Stalder J., Idrizi E., Day C.L. Combined use of Mycobacterium tuberculosis-specific CD4 and CD8 T-cell responses is a powerful diagnostic tool of active tuberculosis. Clin. Infect. Dis. 2015;60:432–437. doi: 10.1093/cid/ciu795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brennan P.J. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2003;83:91–97. doi: 10.1016/s1472-9792(02)00089-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material