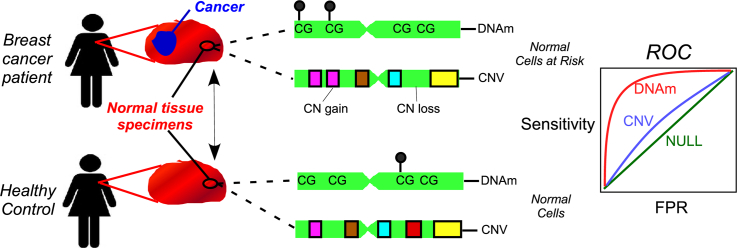
Abstract
Normal tissue at risk of neoplastic transformation is characterized by somatic mutations, copy-number variation and DNA methylation changes. It is unclear however, which type of alteration may be more informative of cancer risk. We analyzed genome-wide DNA methylation and copy-number calls from the same DNA assay in a cohort of healthy breast samples and age-matched normal samples collected adjacent to breast cancer. Using statistical methods to adjust for cell type heterogeneity, we show that DNA methylation changes can discriminate normal-adjacent from normal samples better than somatic copy-number variants. We validate this important finding in an independent dataset. These results suggest that DNA methylation alterations in the normal cell of origin may offer better cancer risk prediction and early detection markers than copy-number changes.
Keywords: Cancer, Cancer-risk, Early detection, DNA methylation, Epigenetic, Copy-number, Breast cancer
Graphical Abstract
Highlights
-
•
Copy-number and DNA methylation changes in normal-adjacent breast tissue become enriched in the matched breast cancer.
-
•
Unlike copy-number changes, DNA methylation can discriminate normal-adjacent from the normal tissue of cancer-free women.
-
•
Epigenetic alterations may be more suitable as cancer risk biomarkers.
Most cancers arise because of molecular alterations that accrue in normal cells as a result of aging and exposure to environmental risk factors. Molecular alterations include those affecting a cell's hardware, commonly termed “genetic” (e.g. mutations and gene duplications), as well as those affecting a cell's software (termed “epigenetic”). An example of an epigenetic change is DNA methylation, a chemical tag which controls the activity of nearby genes. The relative significance of genetic vs. epigenetic changes in the development of cancer is unknown. Our study compares DNA methylation to gene copy number changes in normal cells adjacent to breast cancer to normal cells from healthy women, and concludes that DNA methylation can better predict cancer status. This suggests that epigenetic alterations may be more suitable for developing cancer risk biomarkers.
1. Introduction
Throughout life, normal cells acquire somatic alterations in the genome and epigenome, both of which are thought to contribute to the onset of neoplasia and cancer [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11]. Mapping genetic and epigenetic changes in normal tissue at risk of neoplastic transformation is therefore critically important for understanding oncogenesis, identifying early causal drivers and for cancer risk prediction [12]. Although a number of studies have been able to link somatic mutations and copy-number-variants (CNVs) in whole blood to the future risk of hematological and solid cancers [2,4, 5, 6,13,14], analogous results for somatic alterations in the epithelial cell of origin of solid cancers have remained elusive. Indeed, identifying somatic mutations in normal tissue is technically challenging [12,15,16], with only a couple of studies having been able to associate epithelial cancer risk to somatic mutations in normal (epithelial) tissue [17,18]. In contrast, DNA methylation (DNAm) changes have been correlated to cancer risk in blood [7,19, 20, 21], are frequently observed in preneoplastic epithelial tissue [22, 23, 24, 25, 26, 27], and in the context of cervical smears have allowed prospective risk prediction of a high-grade intraepithelial neoplasia independently of HPV status [25].
Two recent studies formally compared somatic mutations/CNVs to DNAm changes in their ability to predict prospective risk of gastric and esophageal cancer [17,18]. One study showed that DNAm changes may be a better risk predictor than somatic mutations, specially for gastric cancer [18], whilst the other study showed that both CNVs and DNAm changes were better than somatic mutations at predicting progression of intestinal metaplasia to gastric cancer [17]. Thus, both studies underscore the importance of DNAm changes in carcinogenesis and suggest that epigenetic alterations may be a better molecular cancer risk predictor than genetic changes. However, despite these two studies, the relative importance of genetic and epigenetic alterations for cancer risk prediction remains unclear.
Here we decided to shed further light on this outstanding question. Although comparing different types of molecular alteration as predictors of cancer risk is technically challenging due to the requirement of measuring all relevant molecular profiles in the relevant tissue and in a relatively large number of individuals, several studies have shown the feasibility of using Illumina Methylation 450 k/EPIC beadarrays to obtain high-confidence CNV calls [28, 29, 30], thus allowing at least for an objective comparison between CNV and DNAm. Here we conduct such a comparison in the context of an epithelial cancer using a cohort of 50 normal healthy breast samples, 42 age-matched normal samples collected adjacent to breast cancer, and a total of 305 invasive breast cancers (of which 42 were matched to the normal-adjacent ones), all profiled with Illumina 450 k beadarrays [24]. Since cell type heterogeneity represents a major source of DNAm variation in a complex tissue such as breast, we use recent state-of-the art statistical techniques to rigorously adjust for this major confounder. Using these techniques, as well as an independent validation, we demonstrate that DNAm changes in normal cells are more predictive of breast cancer status than their CNV counterparts.
2. Materials and Methods
2.1. Breast Cancer DNA Methylation Datasets
We analyzed 2 different normal breast and breast cancer tissue datasets, both profiled with the same Illumina Infinium 450 k DNAm technology. The Erlangen set was generated and analyzed by us previously [24], consisting of 50 normal healthy breast samples, 42 age-matched normal-adjacent breast cancer pairs (84 samples in total), and an additional 263 unmatched breast cancers. The clinical characteristics and normalization of the DNAm dataset was described previously [24]. The second “validation” dataset generated Illumina 450 k profiles for 18 normal healthy (from breast reduction surgery) breast samples, as well as 70 normal samples found adjacent to breast cancer [31]. Clinical characteristics and normalization of the Infinium data was described by us previously [24,31].
2.2. Construction and Validation of a Reference DNA Methylation Database for Breast Tissue
We aimed to build a reference DNAm database for breast tissue that would allow us to estimate fractions of epithelial, adipocyte and immune-cells from the DNAm profile of a sample, using the EpiDISH algorithm [32]. To construct the reference database, we used 450 k data representing human mammary epithelial cells (HMECs) from Lowe et al. [33], adipocytes from Nazor et al. [34] and all 7 major immune cell types (neutrophils, eosinophils, monocytes, CD4+ and CD8+ T-cells, B-cells and NK-cells) from Reinius et al. [35]. These 450 k profiles were used in conjunction with an empirical Bayes framework [36] to select differentially methylated CpGs (DMCs) between all 9 cell types, demanding FDR < 0.05 and at least 50% difference in average DNAm between cell types. Cell type specific DMCs were filtered further by demanding that they map to a DNase Hypersensitive Site (DHS), as determined by the NIH Epigenomics Roadmap (if such cell type specific DHS data were available), following a procedure we used previously [32]. This resulted in a reference matrix of 349 DMCs and 9 nine cell types. For an independent sample, cell type fractions for the 9 cell types can be estimated using EpiDISH (using the implementation with Robust Partial Correlations).
We performed three separate validations/tests to ensure that the reference DNAm profiles are representative of epithelial, fat and immune-cells. First, we collected Illumina 450 k data representing these same cell types from independent studies: HMECs from ENCODE [37], adipocytes and blood samples from Slieker et al. and [38]. We constructed 100 in-silico random mixtures of these 3 cell types and compared estimated to true cell-fractions. Second, we applied the reference DNAm profile database and EpiDISH to purified monocytes, T-cells and B-cells from 50 monozygotic twin pairs [39], as this should correctly predict zero fractions for epithelial and adipocytes and near 100% for blood cell types. Third, we applied the reference DNAm profile database and EpiDISH to WGBS data of two IHEC samples enriched for breast epithelial cells [40], as this should predict higher cell-fractions for the epithelial component.
2.3. Identification of DNAm Field Defects
The procedure used to identify epigenetic field defects in normal-adjacent breast tissue was described by us previously [24]. Briefly, we used our iEVORA algorithm to identify differentially variable (DV) and differentially methylated CpGs (DVMCs) between the 50 normal healthy and 42 normal-adjacent samples. The iEVORA algorithm demands genome-wide significance (after correction for multiple testing) at the level of differential variance only, thus defining differentially variable CpGs (DVCs), but subsequently re-ranks DVCs by a t-statistic, in order to favor DVCs where the differential variance is driven by as many outliers as possible. This re-ranking heuristic achieves a good compromise between sensitivity and the type-1 error rate, as demonstrated by us previously [41]. DVMCs were selected using a FDR threshold of 0.001 for differential variability (P-values estimated using Bartlett's DV test, which we stress can also be interpreted as a normality deviation test) and a P-value threshold of 0.05 for the t-statistics. Subsequently, we restrict to hypervariable DVMCs, i.e. the subset exhibiting increased variance in the normal-adjacent samples, as the underlying hypothesis is that samples exhibiting deviations from the normal-state represent those at higher risk of carcinogenic transformation.
An appealing feature of using differential variability statistics to identify DNAm alterations in normal-adjacent samples compared to healthy normals is that the resulting hyperV DVMCs are less likely to be driven by changes in cell type composition compared to randomly selected set of CpGs. To see this, we note that the use of the differential variability statistic favors CpGs (hyperV DVMCs) that show ultra-stable DNAm profiles across the normal healthy samples (i.e. very small variance), with outliers driving increased variance in the normal-adjacent specimens. The ultra-high stability of DNAm across the normal healthy samples means that these CpGs are not markers of underlying cell types (in breast these are mainly epithelial cells, adipocytes and immune cells), since variations in the adipose, epithelial and immune cell fractions dominate the top components of variation across normal samples [24]. To prove the result formally, we used our EpiDISH algorithm [32] and our reference DNAm database for breast tissue to estimate epithelial, adipose and immune-cell fractions in all 50 normal samples from healthy women, demonstrating that the top PC in a PCA correlated with these fractions. We then derived CpGs correlating significantly with the estimated epithelial and adipose fractions, thus defining “cell type” DMCs (ctDMCs). We then compared how the previously selected hyperV DVMCs ranked among the list of ctDMCs (i.e those CpGs correlating most strongly with cell type composition) to demonstrate that hyperV DVMCs are ranked significantly lower than a randomly selected set of 10,000 non-DVMCs. We also compared the ranking of the hyperV DVMCs to all non-DVMCs, which did not alter the conclusions.
2.4. CNV Calling Procedure
We used the following procedure to derive copy number alterations for both the Erlangen and validation Illumina 450 k sets. First, idat files were loaded, background-corrected and normalized using functions implemented in the minfi package [42]. The returned MethylSet object was then used as input to the conumee package [43], to infer CNV states. Briefly, conumee performs the inference in 3-steps: (i) background corrected intensity values of the “methylated” and “unmethylated” channels are added, and the log2-ratio of probe intensities of the query sample (this includes any sample, be it normal, normal-adjacent or cancer) to the average over all normal healthy samples is calculated, (ii) the median log2-ratio of probes within predefined genomic bins defines the bin-intensity value, and the bin intensity values are then shifted to minimize the median absolute deviation of all bin intensities from zero to determine the copy-number neutral state, (iii) segmentation is performed using the circular binary segmentation (CBS) algorithm implemented in the DNAcopy package [44]. For calling CN gain or loss, we used sample-specific thresholds instead of the widely used cutoffs (±0.1), in order to reduce the bias caused by cell type heterogeneity. The sample-specific threshold for CN gain/loss is determined automatically by analyzing the distribution of all shifted bin intensity values. For normal-adjacent samples, the median of the log2 ratio + 2σ or +6σ was computed for the 90% of central bins (ordered by their log ratios) to call gains and amplifications, respectively. The median of the log2 ratio − 2.5σ or −7σ was used to call losses and deletions, respectively. For cancer samples, the median of the log2 ratio + 2σ or + 6σ was computed for the 50% of the central bins (ordered by their log ratios) to call gains and amplifications, respectively. The median of the log2 ratio − 2.5σ or −7σ was used to call losses and deletions, respectively. All these thresholds for calling gains, losses, amplifications and deletions, have been extensively validated [45]. Thus, using these thresholds, the copy number state of each segment falls into five categories: deletion, loss, neutral, gain, amplification. To assign copy number states to genes we mapped segments to genes and assigned states according to the procedure used and validated by the METABRIC consortium [45].
The procedure described above to obtain CNV calls uses sample-specific thresholds, which directly accounts for the proportion of non-neoplastic cells in the sample [45]. In the case of normal breast tissue adjacent to breast cancer, non-neoplastic cells will include stromal cells like adipocytes and immune-cells, and the sample-specific threshold should therefore also automatically adjust for variations in the adipose and immune-cell content. To check this, we used the previously estimated epithelial, adipocyte and immune cell fractions (from EpiDISH algorithm) in the normal-adjacent samples, and also computed for each sample, a genomic instability index (GII), measured as the fraction of the genome that is altered [46]. Finally, we observed that the GII and the epithelial purity index from EpiDISH algorithm were not correlated, indicating that our CNV calls have adjusted reasonably well for variations in epithelial cell purity.
In order to assess how the results depend on the segmentation algorithm used, we also called CN-states using the cnAnalysis450k R-package with standard default parameter settings [47].
2.5. Differential Copy-Number Alteration Analysis
Differential CN analysis between normal-healthy and normal-adjacent tissue was performed using a statistical test for differences in binomial proportions, as given by the prop.test function of R. To correct for multiple testing we used a sample-relabeling strategy whereby for each of 100 distinct permutations of phenotype labels we counted the number of genes with P-values as significant or more than the observed ones. These numbers were averaged over the 100 permutations and compared to the observed number of genes passing the same significance P-values.
2.6. Identification of CNV Field Defects
Since the differential CN analysis between normal-healthy and normal-adjacent tissue did not result in genome-wide significance, we adopted a feature selection heuristic that mimics the feature selection step in the iEVORA algorithm. Specifically, we identified genes that exhibited no CN-change across the 50 normal healthy samples (thus being ultra-stable and of zero variance), but which exhibited at least 1 CN alteration across the 42 normal adjacent samples. We verified that there significantly more genes exhibiting this type of pattern, than genes exhibiting the reverse pattern with no CN-change across the 42 normal-adjacent samples and with at least 1 CN alteration across the 50 normal healthy ones. This is the feature selection procedure we implemented when constructing and evaluating risk predictors from the CN-state data.
2.7. Computation of Cancer Risk Scores and Prediction Using AUC Analysis
2.7.1. Internal Cross-Validation
We used a five-fold cross validation strategy and an adaptive-index algorithm [48] to obtain cancer-risk prediction estimates in the Erlangen set. Briefly, the 50 normal (N) and 42 normal-adjacent (NADJ) samples were split into 5 bags, with 4 bags containing 10 Ns and 8 NADJs and one final bag containing 10 Ns and 10 NADJs. At each fold, 4 bags were used for training and feature selection, with 1 bag left as blind test set and for model selection. In the case of DNAm, at each fold we applied iEVORA with FDR(Bartlett-test) < 0.001 and P(t-test) < 0.05 thresholds to a training set consisting of the 4-bags, selecting hypervariable DVMCs (hyperV DVMCs). With these hyperV DVMCs we then estimated a “risk-score” in the leave-one-out (LOO) bag. The risk-score for each sample in the LOO bag was obtained as the fraction of DVMCs exhibiting a significant deviation in DNAm in that sample compared to the normal samples, i.e.
where I is an indicator function with a value 1 if the corresponding beta-value βcs is unlikely to have been derived from a normal with mean μ and standard deviation σ, as estimated across the normal (N) samples, and where the summation is over the DVMCs selected in the training set of 4-bags. The significance of the deviation was determined by computing the z-score of the DNAm value in the sample relative to a Gaussian approximating the distribution of DNAm values in the normal samples. Thus, the risk score depends on two parameters: (i) the specific top-number of DVMCs used to average the score over, and (ii) the significance threshold itself. We allowed these two parameters to vary, defining a grid, for each point in the grid obtaining a risk score. In the case of the significance threshold (pvth), we considered the following values: 1e-5, 5e-5, 1e-4, 5e-4, 0.001, 0.005, 0.01. In the case of the number of top-ranked DVMCs to consider (ntop), we allowed ntop to vary in units of 50 CpGs, starting from 50 and ending at the smallest possible value across the 5 partitions (recall that for each fold, we obtain a different set and number of significant DVMCs). Finally, for each pvth and ntop value, we combine the risk-scores for each LOO-bag over the 5 CV folds, thus allowing us to derive an unbiased measure of discrimination accuracy in blinded samples, and to determine which model (parameter choices) generalizes best. As a measure of discrimination accuracy we used the Area Under the Receiver Operator Characteristic Curve (AUC). This identified ntop = 469 and pvth = 0.001 as an optimal parameter combination.
In the case of CNV, the risk-score for each sample in the LOO bag was obtained as the fraction of selected genes with CN aberrations. The genes were selected based on their frequency of CN alteration (gain/loss) across all normal-adjacent samples of a training set (4 bags) and requiring no CN-change across the normal-healthy ones. Thus, the risk score depends on one parameter, which is the frequency-threshold of CN alteration, i.e. the minimum number of samples exhibiting a CN alteration. For each fold, we varied the frequency from 1 to 10 to select different numbers of genes and generating corresponding risk-scores for the LOO bag. Finally, for each frequency value, we combined the risk-scores for each LOO-bag over the 5 CV folds to subsequently derive an AUC. This identified the optimal parameter to be 4.
2.7.2. External Validation
In order to validate our risk prediction model we used the model with ntop = 469 and pvth = 0.001 with the 469 top-ranked hyperV DVMCs selected from the full training set. Equivalently, one can combine the 5 derived classifiers using an ensemble classifier approach, which leads to near identical results. To obtain risk scores in the external validation set [29], we applied the above model to these external samples, deriving the AUC plus 95% confidence interval. In the case of CNV, we applied the risk prediction model with genes exhibiting at least 4 CN gains or losses in the training (Erlangen) set.
2.7.3. Some Notes
(a) When estimating the risk score as the fraction of hyperV DVMCs exhibiting a significant deviation from the normal state, there could be ties between samples, specially if deviations are infrequent events. To resolve these matches we used the average of the −log10[P-values] to favor samples with higher significance levels. Specifically, for samples with a tied risk-score, R, we computed these averages, rescaling these average values to be between 0 and 1-ε (ε a very small number e.g. 1e-6), denoting these values by λ. For these samples we then define new risk scores using the formula: R' = (Ru – R) λ + R, where Ru is the closest risk-value to R satisfying Ru > R. For λ = 0, R' = R, and for λ = 1- ε, R' = Ru – ε(Ru - R) < Ru, as required. ([60b]) When estimating the risk score in the LOO bags or in the external validation sets, we compute deviations relative to the normal samples from the LOO bags or external validation set. Thus, our classifiers are not single-sample classifiers, and our procedure merely validates selected features. This is justified since the aim of our study is a comparison of the validity and generalizability of the features (in relation to cancer risk) selected from DNAm with those derived from CNV data. ([60c]) In the case of copy-number, we performed the same risk prediction analysis described above, but for 4 separate procedures designed to test the robustness of the conclusions to the process of segmentation and CNV-calling. In one case, we performed the analysis described above but for genomic bins (instead of genes), as defined by the conumee package. In the second case, we performed the same analysis but at the level of individual probes (ie using the log2 of the intensity (I) ratio probe values (I=U + M)). In the third case, we used the segmentation algorithm from the cnAnalysis450k R-package [47] to obtain CN-state calls. In the fourth case, we used again probe-level data but this time not calling CN-states, but running an Elastic Net logistic regression classifier [49] on the log2 I ratio values (I=U + M).
2.8. Further Details on the Elastic Net Classifier Implementation
We used a nested cross-validation strategy to obtain cancer-risk prediction estimates in the Erlangen dataset based on CN log2(I) ratios. Briefly, the 10 normal (N) and 42 normal-adjacent (NADJ) samples were split into 5 bags, with 4 bags containing 10 Ns and 8 NADJs and one final bag containing 10 Ns and 10 NADJs. We used the elastic net model implemented in glmnet R-package for feature selection and risk score estimation [49]. At each fold, 4 bags were used for training to determine which model (parameter choices) generalizes best, with 1 bag left as blind set. The parameter tuning process is based on the nested cross-validation using the cv.glmnet function in glmnet R package, with a line search of alpha starting from 0.1 to 0.9 with 0.05 increase each time. The risk-score for each sample in the LOO bag was obtained using the best model trained in the 4 bags. We found the 5 best models use the alpha 0.45, 0.4, 0.8, 0.65, 0.7 separately as the optimal parameter choice. We combined the risk-scores for each LOO-bag over the 5 CV folds, thus allowing us to derive an unbiased measure of discrimination accuracy in blinded samples. When estimating the risk score in the external validation set, we used the average of coefficient vectors for all 5 models in the training dataset.
2.9. Data Availability
All data analyzed in this manuscript is already publicly available from GEO (www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE69914 and GSE67919, or from the TCGA data portal (https://gdc.cancer.gov).
3. Results
3.1. DNA Methylation Outliers in Normal-Adjacent Samples Mark Changes in Epithelial Cells
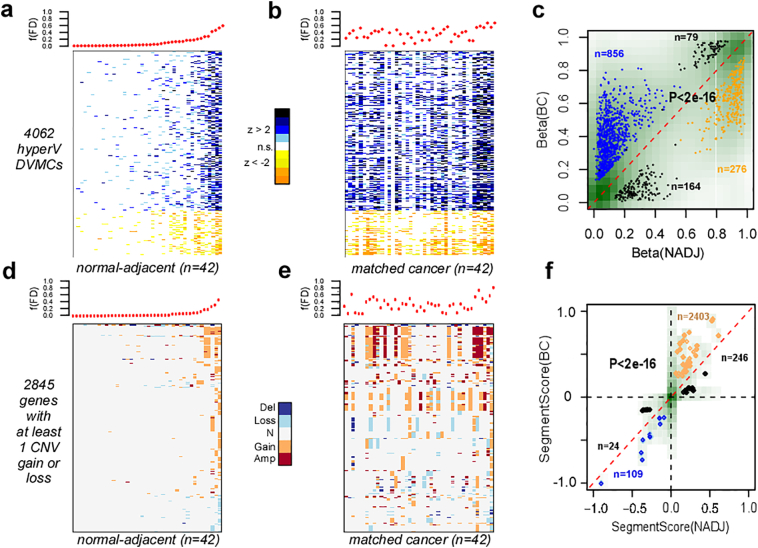
Previously, we used Illumina Infinium 450 k DNAm beadarrays to profile the methylation state of approximately 480,000 CpGs in normal breast tissue from healthy women (n = 50), in the normal breast tissue adjacent to breast cancer (n = 42) and in a total of 305 invasive breast cancers [24]. As shown by us, the normal adjacent samples could not be discriminated from the normal healthy ones using a feature selection paradigm based on differential methylation, as none of the top-ranked differentially methylated CpGs (DMCs) passed genome-wide significance levels [24]. However, using an entirely different feature selection paradigm based on differential DNAm variance, we identified many differentially variable and methylated CpGs (DVMCs) between the two normal tissue types [24]. Importantly, we demonstrated that most DVMCs exhibited increased DNAm variance (termed hyperV DVMCs), in the normal adjacent tissue compared to normal healthy (Fig.1a, Materials & Methods). These hyperV DVMCs may mark epigenetic field defects, as they are more frequently altered across the matched breast cancers (Fig.1b) and were specifically enriched in the breast cancer matched to the given normal-adjacent tissue (Fig.1c).
Fig. 1.
The aberrant CNV and DNAm landscape in normal cells at risk of neoplastic transformation. a) Top panel displays the fraction of “field defects (FD)” f(FD) for all 42 normal-adjacent samples from the Erlangen set, with samples ranked in increasing order. Lower panel is a corresponding heatmap displaying the significance, i.e. z-scores, of DNAm changes relative to the normal healthy breast samples and over the 4062 hypervariable (hyperV) differentially variably and differentially methylated (DVMCs), over which the f(FD) is computed. The z-score is computed relative to the 50 normal healthy samples, and measures the standardized deviation of a sample's DNAm value from a Gaussian with mean and standard deviation as estimated over the 50 normal healthy samples. f(FD) is computed as the fraction of the 4062 DVMCs which exhibit a significant z-score deviation (using a P-value threshold of 0.001). b) Top panel displays the f(FD) for the 42 matched breast cancers, with the women ordered as in a). Lower panel is the corresponding heatmap displaying the significance of DNAm alterations of the same hyperV DVMCs in the breast cancers compared to normal healthy breast tissue. c) Scatterplot of the DNAm values for the 4062 DVMCs in the normal-adjacent sample (x-axis) vs. the corresponding value in the matched breast cancer (y-axis) for all 42 women. Density of data points is displayed in green with darkgreen representing denser regions. The hypermethylated (hypomethylated) DVMCs in normal-adjacent tissue compared to healthy normal, for which the DNAm value in breast cancer was higher by 0.1 (or lower by 0.1) than in the matched normal-adjacent sample are indicated in blue (orange). Data points hypomethylated (hypermethylated) in normal-adjacent tissue relative to normal-healthy but which exhibited significant hypermethylation (hypomethylaton) in cancer are indicated in black. P-value is from a one-tailed Fisher's exact test. d-e) As a-b), but now for the 2845 genes that exhibit at least 1 copy gain or loss across the 42 normal-adjacent samples compared to healthy breast, with f(FD) now defined as the fraction of genes exhibiting a gain or loss across the 2845 genes. d As c), but now plotting the segment value of the gene in normal-adjacent tissue (x-axis) compared to its value in the matched breast cancer (y-axis). Because several genes may map to the same segment, the data points reflect segments rather than genes, hence why there appear to be less data points than expected.
However, breast is a complex tissue made up primarily of epithelial, adipose/stromal and immune cells. Thus, it is important to establish that the DNAm outlier events (i.e. the hyperV DVMCs) are not driven by changes in the stromal milieu, but instead mark DNAm changes in the epithelial compartment. To address this challenge, we first sought to estimate the fractions of epithelial, adipose and immune cells in each sample. To this end, we constructed a DNAm reference matrix consisting of DNAm profiles of pure breast epithelial, adipose and all major immune cell subtypes (CD4 + T-cells, CD8 + T-cells, Natural-Killer Cells, B-cells, Monocytes, Neutrophils and Eosinophils) (Materials & Methods). The reference matrix was defined over a set of 349 CpGs which were highly discriminative of the underlying cell subtypes (Supplementary Table 1). This reference matrix can then be used in conjunction with the EpiDISH algorithm to obtain sample-specific cell type fraction estimates [32]. We validated the reference matrix using in-silico mixtures of independently generated DNAm profiles representing pure breast, fat and immune cell subtypes (Fig.S1a). As further validation, we also applied it to Blueprint Illumina 450 k data representing purified T-cells, B-cells and Monocytes [39], as well as whole-genome bisulfite sequencing (WGBS) data from the International Human Epigenome Consortium (IHEC) [50] (Fig.S1b).
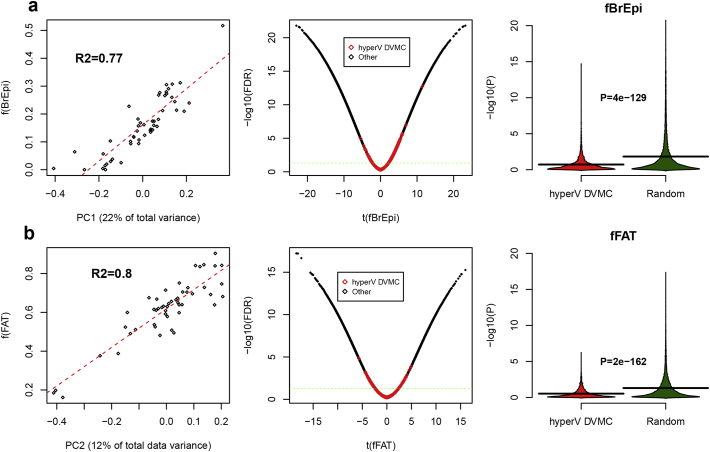
Having validated the reference DNAm matrix, we next applied it to our DNAm dataset of breast samples, estimating sample-specific fractions of epithelial, adipose and immune cells. As assessed over the 50 normal healthy samples, the estimated fraction of epithelial and adipose cells correlated fairly well with the top two principal components (R2 values ~0.8, Fig.2a- [60b]), thus demonstrating that the epithelial-fat ratio is the main source of DNAm variation in breast tissue. Finally, in order to demonstrate that our DNAm outliers (hyperV DVMCs) are not driven by alterations in these cell type fractions, we ranked all CpGs according to their strength of association with these fractions, which confirmed that hyperV DVMCs were significantly underenriched among the most highly correlated features (Fig.2a- [60b]). More formally, we compared the correlation significance P-values of the hyperV DVMCs to those of 10,000 randomly selected non-DVMCs, which confirmed that hyperV DVMCs were significantly less correlated with epithelial or fat content than non-DVMCs (Fig.2a- [60b]). Thus, the DNAm outliers defined by hyperV DVMCs are not caused by changes in the epithelial-fat ratio, and most likely reflect alterations in the epithelial compartment of the breast tissue samples.
Fig. 2.
DNAm outliers (hypervariable DVMCs) in normal-adjacent breast are not driven by changes in the epithelial-adipose ratio. a) Left panel is a scatterplot between the top-PC (PC1, x-axis) against the estimated fraction of breast epithelial cells for the 50 normal healthy breast samples from the Erlangen set [24], as derived from Illumina 450k DNAm data. Linear regression and R2 values are given, demonstrating the strong correlation. Middle panel is a volcano-type plot of the t-statistic of a linear regression of CpGs DNAm profiles against the estimated breast epithelial cell fraction over the same 50 normal healthy samples, for all ~450k CpGs on the array, with the y-axis labeling the significance level of the t-statistic (−log10(FDR)). Green dashed line represents FDR=0.05. Plot shows how hyperV DVMCs (indicated in red) are underenriched among CpGs most strongly associated with variations in the breast epithelial fraction. Right panel compares the significance levels (y-axis) of the hyperV DVMCs against a randomly selected set of 10,000 non-DVMCs. P-value is from a one-tailed Wilcoxon rank sum test, demonstrating that hyperV DVMCs are less associated with variations in the breast epithelial fraction than a random set of CpGs. b) As a), but now for the estimated adipose/fat cell type fractions. The fraction of breast epithelial cells was estimated using the EpiDISH algorithm using robust partial correlations [32] and an extension of the reference DNAm database used in [24]. This reference database included reference DNAm profiles for breast epithelial cells, adipocytes and 7 blood cell subtypes (CD4+ T-cells, CD8+ T-cells, NK-cells, B-cells, Neutrophils, Eosinophils, Monocytes).
3.2. Differential Copy-Number Analysis Between Normal-Adjacent and Normal Tissue Reveals no Genome-Wide Significance
Next, we asked if CNVs differ between normal-adjacent and normal healthy tissue. Since Illumina 450 k data can also be used to derive CNV profiles [28], we obtained CNV calls in the samples and from the same DNA-assay, using a previously validated procedure designed to automatically adjust for cell type heterogeneity (Materials & Methods). Validating the adjustment procedure, the genomic instability index (GII), which reflects the overall amount of aberrant CNV in a normal-adjacent sample, did not correlate with the estimated fraction of breast epithelial cells (Fig.S2). Further validating the procedure, the derived CNV landscape across our 305 invasive breast cancers was highly similar to those derived using SNP-based technologies in the breast cancer TCGA [51] and METABRIC [45] cohorts, and was able to detect known ~500 kb amplicons and deletions (Figs.S3-S5). However, there were also differences in the derived CNV landscapes, which we attribute to differential probe representation between the 450 k and Affymetrix SNP 6.0 arrays used in the TCGA/METABRIC: for instance, frequent loss of a 10 Mb region on 17q containing 3 tumor suppressor genes (AXIN2, BRIP1, CLTC) (Fig.S3) was not observed with SNP arrays, probably due to a sparser representation (3099 SNP probes compared to 9857,450 k probes in this region). As a final check of our CNV-calling procedure, we observed good agreement of estimated frequencies of chromosome 1q gain and chromosome 16q heterozygous loss, with those reported previously (Fig.S6), thus validating our thresholds for single-copy gains and losses.
Having established that our CNV calling procedure is accurate, we next used a statistical procedure that tests for differences in binomial proportions (a well-known Chi-Square test, Materials & Methods), to determine if frequencies of CN gain or loss differ significantly between the 42 normal-adjacent and the 50 normal-healthy tissues. In the case of CN gain, this analysis was done for a total of 4269 genes exhibiting at least 1 CN gain across all 92 samples. Only 147 genes passed an unadjusted P-value threshold of 0.05, with the smallest P-value being 0.019, which corresponded to genes exhibiting no CN gain across the normal samples but 6 gains across the 42 normal-adjacent ones. To assess the overall statistical significance of these 147 genes, we permuted the phenotype-labels a 100 times and recomputed P-values, revealing that only once did more genes pass the same threshold. In contrast to gains, we did not observe any genome-wide significance for differences in the frequency of CN loss between normal and normal-adjacent tissue. Thus, overall, the differential CN analysis did not reveal genome-wide significance, with only a very marginal effect for gains, driven by genes with no CN alteration across normal healthy samples and at least 6 gains across the normal-adjacent ones.
3.3. CNV Field Defects are Enriched in the Adjacent Breast Cancer
The previous differential CNV analysis mirrors the standard differential methylation analysis in that there is no, or very weak, genome-wide statistical significance. However, in the case of DNAm, we observed that many more CpGs were hypervariable in the normal-adjacent breast compared to the normal-healthy tissue [24]. A similar pattern was also evident at the CN-level: a total of 2845 genes exhibited no CN-change across the normal samples with at least 1 CN-change across the 42 normal-adjacent tissues (Fig.1d, Supplementary Table 2), in contrast to 1295 genes exhibiting a reverse pattern (i.e. no CN-change in any of the 42 normal-adjacent samples, but at least 1 CN-change across the 50 normals) (Fig.S7). Thus, based on this, we decided to investigate the pattern of CN alteration of the 2845 genes in the matched breast cancers. As with the hyperV DVMCs, we observed an increase in the frequency of alteration in the invasive breast cancers (Fig.1e). Using the matched 42 breast cancers, we observed that CN-gains in normal-adjacent tissue exhibited a preference for a higher level gain in the matched breast cancer, a pattern also present, but less evident, for CN losses (Fig.1f). This indicates that CN changes in the normal-adjacent cells become more aggravated in the adjacent breast cancer. We note that this pattern of enrichment is similar to that seen at the DNAm level (Fig.1c). Of note, the frequency of CN alteration over the 2845 genes correlated well with the corresponding frequency of DNAm field defects, although this correspondence was only evident for the 4 to 5 samples carrying the largest fractions of alteration (SI Fig. S8).
3.4. DNA Methylation Changes Discriminate Normal-Adjacent from Normal Samples Better than Copy Number Variations
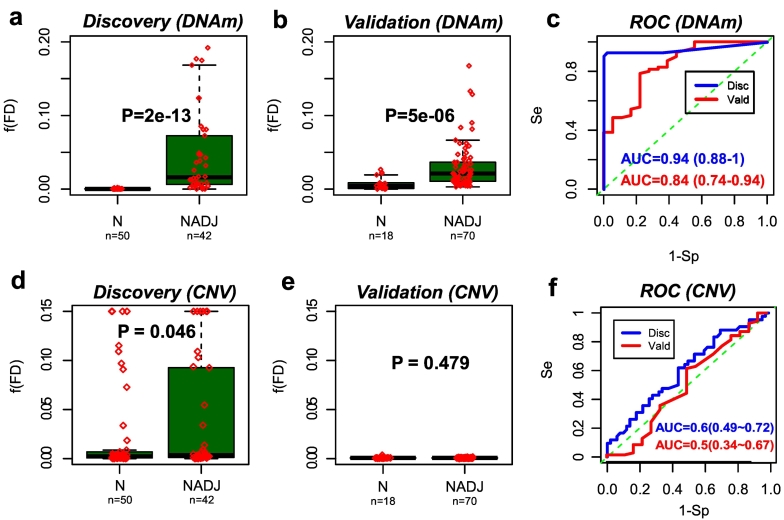
Having identified DNAm and CN alterations in normal-adjacent tissue which become enriched in the matched breast cancers, we next asked which type of alteration better discriminates the 42 normal-adjacent samples (representing normal cells at risk of neoplastic transformation) from the 50 normal healthy ones. To address this, we built predictors of cancer-risk, separately for CNVs and DNAm changes, using a five-fold cross-validation strategy (Materials & Methods). Briefly, for each fold, predictors were developed using a training set and sample-specific risk scores reflecting the overall load of CNV or DNAm alteration over selected loci, were computed in the blind test set. Finally, risk scores were combined over the five folds to give an unbiased estimate of the discrimination accuracy as given by the Area Under the Curve (AUC). We observed that DNAm changes achieved a higher level of discrimination accuracy than CNVs: AUC = 0.94 (95% CI: 0.88–1) for DNAm and AUC = 0.60 (95% CI: 0.49–0.72) for CNVs (Fig. 3a,c,d,f, Figs.S9-10). In fact, for CNV the AUC was not significantly above 0.5.
Fig. 3.
DNAm patterns predict normal-adjacent status better than CNV a.) Boxplot of the fraction of DNAm field defects, [60d](FD), in the Erlangen discovery set, between the 50 normal breast samples from healthy women and the 42 normal-adjacent samples, as assessed using a 5-fold cross-validation and an adaptive index algorithm. P-value is from a one-tailed Wilcoxon rank sum test [60b].) Boxplot of the fraction of DNAm field defects, [60d](FD), in the validation set, between 18 normal breast samples from healthy women and 70 normal-adjacent samples, as assessed using the optimal adaptive index classifier as inferred from the Erlangen discovery set. P-value is from a one-tailed Wilcoxon rank sum test [60c].) Corresponding ROC curves and AUC values plus their 95% confidence intervals. d- [60d]) Exactly as [a-c]), but now for the field defects inferred from CNV data.
Given the limited size of our Erlangen dataset (n = 92), it is critically important to validate the above result in independent data to support its significance. Using the risk predictors derived in the Erlangen set, we thus estimated risk-scores in an independent Illumina 450 k dataset encompassing 18 normal healthy and 70 normal-adjacent breast samples [31]. This confirmed a statistically significant discrimination in the case of DNAm (AUC = 0.84 (95% CI: 0.74–0.94)), while also confirming non-significance in the case of CNV (AUC = 0.50 (95% CI: 0.34–0.67)) (Fig. 3b,c,e,f).
In order to confirm that the difference in discrimination accuracy is not the result of overly stringent thresholds used in the CN segmentation algorithm, nor dependent on the segmentation method itself, we repeated the CN-analysis in 3 different ways: (i) at the probe-level, (ii) at the level of genomic bins, as defined in the conumee package [52] and (iii) using an entirely different copy-number and segmentation package (cnAnalysis450k) [47]. All three analyses confirmed that it was not possible to construct a CNV-based risk classifier that would validate strongly in our independent dataset (Fig.S11). Associations, if any, were only marginal (Fig.S11). We also performed the CN-based analysis at the level of individual probes using a powerful Elastic Net classifier [49,53], which resulted in a negative validation (Fig. S12). Thus, the difference in predictive ability between the DNAm and CNV-based classifiers is not an artefact of the segmentation algorithm or of the parameter choices used in these algorithms. Conversely, to demonstrate the importance of the feature selection framework used in iEVORA in the case of DNAm data, we trained an Elastic Net classifier on the DNAm data using the same 5-fold cross-validation procedure as in the CN-case. This did not result in a consistently significant AUC across both discovery and validation sets (Fig. S13). Our analyses therefore attribute the difference in classification performance to the biological significance of the DNAm outliers in the normal-adjacent samples, and tthe differential variance feature selection algorithm which can robustly identify such outliers.
3.5. The Hyper DVMCs are not Driven by Genomic Loss or Deletions
Because of the nature of the DNAm-assay, genomic regions that are lost or deleted could result in artefactual shifts in DNAm of probes mapping to these regions [28]. Specifically, if the U and M intensity values are not significantly above background, as they would be for probes in deleted regions, DNAm beta-values might hover around 0.5, resulting in hypermethylation if the probe is normally unmethylated, or hypomethylation if the probe is normally methylated. Although our classification analysis above strongly suggests that hyperV DVMCs do not fall within deleted regions (as otherwise the CNV-based risk predictor would perform as well as the DNAm-based one), we sought to obtain independent confirmation of this. The 4062 hyperV DVMCs mapped to a total of 1768 genes, of which 1681 had reliable CN calls. We verified that none of these 1681 genes exhibited a genomic deletion (2-copy loss) in any of the 42 normal-adjacent samples, thus confirming that their aberrant DNAm is not a CN-artefact. Moreover, the fraction of DNAm alterations attributable to a 1-copy loss was very small, exhibiting a maximum per sample of 6% and with 34/42 (81%) normal-adjacent samples exhibiting no single CN-loss at any of the hyperV DVMC probes (Fig. S14).
4. Discussion
The results presented here are relevant to one of the most pressing questions in oncogenesis, namely, what is the relative role of genetic versus epigenetic alterations in the development of cancer. Both types of alteration are seen in normal cells as a function of age and other major risk factors, and their frequency increases in cancer cells themselves. Although for technical reasons we did not consider somatic mutations, we did compare CNVs to DNAm changes in normal breast samples. We found that although both CNV and DNAm alterations seen in normal-adjacent samples become enriched in their matched cancers, that only the epigenetic changes could significantly discriminate normal adjacent from normal healthy tissue. This was demonstrated using a rigorous cross-validation strategy in the discovery set, and further validated in an independent cohort.
Importantly, we verified that the difference in performance between the DNAm and CNV-based predictors was not the consequence of stringent parameter choices when implementing the CN segmentation algorithms, as results were largely unchanged if CN-analyses had been performed at the level of probes, genomic bins or using an altogether different segmentation method. In fact, the analyses presented here confirm the biological and predictive significance of the DNAm outliers in the normal-adjacent tissue, since training a powerful elastic net classifier (which by design does not identify DNAm outliers) on the DNAm data did not result in a positive validation (Fig.S13). Moreover, the feature selection step implemented on the CN-data was designed to mimic the feature selection step in iEVORA, yet the corresponding CN-based predictors failed to validate consistently across the discovery and validation sets (Fig.3f). Further attesting to the greater biological and predictive significance of the DNAm outliers, we observed that the corresponding fraction of DNAm field defects was more variable across the 42 normal-adjacent samples, enabling more of these to be discriminated from the normal healthy ones (Fig.1a,d). Intriguingly, while there was concordance between the 4 or 5 samples with the highest DNAm and CNA field defect loads (Fig.S8), the specific CNAs in these samples were generally not representative for the rest of the normal-adjacent samples. Thus, we conclude that the improved prediction derived from the DNAm data is driven by the biological significance of the DNAm outliers, and the differential variance feature selection step in iEVORA, that allows these outliers to be identified.
While this important finding does not imply that DNAm changes are functionally more important in the development of cancer, it clearly indicates that epigenetic changes might represent more relevant cancer risk biomarkers. For instance, by measuring DNAm at the hyperV DVMCs in cell-free or circulating DNA it may be possible to develop non-invasive early detection or risk prediction tests assuming enough precursor cell DNA can be captured [54, 55, 56, 57].
The data presented here is also consistent with other studies suggesting that DNAm alterations may play a more causative role than CNVs in the earliest stages of carcinogenesis. First, risk prediction of an epithelial carcinoma (specifically, high grade cervical intraepithelial neoplasia), has been shown to be possible with DNAm patterns in the cell of origin, and using a differential variability feature selection algorithm similar to the one used here [25]. Second, age-associated DNAm alterations preferentially target developmental transcription factors (TFs), and as shown recently by us, tissue-specific transcription factors are also preferentially silenced in the corresponding cancer-type, with promoter hypermethylation emerging as the dominant associative mechanism [58,59]. Indeed, many of the hyperV DVMCs map to binding sites of developmental TFs, suggesting that deregulation of TF-binding via DNAm changes at the regulatory elements might indeed be an early event that contributes to oncogenesis. In contrast, tissue-specific TFs silenced in the corresponding cancer type were not enriched for copy-number deletions [59]. Third, two recent studies have confirmed the importance of DNAm alterations as predictors of cancer risk, one in gastric cancer [17] and another in both gastric and esophageal cancer [18]. Although the former study found that somatic CNAs can predict the risk of progression of an intestinal metaplasia to gastric cancer as well as DNAm, a detailed comparative analysis was not performed. The second study compared DNAm to somatic mutations, concluding that DNAm alterations may be important predictors of cancer risk in the stomach, but not so in the esophagus. The findings obtained in gastric cancer are consistent between the two studies, indicating that DNAm changes are more reliable indicators of cancer risk than somatic mutations. Our findings are in line with these two studies in that DNAm alterations are indicative of cancer-risk. We stress however that our comparative study between DNAm and CNV was restricted to breast cancer, and therefore it is entirely plausible that the cancer risk prediction potential of CN-changes may be very different in other cancer types, similar to what has been observed for somatic mutations [18].
We acknowledge that the results presented here need to be interpreted with caution, as our study has a number of other additional limitations. One caveat is that the improved prediction performance of DNAm over CNVs could be due to the use of a technology which was designed to measure DNAm and not CNV. However, we and others have clearly demonstrated that the intensity values (i.e. the sum of methylated and unmethylated intensities) provided by the Illumina 450 k technology are perfectly suitable to detect both large-scale aberrations as well as small scale amplicons and deletions [28,47]. Moreover, the ability to detect DNAm and CNV from the same DNA sample can be seen as a succinct technical advantage of the approach taken here, since technological biases and confounders (e.g. signal distribution, dynamic range and background signal) are accounted for by using the same assay for two data-types. On the other hand, the reduced genomic coverage of the 450 k beadarray, which is limited to approximately 480,000 CpGs, imposes a major limitation, as it has a sparse representation for regions with low CpG density. The technology is also unable to measure signals related to methylation or copy number gains at repeat elements, which is a good proxy for genome stability. Thus, overall, we stress caution when extrapolating the results obtained here to those that we would obtain using whole-genome profiling.
Another limitation is that our study was not of a prospective nature. For epithelial cancers it is technically and logistically challenging to set-up prospective studies due to the need to collect the cell of origin in advance of diagnosis, which is generally not easily accessible. One exception is cervical cancer, and a previous study showed that DNA methylation profiles measured in cytologically normal cervical samples could discriminate women who developed a cervical intraepithelial neoplasia of grade 2 or higher (CIN2+) within 3 years from those who did not [25]. This is important, because we observe that the patterns of DNAm in normal cervix at risk of neoplastic transformation are similar to those seen in the normal breast tissue found adjacent to breast cancer [24], in both cases with hypervariable DVMCs/DVCs mapping preferentially to targets of the polycomb-repressive complex PRC2. Thus, using normal-adjacent samples may indeed be a valuable strategy to identify cancer risk biomarkers. Of note, the normal-adjacent breast samples analyzed here were also taken at a wide margin, specifically at least 3 cm away from the invasive cancer boundary, which means that contamination of normal-adjacent samples by neighboring cancer cells is unlikely to explain the 20 to 30% methylation differences seen among the hyperV DVMCs. In this regard, we also showed, by adjusting for the epithelial-adipose ratio of the breast samples, that these DNAm outliers are not artefacts of changes in this ratio. Indeed, the differential variability feature selection algorithm we used here identifies CpGs that exhibit fairly stable DNAm values across all the healthy normal breast samples, and this is only possible if they are not differentially methylated between the major cell types in breast tissue (epithelial and adipose cells), as shifts in the epithelial-adipose ratio drive most of the DNAm variation across the normal healthy samples.
In summary, our analysis suggests that DNAm and CNV alterations in normal cells adjacent to breast cancer are enriched in the matched tumors, but that only DNAm changes can discriminate the normal-adjacent from normal-healthy samples. Thus, epigenetic alterations may constitute more relevant cancer risk biomarkers, which supports a model of oncogenesis whereby epigenetic alterations play a more fundamental role in the earliest stages of cancer development.
The following are the supplementary data related to this article.
Supplementary material
Acknowledgements & Funding Sources
This work was supported by the Eve Appeal, NSFC (National Science Foundation of China) grants, grant numbers 31571359 and 31401120 and by a Royal Society Newton Advanced Fellowship (NAF project number: 522438, NAF award number: 164914). The authors are supported by the European Union's Horizon 2020 Programme (H2020/2014-2020) under grant agreement number 634570 (Project FORECEE: www.forecee.eu/) and MW also receives support from the European Research Council (ERC Advanced Grant ERC-BRCA) and the National Institute for Health Research (NIHR) University College London Hospitals (UCLH) Biomedical Research Centre.
Declaration of Interests
The authors declare that they have no competing interests.
Author Contributions
Manuscript was conceived and written by AET with contributions from GY and MW. Statistical analyses were performed by GY and AET.
References
- 1.Feinberg A.P., Ohlsson R., Henikoff S. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 2.Genovese G., Kahler A.K., Handsaker R.E., Lindberg J., Rose S.A., Bakhoum S.F. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Issa J.P. Epigenetic variation and cellular Darwinism. Nat. Genet. 2011;43:724–726. doi: 10.1038/ng.897. [DOI] [PubMed] [Google Scholar]
- 4.Jacobs K.B., Yeager M., Zhou W., Wacholder S., Wang Z., Rodriguez-Santiago B. Detectable clonal mosaicism and its relationship to aging and cancer. Nat. Genet. 2012;44:651–658. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P.V., Mar B.G. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laurie C.C., Laurie C.A., Rice K., Doheny K.F., Zelnick L.R., McHugh C.P. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat. Genet. 2012;44:642–650. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levine M.E., Hosgood H.D., Chen B., Absher D., Assimes T., Horvath S. DNA methylation age of blood predicts future onset of lung cancer in the women's health initiative. Aging. 2015;7:690–700. doi: 10.18632/aging.100809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Severi G., Southey M.C., English D.R., Jung C.H., Lonie A., McLean C. Epigenome-wide methylation in DNA from peripheral blood as a marker of risk for breast cancer. Breast Cancer Res. Treat. 2014;148:665–673. doi: 10.1007/s10549-014-3209-y. [DOI] [PubMed] [Google Scholar]
- 9.Teschendorff A.E., Menon U., Gentry-Maharaj A., Ramus S.J., Weisenberger D.J., Shen H. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20:440–446. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomasetti C., Vogelstein B., Parmigiani G. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc. Natl. Acad. Sci. U. S. A. 2013;110:1999–2004. doi: 10.1073/pnas.1221068110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Z., Wong A., Kuh D., Paul D.S., Rakyan V.K., Leslie R.D. Correlation of an epigenetic mitotic clock with cancer risk. Genome Biol. 2016;17:205. doi: 10.1186/s13059-016-1064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spira A., Yurgelun M.B., Alexandrov L., Rao A., Bejar R., Polyak K. Precancer atlas to drive precision prevention trials. Cancer Res. 2017;77:1510–1541. doi: 10.1158/0008-5472.CAN-16-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dumanski J.P., Rasi C., Lonn M., Davies H., Ingelsson M., Giedraitis V. Mutagenesis. Smoking is associated with mosaic loss of chromosome Y. Science. 2015;347:81–83. doi: 10.1126/science.1262092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsberg L.A., Rasi C., Malmqvist N., Davies H., Pasupulati S., Pakalapati G. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat. Genet. 2014;46:624–628. doi: 10.1038/ng.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper C.S., Eeles R., Wedge D.C., Van Loo P., Gundem G., Alexandrov L.B. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat. Genet. 2015;47:367–372. doi: 10.1038/ng.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoang M.L., Kinde I., Tomasetti C., McMahon K.W., Rosenquist T.A., Grollman A.P. Genome-wide quantification of rare somatic mutations in normal human tissues using massively parallel sequencing. Proc. Natl. Acad. Sci. U. S. A. 2016;113:9846–9851. doi: 10.1073/pnas.1607794113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang K.K., Ramnarayanan K., Zhu F., Srivastava S., Xu C., Tan A.L.K. Genomic and epigenomic profiling of high-risk intestinal metaplasia reveals molecular determinants of progression to gastric Cancer. Cancer Cell. 2018;33:137–150. doi: 10.1016/j.ccell.2017.11.018. (e135) [DOI] [PubMed] [Google Scholar]
- 18.Yamashita S., Kishino T., Takahashi T., Shimazu T., Charvat H., Kakugawa Y. Genetic and epigenetic alterations in normal tissues have differential impacts on cancer risk among tissues. Proc. Natl. Acad. Sci. U. S. A. 2018;115:1328–1333. doi: 10.1073/pnas.1717340115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baglietto L., Ponzi E., Haycock P., Hodge A., Bianca Assumma M., Jung C.H. DNA methylation changes measured in pre-diagnostic peripheral blood samples are associated with smoking and lung cancer risk. Int. J. Cancer. 2017;140:50–61. doi: 10.1002/ijc.30431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fasanelli F., Baglietto L., Ponzi E., Guida F., Campanella G., Johansson M. Hypomethylation of smoking-related genes is associated with future lung cancer in four prospective cohorts. Nat. Commun. 2015;6 doi: 10.1038/ncomms10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Z., Bolick S.C., DeRoo L.A., Weinberg C.R., Sandler D.P., Taylor J.A. Epigenome-wide association study of breast cancer using prospectively collected sister study samples. J. Natl. Cancer Inst. 2013;105:694–700. doi: 10.1093/jnci/djt045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernstein C., Nfonsam V., Prasad A.R., Bernstein H. Epigenetic field defects in progression to cancer. World J. Gastrointest. Oncol. 2013;5:43–49. doi: 10.4251/wjgo.v5.i3.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones A., Teschendorff A.E., Li Q., Hayward J.D., Kannan A., Mould T. Role of DNA methylation and epigenetic silencing of HAND2 in endometrial cancer development. PLoS Med. 2013;10 doi: 10.1371/journal.pmed.1001551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teschendorff A.E., Gao Y., Jones A., Ruebner M., Beckmann M.W., Wachter D.L. DNA methylation outliers in normal breast tissue identify field defects that are enriched in cancer. Nat. Commun. 2016;7 doi: 10.1038/ncomms10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teschendorff A.E., Jones A., Fiegl H., Sargent A., Zhuang J.J., Kitchener H.C. Epigenetic variability in cells of normal cytology is associated with the risk of future morphological transformation. Genome Med. 2012;4:24. doi: 10.1186/gm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ushijima T. Epigenetic field for cancerization. J. Biochem. Mol. Biol. 2007;40:142–150. doi: 10.5483/bmbrep.2007.40.2.142. [DOI] [PubMed] [Google Scholar]
- 27.Ushijima T., Hattori N. Molecular pathways: involvement of helicobacter pylori-triggered inflammation in the formation of an epigenetic field defect, and its usefulness as cancer risk and exposure markers. Clin. Cancer Res. 2012;18:923–929. doi: 10.1158/1078-0432.CCR-11-2011. [DOI] [PubMed] [Google Scholar]
- 28.Feber A., Guilhamon P., Lechner M., Fenton T., Wilson G.A., Thirlwell C. Using high-density DNA methylation arrays to profile copy number alterations. Genome Biol. 2014;15:R30. doi: 10.1186/gb-2014-15-2-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knoll M., Debus J., Abdollahi A. cnAnalysis450k: an R package for comparative analysis of 450k/EPIC illumina methylation array derived copy number data. Bioinformatics. 2017;33:2266–2272. doi: 10.1093/bioinformatics/btx156. [DOI] [PubMed] [Google Scholar]
- 30.Marzouka N.A., Nordlund J., Backlin C.L., Lonnerholm G., Syvanen A.C., Carlsson Almlof J. CopyNumber450kCancer: baseline correction for accurate copy number calling from the 450k methylation array. Bioinformatics. 2016;32:1080–1082. doi: 10.1093/bioinformatics/btv652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hair B.Y., Xu Z., Kirk E.L., Harlid S., Sandhu R., Robinson W.R. Body mass index associated with genome-wide methylation in breast tissue. Breast Cancer Res. Treat. 2015;151:453–463. doi: 10.1007/s10549-015-3401-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teschendorff A.E., Breeze C.E., Zheng S.C., Beck S. A comparison of reference-based algorithms for correcting cell-type heterogeneity in epigenome-wide association studies. BMC Bioinformatics. 2017;18:105. doi: 10.1186/s12859-017-1511-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowe R., Overhoff M.G., Ramagopalan S.V., Garbe J.C., Koh J., Stampfer M.R. The senescent methylome and its relationship with cancer, ageing and germline genetic variation in humans. Genome Biol. 2015;16:194. doi: 10.1186/s13059-015-0748-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nazor K.L., Altun G., Lynch C., Tran H., Harness J.V., Slavin I. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10:620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reinius L.E., Acevedo N., Joerink M., Pershagen G., Dahlen S.E., Greco D. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. (Article3) [DOI] [PubMed] [Google Scholar]
- 37.Gerstein M.B., Kundaje A., Hariharan M., Landt S.G., Yan K.K., Cheng C. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slieker R.C., Bos S.D., Goeman J.J., Bovee J.V., Talens R.P., van der Breggen R. Identification and systematic annotation of tissue-specific differentially methylated regions using the Illumina 450k array. Epigenetics Chromatin. 2013;6:26. doi: 10.1186/1756-8935-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paul D.S., Teschendorff A.E., Dang M.A., Lowe R., Hawa M.I., Ecker S. Increased DNA methylation variability in type 1 diabetes across three immune effector cell types. Nat. Commun. 2016;7 doi: 10.1038/ncomms13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stunnenberg H.G., International Human Epigenome, Hirst M. The international human epigenome consortium: a blueprint for scientific collaboration and discovery. Cell. 2016;167:1897. doi: 10.1016/j.cell.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 41.Teschendorff A.E., Jones A., Widschwendter M. Stochastic epigenetic outliers can define field defects in cancer. BMC Bioinformatics. 2016;17:178. doi: 10.1186/s12859-016-1056-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hovestadt V., Zapatka M. 2017. Conumee: enhanced copy-number variation analysis using illumina DNA methylation arrays. (1.9.0 edn (CRAN, 2017)) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olshen A.B., Venkatraman E.S., Lucito R., Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 45.Curtis C., Shah S.P., Chin S.F., Turashvili G., Rueda O.M., Dunning M.J. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chin S.F., Teschendorff A.E., Marioni J.C., Wang Y., Barbosa-Morais N.L., Thorne N.P. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol. 2007;8:R215. doi: 10.1186/gb-2007-8-10-r215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knoll M., Debus J., Abdollahi A. cnAnalysis450k: an R package for comparative analysis of 450k/EPIC illumina methylation array derived copy number data. Bioinformatics. 2017;33:2266–2272. doi: 10.1093/bioinformatics/btx156. [DOI] [PubMed] [Google Scholar]
- 48.Tian L., Tibshirani R. Adaptive index models for marker-based risk stratification. Biostatistics. 2011;12:68–86. doi: 10.1093/biostatistics/kxq047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Friedman J., Hastie T., Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 50.Stunnenberg H.G., International Human Epigenome, Hirst M. The international human epigenome consortium: a blueprint for scientific collaboration and discovery. Cell. 2016;167:1145–1149. doi: 10.1016/j.cell.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 51.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hovestadt V., Zapatka M. 2017. Conumee: Enhanced Copy-Number Variation Analysis Using Illumina DNA Methylation Arrays. (CRAN) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waldmann P., Meszaros G., Gredler B., Fuerst C., Solkner J. Evaluation of the lasso and the elastic net in genome-wide association studies. Front. Genet. 2013;4:270. doi: 10.3389/fgene.2013.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo S., Diep D., Plongthongkum N., Fung H.L., Zhang K., Zhang K. Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nat. Genet. 2017;49:635–642. doi: 10.1038/ng.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lehmann-Werman R., Neiman D., Zemmour H., Moss J., Magenheim J., Vaknin-Dembinsky A. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E1826–1834. doi: 10.1073/pnas.1519286113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Widschwendter M., Evans I., Jones A., Ghazali S., Reisel D., Ryan A. Methylation patterns in serum DNA for early identification of disseminated breast cancer. Genome Med. 2017;9:115. doi: 10.1186/s13073-017-0499-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Widschwendter M., Zikan M., Wahl B., Lempiainen H., Paprotka T., Evans I. The potential of circulating tumor DNA methylation analysis for the early detection and management of ovarian cancer. Genome Med. 2017;9:116. doi: 10.1186/s13073-017-0500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y., Widschwendter M., Teschendorff A.E. Systems-epigenomics inference of transcription factor activity implicates aryl-hydrocarbon-receptor inactivation as a key event in lung cancer development. Genome Biol. 2017;18:236. doi: 10.1186/s13059-017-1366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Teschendorff A.E., Zheng S.C., Feber A., Yang Z., Beck S., Widschwendter M. The multi-omic landscape of transcription factor inactivation in cancer. Genome Med. 2016;8:89. doi: 10.1186/s13073-016-0342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
All data analyzed in this manuscript is already publicly available from GEO (www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE69914 and GSE67919, or from the TCGA data portal (https://gdc.cancer.gov).