Abstract
Mechanisms to elicit antiviral immunity, a natural host response to viral pathogen challenge, are of imminent relevance to cancer immunotherapy. ‘Oncolytic’ viruses, naturally existing or genetically engineered viral agents with cell type-specific propagation in malignant cells, were ostensibly conceived for their tumor cytotoxic properties. Yet, their true therapeutic value may rest in their ability to provoke antiviral signals that engage antitumor immune responses within the immunosuppressive tumor microenvironment. Coopting such ‘oncolytic’ viral agents to instigate antitumor immunity is not an easy feat. During co-evolution with their hosts, viruses acquired sophisticated strategies to block inflammatory signals, intercept innate antiviral interferon responses and prevent antiviral effector responses, e.g. by interfering with antigen presentation and T cell costimulation. The resulting struggle of host innate inflammatory and antiviral responses vs. viral immune evasion and suppression, determines the potential for antitumor immunity to occur. Moreover, paradigms of early host:virus relations established in normal immunocompetent organisms may not hold in the profoundly immunosuppressive tumor microenvironment. In this review, we explain the mechanisms of recombinant non-pathogenic poliovirus, PVSRIPO, which is currently in Phase-1 clinical trials against recurrent glioblastoma. We focus on an unusual host:virus relationship, defined by the simple and cytotoxic replication strategy of poliovirus, which generates inflammatory perturbations conducive to tumor antigen-specific immune priming.
Keywords: Poliovirus, internal ribosomal entry site, CD155, interferon, neutrophils, cancer immunotherapy
INTRODUCTION
Viruses, from a clinical translational perspective, are exceedingly complex biological agents, that unfold a range of disparate activities in their target tissues. The much-touted ability of many viruses to attach to, enter, replicate in, and kill high-passage clonal cancer cell lines in vitro often yields efficacy in immunocompromised rodent tumor models. These properties, embodied in the adjective ‘oncolytic’, do not constitute a sufficient rationale for clinical application. Rather, it is the full range of virus:host relations that determine the response to tumor targeting with viruses. Naturally, this range is not limited to unilateral (‘oncolytic’) viral tumor cytopathogenicity, but encompasses an elaborate, multilayered relationship with the host’s immune system. First and foremost, viruses administered to cancer patients encounter an onslaught of innate antiviral and host inflammatory processes that determine the virus’ fate and define the potential for therapeutic outcomes. The central questions addressed in this review are: (1) can safe viral agents be derived, whose range of actions is not thwarted by the innate antiviral response; and (2) can the interplay between viral tumor cytotoxicity and the resulting innate host inflammatory response generate conditions conducive to provoking antitumor immunity?
This review centers on PVSRIPO, the live attenuated (Sabin) type 1 poliovirus vaccine containing a foreign internal ribosomal entry site (IRES) of human rhinovirus type 2 (1). This IRES substitution in PVSRIPO is associated with profound neuro-attenuation, manifest as the inability of PVSRIPO to cause polio- or meningo-encephalomyelitis after intracerebral inoculation in old-world primates that serve as WHO-standard model organisms for studies of poliovirus neurovirulence (2), or in human subjects (3). This constitutes the basis for PVSRIPO biosafety. The primary rationale for pursuing PVSRIPO for clinical application is poliovirus’ intriguing tropism for CD155, an immune checkpoint molecule virtually universally expressed in malignant cells of solid neoplasia, as well as in myeloid and endothelial cells (4). Enteroviruses (poliovirus is their flagship member) have a peculiar, unusually simple relationship with the host innate antiviral interferon (IFN) response system. We believe that this is a decisive factor in the immunogenic mechanisms of PVSRIPO (5). Lastly, tumor infection with PVSRIPO elicits a range of acute inflammatory events leading to profuse immune cell invasion, resulting in an immune-engaged tumor microenvironment. These events resemble classic cascades of the inflammatory response to pathogen challenge and are highly desirable in the cancer immunotherapy context (6).
SAFETY OF THE RECOMBINANT POLIO:RHINOVIRUS CHIMERA, PVSRIPO
In humans, productive poliovirus replication is restricted to two sites: the gastrointestinal tract and its associated lymphatic structures, and medullary/spinal cord motor neurons (7, 8). These reservoirs for poliovirus propagation coincide with CD155 expression (9, 10). Poliovirus propagation in the gastrointestinal tract does not produce overt enteric pathology and is generally clinically silent; tropism for lower motor neurons is the reason for the pathognomonic signature of poliovirus -paralytic poliomyelitis- and an obvious grave concern for biosafety considerations.
Therefore, clinically safe, replication-competent polioviruses require complete ablation of their neurovirulent potential. The desirable safety characteristics for an ‘oncolytic’ poliovirus construct essentially are identical to the demands on a safe live-attenuated poliovirus vaccine: (1) retain undiminished propagation in neoplastic cells (or mitotically active gastrointestinal epithelial cells); (2) high level of intrinsic neuro-attenuation; (3) stability of the neuro-attenuated genotype. Neuro-attenuation of poliovirus has been famously achieved with the Sabin vaccine strains, which attain at least 2 of these 3 desirable characteristics (11). The poliovirus (Sabin) vaccines were derived through variable passage in diverse explant cell cultures of several old-world primate species and/or through selection with non-human primate neurovirulence testing (12). Sequencing of their genomes revealed a major safety mechanism of each of the 3 Sabin vaccine strains: single point mutations in a critical area of the viral IRES [stem loop domain V; (13)]. Incidences of poliovirus (Sabin) vaccine-associated poliomyelitis in vaccine recipients are associated with genetic reversion of these single point IRES mutations to the wild type sequence (14, 15).
The IRES is a highly structured, essential ~480 nucleotide (nt) feature in the 5′ untranslated region of the polioviral genome, responsible for attracting host translation factors and, thus, ribosomes to viral RNA (16, 17). Stem loop domain V, harboring all 3 attenuating poliovirus (Sabin) IRES point mutations, is the main structure involved in recruiting the ‘ribosome-adaptor’ of the mammalian translation apparatus, the translation initiation helicase composed of eukaryotic initiation factors 4G (eIF4G):eIF4A:eIF4B (18, 19). Neuro-attenuation of the poliovirus (Sabin) vaccine strains likely is due to cell type-specific defects in recruiting eIF4G:4A:4B and, hence, ribosomes in neurons (18–20). The extreme genetic austerity of poliovirus (a genome of ~7,400 nt encodes for 7 non-structural proteins involved in RNA replication) is the reason for an exceedingly simple life cycle. Poliovirus:host relations are shaped by the virus’ ability to direct protein synthesis at incoming genomes immediately upon host cell entry. Therefore, the capacity for instantly recruiting ribosomes to incoming viral RNA dictates all subsequent steps in the virus life cycle and explains the neuro-attenuation phenotype achieved by single IRES point mutations in the poliovirus (Sabin) vaccines.
In PVSRIPO, instead of the single attenuating point mutation at nt 480 in stem loop domain V of the IRES of its precursor poliovirus type 1 (Sabin), the entire IRES is swapped with its counterpart from human rhinovirus type 2. Compared to poliovirus type 1 (Sabin), this improves one and adds the 3rd demand on a safe, replication-competent poliovirus: (1) PVSRIPO retains high cytolytic potential in mitotically active cells, e.g. neoplastic cells, due to unhinged protein kinase C (PKC)-Raf-ERK1/2-MNK signaling to protein synthesis machinery (21–27); (2) the genetic footprint for neuro-attenuation is much sturdier and mediates an elevated level of ‘neuronal incompetence’ (28–31); (3) PVSRIPO exhibits genetic stability upon serial propagation, e.g. in intended target tissues for PVSRIPO (29) or during manufacturing through serial passage in Vero (Chlorocebus Sabaeus kidney) cells. The reason for PVSRIPO’s improved genetic stability is the distinct origin of its IRES. The type 1 poliovirus (Sabin) IRES acquired its artificial point mutation after selection in tissue culture/non-human primates (the reason for this is unknown); this mutation does not occur in circulating wild-type poliovirus strains, i.e. it has been selected against through thousands of years of poliovirus:human co-speciation and co-evolution. Thus, it represents an artificially imposed impediment to viral fitness that is subject to reversion. In contrast, PVSRIPO contains the intact, fully functional, evolutionarily optimized IRES of human rhinovirus type 2. The rhinoviral IRES in PVSRIPO functions precisely as it has evolved to, whereas the Sabin IRESes represent artificial, ‘tainted’ versions of their ideal form.
TARGETING THE CD155 IMMUNE CHECKPOINT
Of all aspects dictating the virus:host relationship, few are more important than those that determine host cell type-specific tropism, i.e. host cell receptors mediating viral attachment and entry. This is true for the subject of this review, poliovirus, whose status as one of the great scourges of humanity is due to its receptor choice. The limitations imposed on viruses by their target tropism inherently restrict their potential clinical utility, as viruses evolved to target host cell receptors in their preferred replication sites in their host’s organism rather than neoplastic disease.
Given the role of receptor specificity in determining host:virus interactions, it is intuitive that tropism is critical when attempting to target malignancy with viruses. The poliovirus receptor [PVR (32); a.k.a. Nectin-like molecule 5 (Necl5) (4); CD155 (33)] was discovered through a monoclonal antibody that blocks poliovirus infection of HeLa cells (34). CD155 is necessary and sufficient to convey susceptibility to poliovirus (32). Only humans and old-world primates express a functional poliovirus receptor and are susceptible to paralytic poliomyelitis (Chimpanzees are more susceptible than old world primate species that are phylogenetically more distant to humans); resistance of new world non-human primate species to poliovirus was linked to sequence differences in their CD155 gene (35). In addition to defining the essentially human-only host range, CD155 is the principal determinant of poliovirus’ tightly restricted tissue and cell tropism in the gastrointestinal tract and its associated lymphatic structure and in lower motor neurons (7, 8). In addition to these sites of active poliovirus propagation, CD155 was detected on cells of monocytic lineage (monocytes, macrophages, dendritic cells) (36–38) and on endothelial cells (39, 40). Both, monocytic lineage and endothelial cells are susceptible to poliovirus infection in vitro, but likely are not significant reservoirs for viral propagation in vivo. For example, studies in CD155-transgenic mice revealed high levels of CD155 expression in kidneys (41), likely due to the large endothelial compartment in this organ; yet, there is no poliovirus replication in the kidneys of CD155-transgenic mice, or infected chimpanzees (7, 8). A potential function for monocytic-lineage cell infection for poliovirus susceptibility or in the pathogenesis of paralytic poliomyelitis is unknown. Infection of CNS endothelial cells may facilitate crossing of the blood-brain-barrier and may be a prelude to CNS invasion (42).
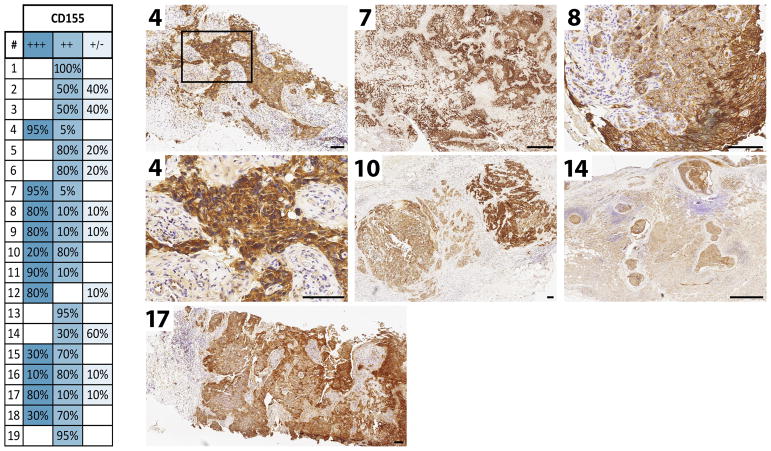
Amongst its relatives within the nectin/nectin-like family of cell adhesion molecules, CD155 stands out because of its intricate relation with cancer (4). CD155 expression is high in virtually all solid neoplasias (4), except Burkitt lymphoma, where CD155 is transcriptionally silenced, possibly due to Epstein Barr virus infection (43). We recently described validation of an immunohistochemistry protocol for reliable detection of CD155 expression in formaldehyde-fixed, paraffin-embedded tumor tissue sections [in glioblastoma; (9)]. Utilization of this assay in 19 samples of triple-negative breast cancer is shown in Figure 1. The mechanisms underlying CD155 upregulation in neoplasia are poorly understood, but have been linked to disruption of normal tissue architecture (44) or the DNA damage response (45). CD155 recently emerged as an immune checkpoint, due to its high-affinity binding to T cell immunoreceptor with Ig and ITIM domains (TIGIT) on natural killer- and T cells; its poorly understood physiological roles therefore may involve modulation of the immune response (46–50). Broadly upregulated CD155 expression in solid neoplasia means that neoplastic cells in such lesions are susceptible to PVSRIPO infection. While CD155 expression is categorically required for mediating tumor cell susceptibility to PVSRIPO, CD155 expression levels may not influence targeting by PVSRIPO (9), as the number of receptors required for productive infection of host cells likely is extremely low (51).
Figure 1.
Immunohistochemistry for the PV receptor (CD155) in 19 triple-negative breast cancer cases. For methods and detailed information on the assay, expression scoring, etc. see Chandramohan et al. (9). The table summarizes expression scores in the 19 cases. The numbered panels show IHC results for the corresponding cases in the table. Size bars represent 1 mm (7, 14) or 100 μm (4, 8, 10, 17).
THE INNATE ANTIVIRAL RESPONSE
Arguably, the most important first line of host defense against virus infection is the innate antiviral interferon (IFN) response (52). The appearance of viral signatures engages pattern recognition receptors, which activate a signaling network that results in transcriptional induction of type 1 IFNs (53). Type 1 IFNs, through autocrine and paracrine effects [mediated via the type 1 IFN receptor (IFNAR) and downstream JAK-STAT signaling cascades], set up an anti-viral state characterized by the induction of IFN-stimulated genes (ISG) (54).
Preclinical investigations of viral cancer-targeting agents frequently involve clonal high-passage cancer cell lines that either cannot produce or sense a type 1 IFN response, e.g. due to loss of expression of type 1 IFNs or IFNAR. This choice has been justified with claims that such phenotypes may epitomize the malignant state (55). There is much empirical evidence that refutes these -largely unsubstantiated- claims. Loss of type 1 IFN expression/IFN responsiveness is a well-known effect of tissue culture, even in non-cancerous cells. The Vero cell line, immortalized (non-transformed) Chlorocebus Sabaeus kidney cells, is a staple of viral vaccine manufacture (56). Vero cells cannot mount type 1 IFN responses upon virus infection (57), because they lost the type 1 IFN gene cluster during serial passage in tissue culture (58). Therefore, it cannot be assumed that high-passage clonal cancer cell lines accurately represent the IFN status of their tissue of origin.
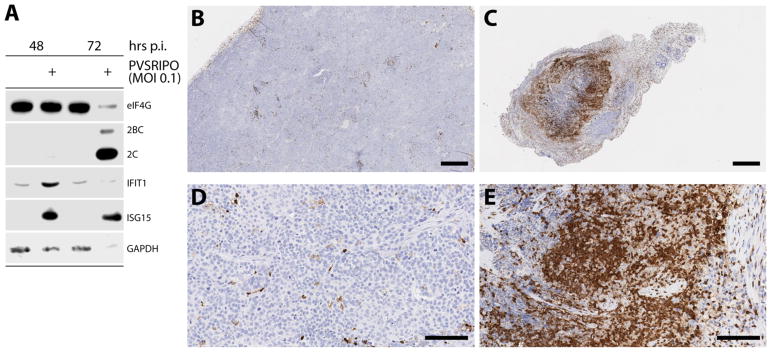
The catastrophic genetic heterogeneity of advanced, treatment-refractory cancers likely gives rise to subsets of tumor cells where the integrity of genes involved in the innate antiviral IFN response is lost. However, claiming the loss of IFN ‘competence’ as a defining characteristic of the malignant state is a gross misrepresentation. This is borne out in studies of the innate antiviral IFN response in primary explant patient tumor cells, prior to extended passage in tissue culture, e.g. in glioblastoma (59). Also, even many clonal high-passage cancer cell lines frequently exhibit normal type I IFN responses upon virus infection. A case in point is the SUM149 triple negative inflammatory breast cancer cell line used in our studies (Fig. 2) (6).
Figure 2.
PVSRIPO replication induces type I IFN responses and profound neutrophil invasion in a breast cancer xenotransplantation model (SUM149) (6). A. Infection of SUM149 (triple negative breast cancer) cells with PVSRIPO at a multiplicity of infection (MOI) of 0.1 causes prototypical signs of active viral translation (viral proteins 2BC/2C), viral cytopathogenic damage (eIF4G cleavage), and the type 1 IFN response (induction of ISGs IFIT1 and ISG15). At later intervals post infection (72 hrs), there is global loss of the proteome (see the GAPDH loading control) due to sweeping destruction of cancer cells. B–E. Immunohistochemistry for CD11b, a marker of myeloid cells, in SUM149 xenotransplanted tumors in athymic mice. These cells were determined to be largely neutrophils via flow cytometry [CD11b+, F4/80 negative, Ly6C+, Ly6G+ (6)]. Tumors in mock-treated animals (B; see detail at higher magnification in D) contain minimal neutrophil infiltrates. In contrast, PVSRIPO-treated tumors exhibit massive neutrophil invasion (C; see detail at higher magnification in E). For methods and detailed information on the assay, the xenotransplantation model, etc. see Holl et al. (6). Size bars represent 500 μm (B, C) or 100 μm (D, E).
The innate antiviral IFN response can represent a formidable barrier to achieving any biologically significant activities with viral agents, as it can thwart the infectious cascade prior to any such activities taking hold. However, inhibition or attenuation of antiviral IFN responses to overcome this barrier may not be therapeutically logical, as the immunotherapy benefit of oncolytic viruses may depend on such IFN responses (60). With PVSRIPO, we are exploiting the unique and remarkable relative IFN insensitivity of poliovirus. Poliovirus infection can proceed to full cytopathogenicity, even in cells that were pre-treated with type 1 IFNs and exhibit productive type 1 IFN responses (61). This is evident with PVSRIPO infection of SUM149 cells, where at multiplicities of infection of 0.1 (implying multiple rounds of infection in the culture in the presence of an active type 1 IFN response), complete cytolysis as indicated by eIF4G cleavage and decrease in total protein is achieved (Fig. 2) (6). As with other aspects of the poliovirus:host relationship, this phenotype is mostly due to the exceedingly simple (and cytotoxic) viral life cycle. The relative IFN-insensitivity phenotype of poliovirus was linked to the actions of a highly cytotoxic viral protease, 2Apro (61), which is responsible for cleavage of eIF4G in infected cells (Fig. 2) (62). Immediate early eIF4G cleavage upon poliovirus/PVSRIPO entry shuts off host protein synthesis, providing an enormous advantage to the virus, by eliminating host cell protein synthesis and favoring selective viral translation (62). This highly cytotoxic program can succeed in the presence of an active type 1 IFN response (61), explaining why with PVSRIPO, stimulation of the innate antiviral IFN response and tumor cell killing can occur in the same cell (Fig. 2). This means that upon PVSRIPO infection, the tumor cell proteome is released in the presence of potent pathogen-associated molecular patterns [PAMP; viral double-stranded RNA; (63)], danger-associated molecular patterns (DAMP) and an active type 1 IFN response (Fig. 2).
THE HOST INNATE INFLAMMATORY RESPONSE TO PVSRIPO TUMOR INFECTION
PVSRIPO must be inoculated intratumorally to unfold its range of activities leading up to antitumor immunity (64). Systemic administration of any tumor-targeting viral agent faces high hurdles, e.g. the blood brain barrier, circulating serum neutralizing antibodies, complement, a failure to reach the intended target and/or reach critical concentration in the intended target. Since all ‘oncolytic’ viruses are naturally devoid of pathogenicity or attenuated versions of pathogenic precursors, their ability to home to the intended target may be much reduced. After intratumoral virus administration: what are the host:virus interactions that determine the response to tumor infection?
As intimated above, the primary goal of oncolytic virus therapy is to generate new antitumor immune populations that persist after the virus is cleared from the host, thereby producing lasting antitumor immunity, capable of restricting tumor progression and distant metastatic lesions. Our empirical findings indicate that intratumoral infusion of PVSRIPO may elicit a highly intriguing mix of early innate antiviral activation, pro-inflammatory stimulation and immune cell invasion that may provide an ideal platform/context for generating antitumor immunity.
The initial cellular immune response to pathogen infection is mediated by neutrophils and macrophages, which directly destroy pathogens, kill pathogen-infected cells, and recruit antigen presenting cells and adaptive immune cells to generate anti-pathogen memory (65). Thus, phagocytes function not only as an effector arm of the immune system, but also a critical mediator of adaptive immune responses. Therefore, towards the development of productive 8 antitumor immunity, the engagement and activation of phagocytes holds potential to elicit multilayered innate and adaptive responses.
In breast and prostate cancer xenotransplantation models, single intratumoral administration of PVSRIPO led to rapid production of chemokines and cytokines one day after therapy (6). Analysis of tumor immune infiltrates following therapy revealed rampant neutrophil influx 1–2 days after treatment (Fig. 2) (6). This was reminiscent of immune cell infiltrations that had been described, but not defined, in earlier studies of PVSRIPO treatment in mouse xenotransplantation models with human malignant glioma models (27, 29, 66). Neutrophil tumor invasion was associated with the production of type I IFNs and markers of neutrophil activity including myeloperoxidase (MPO), H2O2, and inducible nitric oxid synthase (iNOS) (6).
Intriguingly, the response to PVSRIPO tumor infection was dominated by neutrophils and not macrophages; which may indicate a DAMP response (67), along with pro-inflammatory cytokines/chemokines that recruit neutrophils and induce endothelial adhesion proteins involved in leukocyte transendothelial migration. Indeed, several key chemokines known to recruit neutrophils (e.g. CCL3, CCL4, CCL5, CSF3, IL-6, etc.) were observed in PVSRIPO-treated xenografts (6). The source of these cytokines is the subject of ongoing investigations; however, it is likely that tissue-resident macrophages and endothelial cells likely participate in neutrophil recruitment (68).
Neutrophil invasion in the respiratory tract is a hallmark of rhinovirus infection (69–74) (the histopathologic and inflammatory response to poliovirus infection in the gastrointestinal tract is unknown). Rhinoviruses and polioviruses are part of the Enterovirus genus in the Picornavirus family; the recombinant IRES in PVSRIPO is that of human rhinovirus type 2. Since neutrophil invasion of the respiratory tract occurs with natural infection, direct administration of large quantities of virus directly into tumors, as in our pre-clinical rodent tumor models (Fig. 2) (6), where susceptible host cells abound, may exacerbate the inflammatory response and neutrophil infiltration.
Such neutrophil engagement may lead to direct tumor cell killing via the release of cytotoxic proteins and reactive oxygen species as well as indirect killing through the production of TNF-α (6). Perhaps more importantly, neutrophils may be involved in mediating a transition to adaptive antitumor immunity following PVSRIPO therapy through the production of pro-inflammatory cytokines (75).
CONCLUSIONS
Viruses could play a valuable role in cancer immunotherapy regimens in the future. This will require diverting focus away from attempts to maximize cytotoxicity in malignant cells, which defined the field for decades, and better efforts to refine our understanding of the host:virus immune interface. Intricate host innate antiviral response systems compete with elaborate immune evasion and suppression strategies adopted by viruses to out-smart their hosts. Developing efficacious viral cancer immunotherapy approaches will only be possible when fully taking this complexity into account. In the emerging cancer immunotherapy landscape, viral targeting strategies likely will need to be combined with other modalities. Immunogenic mechanisms outlined in this review, e.g. eliciting type 1 IFN responses in infected cancer cells, or recruiting immune cell invasion into notoriously immunologically ‘cold’ neoplasias, are sought-after principles with broad appeal for combination with other forms of immunotherapy.
Acknowledgments
We thank Eda K. Holl, Sarah Sammons, Kimberly L Blackwell and E. Shelley Hwang for their help with procuring and processing patient breast cancer tissues; Eda K. Holl for her work on rodent breast cancer models; Vidya Chandramohan, Jeffrey D. Bryant, Roger E. McLendon and Darell D. Bigner for their work on the immunohistochemistry of CD155 and contributions to developing PVSRIPO; and Michael C. Brown for editing this text. This work was supported by PHS grants CA124756 and CA190991 (MG), grants from the Lefkofsky Family Foundation and from Hope & Gavin Wolfe (MG); Department of Defense (DoD) breast cancer research program (BCRP) award W81XWH-16-1-0354 (SKN) and discretionary support from the Department of Surgery (SKN).
Footnotes
DISCLOSURE STATEMENT
MG and SKN own intellectual property related to this research, which has been licensed to Istari Oncology, Inc. MG is a co-founder and equity holder in Istari Oncology, Inc.
LITERATURE CITED
- 1.Gromeier M, Alexander L, Wimmer E. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc Natl Acad Sci U S A. 1996;93:2370–5. doi: 10.1073/pnas.93.6.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dobrikova EY, Goetz C, Walters RW, Lawson SK, Peggins JO, Muszynski K, Ruppel S, Poole K, Giardina SL, Vela EM, Estep JE, Gromeier M. Attenuation of neurovirulence, biodistribution, and shedding of a poliovirus:rhinovirus chimera after intrathalamic inoculation in Macaca fascicularis. J Virol. 2012;86:2750–9. doi: 10.1128/JVI.06427-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desjardins A, Sampson JH, Peters K, Ranjan T, Vlahovic G, Threatt S, Herndon JI, Boulton S, Lally-Goss D, McSherry F, Friedman A, Friedman H, Bigner D, Gromeier M. Phase I study of an Oncolytic polio/rhinovirus recombinant (PVSRIPO) against recurrent glioblastoma. SNO Annual Meeting; San Francisco, CA: Neuro-Oncology; 2013. p. 107. [Google Scholar]
- 4.Takai Y, Miyoshi J, Ikeda W, Ogita H. Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat Rev Mol Cell Biol. 2008;9:603–15. doi: 10.1038/nrm2457. [DOI] [PubMed] [Google Scholar]
- 5.Brown MC, Gromeier M. Cytotoxic and immunogenic mechanisms of recombinant oncolytic poliovirus. Curr Opin Virol. 2015;13:81–5. doi: 10.1016/j.coviro.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holl EK, Brown MC, Boczkowski D, McNamara MA, George DJ, Bigner DD, Gromeier M, Nair SK. Recombinant oncolytic poliovirus, PVSRIPO, has potent cytotoxic and innate inflammatory effects, mediating therapy in human breast and prostate cancer xenograft models. Oncotarget. 2016;7:79828–41. doi: 10.18632/oncotarget.12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bodian D. Emerging concept of poliomyelitis infection. Science. 1955;122:105–8. doi: 10.1126/science.122.3159.105. [DOI] [PubMed] [Google Scholar]
- 8.Sabin AB. Pathogenesis of poliomyelitis; reappraisal in the light of new data. Science. 1956;123:1151–7. doi: 10.1126/science.123.3209.1151. [DOI] [PubMed] [Google Scholar]
- 9.Chandramohan V, Bryant J, Piao H, Keir S, Lipp E, Lefaivre M, Perkinson K, Bigner DD, Gromeier M, McLendon RE. Validation of an Immunohistochemistry Assay for Detection of CD155, the Poliovirus Receptor, in Malignant Gliomas. Archives of Pathology and Laboratory Medicine. 2017 doi: 10.5858/arpa.2016-0580-OA. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwasaki A, Welker R, Mueller S, Linehan M, Nomoto A, Wimmer E. Immunofluorescence analysis of poliovirus receptor expression in Peyer’s patches of humans, primates, and CD155 transgenic mice: implications for poliovirus infection. J Infect Dis. 2002;186:585–92. doi: 10.1086/342682. [DOI] [PubMed] [Google Scholar]
- 11.Sabin AB, Boulger LR. History of Sabin attenuated poliovirus oral live vaccine strains. Journal of Biological Standardization. 1973;1:115–8. [Google Scholar]
- 12.Sabin AB. Properties and behavior of orally administered attenuated poliovirus vaccine. J Am Med Assoc. 1957;164:1216–23. doi: 10.1001/jama.1957.62980110008008. [DOI] [PubMed] [Google Scholar]
- 13.Wimmer E, Hellen CU, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 14.Evans DM, Dunn G, Minor PD, Schild GC, Cann AJ, Stanway G, Almond JW, Currey K, Maizel JV., Jr Increased neurovirulence associated with a single nucleotide change in a noncoding region of the Sabin type 3 poliovaccine genome. Nature. 1985;314:548–50. doi: 10.1038/314548a0. [DOI] [PubMed] [Google Scholar]
- 15.Georgescu MM, Balanant J, Macadam A, Otelea D, Combiescu M, Combiescu AA, Crainic R, Delpeyroux F. Evolution of the Sabin type 1 poliovirus in humans: characterization of strains isolated from patients with vaccine-associated paralytic poliomyelitis. J Virol. 1997;71:7758–68. doi: 10.1128/jvi.71.10.7758-7768.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–43. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–5. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 18.de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CU. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc Natl Acad Sci U S A. 2009;106:9197–202. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sweeney TR, Abaeva IS, Pestova TV, Hellen CU. The mechanism of translation initiation on Type 1 picornavirus IRESs. EMBO J. 2014;33:76–92. doi: 10.1002/embj.201386124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ochs K, Zeller A, Saleh L, Bassili G, Song Y, Sonntag A, Niepmann M. Impaired binding of standard initiation factors mediates poliovirus translation attenuation. J Virol. 2003;77:115–22. doi: 10.1128/JVI.77.1.115-122.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown MC, Bryant JD, Dobrikova EY, Shveygert M, Bradrick SS, Chandramohan V, Bigner DD, Gromeier M. Induction of viral, 7-methyl-guanosine cap-independent translation and oncolysis by mitogen-activated protein kinase-interacting kinase-mediated effects on the serine/arginine-rich protein kinase. J Virol. 2014;88:13135–48. doi: 10.1128/JVI.01883-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown MC, Dobrikov MI, Gromeier M. Mitogen-activated protein kinase-interacting kinase regulates mTOR/AKT signaling and controls the serine/arginine-rich protein kinase-responsive type 1 internal ribosome entry site-mediated translation and viral oncolysis. J Virol. 2014;88:13149–60. doi: 10.1128/JVI.01884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown MC, Gromeier M. Oncolytic immunotherapy through tumor-specific translation and cytotoxicity of poliovirus. Discov Med. 2015;19:359–65. [PMC free article] [PubMed] [Google Scholar]
- 24.Brown MC, Gromeier M. MNK Controls mTORC1:Substrate Association through Regulation of TELO2 Binding with mTORC1. Cell Rep. 2017;18:1444–57. doi: 10.1016/j.celrep.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dobrikov M, Dobrikova E, Shveygert M, Gromeier M. Phosphorylation of eukaryotic translation initiation factor 4G1 (eIF4G1) by protein kinase C{alpha} regulates eIF4G1 binding to Mnk1. Mol Cell Biol. 2011;31:2947–59. doi: 10.1128/MCB.05589-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobrikov MI, Dobrikova EY, Gromeier M. Dynamic Regulation of the Translation Initiation Helicase Complex by Mitogenic Signal Transduction to Eukaryotic Translation Initiation Factor 4G. Mol Cell Biol. 2013;33:937–46. doi: 10.1128/MCB.01441-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goetz C, Everson RG, Zhang LC, Gromeier M. MAPK signal-integrating kinase controls cap-independent translation and cell type-specific cytotoxicity of an oncolytic poliovirus. Mol Ther. 2010;18:1937–46. doi: 10.1038/mt.2010.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell SA, Lin J, Dobrikova EY, Gromeier M. Genetic determinants of cell type-specific poliovirus propagation in HEK 293 cells. J Virol. 2005;79:6281–90. doi: 10.1128/JVI.79.10.6281-6290.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dobrikova EY, Broadt T, Poiley-Nelson J, Yang X, Soman G, Giardina S, Harris R, Gromeier M. Recombinant oncolytic poliovirus eliminates glioma in vivo without genetic adaptation to a pathogenic phenotype. Mol Ther. 2008;16:1865–72. doi: 10.1038/mt.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merrill MK, Dobrikova EY, Gromeier M. Cell-type-specific repression of internal ribosome entry site activity by double-stranded RNA-binding protein 76. J Virol. 2006;80:3147–56. doi: 10.1128/JVI.80.7.3147-3156.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merrill MK, Gromeier M. The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the rhinovirus type 2 internal ribosome entry site. J Virol. 2006;80:6936–42. doi: 10.1128/JVI.00243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendelsohn CL, Wimmer E, Racaniello VR. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56:855–65. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 33.Kishimoto T, Kikutani H, Von dem Borne AEGK, et al., editors. Leucocyte typing VI: white cell differentiation antigens. New York: Garland Publishing Inc; 1997. [Google Scholar]
- 34.Nobis P, Zibirre R, Meyer G, Kuhne J, Warnecke G, Koch G. Production of a monoclonal antibody against an epitope on HeLa cells that is the functional poliovirus binding site. J Gen Virol. 1985;66(Pt 12):2563–9. doi: 10.1099/0022-1317-66-12-2563. [DOI] [PubMed] [Google Scholar]
- 35.Khan S, Peng X, Yin J, Zhang P, Wimmer E. Characterization of the New World monkey homologues of human poliovirus receptor CD155. J Virol. 2008;82:7167–79. doi: 10.1128/JVI.02664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freistadt MS, Eberle KE. Hematopoietic cells from CD155-transgenic mice express CD155 and support poliovirus replication ex vivo. Microb Pathog. 2000;29:203–12. doi: 10.1006/mpat.2000.0386. [DOI] [PubMed] [Google Scholar]
- 37.Freistadt MS, Fleit HB, Wimmer E. Poliovirus receptor on human blood cells: a possible extraneural site of poliovirus replication. Virology. 1993;195:798–803. doi: 10.1006/viro.1993.1433. [DOI] [PubMed] [Google Scholar]
- 38.Wahid R, Cannon MJ, Chow M. Dendritic cells and macrophages are productively infected by poliovirus. J Virol. 2005;79:401–9. doi: 10.1128/JVI.79.1.401-409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manes TD, Pober JS. Identification of endothelial cell junctional proteins and lymphocyte receptors involved in transendothelial migration of human effector memory CD4+ T cells. J Immunol. 2011;186:1763–8. doi: 10.4049/jimmunol.1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reymond N, Imbert AM, Devilard E, Fabre S, Chabannon C, Xerri L, Farnarier C, Cantoni C, Bottino C, Moretta A, Dubreuil P, Lopez M. DNAM-1 and PVR regulate monocyte migration through endothelial junctions. J Exp Med. 2004;199:1331–41. doi: 10.1084/jem.20032206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren S, Rollins BJ. Cyclin C/cdk3 promotes Rb-dependent G0 exit. Cell. 2004;117:239–51. doi: 10.1016/s0092-8674(04)00300-9. [DOI] [PubMed] [Google Scholar]
- 42.Blinzinger K, Simon J, Magrath D, Boulger L. Poliovirus crystals within the endoplasmic reticulum of endothelial and mononuclear cells in the monkey spinal cord. Science. 1969;163:1336–7. doi: 10.1126/science.163.3873.1336. [DOI] [PubMed] [Google Scholar]
- 43.Solecki D, Schwarz S, Wimmer E, Lipp M, Bernhardt G. The promoters for human and monkey poliovirus receptors. Requirements for basic and cell type-specific activity. J Biol Chem. 1997;272:5579–86. doi: 10.1074/jbc.272.9.5579. [DOI] [PubMed] [Google Scholar]
- 44.Erickson BM, Thompson NL, Hixson DC. Tightly regulated induction of the adhesion molecule necl-5/CD155 during rat liver regeneration and acute liver injury. Hepatology. 2006;43:325–34. doi: 10.1002/hep.21021. [DOI] [PubMed] [Google Scholar]
- 45.Cerboni C, Fionda C, Soriani A, Zingoni A, Doria M, Cippitelli M, Santoni A. The DNA Damage Response: A Common Pathway in the Regulation of NKG2D and DNAM-1 Ligand Expression in Normal, Infected, and Cancer Cells. Front Immunol. 2014;4:508. doi: 10.3389/fimmu.2013.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blake SJ, Stannard K, Liu J, Allen S, Yong MC, Mittal D, Aguilera AR, Miles JJ, Lutzky VP, de Andrade LF, Martinet L, Colonna M, Takeda K, Kuhnel F, Gurlevik E, Bernhardt G, Teng MW, Smyth MJ. Suppression of Metastases Using a New Lymphocyte Checkpoint Target for Cancer Immunotherapy. Cancer Discov. 2016;6:446–59. doi: 10.1158/2159-8290.CD-15-0944. [DOI] [PubMed] [Google Scholar]
- 47.Chan CJ, Martinet L, Gilfillan S, Souza-Fonseca-Guimaraes F, Chow MT, Town L, Ritchie DS, Colonna M, Andrews DM, Smyth MJ. The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions. Nat Immunol. 2014;15:431–8. doi: 10.1038/ni.2850. [DOI] [PubMed] [Google Scholar]
- 48.Dougall WC, Kurtulus S, Smyth MJ, Anderson AC. TIGIT and CD96: new checkpoint receptor targets for cancer immunotherapy. Immunol Rev. 2017;276:112–20. doi: 10.1111/imr.12518. [DOI] [PubMed] [Google Scholar]
- 49.Martinet L, Smyth MJ. Balancing natural killer cell activation through paired receptors. Nat Rev Immunol. 2015;15:243–54. doi: 10.1038/nri3799. [DOI] [PubMed] [Google Scholar]
- 50.Stanietsky N, Simic H, Arapovic J, Toporik A, Levy O, Novik A, Levine Z, Beiman M, Dassa L, Achdout H, Stern-Ginossar N, Tsukerman P, Jonjic S, Mandelboim O. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc Natl Acad Sci U S A. 2009;106:17858–63. doi: 10.1073/pnas.0903474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Solecki D, Gromeier M, Harber J, Bernhardt G, Wimmer E. Poliovirus and its cellular receptor: a molecular genetic dissection of a virus/receptor affinity interaction. J Mol Recognit. 1998;11:2–9. doi: 10.1002/(SICI)1099-1352(199812)11:1/6<2::AID-JMR380>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 52.Katze MG, He Y, Gale M., Jr Viruses and interferon: a fight for supremacy. Nat Rev Immunol. 2002;2:675–87. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 53.Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–45. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stojdl DF, Lichty B, Knowles S, Marius R, Atkins H, Sonenberg N, Bell JC. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat Med. 2000;6:821–5. doi: 10.1038/77558. [DOI] [PubMed] [Google Scholar]
- 56.Barrett PN, Mundt W, Kistner O, Howard MK. Vero cell platform in vaccine production: moving towards cell culture-based viral vaccines. Expert Rev Vaccines. 2009;8:607–18. doi: 10.1586/erv.09.19. [DOI] [PubMed] [Google Scholar]
- 57.Desmyter J, Melnick JL, Rawls WE. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero) J Virol. 1968;2:955–61. doi: 10.1128/jvi.2.10.955-961.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Osada N, Kohara A, Yamaji T, Hirayama N, Kasai F, Sekizuka T, Kuroda M, Hanada K. The genome landscape of the african green monkey kidney-derived vero cell line. DNA Res. 2014;21:673–83. doi: 10.1093/dnares/dsu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Glas M, Coch C, Trageser D, Dassler J, Simon M, Koch P, Mertens J, Quandel T, Gorris R, Reinartz R, Wieland A, Von Lehe M, Pusch A, Roy K, Schlee M, Neumann H, Fimmers R, Herrlinger U, Brustle O, Hartmann G, Besch R, Scheffler B. Targeting the cytosolic innate immune receptors RIG-I and MDA5 effectively counteracts cancer cell heterogeneity in glioblastoma. Stem Cells. 2013;31:1064–74. doi: 10.1002/stem.1350. [DOI] [PubMed] [Google Scholar]
- 60.Zamarin D, Holmgaard RB, Subudhi SK, Park JS, Mansour M, Palese P, Merghoub T, Wolchok JD, Allison JP. Localized oncolytic virotherapy overcomes systemic tumor resistance to immune checkpoint blockade immunotherapy. Sci Transl Med. 2014;6:226ra32. doi: 10.1126/scitranslmed.3008095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morrison JM, Racaniello VR. Proteinase 2Apro is essential for enterovirus replication in type I interferon-treated cells. J Virol. 2009;83:4412–22. doi: 10.1128/JVI.02177-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Etchison D, Milburn SC, Edery I, Sonenberg N, Hershey JW. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem. 1982;257:14806–10. [PubMed] [Google Scholar]
- 63.Richards OC, Martin SC, Jense HG, Ehrenfeld E. Structure of poliovirus replicative intermediate RNA. Electron microscope analysis of RNA cross-linked in vivo with psoralen derivative. J Mol Biol. 1984;173:325–40. doi: 10.1016/0022-2836(84)90124-4. [DOI] [PubMed] [Google Scholar]
- 64.Brown MC, Dobrikova EY, Dobrikov MI, Walton RW, Gemberling SL, Nair SK, Desjardins A, Sampson JH, Friedman HS, Friedman AH, Tyler DS, Bigner DD, Gromeier M. Oncolytic polio virotherapy of cancer. Cancer. 2014;120:3277–86. doi: 10.1002/cncr.28862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. 2013;13:159–75. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- 66.Gromeier M, Lachmann S, Rosenfeld MR, Gutin PH, Wimmer E. Intergeneric poliovirus recombinants for the treatment of malignant glioma. Proc Natl Acad Sci U S A. 2000;97:6803–8. doi: 10.1073/pnas.97.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pittman K, Kubes P. Damage-associated molecular patterns control neutrophil recruitment. J Innate Immun. 2013;5:315–23. doi: 10.1159/000347132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim ND, Luster AD. The role of tissue resident cells in neutrophil recruitment. Trends Immunol. 2015;36:547–55. doi: 10.1016/j.it.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fleming HE, Little FF, Schnurr D, Avila PC, Wong H, Liu J, Yagi S, Boushey HA. Rhinovirus-16 colds in healthy and in asthmatic subjects: similar changes in upper and lower airways. Am J Respir Crit Care Med. 1999;160:100–8. doi: 10.1164/ajrccm.160.1.9808074. [DOI] [PubMed] [Google Scholar]
- 70.Graham D, Henderson F, House D. Neutrophil influx measured in nasal lavages of humans exposed to ozone. Arch Environ Health. 1988;43:228–33. doi: 10.1080/00039896.1988.9934938. [DOI] [PubMed] [Google Scholar]
- 71.Jarjour NN, Gern JE, Kelly EA, Swenson CA, Dick CR, Busse WW. The effect of an experimental rhinovirus 16 infection on bronchial lavage neutrophils. J Allergy Clin Immunol. 2000;105:1169–77. doi: 10.1067/mai.2000.106376. [DOI] [PubMed] [Google Scholar]
- 72.Mallia P, Message SD, Contoli M, Gray KK, Telcian A, Laza-Stanca V, Papi A, Stanciu LA, Elkin S, Kon OM, Johnson M, Johnston SL. Neutrophil adhesion molecules in experimental rhinovirus infection in COPD. Respir Res. 2013;14:72. doi: 10.1186/1465-9921-14-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Naclerio RM, Proud D, Lichtenstein LM, Kagey-Sobotka A, Hendley JO, Sorrentino J, Gwaltney JM. Kinins are generated during experimental rhinovirus colds. J Infect Dis. 1988;157:133–42. doi: 10.1093/infdis/157.1.133. [DOI] [PubMed] [Google Scholar]
- 74.Zhu J, Message SD, Qiu Y, Mallia P, Kebadze T, Contoli M, Ward CK, Barnathan ES, Mascelli MA, Kon OM, Papi A, Stanciu LA, Jeffery PK, Johnston SL. Airway inflammation and illness severity in response to experimental rhinovirus infection in asthma. Chest. 2014;145:1219–29. doi: 10.1378/chest.13-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Deniset JF, Kubes P. Recent advances in understanding neutrophils. F1000Res. 2016;5:2912. doi: 10.12688/f1000research.9691.1. [DOI] [PMC free article] [PubMed] [Google Scholar]