Abstract
Treatment-induced neuroendocrine prostate cancer (t-NEPC) is an aggressive subtype of prostate cancer (PCa) that becomes more prevalent when hormonal therapy, chemotherapy, or radiation therapy is applied to patients with metastatic prostate adenocarcinoma (AdPC). How AdPC cells survive these anti-cancer therapies and progress into t-NEPC remains unclear. By comparing the whole transcriptomes between AdPC and t-NEPC, we identified Bif-1, an apoptosis-associated gene, which undergoes alternative RNA splicing in t-NEPC. We found that while Bif-1a is the predominant variant of the Bif-1 gene in AdPC, two neural-specific variants, Bif-1b and Bif-1c, are highly expressed in t-NEPC patients, patient derived xenografts, and cell models. The neural-specific RNA splicing factor, SRRM4, promotes Bif-1b and Bif-1c splicing, and the expression of SRRM4 in tumors is strongly associated with Bif-1b/-1c levels. Furthermore, we showed that Bif-1a is pro-apoptotic, while Bif-1b and Bif-1c are anti-apoptotic in PCa cells under camptothecin and UV light irritation treatments. Taken together, our data indicate that SRRM4 regulates alternative RNA splicing of the Bif-1 gene that enables PCa cells resistant to apoptotic stimuli under anti-cancer therapies, and may contribute to AdPC progression into t-NEPC.
Keywords: Bif-1, SRRM4, Alternative RNA splicing, Neuroendocrine prostate cancer, Apoptosis
Abbreviations: PCa, prostate cancer; t-NEPC, treatment-induced neuroendocrine prostate cancer; Bif-1, Bax-interacting factor 1; Bax, BCL-2 Associated X Protein; AdPC, prostate adenocarcinoma; SRRM4, Serine/Arginine Repetitive Matrix 4; NE, neuroendocrine; RISH, RNA in situ hybridization; SCNC, small cell neuroendocrine carcinoma; CPT, camptothecin; N-BAR, N-terminal Bin-amphiphysin-Rvs
Highlights
-
•
Alternative RNA splicing of the apoptosis-related gene, Bif-1, is associated with the development of t-NEPC.
-
•
SRRM4 regulates alternative RNA splicing of the Bif-1 gene.
-
•
Bif-1a in AdPC cells is pro-apoptotic, while neural Bif-1 variants, Bif-1b/-1c, enable tumor cells resistant to apoptosis.
Treatment-induced neuroendocrine prostate cancer (t-NEPC) is an aggressive subtype of castration-resistant prostate cancer. It becomes more prevalent when more potent androgen receptor inhibitors are applied to patients. However, mechanisms by which t-NEPC develops remain unclear. Here we report alternative RNA splicing of the apoptosis-related gene, Bif-1, may contribute to t-NEPC establishment. We show that the expression of neural Bif-1 variants is upregulated in t-NEPC, and confers tumor cells resistance to apoptotic stimuli. We propose that tumor cells have to first develop mechanisms to counteract therapy-induced cell death before they can undergo neuroendocrine differentiation for t-NEPC.
1. Introduction
While primary (de novo) neuroendocrine prostate cancer (NEPC) is extremely rare, accounting for ~1% of all prostate cancers (PCa) [9], treatment-induced NEPC (t-NEPC) is more prevalent in patients who have a history of adenocarcinoma (AdPC), and have received single or multiple rounds of hormonal therapy, radiation therapy, or chemotherapy [24]. T-NEPC is responsible for approximately 25% of PCa-related deaths [2,22,32]. Because t-NEPC is highly aggressive and metastatic, once a diagnosis is confirmed patient survival is only ~7 months [34]. Beyond systematic chemotherapy, no targeted therapy is currently available due to our limited knowledge on the molecular underpinning of t-NEPC development.
At this time, a consensus on how AdPC is transformed into t-NEPC has not been reached. However, multiple hypotheses have been proposed including that t-NEPC originates from: i) PCa stem-like cells that retain traits of resistance to apoptosis under anti-cancer therapy, self-renewal, and invasion [29]; ii) AdPC cells that undergo NE differentiation followed by t-NEPC tumorigenesis [5,20,39]; or iii) benign prostatic neuroendocrine cells that acquire tumorigenesis capacity [26]. Whole-exome sequencing had revealed that t-NEPC and AdPC cells in patients have similar genetic mutation landscapes, including the distribution of non-silent point mutations, polyploidy, and somatic copy-number genomic burden [5,20]. These findings support that t-NEPC is likely derived from AdPC.
Because AdPC and t-NEPC share similar genomic features but have dramatically different transcriptomes, we hypothesized that alternative RNA splicing may play a key role in AdPC progression to t-NEPC. Through analyzing published whole-transcriptome sequencing data sets from two patient cohorts [4,13] and paying particular interests in identifying RNA splicing events that are unique to t-NEPC, we have identified a t-NEPC specific RNA splicing signature that is predominantly controlled by the RNA splicing factor, SRRM4 [19]. We have further demonstrated that SRRM4 not only induces a NEPC transcriptome and neuroendocrine (NE)-like morphology to AdPC cells, but more importantly transforms AdPC cells into NEPC xenografts [19]. We have subcutaneously inoculated LNCaP cells that overexpress SRRM4 into castrated nude mice continuously for five passages over 18 months, and generated a series of t-NEPC xenograft models, called LnNE [18]. These LnNE tumors express strong NE markers and present with NEPC morphology. Tumors at later passages grow more aggressively and become androgen-insensitive and PSA negative [18]. These features mimic the characteristics of AdPC progression to t-NEPC in patients. Together, these findings highlight that SRRM4 is a clinically relevant driver gene of t-NEPC by regulating alternative RNA splicing of multiple genes. Therefore, further characterization of these SRRM4 target genes would help understand how AdPC progresses into t-NEPC in patients.
Since t-NEPC develops after patients are given anti-cancer therapies for AdPC, we speculate that the AdPC cells that give rise to t-NEPC should have phenotypes that render them resistant to therapy-induced cell death. Through investigating the SRRM4 transcriptome, we have found that an apoptotic-associated gene, Bax interacting factor-1 (Bif-1), undergoes alternative RNA splicing in t-NEPC. There are three major splice variants of the Bif-1 gene in human cells [6,23,25]. Inclusions of exons 6s and 7, or exons 6l and 7, into Bif-1a (NM_016009.4) mRNA give rise to Bif-1b (NP_001193581) and Bif-1c (XP_006710735), respectively. Most studies on the Bif-1 gene have been focused on Bif-1a, since it is the predominant variant in non-neuronal cells [6,30]. Bif-1a and Bax are pro-apoptotic proteins that are localized in the cytosol in their inactive forms. When activated by apoptotic stimuli, they are translocated to the mitochondrial membrane for mitochondria-mediated apoptosis [6]. Bif-1a was reported to either activate Bax to promote mitochondrial outer membrane permeabilization [8,30], or self-oligomerize independent of Bax to form pores on the mitochondrial membrane and cause apoptosis [28]. In neuronal cells, Bif-1b and Bif-1c are the predominant forms of the Bif-1 gene, and their cellular functions are relatively unclear. However, Morrison et al. reported that Bif-1 in neuronal cells promotes cell viability and maintains mitochondrial morphology [33]. Together, these findings implied that Bif-1 splice variants may have opposite effects to cell apoptosis. Therefore, this study aimed to characterize the function of alternative RNA splicing of the Bif-1 gene in PCa cells during t-NEPC development.
2. Materials and Methods
2.1. Ethics Approval and Consent to Participate
Studies involving human data or human tissues were approved by the Research Ethics Committee of the University of British Columbia (H09-01628) and performed according to relevant guidelines and regulations expressed in the Declaration of Helsiniki. Informed consent was obtained from all participating individuals.
2.2. RNA-seq Data from PCa Patients
Whole transcriptome sequencing data from two Vancouver Prostate Centre (VPC) and Beltran patient cohorts as well as the LnNE tumor models are from our previous publications [4,13,18].
2.3. Tissue Microarrays (TMAs)
Prostate tumor samples were extracted from the VPC tissue bank and used to build a castration-resistant PCa TMA, as previously reported [17,36]. This TMA contains 64 tissue cores from 32 patients who had received hormonal therapy, chemotherapy or radiotherapy. The recurred tumors were removed by transurethral resection prostatectomy to relieve lower urinary tract symptoms. This TMA also contains two brain tissue cores from donors.
2.4. RNA In Situ Hybridization (RISH) and Immunohistochemistry (IHC)
The RISH probes targeting the 952–1003 bp of NM_001206652.1 for both Bif-1b and -1c variants, and the 496–835 bp of NM_194286.3 for SRRM4 were designed by Advanced Cell Diagnostic (Hayward, USA). A probe targeting the dapB gene of bacteria was used as a negative control probe. RISH assays were performed using the BaseScope™ assay kit following manufacture's instruction. IHC was performed by Ventana Discovery XT (Ventana) using a DAB MAP kit, as previously reported [17,36]. A Leica SCN400 scanner to form digital images scanned all stained slides.
Positive RISH signals were presented as red dots under 40 × magnification. RISH signals were scored as 0 if no positive signal; 1 if RISH signals were positive in ≤ 20% of all cells within a core; and 2 if RISH signals were positive in > 20% of the cells in the entire tissue core. RISH positive cells with a score of 1 usually have ≤ 2 RISH dots/cell, and RISH positive cells with a score of 2 usually have multiple dots that can merge into dot clusters. IHC scores of CHGA, SYP, CD56, AR, and PSA were calculated by IHC signal intensity (no, low, medium, and high as 0–3) multiplied by the percentage of positive cells (0–100%). IHC scores ≥ 0.3 were considered to be positive.
The histology of castration-resistant tumors is classified either as AdPC, small cell neuroendocrine carcinoma (SCNC), or AdPC with abundant neuroendocrine cells (AdNC). AdPC contains tumor cells that form glandular structures. Comparing to benign prostate glands, AdPC glands are smaller, more compacted, and homogeneous. AdPC cells are large, with vesicular nuclei and prominent nucleoli. AdPC have rare NE cells. SCNC contain only NE tumor cell populations that grow as solid sheets, cords, or individual cells without glandular formation. The tumor cells exhibit NE features including hyperchromatic nuclei, finely granular and homogenous chromatin pattern, and no nucleoli. Cells have scant cytoplasm and high nucleus:cytoplasm ratios. SCNC often contain areas of necrosis and crush artifact. Mitotic and apoptotic figures are frequent. AdNC are more histologically similar to AdPC than to SCNC, but cannot be classified as typical AdPC or SCNC. These tumors contain mixed cell populations with a large number (> 10%) of NE cells.
2.5. PCa Cell Models and Patient Derived Xenografts
LNCaP, 22RV-1, PC-3, DU145, NCI-H660 PCa cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Dr. Rennie from the Vancouver Prostate Centre generously provided C4-2 and 293 T cell lines. LNCaP95 cells were a kind gift from Dr. Alan Meeker of Johns Hopkins University. Cell culture conditions were described before [14, 15, 16, 17,19,21]. AdPC and NEPC patient derived xenografts (PDXs) were previously reported [20] and shared by Dr. Yuzhuo Wang from Vancouver Prostate Centre.
2.6. PCR and Immunoblotting Assays
Real-time qPCR and immunoblotting arrays were performed as previously reported [21]. Information on primers and antibodies is listed in Tables S1 and 2, respectively. Experiments were repeated at least three times.
2.7. siRNA and DNA Transient Transfection
Cells were transfected with control siRNA (Dharmacon) and siRNA SMARTpool targeting total Bif-1 (#L-017086-00-0005, Dharmacon) and SRRM4 (#L-019322-02-0005, Dharmacon) using Lipofectamine RNAiMAX (Invitrogen). Lipofectamine 3000 (Invitrogen) and SuperFect Transfection Reagent (QIAGEN) were used for plasmid transfection. Expression vectors for SRRM4, PTB, ASF/SF2, PSF, U2AF65, hnRNP A1 were previously described [14,21].
2.8. In Vivo RNA Binding Assays
In vivo RNA binding assays were performed as we reported previously [14,19,21]. Briefly, LNCaP cells were transfected with Flag-tagged SRRM4 plasmid, followed by formaldehyde treatment to crosslink SRRM4 with its RNA substrates. After immunoprecipitation of SRRM4, eluted RNA fragments were used as templates to measure the enrichment of SRRM4 binding sites at the Bif-1 gene. Data were from three replicated experiments.
2.9. Construction of Expression Plasmids and PCa Cell Lines by Lentiviral Approaches
Construction of lentivirus vector followed the procedure as we reported [17,19]. The pEGFP-N2-Bif-1a plasmid was kindly provided by Dr. Hong-Gang Wang (Penn State University College of Medicine), and was used as the template to clone the cDNA of Bif-1a, -1b, and -1c into the pCMV2 for transient transfection, and pDONR221 and pFUGWBW vectors for lentivirus infections as we reported [35]. LNCaP, PC-3 and C4-2 cells were infected with lentivirus encoding control or Bif-1 variants followed by blasticidin selection. Sanger sequencing was used to validate all expression vectors.
2.10. Statistics
Statistical analysis was performed using the GraphPad Prism 5.01 software (GraphPad Software, CA, USA). Differences between the two groups were compared by unpaired student t-test. One-way ANOVA followed by a Newman-Keuls multiple comparison test was used to compare differences among multiple groups. The proportion of Bif-1b/-1c RISH scores among different tumor groups was compared based on Fisher exact test. Person correlation analysis was employed to test the correlation between RISH scores and IHC scores, and between RISH scores and number of positive NE markers. The sensitivity and specificity of Bif-1b/-1c to detect t-NEPC were calculated as described [1]. The levels of significance were set at p < 0.05 as *, p < 0.01 as ** and p < 0.001 as ***.
3. Results
3.1. Whole-transcriptome Sequencing Identifies Alternative RNA Splicing of the Bif-1 Gene in t-NEPC
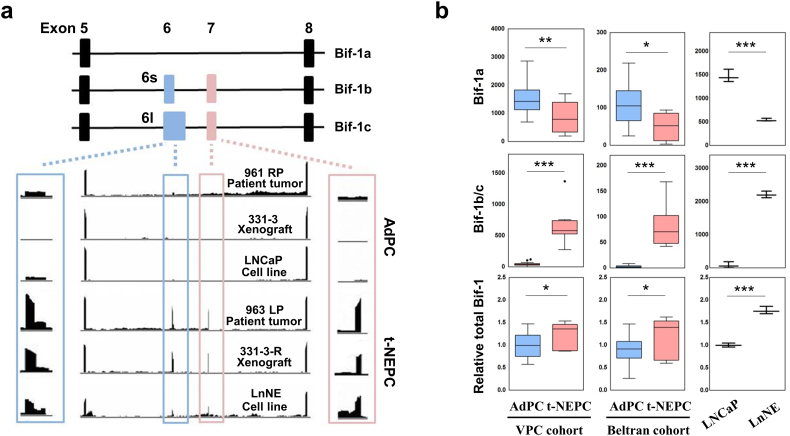
Three whole-transcriptome sequencing data sets all indicated that the neural-specific variants of Bif-1b and Bif-1c were expressed at extremely low levels in AdPC, but were dramatically upregulated in t-NEPC (Fig. 1a). These data are from: i) the VPC cohort that contains 31 AdPC and 7 NEPC samples [20]; ii) the Beltran cohort that contains 30 AdPC and 6 NEPC patient samples [4]; and 3) triplicate RNA samples from LNCaP and LnNE cell models [19]. In both the VPC and Beltran patient cohorts, there is a 42–52% decrease in Bif-1a mRNA levels in t-NEPC comparing to AdPC patient samples. However, Bif-1b and Bif-1c mRNA levels in t-NEPC are 16–31 fold of that in AdPC, resulting in moderate increases of total Bif-1 mRNA levels in t-NEPC (p < 0.05) (Fig. 1b). Consistent results were found in the LnNE model. There is a 63% reduction of Bif-1a, and 28 fold induction of Bif-1b and Bif-1c expression in LnNE cells compared to that in LNCaP cells, resulting in a 1.8 fold induction of total Bif-1 mRNA. Stronger changes in alternative RNA splicing of the Bif-1 gene in the LnNE model may be explained by the fact that this model represents one sub-population of PCa cells in t-NEPC. Together, these RNA sequencing results indicated that the upregulation of Bif-1b and Bif-1c is accompanied by the downregulation of Bif-1a during AdPC progression to t-NEPC. These data suggest that alternative RNA splicing of the Bif-1 gene may contribute to t-NEPC development.
Fig. 1.
RNA Sequencing data show alternative RNA splicing of the Bif-1 gene in t-NEPC.
(a) Illustration of three spliced variants of the Bif-1 gene and the position of exons - Bif-1a (non-neural variant), and Bif-1b and Bif-1c (neural-specific variants). Bif-1b contains a shorter version of exon 6 (6s), whereas Bif-1c contains a longer version of exon 6 (6l). Integrative Genomics Viewer (IGV) was used to visualize the coverage of Bif-1 variants in AdPC and t-NEPC patient tumors, patient-derived xenografts (PDXs) and cell models.
(b) RNA-seq results showed the expression of each Bif-1 variants and total Bif-1 gene expression in AdPC and t-NEPC patient tumor samples, PDXs or cell lines. Student t-test ***denotes p < 0.001, **denotes p < 0.01 and *denotes p < 0.05; AdPC, prostate adenocarcinoma; t-NEPC, treatment-related neuroendocrine prostate cancer; VPC, Vancouver Prostate Centre.
3.2. Bif-1b and Bif-1c Expression is Highly Correlated With t-NEPC in Patients
Currently, there are no Bif-1 antibodies that recognize Bif-1b and Bif-1c specifically in IHC. When compared to Bif-1a, Bif-1b and Bif-1c have an additional 21 and 37 amino acids in sequence, respectively. To confirm our RNA-sequencing results and study the expression of Bif-1b and Bif-1c in association with t-NEPC, we developed the RISH technique using a probe targeting the junction of exons 7 and 8, which is shared by both Bif-1b and Bif-1c. Several control experiments were performed to validate the specificity of the probe (Figs. S1–2). RISH assays were performed: 1) on Bif-1b/Bif-1c positive tissues from NCI-H660 and LnNE xenografts, and human brain; 2) on Bif-1b/-1c negative tissues from LNCaP adenocarcinoma xenograft; and 3) by using a negative control RNA probe that does not cross-react to any human RNA. RISH signals for Bif-1b/-1c were seen as red dots or dot clusters.
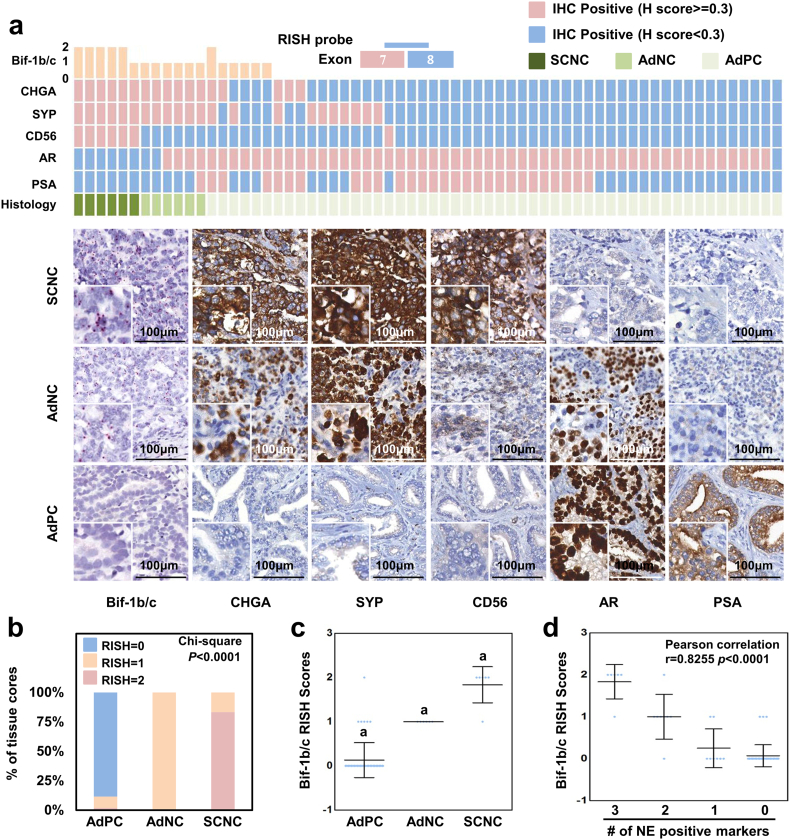
Among castration-resistance tumors, Bif-1b/-1c was detected in 28% (18/64) of tissue cores (Fig. 2a). There were six SCNC cores that were CHGA, SYP and CD56 triple positive, and AR and PSA double negative. All of these cores were Bif-1b/-1c positive, and five of them had RISH scores of 2. There were 6 AdNC tissue cores that were all CHGA and SYP positive. Three of these cores were AR positive, and one was AR and PSA double positive. These AdNC cores showed mixed SCNC and AdPC phenotypes, suggesting an ongoing transition from AdPC to SCNC. We observed that all AdNC cores were Bif-1b/-1c positive. Bif-1b/-1c were also expressed in six AR positive AdPC tissue cores, among which three expressed at least one NE marker.
Fig. 2.
The expression of neural-specific Bif-1 variants correlate with t-NEPC.
(a) A RISH probe targeting the junction between exons 7 and 8 is unique to neural-specific variants of the Bif-1 gene and is applied on castration-resistant PCa TMA. IHC assays using CHGA, SYP, CD56, AR and PSA antibodies were performed on the TMA. RISH and IHC scores as well as tumor histology were evaluated as described in the Methods section. Each column represents one of the 64 tissue cores from 32 patients. IHC scores ≥ 0.3 are considered as positive. Representative RISH and IHC images are presented.
(b–c) Castration-resistant tumor cores were grouped according their histology into SCNC, AdNC and AdPC. Distributions of RISH scores of neural-specific Bif-1 variants in each tumor group are plotted (“a” means significant difference with the other two groups, p < 0.001).
(d) Scattered plots show RISH scores of neural-specific Bif-1 variants in association with the numbers of positive NE markers by Pearson's Chi-square test.
Statistical analyses indicated that Bif-1b/-1c expression is increased in SCNC and AdNC (Fig. 2b–c). The Bif-1b/-1c expression is positively correlated not only with IHC scores of CHGA, SYP and CD56 (Pearson correlation r = 0.7977, 07903 and 0.6993, respectively; p < 0.0001), but also with the numbers of NE markers (r = 0.8255, p < 0.0001) (Table 1 & Fig. 2d). Moreover, Bif-1b/-1c expression is negatively, but weakly, correlated with AR (r = −0.4752, p < 0.0001) and PSA (r = −0.3335, p = 0.0071) (Table 1). If NEPC is defined as SCNC and AdNC, the sensitivity of Bif-1b/-1c to detect t-NEPC is 1.00 (95%CI: 0.923–1.00) and the specificity is 0.885 (95%CI: 0.766–0.957) (Table 2), indicating that all NEPC are Bif-1b/-1c positive, and approximately 12% Bif-1b/-1c positive tumors are not NEPC. However, it remains to be determined whether these 12% Bif-1b/-1c positive tumors will later develop into t-NEPC. Together, these results indicate that Bif-1b/-1c expression is highly correlated with t-NEPC development.
Table 1.
The correlation of Bif-1b/-1c with the markers for AdPC and NEPC. Bif-1b/-1c expression is positively correlated with IHC scores of CHGA, SYP and CD56, and negatively correlated with that of AR and PSA (Person correlation analysis).
| Correlation with Bif-1b/-1c | Pearson r value | p value |
|---|---|---|
| CHG A | 0.7977 | <0.0001 |
| SYP | 0.7903 | <0.0001 |
| CD56 | 0.6993 | <0.0001 |
| AR | −0.4752 | <0.0001 |
| PSA | −0.3335 | 0.0071 |
Table 2.
The sensitivity and specificity of Bif-1b/-1c to detect t-NEPC. If t-NEPC here is defined as SCNC and AdNC, Bif-1b/-1c has high sensitivity and specificity to detect t-NEPC (Fisher exact test p < 0.0001).
| Bif-1b/-1c | AdPC | t-NEPC | Sensitivity | Specificity |
|---|---|---|---|---|
| + | 6 | 12 | 1 (0.923–1) | 0.885 (0.766–0.957) |
| − | 46 | 0 |
3.3. Bif-1b and Bif-1c Expression Correlate With NEPC PDXs and Cell Models
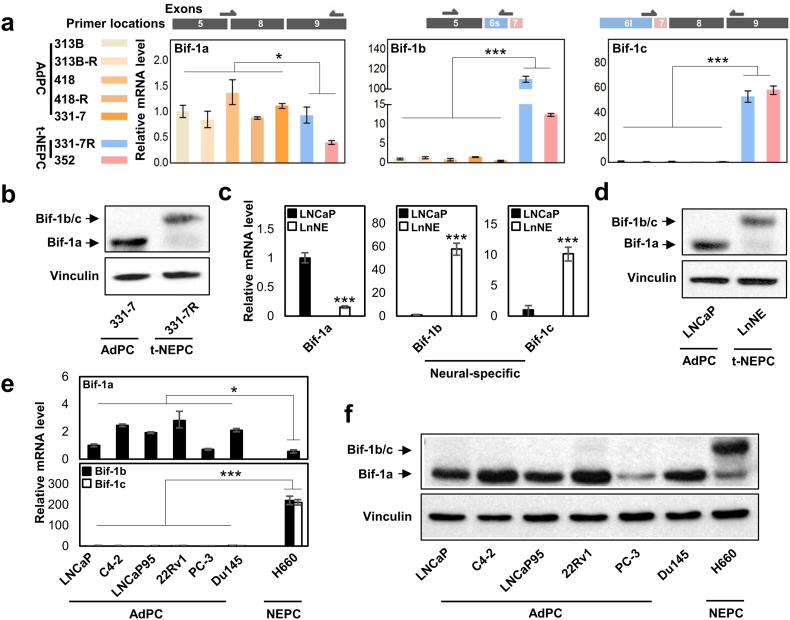
We have designed real-time PCR primers that target exon junctions specific to the three Bif-1 variants (Fig. 3a). Consistent with RNA sequencing results from t-NEPC patients, the mRNA levels of Bif-1a are downregulated, while Bif-1b and Bif-1c levels are dramatically upregulated in t-NEPC PDXs. Particularly, the 331-7R PDX was derived from 331-7 that presented with typical AdPC histology, but developed into t-NEPC by castration surgery. The conversion of Bif-1a to Bif-1b/-1c proteins was further validated by immunoblotting assays with a Bif-1 antibody against all Bif-1 variants (Fig. 3b). Real-time PCR assays showed that reduced Bif-1a and enhanced Bif-1b/-1c in both mRNA and protein levels in our LnNE model (Fig. 3c–d). Furthermore, both Bif-1b and Bif-1c mRNA and protein levels are at extremely low levels in all commonly used AdPC cell lines (Fig. 3e–f), but are highly expressed in the well established NEPC cell model, NCI-H660. Collectively, our results indicated that the neural-specific Bif-1b and Bif-1c variant expression by alternative RNA splicing of the Bif-1 gene is dramatically upregulated in t-NEPC PDXs and cell models, similar to the RNA sequencing findings from patient tumors.
Fig. 3.
Bif-1b and Bif-1c expression in t-NEPC PDXs and cell models.
(a) Primers were designed to detect Bif-1a, Bif-1b and Bif-1c specifically as shown. MRNA levels of each Bif-1 variant were measured in seven PDXs.
(b) Protein lyses extracted from 331-7 (AdPC) and its paired 331-7R (t-NEPC) were used to measure Bif-1 protein levels by immunoblotting.
(c–d) Total RNA and whole cell lysates were extracted from LNCaP and LnNE cell models to measured Bif-1 splice variants.
(e–f) The expressions of Bif-1 splice variants in AdPC and NEPC cell lines were measured by real-time PCR and immunoblotting assays.
Experiments were repeated at least three times. Only one set of the representative immunoblots is shown. Statistical analyses were performed by unpaired student's t-test with (*denotes p < 0.05 and ***denotes p < 0.001).
3.4. SRRM4 Regulates Alternative RNA Splicing of the Bif-1 Gene in PCa Cells
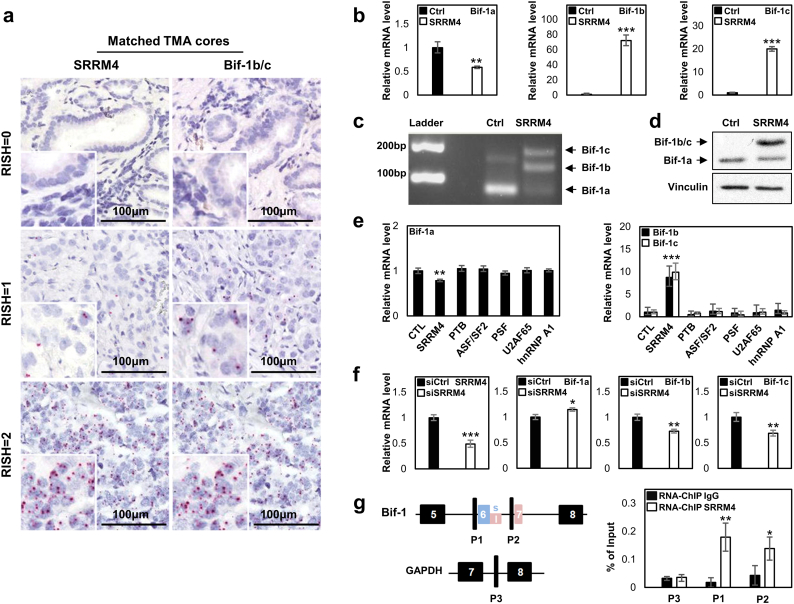
Because Bif-1b/-1c were identified from the t-NEPC specific RNA splicing signature that is predominantly controlled by SRRM4, we performed RISH assays to evaluate the association of Bif-1b/-1c with SRRM4 expression in t-NEPC patients (Table 3 & Fig. 4a). In the castration-resistant TMA, matched tissue cores showed a strong positive correlation between SRRM4 and Bif-1b/-1c expression (Pearson correlation r = 0.9199, p < 0.0001). Among the 6 SCNC cores, five had both SRRM4 and Bif-1b/-1c RISH scores of 2 and one had a score of 1 (Table S3). All 6 AdNC tissue cores were SRRM4 and Bif-1b/-1c positive, and 46 out of the 52 AdPC scores were both SRRM4 and Bif-1b/-1c negative.
Table 3.
The association of Bif-1b/-1c and SRRM4 in t-NEPC patients. Bif-1b/-1c expression is positively correlated with RISH scores of SRRM4 (Pearson correlation r = 0.9292 p < 0.0001).
| Bif-1b/-1c |
||||
|---|---|---|---|---|
| RISH Score | 0 | 1 | 2 | |
| SRRM4 | 0 | 46 | 1 | 0 |
| 1 | 0 | 9 | 1 | |
| 2 | 0 | 2 | 5 | |
Fig. 4.
SRRM4 regulates alternative RNA splicing of the Bif-1 gene.
(a) Matched tissue cores detected SRRM4 and neural-specific Bif-1 variants in PCa tumor samples.
(b) Real-time qPCR measured mRNA levels of Bif-1 variants in DU145 cells stably expressing control (Ctrl) or SRRM4.
(c) A representative image showed results from regular PCR to detect all Bif-1 variants in DU145 cells stably expressing Ctrl or SRRM4.
(d) Protein lyses from DU145 cells stably expressing Ctrl or SRRM4 were used to measure Bif-1 protein levels by immunoblotting assays.
(e) LNCaP cells were transiently transfected with six RNA splicing factors. Total RNA was extracted to measure the mRNA levels of Bif-1 variants by real-time PCR.
(f) VCaP cells with SRRM4 endogenous expression were transfected with Ctrl or SRRM4 siRNA for 48 h. Total RNA was extracted to measure the mRNA levels of SRRM4 and Bif-1 variants by real-time PCR.
(g) LNCaP cells were transfected with Flag-SRRM4 plasmids. In vivo RNA binding assays were performed using Flag antibody. Eluted RNA fragments were used as templates to perform real-time PCR to measure SRRM4 recruitment to the indicated region. Experiments were repeated three times, and one-way ANOVA or unpaired t-test were performed with *denotes p < 0.05, **denotes p < 0.01 and ***denotes p < 0.001.
To investigate whether SRRM4 regulates Bif-1 gene splicing, SRRM4 was introduced to DU145 cells. Real-time PCR showed a 50% reduction of Bif-1a, and 70 and 20 fold induction of Bif-1b and Bif-1c mRNA levels in these cells (Fig. 4b). These expression changes were also supported by reverse transcription PCR using a pair of primers at exons 5 and 8 that amplified all splice variants of the Bif-1 gene (Fig. 4c), and was further validated by immunoblotting with the Bif-1 antibody (Fig. 4d). We also introduced SRRM4 and five other RNA splicing factors into LNCaP cells and observed that only SRRM4 can promote Bif-1 RNA splicing (Fig. 4e). Furthermore, SRRM4 knockdown by siRNA was accompanied with an upregulation of Bif-1a (p < 0.05) and a downregulation of Bif-1b and Bif-1c (p < 0.01) in SRRM4 positive VCaP cells (Fig. 4f). Lastly, in vivo RNA binding assays were performed on LNCaP cells transfected with SRRM4. SRRM4 was recruited to the regions next to the 3′ splice sites of Bif-1 intron 5 and intron 6 (designated as P1 and P2, respectively), but not the control region (designated as P3) in the GAPDH gene (Fig. 4g). These results indicated that SRRM4 recruitment to the Bif-1 pre-mRNA promotes alternative RNA splicing to generate neural variants of Bif-1b and Bif-1c.
3.5. Bif-1b and Bif-1c Have Opposite Effects to Bif-1a to PCa Cell Apoptosis
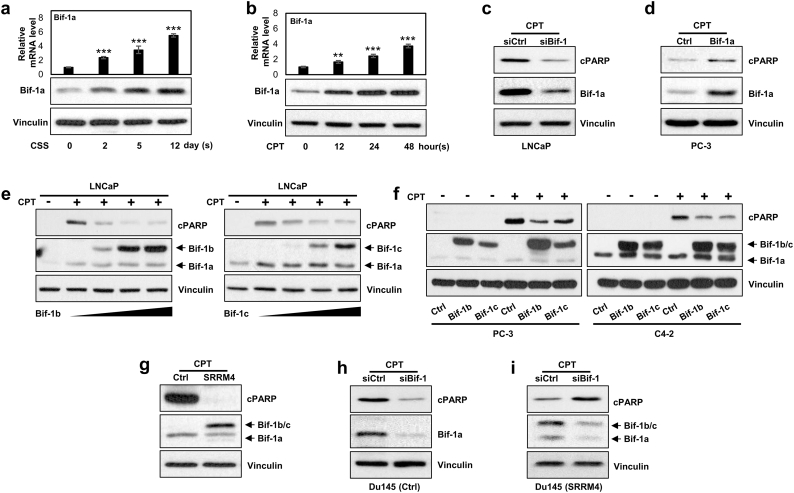
To assess the impacts of Bif-1 splice variants on PCa cell apoptosis, we first treated LNCaP cells with either androgen deprivation for 0–12 days or 0.2 μM of camptothecin (CPT) for 0–48 h. LNCaP cells express only Bif-1a, which can be upregulated by either androgen deprivation or CPT in a time-dependent manner (Fig. 5a–b). Bif-1a expression was also induced by UV light irritation (Fig. S3a). We observed that these treatments also induced neuroendocrine differentiation of PCa cells, shown by reduced luminal epithelial markers and increased neuroendocrine markers (Fig. S3b). However, SRRM4 was not induced by any treatment in these cells. To confirm whether Bif-1a is pro-apoptotic, as reported in other cell contexts, we showed that Bif-1a RNA depletion in LNCaP (Bif-1a high cell line (Fig. 3f)) cells resulted in less apoptotic responses, while overexpression of Bif-1a in PC-3 cells (Bif-1a low cell line) resulted in enhanced apoptotic responses under CPT treatment as shown by cleave PARP1 (cPARP) levels (Fig. 5c–d). These results indicate that Bif-1a expression is upregulated by stress conditions, and can sensitize stress-induced PCa cell apoptosis.
Fig. 5.
Bif-1b and Bif-1c have opposite effects to Bif-1a in PCa cell apoptosis.
(a–b) LNCaP cells were treated with 10% charcoal-stripped serum (CSS) for 0–12 days or treated with CPT for 0–48 h. Total RNA and whole cell protein were extracted to measure Bif-1a mRNA and protein levels, respectively. One-way ANOVA followed by a Newman-Keuls multiple comparison test was used (**denotes p < 0.01 and ***denotes p < 0.001).
(c) LNCaP cells were transfected with control or Bif-1 siRNA for 24 h, and then treated with 8 μM CPT for 24 h. Cleaved PARP, Bif-1 and vinculin expression were measured by immunoblotting.
(d) PC-3 cells were transfected with control or Bif-1a expression vector, and treated with 8 μM CPT for 48 h. Cleaved PARP, Bif-1 and vinculin expression were measured by immunoblotting.
(e–f) LNCaP cells were transiently transfected with 0, 2, 4, or 6 μg of Bif-1b or Bif-1c vector, and then treated with or without 8 μM CPT for 24 h in (e); C4-2 and PC-3 cells were infected by lentivirus encoding control, Bif-1b or Bif-1c (f). After blasticydin selection, cells were treated with 8 μM CPT for 24 h to C4-2 cell or 48 h to PC-3 cells. Cleaved PARP, Bif-1 and vinculin expression were measured by immunoblotting.
(g–i) DU145 cells were infected with by lentivirus encoding control or SRRM4. After blasticydin selection, cells were treated with 8 μM CPT for 24 h in (g). DU145 control (h) or DU145(SRRM4) (i) cells were transfected with Bif-1 siRNA and treated with 16 μM CPT for 48 h. Cleaved PARP, Bif-1 and vinculin expression were measured by immunoblotting.
When LNCaP cells were transfected with increasing doses of Bif-1b or Bif-1c expression vectors and then treated with CPT, cell apoptosis responses were alleviated (Fig. 5e). Similar results were also observed in PC3, C4-2 and LNCaP cells that enhanced Bif-1b or Bif-1c expression reduced cPARP levels in cells under CPT or UV light exposure (Figs. 5f & S4). The opposite effects of Bif-1 splice variants to PCa cell apoptosis were further validated in DU145 cells (Fig. 5g–i). The control DU145 cells only expressed Bif-1a variant. When these cells were transfected with either control or Bif-1 siRNA and treated with CPT, cPARP levels decreased. In contrast, Bif-1b/-1c are the predominant splice variants of the Bif-1 gene in SRRM4 overexpressed DU145 cells (a NEPC cell model (Fig. S5)). RNA depletion of the Bif-1 gene increased cPARP levels. Together, these studies indicated that Bif-1a and Bif-1b/-1c have opposite effects on PCa cell apoptosis.
4. Discussion
T-NEPC is a clinical presentation of tumor plasticity when tumor cells encountering anti-cancer therapies. To survive hormonal, radiation or chemical therapies, tumor cells have to first development mechanisms to become resistant to therapy-induced cell death before they can undergo neuroendocrine differentiation and subsequent NEPC establishment. In this context, our study demonstrated that alternative RNA splicing of the Bif-1 gene converts pro-apoptotic Bif-1a into anti-apoptotic Bif-1b and Bif-1c variants, and that this process is controlled by the neural-specific splicing factor SRRM4, which had been previously demonstrated a driver gene of t-NEPC. These findings highlight how alternative RNA splicing of apoptosis-associated genes, such as Bif-1, controls cell destination and can play a critical role during the development of therapy-resistant diseases. Whether interrupting these splicing processes would provide new avenues to treat t-NEPC warrants further investigation.
How alternative RNA splicing regulates the apoptotic property of Bif-1 splice variants remains to be determined. Bif-1 proteins have an N-BAR (Bin-amphiphysin-Rvs) domain in their N-terminus and a Src homology 3 (SH3) domain at their C-terminus. While the SH3 domain is shared among all Bif-1 members, Bif-1b and Bif-1c have altered N-BAR domains due to alternative RNA splicing. The Bif-1a N-BAR domain has been demonstrated to promote cellular apoptosis through multiple mechanisms. This domain mediates Bif-1a's association with the mitochondrial membrane to affect mitochondrial outer membrane permeabilization and promote mitochondria-mediated apoptosis [8]. This Bif-1a recruitment to the mitochondrial membrane is further strengthened by apoptotic stimuli [6,11]. Additionally, the N-BAR domain is also responsible for its protein interactions with and activation of Bax, a member of the Bcl-2 family with pro-apoptotic activities [8]. Furthermore, Bif-1 uses the N-BAR domain to interact with the intracellular membrane and induce membrane curvature to control the formation of autophagosome in response to nutrition deprivation [31]. These findings led us to propose that altered conformation of the N-BAR domain in Bif-1b and Bif-1c by RNA splicing may abolish Bif-1 recruitment to mitochondria or intracellular membrane, or Bif-1 protein interaction with Bax, resulting in protection of cells from therapy-induced cell death. This hypothesis is supported by previous studies showing that SRRM4 regulated RNA splicing during neurogenesis targets “microexons”, which alter the protein interaction domains of the targeted genes to influence their functions [10].
Alternative RNA splicing of the Bif-1 gene by SRRM4 further supports the idea that SRRM4 is a t-NEPC driver gene. SRRM4 was identified by its functions in regulating a t-NEPC specific RNA splicing signature in patient samples [19]. Upregulation of SRRM4 expression levels in t-NEPC had also been demonstrated by using RNA-seq and gene microarray analyses [5,37]. Because SRRM4 antibodies for IHC are not available, we have recently developed RNA in situ hybridization techniques to demonstrate that SRRM4 expression has high sensitivity and specificity to detect t-NEPC in castration-resistant prostate tumors (unpublished data; Y Li et al.). SRRM4 upregulation was also observed in AdPC with overt NE biomarker expression and NE-like morphological changes. RNA splicing of the Bif-1 gene by SRRM4 supports the multi-functional properties of SRRM4 in driving t-NEPC development. SRRM4 inhibit the expression of REST [19] and FoxA1 [12] transcription factors that can promote neuroendocrine differentiation of AdPC cells. SRRM4 can also regulate RNA splicing of MEAF6 histone acetyltransferase to stimulate PCa cell proliferation and invasion [14]. In this study, we further demonstrated that SRRM4 confers anti-apoptotic properties of PCa cells when under anti-cancer therapies through Bif-1 gene splicing.
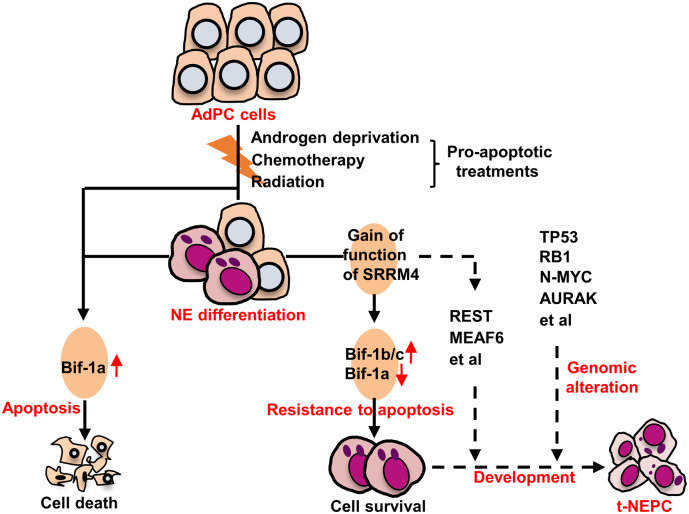
Our studies revealed that not only androgen deprivation, but also camptothecin and UV light irritation induced neuroendocrine differentiation of LNCaP AdPC cells (Fig. S3b). In addition, AdPC cells were reported to acquire NE marker expression and NE-like morphology by cAMP, IL6, hypoxia, and radiation treatments [3,7,27,38]. These findings indicate that there are multiple signaling pathways that promote AdPC cell differentiation toward NE lineage. However, gaining anti-apoptotic ability of cancer cells under various anti-cancer therapies will be the initial step that permits t-NEPC development, during which process SRRM4 regulated Bif-1 splicing may play an important role (Fig. 6).
Fig. 6.
A schematic diagram proposes a model of SRRM4 and Bif-1 in promoting t-NEPC development.
In conclusion, upregulation of the neural-specific RNA variants of the Bif-1 gene confers PCa cell resistant to therapy-induced apoptosis during AdPC progression to t-NEPC.
Acknowledgments
Acknowledgements
None.
Funding Sources
This work is supported by a CIHR operating grant (MOP-137007, PJT-156150) to XD, National Natural Science Foundation of China grants to YT (#81571432) and XC (#81770693), and visiting scholarships from China Scholar Council to YG (#201606370204), YL (#201306231021) and ZL (#201606375104).
Competing Interests
The authors declare that they have no competing interests.
Authors' contributions
Design and study concept: YG, XD. Data collection: YG, YL, ZL, NX. Data analysis and interpretation: YG, YL, JH. Material support: AL, YT, XC. Manuscript writing: YG, JL, XD. All authors read and approved the manuscript.
Disclosure Statement
The authors have nothing to disclose.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.05.002.
Contributor Information
Yu Gan, Email: gywangaoyiwoaini@126.com.
Yinan Li, Email: yli@prostatecentre.com.
Zhi Long, Email: zlong@prostatecentre.com.
Ahn R. Lee, Email: alee@prostatecentre.com.
Ning Xie, Email: nxie@prostatecentre.com.
Jessica M. Lovnicki, Email: jlovnicki@prostatecentre.com.
Yuxin Tang, Email: mmcct@126.com.
Xiang Chen, Email: cxiang1007@126.com.
Jiaoti Huang, Email: jiaoti.huang@duke.edu.
Xuesen Dong, Email: xdong@prostatecentre.com.
Appendix A. Supplementary Data
Supplementary material
References
- 1.Altman D.G., Bland J.M. Diagnostic tests. 1: sensitivity and specificity. BMJ. 1994;308:1552. doi: 10.1136/bmj.308.6943.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aparicio A., Logothetis C.J., Maity S.N. Understanding the lethal variant of prostate cancer: power of examining extremes. Cancer Discov. 2011;1:466–468. doi: 10.1158/2159-8290.CD-11-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bang Y.J., Pirnia F., Fang W.G., Kang W.K., Sartor O., Whitesell L. Terminal neuroendocrine differentiation of human prostate carcinoma cells in response to increased intracellular cyclic AMP. Proc Natl Acad Sci U S A. 1994;91:5330–5334. doi: 10.1073/pnas.91.12.5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beltran H., Rickman D.S., Park K., Chae S.S., Sboner A., Macdonald T.Y. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1:487–495. doi: 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beltran H., Prandi D., Mosquera J.M., Benelli M., Puca L., Cyrta J. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuddeback S.M., Yamaguchi H., Komatsu K., Miyashita T., Yamada M., Wu C. Molecular cloning and characterization of Bif-1. A novel Src homology 3 domain-containing protein that associates with Bax. J Biol Chem. 2001;276:20559–20565. doi: 10.1074/jbc.M101527200. [DOI] [PubMed] [Google Scholar]
- 7.Deng X., Elzey B.D., Poulson J.M., Morrison W.B., Ko S.C., Hahn N.M. Ionizing radiation induces neuroendocrine differentiation of prostate cancer cells in vitro, in vivo and in prostate cancer patients. Am J Cancer Res. 2011;1:834–844. [PMC free article] [PubMed] [Google Scholar]
- 8.Etxebarria A., Terrones O., Yamaguchi H., Landajuela A., Landeta O., Antonsson B. Endophilin B1/Bif-1 stimulates BAX activation independently from its capacity to produce large scale membrane morphological rearrangements. J Biol Chem. 2009;284:4200–4212. doi: 10.1074/jbc.M808050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helpap B., Kollermann J., Oehler U. Neuroendocrine differentiation in prostatic carcinomas: histogenesis, biology, clinical relevance, and future therapeutical perspectives. Urol Int. 1999;62:133–138. doi: 10.1159/000030376. [DOI] [PubMed] [Google Scholar]
- 10.Irimia M., Weatheritt R.J., Ellis J.D., Parikshak N.N., Gonatopoulos-Pournatzis T., Babor M. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 2014;159:1511–1523. doi: 10.1016/j.cell.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karbowski M., Jeong S.Y., Youle R.J. Endophilin B1 is required for the maintenance of mitochondrial morphology. J Cell Biol. 2004;166:1027–1039. doi: 10.1083/jcb.200407046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J., Jin H., Zhao J.C., Yang Y.A., Li Y., Yang X. FOXA1 inhibits prostate cancer neuroendocrine differentiation. Oncogene. 2017;36:4072–4080. doi: 10.1038/onc.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lapuk A.V., Wu C., Wyatt A.W., Mcpherson A., Mcconeghy B.J., Brahmbhatt S. From sequence to molecular pathology, and a mechanism driving the neuroendocrine phenotype in prostate cancer. J Pathol. 2012;227:286–297. doi: 10.1002/path.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee A.R., Li Y., Xie N., Gleave M.E., Cox M.E., Collins C.C. Alternative RNA splicing of the MEAF6 gene facilitates neuroendocrine prostate cancer progression. Oncotarget. 2017;8:27966–27975. doi: 10.18632/oncotarget.15854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H., Xie N., Gleave M.E., Dong X. Catalytic inhibitors of DNA topoisomerase II suppress the androgen receptor signaling and prostate cancer progression. Oncotarget. 2015;6:20474–20484. doi: 10.18632/oncotarget.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y., Xie N., Gleave M.E., Rennie P.S., Dong X. AR-v7 protein expression is regulated by protein kinase and phosphatase. Oncotarget. 2015;6:33743–33754. doi: 10.18632/oncotarget.5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H., Xie N., Chen R., Verreault M., Fazli L., Gleave M.E. UGT2B17 expedites progression of castration-resistant prostate cancers by promoting ligand-independent AR signaling. Cancer Res. 2016;76:6701–6711. doi: 10.1158/0008-5472.CAN-16-1518. [DOI] [PubMed] [Google Scholar]
- 18.Li Y., Chen R., Bowden M., Mo F., Lin Y.Y., Gleave M. Establishment of a neuroendocrine prostate cancer model driven by the RNA splicing factor SRRM4. Oncotarget. 2017;8:66878–66888. doi: 10.18632/oncotarget.19916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y., Donmez N., Sahinalp C., Xie N., Wang Y., Xue H. SRRM4 drives neuroendocrine transdifferentiation of prostate adenocarcinoma under androgen receptor pathway inhibition. Eur Urol. 2017;71:68–78. doi: 10.1016/j.eururo.2016.04.028. [DOI] [PubMed] [Google Scholar]
- 20.Lin D., Wyatt A.W., Xue H., Wang Y., Dong X., Haegert A. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res. 2014;74:1272–1283. doi: 10.1158/0008-5472.CAN-13-2921-T. [DOI] [PubMed] [Google Scholar]
- 21.Liu L.L., Xie N., Sun S., Plymate S., Mostaghel E., Dong X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene. 2014;33:3140–3150. doi: 10.1038/onc.2013.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lotan T.L., Gupta N.S., Wang W., Toubaji A., Haffner M.C., Chaux A. ERG gene rearrangements are common in prostatic small cell carcinomas. Mod Pathol. 2011;24:820–828. doi: 10.1038/modpathol.2011.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modregger J., Schmidt A.A., Ritter B., Huttner W.B., Plomann M. Characterization of Endophilin B1b, a brain-specific membrane-associated lysophosphatidic acid acyl transferase with properties distinct from endophilin A1. J Biol Chem. 2003;278:4160–4167. doi: 10.1074/jbc.M208568200. [DOI] [PubMed] [Google Scholar]
- 24.Parimi V., Goyal R., Poropatich K., Yang X.J. Neuroendocrine differentiation of prostate cancer: a review. Am J Clin Exp Urol. 2014;2:273–285. [PMC free article] [PubMed] [Google Scholar]
- 25.Pierrat B., Simonen M., Cueto M., Mestan J., Ferrigno P., Heim J. SH3GLB, a new endophilin-related protein family featuring an SH3 domain. Genomics. 2001;71:222–234. doi: 10.1006/geno.2000.6378. [DOI] [PubMed] [Google Scholar]
- 26.Priemer D.S., Montironi R., Wang L., Williamson S.R., Lopez-Beltran A., Cheng L. Neuroendocrine tumors of the prostate: emerging insights from molecular data and updates to the 2016 World Health Organization Classification. Endocr Pathol. 2016;27:123–135. doi: 10.1007/s12022-016-9421-z. [DOI] [PubMed] [Google Scholar]
- 27.Qi J., Nakayama K., Cardiff R.D., Borowsky A.D., Kaul K., Williams R. Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors. Cancer Cell. 2010;18:23–38. doi: 10.1016/j.ccr.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rostovtseva T.K., Boukari H., Antignani A., Shiu B., Banerjee S., Neutzner A. Bax activates endophilin B1 oligomerization and lipid membrane vesiculation. J Biol Chem. 2009;284:34390–34399. doi: 10.1074/jbc.M109.021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith B.A., Sokolov A., Uzunangelov V., Baertsch R., Newton Y., Graim K. A basal stem cell signature identifies aggressive prostate cancer phenotypes. Proc Natl Acad Sci U S A. 2015;112:E6544–E6552. doi: 10.1073/pnas.1518007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi Y., Karbowski M., Yamaguchi H., Kazi A., Wu J., Sebti S.M. Loss of Bif-1 suppresses Bax/Bak conformational change and mitochondrial apoptosis. Mol Cell Biol. 2005;25:9369–9382. doi: 10.1128/MCB.25.21.9369-9382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi Y., Coppola D., Matsushita N., Cualing H.D., Sun M., Sato Y. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terry S., Beltran H. The many faces of neuroendocrine differentiation in prostate cancer progression. Front Oncol. 2014;4:60. doi: 10.3389/fonc.2014.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang D.B., Uo T., Kinoshita C., Sopher B.L., Lee R.J., Murphy S.P. Bax interacting factor-1 promotes survival and mitochondrial elongation in neurons. J Neurosci. 2014;34:2674–2683. doi: 10.1523/JNEUROSCI.4074-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H.T., Yao Y.H., Li B.G., Tang Y., Chang J.W., Zhang J. Neuroendocrine prostate Cancer (NEPC) progressing from conventional prostatic adenocarcinoma: factors associated with time to development of NEPC and survival from NEPC diagnosis-a systematic review and pooled analysis. J Clin Oncol. 2014;32:3383–3390. doi: 10.1200/JCO.2013.54.3553. [DOI] [PubMed] [Google Scholar]
- 35.Yu Y., Liu L., Xie N., Xue H., Fazli L., Buttyan R. Expression and function of the progesterone receptor in human prostate stroma provide novel insights to cell proliferation control. J Clin Endocrinol Metab. 2013;98:2887–2896. doi: 10.1210/jc.2012-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu Y., Yang O., Fazli L., Rennie P.S., Gleave M.E., Dong X. Progesterone receptor expression during prostate cancer progression suggests a role of this receptor in stromal cell differentiation. Prostate. 2015;75:1043–1050. doi: 10.1002/pros.22988. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X., Coleman I.M., Brown L.G., True L.D., Kollath L., Lucas J.M. SRRM4 expression and the loss of REST activity may promote the emergence of the neuroendocrine phenotype in castration-resistant prostate cancer. Clin Cancer Res. 2015;21:4698–4708. doi: 10.1158/1078-0432.CCR-15-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Y., Liu C., Cui Y., Nadiminty N., Lou W., Gao A.C. Interleukin-6 induces neuroendocrine differentiation (NED) through suppression of RE-1 silencing transcription factor (REST) Prostate. 2014;74:1086–1094. doi: 10.1002/pros.22819. [DOI] [PubMed] [Google Scholar]
- 39.Zou M., Toivanen R., Mitrofanova A., Floch N., Hayati S., Sun Y. Transdifferentiation as a mechanism of treatment resistance in a mouse model of castration-resistant prostate cancer. Cancer Discov. 2017;7:736–749. doi: 10.1158/2159-8290.CD-16-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material