Abstract
Background
Novel non-invasive biomarkers for gastric cancer (GC) are needed, because the present diagnostic methods for GC are either invasive or insensitive and non-specific in clinic. The presence of stable circulating microRNAs (miRNAs) in plasma suggested a promising role as GC biomarkers.
Methods
Based on the quantitative droplet digital PCR (ddPCR), four miRNAs (miR-21, miR-93, miR-106a and miR-106b) related to the presence of GC were identified in plasma from a training cohort of 147 participants and a validation cohort of 28 participants.
Results
All circulating miRNA levels were significantly higher in the plasma of GC patients compared to healthy controls (P < 0.05). Through a combination of four miRNAs by logistic regression model, receiver operating characteristic (ROC) analyses yielded the highest AUC value of 0.887 in discriminating GC patients from healthy volunteers. Furthermore, miR-21, miR-93 and miR-106b levels were significantly related to an advanced TNM stage in GC patients. ROC analyses of the combined miRNA panel also showed the highest AUC value of 0.809 in discriminating GC patients with TNM stage I and II from stage III and IV. Through combining four miRNAs and clinical parameters, a classical random forest model was established in the training stage. In the validation cohort, it correctly discriminated 23 out of 28 samples in the blinded phase (false rate, 17.8%).
Conclusions
Using the ddPCR technique, circulating miR-21, miR-93, miR-106a and miR-106b could be used as diagnostic plasma biomarkers in gastric cancer patients.
Electronic supplementary material
The online version of this article (10.1186/s12885-018-4601-5) contains supplementary material, which is available to authorized users.
Keywords: Gastric cancer, Liquid biopsy, ddPCR, miR-21, miR-93, miR-106a, miR-106b
Background
Gastric cancer is second most common cancer in terms of incidence and mortality in China, according to the most recent cancer statistics [1]. With the improvement of surgical technique, radiotherapy and chemotherapy in recent years, patients in the early stage of GC had a significant increased 5-year survival rate, but the prognosis for advanced GC remains poor [2, 3]. Thus, it is important to diagnose GC in the early stage thus yielding better outcome. Gastroscopy is the gold standard test for GC diagnosis, but it is invasive and couldn’t be frequently used as regular health examination. Carcinoembryonic antigen (CEA), α-fetoprotein (AFP) and carbohydrate antigen 19–9 (CA19–9) are widely used as non-invasive markers in clinical, but their sensitivities and specificities are not enough for early diagnosis of GC [4]. Therefore, novel non-invasive biomarkers with better sensitivities and specificities are urgently needed.
MicroRNAs (miRNAs) are small noncoding RNAs, about 22–24 bases long, that inhibit their target mRNAs translation by inducing mRNA degradation or translational repression [5, 6]. Up to now, there are thousands of miRNAs have been reported to be associated with tumor growth, invasion, metastasis and apoptosis [7]. Several studies have demonstrated that circulating miRNAs can serve as biomarkers for GC diagnosis. For example, miR-223, miR-16 and miR-100 were highly expressed in the serum of GC patients, and positively associated with TNM stage, metastatic status, tumor size and differentiation grade [8]. The level of let-7a expression in the plasma of GC patients was significant lower, and the value of the area under the receiver-operating characteristic curve was 0.879 for the miR-106a/let-7a ratio in GC patients and healthy volunteers [9]. Thus, miRNAs in peripheral blood have great potential for helping early diagnosis of GC.
Although the results of previous studies are promising, their clinical transferability remains uncertain, which mainly due to the lack of uniformity and reproducibility in the criteria for determining the circulating miRNA levels by quantitative real-time PCR (qPCR). Besides, several variables such as sample storage, RNA isolation, PCR inhibitors and normalization could affect final results [10]. The droplet digital PCR (ddPCR) technique is increasingly considered to be the gold standard in the application of liquid biopsy, because it has shown superior precision and sensitivity, being less affected by PCR inhibitors, and unnecessity of internal/external normalization while detecting low concentration of target nucleic acids molecules [11, 12]. In this study, we used the ddPCR technique to explore the circulating miRNA signatures which could be potential biomarkers for GC diagnosis, and discriminating GC patients with different TNM stage. Four miRNAs, miR-21, miR-93, miR-106a and miR-106b, which have been most reported to be closely correlated with GC in tissue and plasma of patients and represent as candidate biomarkers for human GC, were examined by novel technique of ddPCR. [9, 13–15].
Furthermore, without the assist of tissue biopsy and imaging examinations, it would be difficult for clinicians to make diagnosis and tumor staging for GC, because there are many factors could probably influence the results. To improve the precision and accuracy of diagnosing disease, new approaches such as machine learning which is the main technical basis for data mining, provide an effective solution [16]. Several studies have been reported to use machine learning tools for data mining to diagnose disease or predict prognosis [17–19]. In this study, we explored the use of random forest model based learning for GC diagnosis, by using circulating miRNA expressions and clinical parameters such as age, gender, CEA and CA19–9.
Methods
Patients and blood samples
The present study was approved by the ethics committee of Sichuan Provincial People’s Hospital. All participants provided written informed consent form to approve the use of their blood samples for research purposes.
From Sichuan Provincial People’s Hospital, a total of 101 patients with gastric cancer (GC) and 46 healthy volunteers were recruited to the training cohort between January 2017 and June 2017, and a total of 11 patients with GC and 17 healthy volunteers were recruited to the validation cohort between December 2017 and February 2018. For plasma, 5 ml peripheral blood was collected in EDTA tubes, the sampling time was pre-surgery for GC patients, especially. And within 2 h, plasma was separated by centrifugation at 2000×g for 10 min, the supernatant was followed by a second centrifugation at 12000×g for 20 min. Then, the plasma was either stored at − 80 °C or miRNA was extracted immediately.
For patients, GC paraffin-embedded tissue samples were obtained after surgical resection. The clinicopathological classification and staging were determined according to the World Health Organization pathological classification of tumors. The clinical information for GC patients in the training stage is summarized in Table 1. Among the 101 patients included 51 male and 50 female, the median age was 56 years old (range, 35–75 years) and the median tumor size was 3.9 cm (range, 1.0–7.5 cm). There were 16 cases well differentiated, 35 were moderately differentiated and 50 were poorly differentiated. There were 35 cases without lymph node metastasis, 66 cases with lymph node metastasis, 18 cases with distant metastasis and 83 cases without distant metastasis. According to TNM stage classification, 28 cases were categorized as stage I, 13 cases for stage II, 36 cases for stage III and 24 cases for stage IV.
Table 1.
Clinicopathological characteristics of all individuals in the training stage and relationships with circulating miRNAs in the plasma
| Characteristics | miR-21 | miR-93 | miR-106a | miR-106b | |||||
|---|---|---|---|---|---|---|---|---|---|
| Total (%) | Mean ± SD | P value | Mean ± SD | P value | Mean ± SD | P value | Mean ± SD | P value | |
| GC patients | |||||||||
| Gender | |||||||||
| Male | 51(50.5) | 300.2 ± 127.7 | 0.437 | 206.3 ± 87.8 | 0.318 | 41.3 ± 18.7 | 0.404 | 26.3 ± 11.8 | 0.353 |
| Female | 50(49.5) | 300.8 ± 187.5 | 188.6 ± 70.5 | 37.7 ± 16.1 | 24.0 ± 11.1 | ||||
| Age (years) | |||||||||
| ≥ 60 | 45(44.6) | 303.4 ± 187.7 | 0.562 | 203.5 ± 87.0 | 0.696 | 36.9 ± 16.4 | 0.159 | 22.7 ± 11.2 | 0.019* |
| < 60 | 56(55.4) | 298.1 ± 133.9 | 192.7 ± 73.9 | 41.6 ± 18.2 | 27.1 ± 11.4 | ||||
| Tumor size (cm) | |||||||||
| ≥ 5 | 29(28.7) | 358.8 ± 183.5 | 0.029* | 198.9 ± 63.9 | 0.684 | 43.0 ± 14.9 | 0.071 | 26.5 ± 8.8 | 0.165 |
| < 5 | 72(71.3) | 277.0 ± 143.2 | 196.9 ± 85.8 | 38.1 ± 18.3 | 24.6 ± 12.4 | ||||
| Differentiation | |||||||||
| Well | 16(15.8) | 296.4 ± 158.4 | 0.078 | 200.9 ± 86.1 | 0.976 | 39.7 ± 17.4 | 0.601 | 26.7 ± 13.1 | 0.891 |
| Moderate | 35(34.7) | 353.8 ± 187.5 | 196.0 ± 69.2 | 37.9 ± 18.8 | 25.4 ± 12.1 | ||||
| Poor | 50(49.5) | 264.5 ± 127.9 | 197.5 ± 86.0 | 40.6 ± 16.1 | 24.5 ± 10.6 | ||||
| Lymph node metastasis | |||||||||
| Positive | 66(65.3) | 329.1 ± 172.3 | 0.014* | 210.8 ± 80.7 | 0.019* | 40.1 ± 16.7 | 0.438 | 27.9 ± 11.2 | 0.0002* |
| Negative | 35(34.7) | 246.5 ± 115.4 | 172.4 ± 72.7 | 38.5 ± 19.0 | 20.0 ± 10.1 | ||||
| Distant metastasis | |||||||||
| Positive | 18(21.7) | 304.1 ± 122.3 | 0.435 | 218.8 ± 91.4 | 0.371 | 41.4 ± 15.1 | 0.297 | 25.6 ± 10.5 | 0.572 |
| Negative | 83(78.3) | 299.7 ± 166.8 | 192.9 ± 76.9 | 39.1 ± 18.0 | 25.1 ± 11.7 | ||||
| TNM stage | |||||||||
| I | 28(27.7) | 234 ± 119.7 | 0.0062* | 175.8 ± 78.3 | 0.0571 | 37 ± 19.4 | 0.7371 | 19.3 ± 9 | 0.0016* |
| II | 13(12.9) | 241.8 ± 101.5 | 182.5 ± 84.6 | 40.9 ± 15.6 | 21.7 ± 10.9 | ||||
| III | 36(35.6) | 329.5 ± 173.2 | 195.9 ± 66.1 | 39.2 ± 17.7 | 27.8 ± 11.1 | ||||
| IV | 24(23.8) | 366.2 ± 171.2 | 233.4 ± 89.7 | 42.3 ± 16.4 | 29.9 ± 12 | ||||
| Healthy voluteers | |||||||||
| Gender | |||||||||
| Male | 26(56.5) | 142.8 ± 62.8 | 0.475 | 128 ± 56.6 | 0.784 | 29.2 ± 17.1 | 0.33 | 17.4 ± 10.6 | 0.123 |
| Female | 20(43.5) | 172 ± 101.1 | 133.3 ± 63.2 | 25.1 ± 14.7 | 13.1 ± 7.3 | ||||
| Age (years) | |||||||||
| ≥ 60 | 18(39.1) | 158.2 ± 67.2 | 0.632 | 146.7 ± 66.8 | 0.249 | 28.4 ± 21.4 | 0.486 | 16.4 ± 12.2 | 0.726 |
| < 60 | 28(60.9) | 153.7 ± 91.4 | 119.8 ± 51.8 | 26.8 ± 11.9 | 15 ± 7.4 | ||||
*means P-value< 0.05
RNA isolation and reverse transcription
Total circulating miRNA was extracted from 200 μL plasma using the miRNeasy Serum/Plasma Kit (Qiagen) according to the manufacturer’s protocol. In addition, 10 μL of a 1.5 nmol/L solution of the custom synthetic miRNA cel-miR-54-5p was added after the sample was mixed with 1 mL QIAzol Lysis reagent for 5 min. RNA was eluted from spin columns in 40 μL nuclease-free water.
Four circulating human miRNAs (miR-21, miR-93, miR-106a and miR-106b) and one spike-in control miRNA (cel-miR-54-5p) were determinated by TaqMan™ MicroRNA Assays, TaqMan miRNA Reverse Transcription kits (Life Technologies) and miRNA-specific RT primers were used for reverse transcription. For each sample, 3 μL RNA sample was added in a 15 μL reaction mixture using standard protocol. Then, the resulting cDNA was prepared for the droplet digital PCR.
ddPCR workflow
For each ddPCR assay, 3 μL cDNA sample, 10 μL 2× ddPCR supermix for probes (Bio-Rad), 1 μL 20× TaqMan miRNA probe and 6 μL RNase-free Water was added in a 20 μL reaction mixture. Then, the mixture and 70 μL droplet generation oil for probes (Bio-Rad) were respectively loaded into the sample wells and oil wells of a disposable droplet generator cartridge (Bio-Rad). After that, droplets were generated by QX200 droplet generator device (Bio-Rad) and carefully transferred to a 96-well PCR plate (Eppendorf). The cycling conditions were: 95 °C for 10 min, 40 cycles of 95 °C for 15 s and 57 °C for 1 min, and a final step at 98 °C for 10 min. At the end of the PCR reaction, droplets were read in the QX200 droplet reader and analyzed using the Quantasoft™ version 1.7.4 software (Bio-Rad). In addition, a no template control (NTC) was included in every assay. And the spike-in control miRNA was used as an internal calibrator to monitor extraction efficiency.
Statistical analysis
The statistical analyses were performed using the SPSS version 19.0 software. The Mann-Whitney U test was used to compare significant differences in miRNA expression between different groups. Logistic regression was used to develop a combined miRNA panel to diagnose GC with different TNM stage. Receiver operating characteristic (ROC) curves were established to evaluate the capacity of the tested miRNA to discriminate cancer cases in different TNM stage, and its potential use as a diagnostic tool for detecting GC. A p-value of less than 0.05 was considered to be significant.
A total of 147 participants in the training cohort were grouped into the training data set, and 28 participants in the validation cohort were grouped into the testing data set. In the training stage, a classical random forest algorithm in R version 3.4.2 software was used to construct variable selection models for combined four miRNA panel and clinical parameters in this study. Next, using single blind method, we tested the model by using the 28 cases of the testing data set as a prospective validation set, to assess its predictive ability. And we also retrospectively analyzed the 147 cases of the training data set.
Results
Circulating miRNAs in plasma of GC patients versus healthy controls
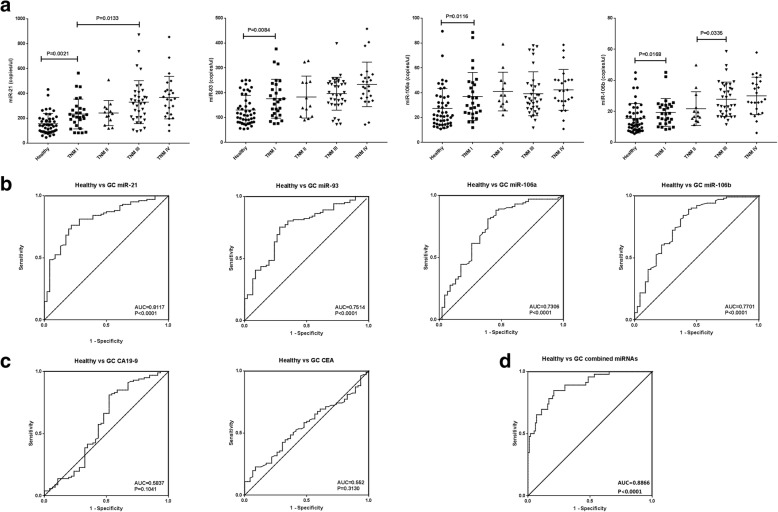
First, we compared the expression levels of four validated miRNAs in plasma from healthy volunteers (n = 46) and GC patients (n = 101) with different TNM stage using ddPCR. All four miRNAs including miR-21, miR-93, miR-106a and miR-106b levels were significantly lower in healthy controls than GC patients with TNM stage I (p = 0.0021, p = 0.0084, p = 0.0116 and p = 0.0168 respectively) (Fig. 1a), as well as TNM stage II, III and IV (Table 2). To evaluate the diagnostic value of the concentrations of these four circulating miRNAs, ROC curve analysis was performed. GC patients with different TNM stage were combined as one group, the area under the curve (AUC) values of miR-21, miR-93, miR-106a and miR-106b were 0.811 (95% confidence interval [CI], 0.739–0.884), 0.751 (95% CI, 0.667–0.836), 0.731 (95% CI, 0.638–0.823) and 0.77 (95% CI, 0.683–0.857), respectively (Fig. 1b). We also detect the CEA and CA19–9 in all 147 participants in the present study, the testing time was pre-surgery for GC patients. The AUC values obtained for CEA and CA19–9 to distinguish the GC patients from the healthy controls were 0.552 (95% CI, 0.456–0.648) and 0.584 (95% CI, 0.473–0.695), respectively (Fig. 1c).
Fig. 1.
Diagnostic value of circulating miRNAs expression signature in discriminating gastric cancer patients from healthy volunteers. a Levels of circulating miR-21, miR-93, miR-106a and miR-106b in plasma of gastric cancer patients and healthy volunteers. The levels of miRNA are presented as copies/μl of PCR reaction. b ROC analysis for individual miRNA. c ROC analysis for the common tumor biomarkers including CEA and CA19–9. d ROC analysis for the combined miRNA panel
Table 2.
Performance of circulating miRNAs for detection of GC with different TNM stages
| miR-21 | miR-93 | miR-106a | miR-106b | |
|---|---|---|---|---|
| P-value | ||||
| Health vs GC TNM I | 0.0021** | 0.0084** | 0.0116* | 0.0168* |
| Health vs GC TNM II | 0.0014** | 0.0263* | 0.0013** | 0.0132* |
| Health vs GC TNM III | < 0.0001** | < 0.0001** | 0.0004** | < 0.0001** |
| Health vs GC TNM IV | < 0.0001** | < 0.0001** | 0.0001** | < 0.0001** |
| GC TNM I VS II | 0.5704 | 0.8142 | 0.2820 | 0.6043 |
| GC TNM I VS III | 0.0133* | 0.1543 | 0.4864 | 0.0018** |
| GC TNM I VS IV | 0.0018** | 0.0102* | 0.1118 | 0.0004** |
| GC TNM II VS III | 0.0791 | 0.4665 | 0.5262 | 0.0335* |
| GC TNM II VS IV | 0.0163* | 0.0952 | 0.6097 | 0.0200* |
| GC TNM III VS IV | 0.2985 | 0.0897 | 0.2164 | 0.3092 |
* means P < 0.05, ** means P < 0.01
Furthermore, through a combination of the expression levels of four validated miRNAs, weighted by the regression coefficient, we developed a miRNA classifier using logistic regression model. It could be used to evaluate the predicted probability of being detected as GC, which was calculated as follows: first, the expression levels of four miRNAs were calculated as miRNA panel score using the following equation: miRNA panel score = 5.218–0.011 × miR-21-0.012 × miR-93-0.037 × miR-106a-0.031 × miR-106b. Then, the predicted probability was calculated by a second equation: predicted probability = EXP (miRNA panel score)/[1 + EXP (miRNA panel score)]. The combination of the four miRNAs exhibited better diagnostic value compared to any individual miRNA, with an AUC of 0.887 (95% CI, 0.83–0.943) (Fig. 1d) by logistic regression analysis, an optimal cut-off point was indicated at 0.315 with a sensitivity of 84.8% and a specificity of 79.2%. These results indicated that the circulating miR-21, miR-93, miR-106a and miR-106b could be considered as more accurate biomarkers than CEA and CA19–9 for GC diagnosis.
Circulating miRNAs in plasma of GC patients with different TNM stage
Although four miRNAs were up-regulated in GC patients with TNM stage II compared with stage I, as well as TNM stage IV compared with stage III, but the differences had no statistically significant (p > 0.05) (Fig. 1a & Table 2). However, miR-21 and miR-106b levels were significantly increased in GC patients with TNM stage III or IV compared with stage I (miR-21:p = 0.0133 and p = 0.0018, miR-106b:p = 0.0018 and p = 0.0004 respectively) (Table 2). miR-106b levels were still significantly increased when compared TNM stage III or IV with stage II in GC patients (p = 0.0335 and p = 0.02 respectively), while it was significant for miR-21 levels only when compared TNM stage IV with stage I in GC patients (p = 0.0163). Moreover, miR-93 and miR-106a levels in GC patients had no significant difference between groups with different TNM stage, except for the miR-93 levels in GC patients with TNM stage I and IV (p = 0.0102).
Furthermore, we combined GC patients with TNM stage I and II as one group, as well as TNM stage III and IV. As expected, the results showed that miR-21 and miR-106b levels were significantly higher in stage III and IV compared to stage I and II (p = 0.0004 and p < 0.0001 respectively), while miR-106a levels still had no significant difference between these two groups (p = 0.3824) (Fig. 2a). However, there was an unexpected significant increase for miR-93 levels in GC patients with stage III and IV compared to stage I and II (p = 0.0218) (Fig. 2a). The AUC values obtained for miR-21, miR-93, miR-106a and miR-106b to distinguish the GC patients with stage I and II from stage III and IV were 0.704 (95% CI, 0.601–0.807), 0.634 (95% CI, 0.52–0.749), 0.552 (95% CI, 0.435–0.668) and 0.736 (95% CI, 0.635–0.836), respectively (Fig. 2b).
Fig. 2.
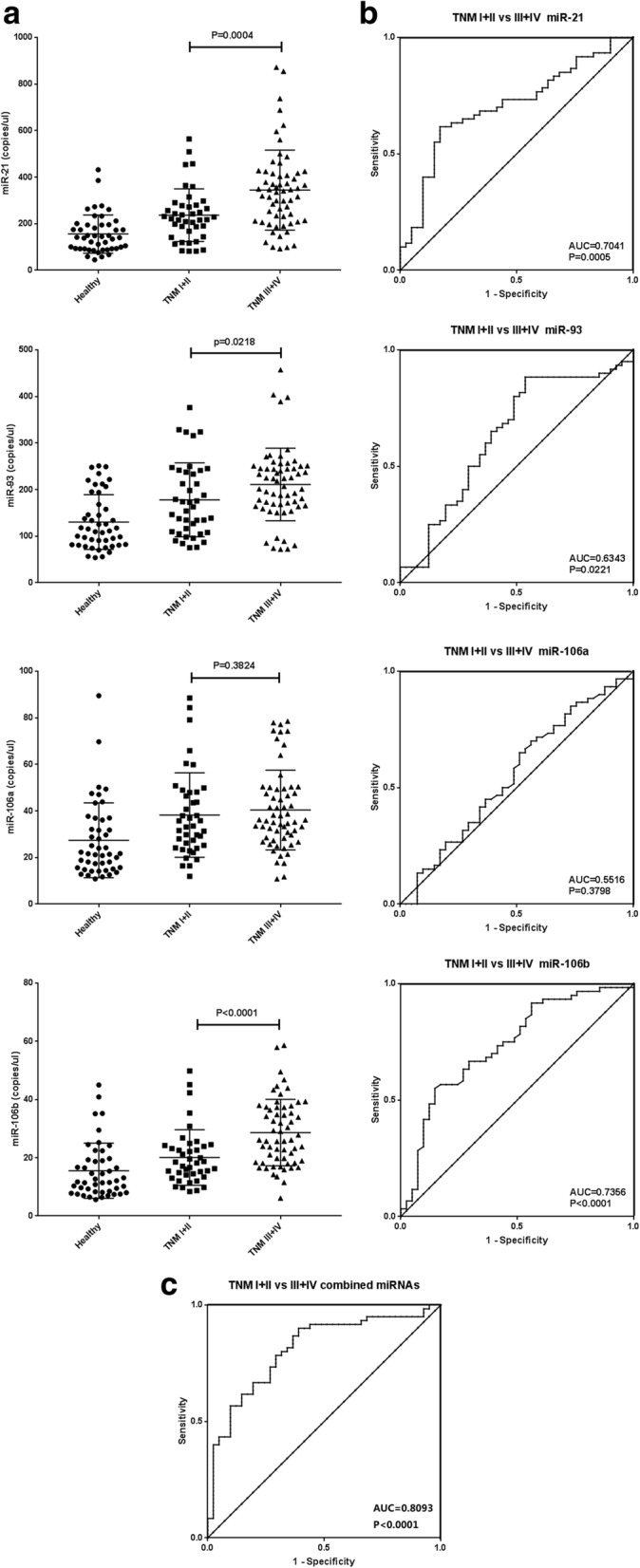
Diagnostic value of circulating miRNAs expression signature in discriminating gastric cancer at different TNM stage. a Levels of circulating miR-21, miR-93, miR-106a and miR-106b in plasma of gastric cancer patients with low TNM stage (stage I and II) and high TNM stage (stage III and IV), and healthy volunteers. The levels of miRNA are presented as copies/μl of PCR reaction. b ROC analysis for individual miRNA. c ROC analysis for the combined miRNA panel
Same as previous analysis, we assigned each patient a risk score which was calculated as follows: First, miRNA panel score = − 2.875 + 0.005 × miR-21 + 0.003 × miR-93-0.034 × miR-106a + 0.115 × miR-106b. Then, risk score = EXP (miRNA panel score)/[1 + EXP (miRNA panel score)]. The combination of the four miRNAs exhibited better capability to discriminate GC with TNM stage I and II from stage III and IV compared to any individual miRNA, with an AUC of 0.809 (95% CI, 0.723–0.896) (Fig. 2c) by logistic regression analysis, an optimal cut-off point was indicated at 0.534 with a sensitivity of 78.3% and a specificity of 70.7%. Taken together, these results demonstrated that circulating miR-21, miR-93 and miR-106b might have a potential diagnostic value for distinguishing GC with different TNM stage.
Correlation between expression levels of circulating miRNA in plasma and clinicopathologic factors in GC patients
Except for the TNM stage, we evaluated whether the levels of four circulating miRNAs are correlated with other clinical characteristics of all GC patients. As it was summarized in Table 1, the expression of miR-21, miR-93, miR-106a and miR-106b didn’t significantly differ between the GC patients based on gender (p = 0.437, p = 0.318, p = 0.404 and p = 0.353 respectively), tumor differentiation (p = 0.078, p = 0.976, p = 0.601 and p = 0.891 respectively) and distant metastasis (p = 0.435, p = 0.371, p = 0.297 and p = 0.572 respectively). However, the levels of miR-21, miR-93 and miR-106b in plasma from the GC patients is significantly related to lymph node metastasis (p = 0.014, p = 0.019 and p = 0.0002 respectively). Moreover, the circulating miR-21 and miR-106b levels are also related to tumor size and age respectively (p = 0.029 and p = 0.019). The results demonstrated the feasibility of these miRNAs for the diagnosis of other clinicopathological characteristics of GC patients.
Random forest model used for GC diagnosis
To evaluate the diagnostic value of circulating miR-21, miR-93, miR-106a and miR-106b combined with other conventional clinical parameters including gender, age, CEA and CA19–9 for GC with different TNM stage, we used a classical random forest algorithm for analysis. A total of 147 participants in the training cohort were grouped as the training data set and used for developing the model. And the other 28 participants in the validation cohort were grouped as the testing data set and used for assessing predictive ability of the model.
In the training stage, using random forest supervised classification algorithm, four microRNAs, three clinical parameters including CEA, age and CA19–9 mostly related to the diagnostic classification were selected (Additional file 1: Figure S1). In the testing stage, we used the developed random forest model to validate both the training data set and the testing data set. It correctly discriminated 147 out of 147 samples in the training data set, and 23 out of 28 samples in the testing data set, which showed 100 and 82.1% accuracy respectively in identifying Healthy volunteers and GC patients with different TNM stage (Table 3), by using the selected variables based on their value cut-offs (Additional file 1: Figure S1). In addition, the most influential factor in this model was miR-21, followed by miR-106b, miR-93, miR-106a, CA19–9, CEA, age and gender (Table 4).
Table 3.
Confusion matrix of the developed random forest model in the testing stage
| Predicted class | ||||
|---|---|---|---|---|
| Healthy | TNM I + II | TNM III + IV | ||
| Training data set | ||||
| Actual class | Healthy | 46 | 0 | 0 |
| TNM I + II | 0 | 41 | 0 | |
| TNM III + IV | 0 | 0 | 60 | |
| Testing data set | ||||
| Actual class | Healthy | 14 | 2 | 1 |
| TNM I + II | 0 | 4 | 1 | |
| TNM III + IV | 1 | 0 | 5 | |
Table 4.
The relative importance of variables in the developed random forest model
| Variables | Mean Decrease Gini |
|---|---|
| Gender | 1.134787 |
| Age | 9.342937 |
| CA19–9 | 11.051689 |
| CEA | 9.918123 |
| miR-21 | 19.143754 |
| miR-93 | 16.274413 |
| miR-106a | 12.627928 |
| miR-106b | 16.550531 |
Discussion
Early diagnosis could greatly improve the survival rates of GC patients. However, the currently used diagnostic methods are either invasive or insensitive, thus limited their application in clinic. In recent years, a number of circulating miRNAs, which are notably stable in the circulation of body fluids [20, 21], are suggested as promising non-invasive diagnostic markers for GC [9, 15, 22–24]. Unfortunately, since circulating miRNAs exist in blood at extremely low concentrations [25], the test results would be made poorly repeatable due to the interference of several variables, such as sample processing protocols, RNA isolation and so on [10, 26]. Most importantly, quantitative real-time PCR is most commonly used but must rely on the use of external calibrators, because it lacks reliable endogenous reference miRNA for normalization of results in plasma or serum. Therefore, the data which produced by a variety of normalization methods in different studies, become non-comparable or difficult to compare. This is a major obstacle for their translation into clinically useful applications [10, 27].
The present study, to our best knowledge, is the first to evaluate the diagnostic value of circulating miRNAs for GC patients using the ddPCR technique. ddPCR is a recently introduced technology which can achieve absolute quantification of nucleic acids based on the principles of sample portioning, end-point PCR and Poisson statistics [28, 29]. Thus, it overcomes the normalization and calibrator issues [30]. Besides, it has shown better precision and sensitivity while detecting low concentration of target nucleic acids molecules [31, 32]. More importantly, ddPCR can tolerate PCR inhibitors which could influence the efficiency of PCR amplification, without affecting the quantitative results of the target [11].
Using ddPCR, we analyzed the levels of circulating miR-21, miR-93, miR-106a and miR-106b in the plasma of GC patients and healthy volunteers. Similar to previous studies [9, 14, 15], we found the significantly increased levels of these miRNAs in GC patients compared with healthy controls, and some miRNAs were associated with advanced TNM stage. ROC curve analysis showed that each miRNA had higher diagnostic sensitivity and specificity than CEA and CA19–9 which were widely used in clinic. Furthermore, through a combination of the expression levels of four validated miRNAs, a patient will be considered to have GC if the predicted probability is higher than the threshold set (0.315 with a sensitivity of 84.8% and specificity of 79.2%) in the model. An AUC of 0.887 (95% CI, 0.83–0.943) and P-values< 0.001 indicate the great potential value of these miRNAs as GC biomarkers.
Based on the results above, we further evaluate the potential use of these miRNAs in discriminating GC with different TNM stage. First, GC patients with TNM stage I and II were combined as one group, as well as stage III and IV, because there was no statistically significant difference between these groups. Then, our results showed that the levels of circulating miR-21, miR-93 and miR-106b in the plasma of GC patients were significantly higher in TNM stage III and IV than stage I and II, except for the miR-106a. As usual, a combination of four miRNAs showed better capability to discriminate GC with different TNM stage. A patient will be considered to have GC with TNM stage III or IV if the risk score is higher than 0.534 (a sensitivity of 78.3% and specificity of 70.7%). ROC analysis also showed an AUC of 0.809 (95% CI, 0.723–0.896) and P-values< 0.001. To our knowledge, this study is the first to demonstrate that these miRNAs might be also used as biomarkers to discriminate GC with TNM stage I and II from stage III and IV.
In the search of possible correlations with clinicopathological features, it was noteworthy that the presence of lymph node metastases was significantly correlated with increased levels of circulating miR-21, miR-93 and miR-106b. Moreover, a high level of circulating miR-21 was significantly related to a bigger tumor size (≥5 cm). These results indicate that these miRNAs might represent biomarkers of tumor aggressiveness, which further improved their value for discriminating GC with different TNM stage. Some studies have reported that high levels of miR-21 expression may induce tumor proliferation, migration and invasion via the downregulation of Noxa or PTEN expressions in GC cells [33, 34]. And miR-93 could promote proliferation and metastasis of GC via targeting TIMP2 or inactivation of the Hippo signaling pathway [35, 36]. In cancer-associated fibroblasts from GC, miR-106b could promote cell migration and invasion by targeting PTEN [37]. And it could also promote cell cycling of GC cells through regulation of p21 and E2F5 target gene expression [38]. These might be the mechanism of its correlation with lymph node metastases and tumor size. However, although it was reported that miR-106a could also regulate invasion and metastasis of GC via targeting TIMP2 [39, 40], and may inhibit extrinsic apoptotic pathway through targeting FAS [41], our results demonstrated that miR-106a expression was not associated with the lymph node metastases and tumor size. Further studies are required.
In clinic, due to the numerous factors that influence the precision and accuracy of diagnosing diseases or predicting of patients’ prognosis, more and more studies are applying machine learning algorithms to medical data, including the detection of GC [20, 42, 43]. There are several algorithms such as random forest, support vector machine and neural networks were commonly used [43, 44]. Here, we chose random forest model since it is easy to interpret, and allowed us to estimate the importance of a variable. After the random forest model was established in the training stage, when we tested the predictive value of this model using the testing data set, our results showed that it correctly discriminated 14 out of 17 healthy volunteers (false rate, 17.6%), 4 out of 5 GC patients with TNM stage I or II (false rate, 20%), and 5 out of 6 GC patients with TNM stage III or IV (false rate, 16.7%). However, the number of cases included in the present study is still far from sufficient to develop a reliable model, and we also didn’t have enough cases to test and validate the model. Further studies with much more cases are urgently required, to improve their application in clinic. Moreover, despite our results and accumulating evidences suggested that circulating miRNAs stably existed in circulation and can indeed be used as biomarkers to identify and monitor a variety of cancers and other diseases, it is still unknown how and why GC causes changes in the levels of these four circulating miRNAs, and whether or how they play roles in physiology. Further studies are also needed.
Conclusions
Overall, the present study demonstrated that by using the ddPCR technique, circulating miR-21, miR-93, miR-106a and miR-106b could be used as diagnostic plasma biomarkers in gastric cancer patients.
Additional files
Figure S1. A random forest model for discriminating healthy volunteers, gastric cancer patients with low TNM stage (stage I and II) and high TNM stage (stage III and IV) G0 represents healthy volunteers; G1 represents GC patients with TNM stage I and II; G2 represents GC patients with TNM stage III and IV. (TIF 624 kb)
Acknowledgments
Funding
This study was supported by the grants from the National Natural Science Foundation of China (No.81172832, 81771723), Sichuan Youth Science and Technology Foundation (No.2013JQ0020), Special Program for Sichuan Youth Science and Technology Innovation (No.2014TD0010), and grant from Health and Family Planning Commission of Sichuan Province (No.110190). The funding body did not have any role in the study design, nor the data collection, analysis and interpretation as well as writing of the manuscript.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AFP
α-fetoprotein
- AUC
Area under the curve.
- CA19–9
Carbohydrate antigen 19–9
- CEA
Carcinoembryonic antigen
- ddPCR
Droplet digital PCR
- GC
Gastric cancer
- miRNA
Microrna
- qPCR
Quantitative real-time PCR
- ROC
Receiver operating characteristic
Authors’ contributions
The authors’ contribution was as following: GPZ: Conceptualization, methodology, formal analysis, investigation, resources, data curation, funding acquisition, writing-original draft, writing–review and editing, and project administration. TJ, YZL and GLH: Validation, formal analysis, investigation, resources, and data curation. CBL and GQL: Conceptualization, formal analysis. GQJ and KW: Conceptualization, methodology, formal analysis. MZY: Conceptualization, methodology, funding acquisition, writing-original draft, writing–review and editing. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the ethics committee of Sichuan Provincial People’s Hospital. All patients provided written informed consent.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12885-018-4601-5) contains supplementary material, which is available to authorized users.
Contributor Information
Gaoping Zhao, Phone: +86-28-87393805, Email: gzhao@uestc.edu.cn.
Tao Jiang, Email: 490397636@qq.com.
Yanzhuo Liu, Email: comelina123@163.com.
Guoli Huai, Email: 1358271315@qq.com.
Chunbin Lan, Email: 191197067@qq.com.
Guiquan Li, Email: 87239657@qq.com.
Guiqing Jia, Email: gqjia2009@163.com.
Kang Wang, Email: 258380648@qq.com.
Maozhu Yang, Phone: +86-28-87393315, Email: 37771782@qq.com.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Hamashima C, Shabana M, Okada K, Okamoto M, Osaki Y. Mortality reduction from gastric cancer by endoscopic and radiographic screening. Cancer Sci. 2015;106:1744–1749. doi: 10.1111/cas.12829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catalano V, Labianca R, Beretta GD, Gatta G, de Braud F, Van Cutsem E. Gastric cancer. Crit Rev Oncol Hematol. 2009;71:127–164. doi: 10.1016/j.critrevonc.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Pasechnikov V, Chukov S, Fedorov E, Kikuste I, Leja M. Gastric cancer: prevention, screening and early diagnosis. World J Gastroenterol. 2014;20:13842–13862. doi: 10.3748/wjg.v20.i38.13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 7.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Wang L, Wu Z, Sun R, Jin H, Ma J, et al. Three dysregulated microRNAs in serum as novel biomarkers for gastric cancer screening. Med Oncol. 2014;31:298. doi: 10.1007/s12032-014-0298-8. [DOI] [PubMed] [Google Scholar]
- 9.Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, et al. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174–1179. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarry J, Schadendorf D, Greenwood C, Spatz A, van Kempen LC. The validity of circulating microRNAs in oncology: five years of challenges and contradictions. Mol Oncol. 2014;8:819–829. doi: 10.1016/j.molonc.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Racki N, Dreo T, Gutierrez-Aguirre I, Blejec A, Ravnikar M. Reverse transcriptase droplet digital PCR shows high resilience to PCR inhibitors from plant, soil and water samples. Plant Methods. 2014;10:42. doi: 10.1186/s13007-014-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campomenosi P, Gini E, Noonan DM, Poli A, D'Antona P, Rotolo N, et al. A comparison between quantitative PCR and droplet digital PCR technologies for circulating microRNA quantification in human lung cancer. BMC Biotechnol. 2016;16:60. doi: 10.1186/s12896-016-0292-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tchernitsa O, Kasajima A, Schäfer R, Kuban RJ, Ungethüm U, Györffy B, et al. Systematic evaluation of the miRNA-ome and its downstream effects on mRNA expression identifies gastric cancer progression. J Pathol. 2010;222:310–319. doi: 10.1002/path.2759. [DOI] [PubMed] [Google Scholar]
- 15.Shiotani A, Murao T, Kimura Y, Matsumoto H, Kamada T, Kusunoki H, et al. Identification of serum miRNAs as novel non-invasive biomarkers for detection of high risk for early gastric cancer. Br J Cancer. 2013;109:2323–2330. doi: 10.1038/bjc.2013.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cios KJ, Moore GW. Uniqueness of medical data mining. Artif Intell Med. 2002;26:1–24. doi: 10.1016/S0933-3657(02)00049-0. [DOI] [PubMed] [Google Scholar]
- 17.Kang J, Schwartz R, Flickinger J, Beriwal S. Machine learning approaches for predicting radiation therapy outcomes: a Clinician's perspective. Int J Radiat Oncol Biol Phys. 2015;93:1127–1135. doi: 10.1016/j.ijrobp.2015.07.2286. [DOI] [PubMed] [Google Scholar]
- 18.Sudharsan B, Peeples M, Shomali M. Hypoglycemia prediction using machine learning models for patients with type 2 diabetes. J Diabetes Sci Technol. 2015;9:86–90. doi: 10.1177/1932296814554260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyaki R, Yoshida S, Tanaka S, Kominami Y, Sanomura Y, Matsuo T, et al. A computer system to be used with laser-based endoscopy for quantitative diagnosis of early gastric cancer. J Clin Gastroenterol. 2015;49:108–115. doi: 10.1097/MCG.0000000000000104. [DOI] [PubMed] [Google Scholar]
- 20.Armand-Labit V, Pradines A. Circulating cell-free microRNAs as clinical cancer biomarkers. Biomol Concepts. 2017;8:61–81. doi: 10.1515/bmc-2017-0002. [DOI] [PubMed] [Google Scholar]
- 21.Wittmann J, Jäck HM. Serum microRNAs as powerful cancer biomarkers. Biochim Biophys Acta. 2010;1806:200–207. doi: 10.1016/j.bbcan.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Shin VY, Ng EK, Chan VW, Kwong A, Chu KM. A three-miRNA signature as promising non-invasive diagnostic marker for gastric cancer. Mol Cancer. 2015;14:202. doi: 10.1186/s12943-015-0473-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou X, Zhu W, Li H, Wen W, Cheng W, Wang F, et al. Diagnostic value of a plasma microRNA signature in gastric cancer: a microRNA expression analysis. Sci Rep. 2015;5:11251. doi: 10.1038/srep11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsujiura M, Komatsu S, Ichikawa D, Shiozaki A, Konishi H, Takeshita H, et al. Circulating miR-18a in plasma contributes to cancer detection and monitoring in patients with gastric cancer. Gastric Cancer. 2015;18:271–279. doi: 10.1007/s10120-014-0363-1. [DOI] [PubMed] [Google Scholar]
- 25.Wang K, Yuan Y, Cho JH, McClarty S, Baxter D, Galas DJ. Comparing the MicroRNA spectrum between serum and plasma. PLoS One. 2012;7:e41561. doi: 10.1371/journal.pone.0041561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sourvinou IS, Markou A, Lianidou ES. Quantification of circulating miRNAs in plasma: effect of preanalytical and analytical parameters on their isolation and stability. J Mol Diagn. 2013;15(6):827–834. doi: 10.1016/j.jmoldx.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 27.Marzi MJ, Montani F, Carletti RM, Dezi F, Dama E, Bonizzi G, et al. Optimization and standardization of circulating MicroRNA detection for clinical application: the miR-test case. Clin Chem. 2016;62:743–754. doi: 10.1373/clinchem.2015.251942. [DOI] [PubMed] [Google Scholar]
- 28.Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem. 2012;84:1003–1011. doi: 10.1021/ac202578x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, Haynes R, et al. The digital MIQE guidelines: minimum information for publication of quantitative digital PCR experiments. Clin Chem. 2013;59:892–902. doi: 10.1373/clinchem.2013.206375. [DOI] [PubMed] [Google Scholar]
- 30.Zhao H, Wilkins K, Damon IK, Li Y. Specific qPCR assays for the detection of orf virus, pseudocowpox virus and bovine papular stomatitis virus. J Virol Methods. 2013;194:229–234. doi: 10.1016/j.jviromet.2013.08.027. [DOI] [PubMed] [Google Scholar]
- 31.Zhao S, Lin H, Chen S, Yang M, Yan Q, Wen C, et al. Sensitive detection of porcine circovirus-2 by droplet digital polymerase chain reaction. J Vet Diagn Investig. 2015;27:784–788. doi: 10.1177/1040638715608358. [DOI] [PubMed] [Google Scholar]
- 32.Brunetto GS, Massoud R, Leibovitch EC, Caruso B, Johnson K, Ohayon J, et al. Digital droplet PCR (ddPCR) for the precise quantification of human T-lymphotropic virus 1 proviral loads in peripheral blood and cerebrospinal fluid of HAM/TSP patients and identification of viral mutations. J Neuro-Oncol. 2014;20:341–351. doi: 10.1007/s13365-014-0249-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun H, Wang P, Zhang Q, He X, Zai G, Wang X, et al. MicroRNA-21 expression is associated with the clinical features of patients with gastric carcinoma and affects the proliferation, invasion and migration of gastric cancer cells by regulating Noxa. Mol Med Rep. 2016;13(3):2701–2707. doi: 10.3892/mmr.2016.4863. [DOI] [PubMed] [Google Scholar]
- 34.Zhang BG, Li JF, Yu BQ, Zhu ZG, Liu BY, Yan M. microRNA-21 promotes tumor proliferation and invasion in gastric cancer by targeting PTEN. Oncol Rep. 2012;27(4):1019–1026. doi: 10.3892/or.2012.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guan H, Li W, Li Y, Wang J, Li Y, Tang Y, et al. MicroRNA-93 promotes proliferation and metastasis of gastric cancer via targeting TIMP2. PLoS One. 2017;12(12):e0189490. doi: 10.1371/journal.pone.0189490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li L, Zhao J, Huang S, Wang Y, Zhu L, Cao Y, et al. MiR-93-5p promotes gastric cancer-cell progression via inactivation of the hippo signaling pathway. Gene. 2018;641:240–247. doi: 10.1016/j.gene.2017.09.071. [DOI] [PubMed] [Google Scholar]
- 37.Yang TS, Yang XH, Chen X, Wang XD, Hua J, Zhou DL, et al. MicroRNA-106b in cancer-associated fibroblasts from gastric cancer promotes cell migration and invasion by targeting PTEN. FEBS Lett. 2014;588(13):2162–2169. doi: 10.1016/j.febslet.2014.04.050. [DOI] [PubMed] [Google Scholar]
- 38.Yao YL, Wu XY, Wu JH, Gu T, Chen L, Gu JH, et al. Effects of microRNA-106 on proliferation of gastric cancer cell through regulating p21 and E2F5. Asian Pac J Cancer Prev. 2013;14(5):2839–2843. doi: 10.7314/APJCP.2013.14.5.2839. [DOI] [PubMed] [Google Scholar]
- 39.Zhu M, Zhang N, He S, Yan R, Zhang J. MicroRNA-106a functions as an oncogene in human gastric cancer and contributes to proliferation and metastasis in vitro and in vivo. Clin Exp Metastasis. 2016;33(5):509–519. doi: 10.1007/s10585-016-9795-9. [DOI] [PubMed] [Google Scholar]
- 40.Zhu M, Zhang N, He S, Lui Y, Lu G, Zhao L. MicroRNA-106a targets TIMP2 to regulate invasion and metastasis of gastric cancer. FEBS Lett. 2014;588(4):600–607. doi: 10.1016/j.febslet.2013.12.028. [DOI] [PubMed] [Google Scholar]
- 41.Wang Z, Liu M, Zhu H, Zhang W, He S, Hu C, et al. miR-106a is frequently upregulated in gastric cancer and inhibits the extrinsic apoptotic pathway by targeting FAS. Mol Carcinog. 2013;52(8):634–646. doi: 10.1002/mc.21899. [DOI] [PubMed] [Google Scholar]
- 42.Liu WZ, White AP, Hallissey MT, Fielding JW. Machine learning techniques in early screening for gastric and oesophageal cancer. Artif Intell Med. 1996;8:327–341. doi: 10.1016/0933-3657(95)00039-9. [DOI] [PubMed] [Google Scholar]
- 43.Li Q, Wang W, Ling X, Wu JG. Detection of gastric cancer with Fourier transform infrared spectroscopy and support vector machine classification. Biomed Res Int. 2013;2013:942427. doi: 10.1155/2013/942427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuruvilla J, Gunavathi K. Lung cancer classification using neural networks for CT images. Comput Methods Prog Biomed. 2014;113:202–209. doi: 10.1016/j.cmpb.2013.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. A random forest model for discriminating healthy volunteers, gastric cancer patients with low TNM stage (stage I and II) and high TNM stage (stage III and IV) G0 represents healthy volunteers; G1 represents GC patients with TNM stage I and II; G2 represents GC patients with TNM stage III and IV. (TIF 624 kb)
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.