Abstract
Preoperative lymph node (LN) status is important for the treatment of bladder cancer (BCa). Here, we report a genomic-clinicopathologic nomogram for preoperatively predicting LN metastasis in BCa. In the discovery stage, 325 BCa patients from TCGA were involved and LN-status-related mRNAs were selected. In the training stage, multivariate logistic regression analysis was used to developed a genomic-clinicopathologic nomogram for preoperative LN metastasis prediction in the training set (SYSMH set, n = 178). In the validation stage, we validated the nomogram using two independent sample sets (SYSUCC set, n = 142; RJH set, n = 104) with respect to its discrimination, calibration and clinical usefulness. As results, we identified five LN-status-related mRNAs, including ADRA1D, COL10A1, DKK2, HIST2H3D and MMP11. Then, a genomic classifier was developed to classify patients into high- and low-risk groups in the training set. Furthermore, a nomogram incorporating the five-mRNA-based classifier, image-based LN status, transurethral resection (TUR) T stage, and TUR lymphovascular invasion (LVI) was constructed in the training set, which performed well in the training and validation sets. Decision curve analysis demonstrated the clinical value of our nomogram. Thus, our genomic-clinicopathologic nomogram shows favorable discriminatory ability and may aid in clinical decision-making, especially for cN-patients.
Keywords: Genomics, Lymphatic metastasis, Nomogram, Urinary bladder neoplasms
Highlights
-
•
A genomic classifier was built to classify bladder cancer patients into high- and low-risk groups of lymph node metastasis.
-
•
A genomic-clinicopathologic nomogram was developed for predicting lymph node metastasis in bladder cancer patients.
-
•
The nomogram shows favorable discrimination and calibration which may aid in clinical decision-making.
Preoperative lymph node status is critical for guiding the management of bladder cancer patients. However, current preoperative nodal staging methods are inaccurate. Here, we proposed a user-friendly tool (nomogram) for the preoperative prediction of lymph node metastasis in bladder cancer, which showed good calibration and discrimination. The favorable predictive accuracy was validated in two external institutes. Thus, our nomogram could be an efficient and valuable tool for individual lymph node metastasis prediction and bladder cancer treatment decision making.
1. Introduction
Bladder cancer (BCa) is a common cancer with globally high mortality (Burger et al., 2013; Antoni et al., 2017). Lymph nodes (LNs) are the most common metastatic sites in BCa. Previous studies suggested that 25%–30% of BCa patients treated with radical cystectomy (RC) and pelvic lymph node dissection (PLND) underwent LN metastasis (Stein et al., 2001; Leissner et al., 2004; Vazina et al., 2004; Abol-Enein et al., 2011; Baltaci et al., 2011; Zehnder et al., 2011; Jensen et al., 2012). Up to 80% of BCa patients with pathologic LN metastasis suffer from recurrence after undergoing RC, while only approximately 30% of BCa patients who are LN-negative (pN0) experience tumor recurrence (Shariat et al., 2006; Stamatakis et al., 2012). In addition, LN-positive (pN1–3) patients have a significantly lower five-year overall survival rate compared with pN0 patients (15%–31% vs. > 60%) (Bassi et al., 1999; Stein et al., 2001; Karl et al., 2009; Zehnder et al., 2014).
Preoperative LN status is critical for BCa treatment decision-making, particularly in helping determine the extent of PLND and the use of neoadjuvant chemotherapy (Kluth et al., 2015; Zargar-Shoshtari et al., 2016). Currently, contrast-enhanced computed tomography (CT) is the standard clinical procedure for preoperatively evaluating the LN stage (McKibben and Woods, 2015). However, the sensitivity of CT at detecting metastatic lesions in the LNs is relative low (31%–45%) (Baltaci et al., 2008; Lodde et al., 2010; Goodfellow et al., 2014). As a consequence, a considerable portion of patients with inaccurate staging may receive inadequate treatment or overtreatment. In particular, understaging might lead to postoperative recurrence or even death (Culp et al., 2014); conversely, overstaging is likely to subject a patient to needless neoadjuvant chemotherapy or unnecessarily extensive PLND. Therefore, there is an urgent need to improve the nodal staging accuracy for the treatment of BCa.
In recent years, gene expression signatures have been used to predict LN metastasis in BCa (Smith et al., 2011; Seiler et al., 2016). However, the models developed in these studies have not yet been used in clinical practice due to limitations. Smith et al. developed a 20-mRNA-based classifier for predicting LN status preoperatively in BCa patients (Smith et al., 2011). The predictive efficacy of the model was modest, with an area under the curve (AUC) of 0.67 in the external validation set. Seiler et al. developed a 51-RNA-based classifier that achieved an AUC of 0.82 for predicting BCa LN metastasis (Seiler et al., 2016), but this study was limited by the lack of external validation. Moreover, the previously developed models were limited to clinically node-negative (cN-) BCa patients, and clinical factors were not incorporated into the predictive models for evaluation.
In the current study, we postulated that inclusive models incorporating a genomic signature and clinicopathologic factors might improve the accuracy of nodal staging. We identified mRNAs that significantly correlated with LN metastasis by mining RNA-SEQ data from the TCGA BLCA project and then developed a multiple-mRNA classifier in our BCa sample set. The genomic classifier was further combined with clinicopathological factors to build an inclusive nomogram for predicting LN status preoperatively. We assessed the predictive accuracy of the nomogram and validated it in two independent sample sets. We also evaluated the predictive efficacy of the nomogram in clinically low-risk subgroups (non-muscle invasive bladder cancer [NMIBC] or cN-).
2. Materials and Methods
2.1. Patients and Clinical Database
In this study, a total of 424 BCa patients who had undergone RC and PLND without preoperative therapy were recruited from three independent cancer centers, including 178 samples from the Sun Yat-sen Memorial Hospital of Sun Yat-sen University (SYSMH) between March 2006 and December 2017, 142 samples from the Sun Yat-sen University Cancer Center (SYSUCC) between April 2002 and September 2017, and 104 samples from the Renji Hospital of Shanghai Jiaotong University School of Medicine (RJH) between June 2013 and July 2017. The inclusion criteria were pT0-4N0-3M0 BCa patients who underwent RC + PLND and those with tumor samples, which were confirmed urothelial carcinoma pathologically. The exclusion criteria were BCa patients who underwent preoperative therapy (either neoadjuvant chemotherapy or radiotherapy) or didn't receive pre-RC transurethral resection of bladder tumor (TURBT) in the three centers, and those with samples with insufficient total RNA or a failed quality control (QC) step. All patients underwent CT or magnetic resonance imaging (MRI) before TURBT. The procedures of PLND and RNA QC were described in the Supplementary Material in detail. The patient recruitment pathway is shown in Supplementary Fig. S1. Clinicopathological data were collected through medical record review and included age, gender, BCa recurrence, image-based tumor size, image-based tumor number, image-based N stage, status of hydronephrosis, TUR T stage, TUR tumor grade, and TUR lymphovascular invasion (LVI). LVI was considered present only if tumor cells were unequivocally presented within or attached to the wall of a vascular or lymphatic space on hematoxylin & eosin stained sections (Cho et al., 2009). As for indeterminate cases and aggressive tumors cases, multiple serial sections were used. The BCa patients were classified using the 2009 TNM staging system (Sobin et al., 2009) and the 2004 WHO classification (Epstein et al., 2004). We defined cases as clinical LN positive (cN+) if pelvic LN > 8 mm or abdominal LN > 10 mm in the maximum short-axis diameter based on CT or MRI (Dorfman et al., 1991; Barentsz et al., 1999). To ensure validity of the pathologic outcomes extraction, all samples were re-assessed by two pathologists (Hong Zen and Lin Wang) while blinded to patient clinicopathological data and the findings of the other reviewer. Interreader reliability measured using the intraclass correlation coefficient was >0.95 for each pathologic characteristic. We used the SYSMH samples as the training set for model development and the SYSUCC and RJH samples as the external validation sets. The institutional review boards of the three centers approved this study and the need to obtain informed consent was waived.
2.2. qRT-PCR
Total RNA from 424 fresh-frozen BCa tissue samples was extracted using RNAiso plus reagent (TaKaRa) according to the manufacturer's instructions, and the expression of BCa LN status-related mRNAs (selected in the discovery stage) was further examined via qRT-PCR. First-strand cDNA was synthesized with PrimeScript™ RT Master Mix (TAKARA) according to the manufacturer's instructions. The qRT-PCR was conducted to examine the expression of the selected mRNAs using SYBR-Green PCR Master Mix (Roche) on a LightCycler 96 Real-Time PCR instrument (Roche). Homo sapiens actin beta (ACTB) was used as an internal reference gene to normalize mRNA levels between different samples for an exact comparison of transcript level. Expression levels of each mRNA were calculated using the −△CT approach (△CT = CT of mRNA – CT of ACTB RNA).
2.3. Source of the TCGA BLCA Project Data
Normalized gene expression data (level 3, RNA-SEQ data from an Illumina HiSeq 2000 platform) were obtained from the UCSC Cancer Genomics Browser (https://genome-cancer.ucsc.edu) on 1 June 2016. The UCSC website contains a detailed description of data normalization. Briefly, the gene expression profile was measured experimentally using the Illumina HiSeq 2000 RNA Sequencing platform at a TCGA Genome Center. Level 3 interpreted data were downloaded from the TCGA Data Portal (https://portal.gdc.cancer.gov/). This dataset provides gene-level transcription estimates as an RSEM normalized count. Genes were mapped onto the human genome coordinates using the UCSC cgData HUGO probeMap.
3. Procedures
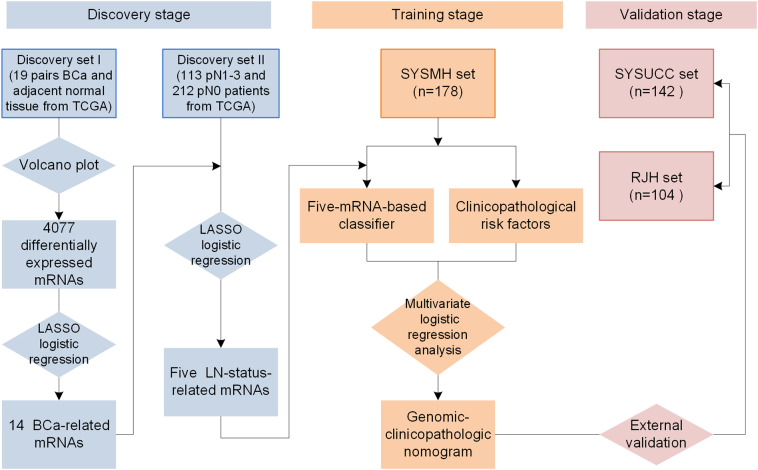
Our study was conducted in three stages: discovery stage, training stage and validation stage. The study flowchart is presented in Fig. 1. In the discovery stage, we used paired samples (tumor/normal) from BCa patients in the TCGA BLCA project as discovery set I (n = 19 pairs); all BCa patients with nodal status information in the TCGA were included in discovery set II (n = 325). Discovery set II included 113 patients with LN-positive disease (pN1–3) and 212 patients with LN-negative disease (pN0). A volcano plot analysis of discovery set I was used to screen for differentially expressed mRNAs, and we then used the least absolute shrinkage and selection operator (LASSO) logistic regression algorithm (Tibshirani, 1996) to identify BCa-associated mRNAs as candidates. Furthermore, the LASSO logistic regression model was used to screen LN status-correlated mRNAs from the candidate mRNAs in discovery set II (see Fig. 2 and Statistical Analysis section).
Fig. 1.
Study flowchart.
BCa = bladder cancer. LASSO = the least absolute shrinkage and selection operator. LN = lymph node. TCGA = the Cancer Genome Atlas. SYSUMH = Sun Yat-sen memorial hospital. SYSUCC = Sun Yat-sen University Cancer Center. RJH = Renji Hospital Shanhai Jiaotong University School of Medicine.
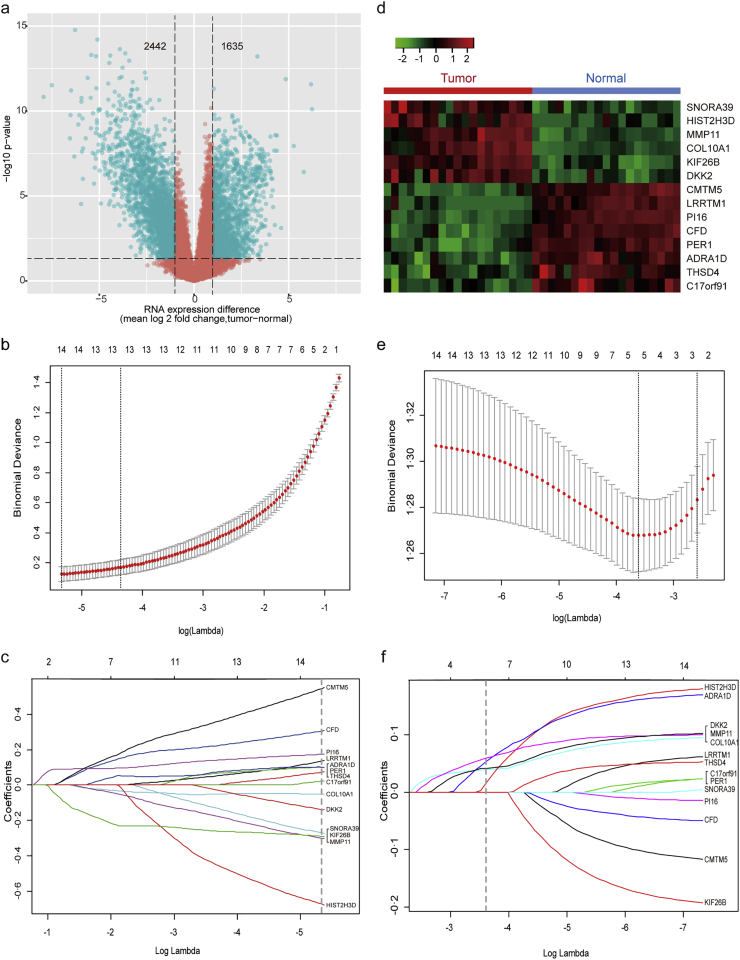
Fig. 2.
Five LN-status-related mRNAs screened in the discovery stage.
(a–d) 14 BCa-related mRNAs screened in discovery set I (n = 19 tumor/normal pairs):
(a) Volcano plot was generated based on absolute fold change (FC) in combination with t-test p-values. The vertical lines represent a two-fold change in expression, and the horizontal line indicates p-value = .05. In comparison of BCa tumor tissues with matched adjacent normal tissues, a total of 4077 differentially expressed mRNAs with statistical significance were identified (green points in the plot). Among them, 1635 mRNAs were up-regulated and 2442 were down-regulated.
(b) Selection of tuning parameter (λ) in the LASSO model via 10-fold cross-validation in discovery set I. Binomial deviances from the LASSO regression's cross-validation procedure were plotted as a function of log(λ). λ is the tuning parameter. Y-axis indicates binomial deviances. The lower x-axis indicates log(λ). Numbers along the upper x-axis represent the average number of predictors. Red dots indicate average deviance values for each model with given λ, and vertical bars through the red dots show the upper and lower values of the deviances. The vertical black lines define the optimal values of λ, where the model provides its best fits to the data. The optimal λ value of 0.0048 with log (λ) = −5.330 was chosen via 10-fold cross-validation based on the minimum criteria.
(c) LASSO coefficients produced by the regression analysis (in B). A vertical line at x-axis with log (λ) = −5.330 were generated based on the minimum criteria in 10-fold cross-validation procedure. The 14 resulting predictors with nonzero coefficients were indicated in the plot.
(d) Hierarchical cluster analysis of 14 BCa-related mRNAs. Red color represents up-regulated expression, and green color indicates down-regulated expression.
(e–f) LASSO logistic regression analysis was used to narrow the 14 BCa-related mRNAs to five LN-status-related ones in discovery set II (n = 325):
(e) Selection of tuning parameter (λ) in the LASSO model via 10-fold cross-validation in discovery set II. The optimal λ value of 0.0270, with log (λ) = −3.613 was chosen based on minimum criteria.
(f) LASSO coefficients produced by the regression analysis (in E). Five LN-status predictors (namely, ADRA1D, COL10A1, DKK2, HIST2H3D and MMP11) with non-zero coefficients were chosen in the LASSO logistic regression model based on the optimal λ value.
In the training stage, a multivariable logistic regression model was used to construct a multi-mRNA-based classifier for predicting LN status in the training (SYSMH) set based on the significant LN status-correlated mRNAs identified in the discovery stage. We then derived the genetic risk scores from the multi-mRNA-based classifier. Receiver operating characteristic (ROC) analysis was performed to investigate the predictive efficiency of the multi-mRNA-based classifier by measuring the AUC. Furthermore, independent validations were conducted using two external samples sets, the SYSUCC and RJH sets. In addition, we evaluated the classification performance of the model in distinguishing BCa patients with a low or high risk of LN metastasis.
To calculate the genetic risk scores reflecting the risk of LN metastasis for each patient, a regression equation was derived using the estimated coefficients in the above multivariable regression model (the five-mRNA classifier). Based on the genetic risk scores computed using the regression formula, we classified BCa patients into low- and high-risk groups based on the optimal risk score cutoff value, which represented the point at which the Youden index (sensitivity + specificity − 1) reached a maximum value in the SYSMH set. Then, we applied the optimal risk score cutoff value to the two independent sample sets (the SYSUCC and RJH sets) to evaluate the classification performance of the model.
To build a genomic-clinicopathologic nomogram, we used a multivariable logistic regression model to identify the clinical risk factors that were significantly correlated with LN status and then combined them with the five-mRNA classifier to construct the inclusive model using the SYSMH set. Then, the performance of the inclusive nomogram was evaluated in the SYSMH set. We also compared the discrimination ability of the nomogram with the genomic classifier and clinicopathologic factors by ROC analysis in all patients.
In the validation stage, the performance of the inclusive nomogram was validated in the independent sample sets, the SYSUCC and RJH sets. The predictive accuracy and discrimination ability of the nomogram were determined using a calibration plot and C-index, respectively. Then, decision curve analysis (DCA) was performed to evaluate the clinical usefulness of the nomogram in three datasets (Vickers et al., 2008; Vickers et al., 2016). DCA was performed by calculating the net benefit for a range of threshold probabilities (Vickers et al., 2008), which place benefits and harms on the same scale. This analysis helps determine whether clinical decision-making based on a model will do more good than harm. DCA provides straightforward information about the clinical value of a model, in contrast to traditional measures such as sensitivity or specificity, which are abstract statistical concepts (Vickers et al., 2016).
Finally, ROC analysis was used to evaluate and compare the discrimination ability of the nomogram with the genomic classifier and clinicopathologic factors in the subgroups diagnosed as clinically low risk (NMIBC or cN-). Based on the risk scores from the inclusive nomogram, we classified clinically low-risk patients into low- and high-risk groups using the optimal risk score cutoff value, which was defined using the approach described above. Then, we defined the optimal risk score cutoff values in the two subgroups (NMIBC or cN-) to evaluate the classification performance of the nomogram.
3.1. Statistical Analysis
Volcano plot analysis was conducted based on absolute fold change (FC > 2.0) in combination with adjusted t-test p values (P < .05). Penalty parameter tuning conducted by 10-fold cross-validation (CV) was used in the LASSO logistic regression. Multivariable logistic regression was used to screen the significant clinicopathologic risk factors and to construct a multi-mRNA-based classifier, and the likelihood ratio test with backward step-wise selection approach was applied.
All the statistical tests were conducted in R statistical software version 3.3.1. The LASSO logistic regression model was performed with the “glmnet” package, and the ROC curves were plotted with the “pROC” package. Hierarchical cluster analysis using the “complete” method with a dissimilarity structure as produced by Euclidean distance was performed with the “R gplots” package. The nomogram and calibration plots were generated with the “rms” package, and the Hosmer-Lemeshow test was performed with the “generalhoslem” package. The DCA was performed with the “dca.R”. A two-sided P value <.05 was considered statistically significant.
3.2. Role of the Funding Source
The sponsors of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author (T-XL) had full access to all the data in the study and had final responsibility for the decision to submit for publication.
3.3. Results
We developed the model for the preoperative prediction of LN metastasis in three stages, including the discovery, training and validation stages. The study flowchart is presented in Fig. 1. In total, we recruited 424 BCa patients who underwent RC and PLND without preoperative therapy from the three centers. We quantitated the expression of the identified mRNAs in all the BCa patients using qRT-PCR. The patients' clinicopathologic characteristics are shown in Table 1. In total, 23.6% (100/424) patients were understaged or overstaged by clinical nodal staging in our study.
Table 1.
Clinicopathological characteristics of patients from three independent centers.
| SYSMH set (n = 178) |
SYSUCC set (n = 142) |
RJH set (n = 104) |
||||
|---|---|---|---|---|---|---|
| pN1–3 (%) | pN0 (%) | pN1–3 (%) | pN0 (%) | pN1–3 (%) | pN0 (%) | |
| Gender | ||||||
| Male | 46 (32%) | 99 (68%) | 39 (33%) | 81 (67%) | 20 (25%) | 60 (75%) |
| Female | 10 (30%) | 23 (70%) | 10 (45%) | 12 (55%) | 7 (29%) | 17 (71%) |
| Age, years | ||||||
| ≤ 65 | 26 (26%) | 75 (74%) | 35 (38%) | 56 (62%) | 11 (27%) | 30 (73%) |
| > 65 | 30 (39%) | 47 (61%) | 14 (27%) | 37 (73%) | 16 (25%) | 47 (75%) |
| Tumor sizea | ||||||
| ≤ 3 cm | 14 (21%) | 54 (79%) | 18 (30%) | 43 (70%) | 15 (22%) | 53 (78%) |
| > 3 cm | 42 (37%) | 68 (63%) | 31 (38%) | 50 (62%) | 12 (33%) | 24 (67%) |
| Number of tumorsa | ||||||
| Single | 43 (38%) | 69 (62%) | 17 (39%) | 27 (61%) | 23 (29%) | 57 (71%) |
| Multiple | 13 (20%) | 53 (80%) | 32 (33%) | 66 (67%) | 4 (17%) | 20 (83%) |
| Clinical N stagea | ||||||
| cN0 | 31 (22%) | 110 (78%) | 33 (28%) | 87 (72%) | 13 (16%) | 72 (84%) |
| cN1–3 | 25 (68%) | 12 (32%) | 16 (73%) | 6 (27%) | 14 (74%) | 5 (26%) |
| Hydronephrosisa | ||||||
| Present | 24 (46%) | 28 (54%) | 25 (64%) | 14 (36%) | 14 (52%) | 13 (48%) |
| Absent | 32 (25%) | 94 (75%) | 24 (23%) | 79 (77%) | 13 (17%) | 64 (83%) |
| TUR T stage | ||||||
| NMIBC | 3 (6%) | 48 (94%) | 1 (4%) | 23 (96%) | 1 (5%) | 20 (95%) |
| MIBC | 53 (42%) | 74 (58%) | 48 (41%) | 70 (59%) | 26 (31%) | 57 (69%) |
| TUR tumor grade | ||||||
| Low | 1 (6%) | 16 (94%) | 0 (0%) | 11 (100%) | 0 (0%) | 4 (100%) |
| High | 55 (34%) | 106 (66%) | 49 (37%) | 82 (63%) | 27 (27%) | 73 (73%) |
| TUR LVI | ||||||
| Present | 24(63%) | 14(27%) | 20 (63%) | 12 (37%) | 13 (76%) | 4 (24%) |
| Absent | 32(23%) | 108(77%) | 29 (26%) | 81 (74%) | 14 (16%) | 73 (84%) |
| Recurrent tumor | ||||||
| Yes | 16 (34%) | 31 (66%) | 19 (53%) | 17 (47%) | 2 (22%) | 7 (78%) |
| No | 40 (31%) | 91 (69%) | 30 (28%) | 76 (62%) | 25 (26%) | 70 (74%) |
Image-based information.
In the discovery stage, volcano plot analysis (FC > 2.0 and P < .05) identified 4077 differentially expressed mRNAs in discovery set I (Fig. 2a). We then used a LASSO logistic regression algorithm to identify 14 BCa-associated mRNAs (Fig. 2c); the optimal tuning parameter λ corresponded to the minimum prediction error selected via the 10-fold CV method (Fig. 2b). With regard to the 14 significant mRNAs, hierarchical cluster analysis successfully separated the 19 pairs of BCa tumor/normal samples into two discrete groups (Fig. 2d). Last, we used a LASSO logistic regression model in discovery set II (n = 325) to screen LN status-correlated mRNAs and selected five mRNAs from the 14 BCa-associated mRNAs. We identified ADRA1D, COL10A1, DKK2, HIST2H3D and MMP11 as significantly correlated with LN status in BCa patients (Fig. 2e and f and Supplementary Table S3). Table 2 lists the relation between expression and LN status for each mRNA.
Table 2.
Univariate association of five-mRNA-based classifier, selected mRNAs and clinicopathological features with lymph status.
| Variable | SYSMH set (n = 178) |
SYSUCC set (n = 142) |
RJH set (n = 104) |
|||
|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |
| Gender (male vs. female) | 0.717 (0.299–1.721) | 0.457 | 1.954 (0.791–4.827) | 0.147 | 1.486 (0.554–3.984) | 0.431 |
| Age (≤65 vs. >65) | 1.287 (0.678–2.442) | 0.440 | 0.804 (0.391–1.652) | 0.553 | 1.277 (0.508–3.210) | 0.604 |
| Tumor size* (≤3 cm vs. >3 cm) | 2.912 (1.401–6.051) | 0.004 | 1.358 (0.671–2.747) | 0.395 | 1.518 (0.613–3.759) | 0.367 |
| Number of tumors* (single vs. multiple) | 0.394 (0.192–0.806) | 0.011 | 1.023 (0.476–2.197) | 0.954 | 0.648 (0.216–1.939) | 0.438 |
| Clinical N stage* (cN0 vs. cN1–3) | 7.392 (3.337–16.376) | <0.0001 | 7.030 (2.534–19.501) | 0.00017 | 15.508 (4.767–50.449) | <0.0001 |
| Hydronephrosis* (absent vs. present) | 2.518 (1.280–4.954) | 0.007 | 5.878 (2.647–13.055) | <0.0001 | 5.302 (2.026–13.874) | 0.001 |
| TUR T stage (NMIBC vs. MIBC) | 11.459 (3.388–38.760) | <0.0001 | 15.771 (2.060–120.751) | 0.008 | 9.123 (1.161–71.665) | 0.036 |
| TUR tumor grade (low vs. high grade) | 8.302 (1.073–64.255) | 0.043 | – | – | – | – |
| TUR LVI (absent vs. present) | 5.786 (2.684–12.473) | <0.0001 | 4.655 (2.026–10.696) | 0.00029 | 16.946 (4.816–59.631) | <0.0001 |
| Recurrent tumor (no vs. yes) | 0.978 (0.472–2.029) | 0.953 | 3.643 (1.686–7.870) | 0.001 | 0.385 (0.045–3.279) | 0.382 |
| ADRA1D (−△CT value) | 1.132 (1.032–1.241) | 0.008 | 0.999 (0.807–1.237) | 0.993 | 1.093 (0.941–1.271) | 0.246 |
| COL10A1 (−△CT value) | 1.136 (1.018–1.268) | 0.023 | 0.788 (0.564–1.101) | 0.162 | 0.890 (0.725–1.093) | 0.267 |
| DKK2 (−△CT value) | 1.133 (1.049–1.223) | 0.001 | 1.599 (1.268–2.018) | <0.0001 | 1.050 (0.897–1.229) | 0.546 |
| HIST2H3D (−△CT value) | 1.153 (1.048–1.268) | 0.003 | 1.029 (0.796–1.330) | 0.828 | 1.040 (0.876–1.234) | 0.657 |
| MMP11 (−△CT value) | 1.399 (1.221–1.603) | <0.0001 | 1.635 (1.328–2.012) | <0.0001 | 1.234 (1.032–1.477) | 0.021 |
| Five-mRNA-classifier (low vs. high risk) | 11.759 (5.486–25.203) | <0.0001 | 7.765 (3.344–18.032) | <0.0001 | 6.481 (2.493–16.853) | 0.00013 |
Image-based information.
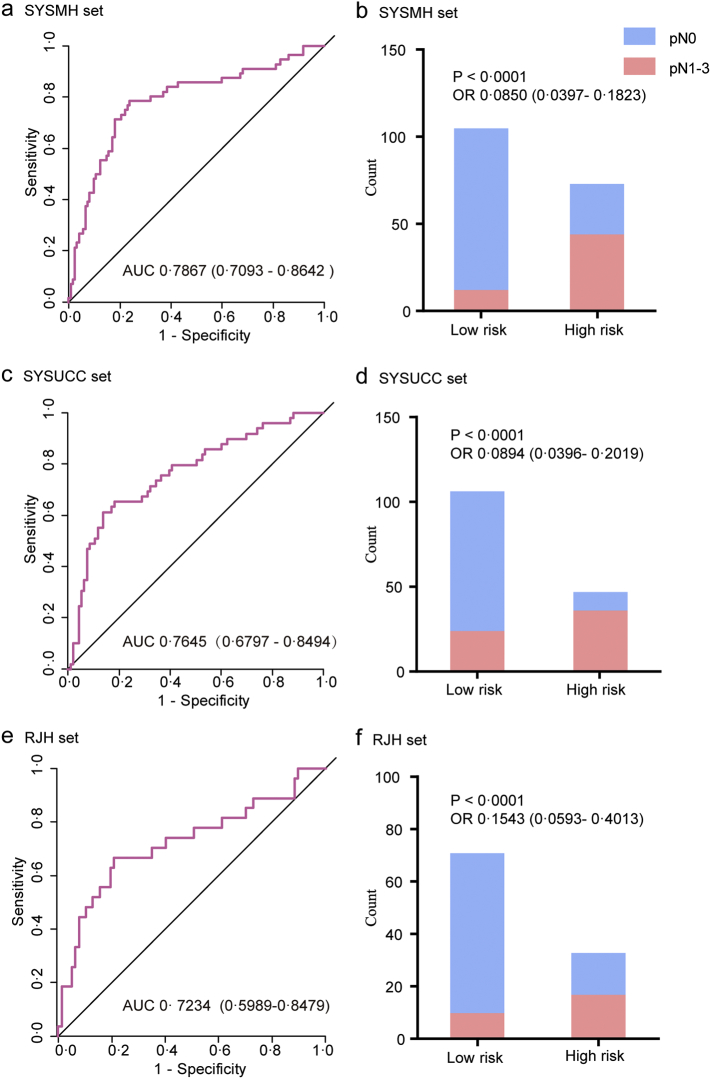
In the training stage, a multivariable logistic regression model was used to construct a five-mRNA-based classifier for predicting LN status in the training set (the SYSMH set, n = 178) based on the findings in the discovery stage. According to this regression model, a risk score for the risk of LN metastasis was calculated for each patient based on their individual expression levels of the five mRNAs: risk score = (5.5735 ∗ ADRA1D expression level) + (0.8713 ∗ COL10A1 expression level) − (0.3087 ∗ DKK2 expression level) + (20.1961 ∗ MMP11 expression level) − (3.5974 ∗ HIST2H3D expression level) − 1.0833. We then derived the genetic risk scores based on the established model. Next, ROC analysis (Fig. 3a) was performed to investigate the predictive efficiency of the classifier. The resulting AUC was 0.7867 (95% CI 0.7093–0.8642), which indicated that our five-mRNA-based classifier had superior predictive efficiency. External validation using the SYSUCC set (n = 142) and the RJH set (n = 104) was performed to validate the predictive efficiency of the classifier, with resulting AUC values of 0.7645 (95% CI 0.6797–0.8494) and 0.7234 (95% CI 0.5989–0.8479), respectively (Fig. 3c and e). Overall, the risk score showed favorable discrimination ability in all three sets.
Fig. 3.
The performance of the models in the training sets (the SYSMH set) and two independent sample sets (the SYSUCC set and the RJH set).
Left panels represent ROC curve analyses for the genomic risk scores derived from the five-mRNA-based classifier; right panels represent high- and low-risk population classifications based on the genomic risk scores. (a, b) the SYSMH set, (c, d) the SYSUCC set, and (e, f) the RJH set. OR (95% CI, chi-square test) and AUC (95% CI) are presented. ROC = receiver operator characteristic. AUC = area under the curve.
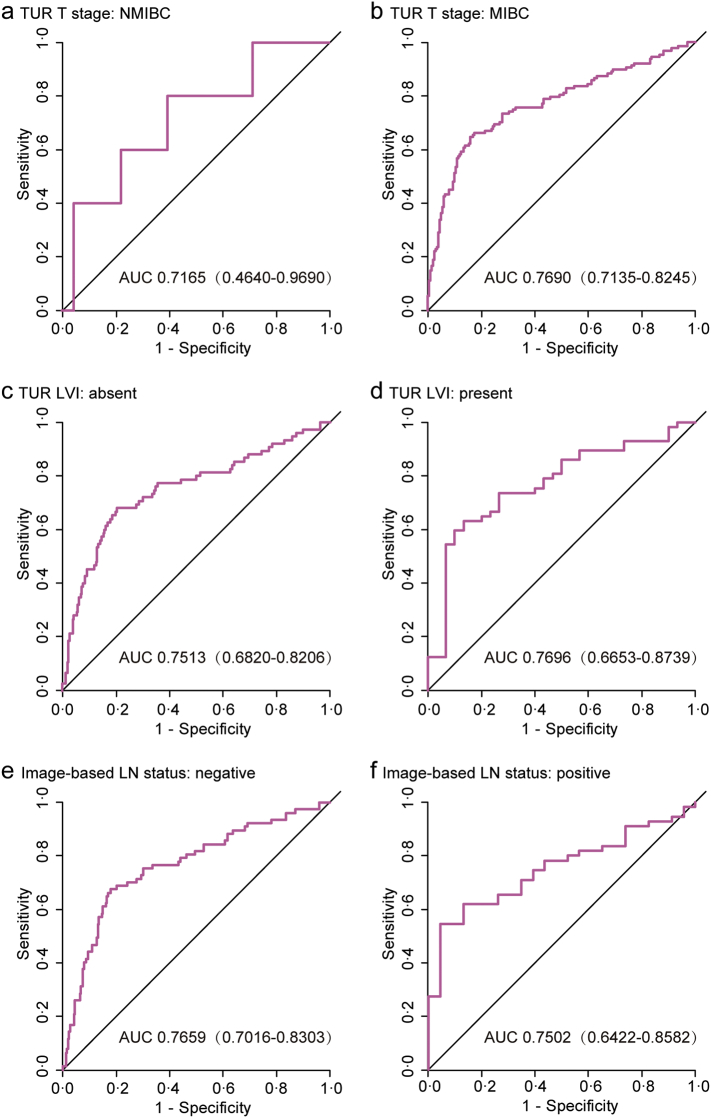
We also evaluated the classification performance of the model in distinguishing BCa patients with a low or high risk of LN metastasis. We defined an optimal risk score cutoff value of −1.005 based on the Youden index in the training set (the SYSMH set). The patients in each set were categorized into low- and high-risk groups according to the cutoff value. Notably, the high-risk group had a higher possibility of harboring LN metastasis in all three sets (Fig. 3b, d and f). When stratified by clinicopathological risk factors, the five-mRNA-based classifier remained a favorable predictive model (Fig. 4 and Supplementary Fig. S2). There is a potential benefit to identifying the high-risk population among those who are clinically considered low risk (NMIBC or cN-) and the low-risk population among those who are clinically characterized as high risk (MIBC or cN+), thus avoiding overstaging or understaging in clinical practice.
Fig. 4.
Stratified ROC curve analyses to evaluate the influences of clinicopathological risk factors on the performance of the five-mRNA-based classifier in all 424 patients with bladder cancer.
(a, b) TUR T stage. (c, d) TUR LVI. (e, f) Image-based LN status. ROC = receiver operator characteristic. AUC = area under the curve. LN = lymph node.
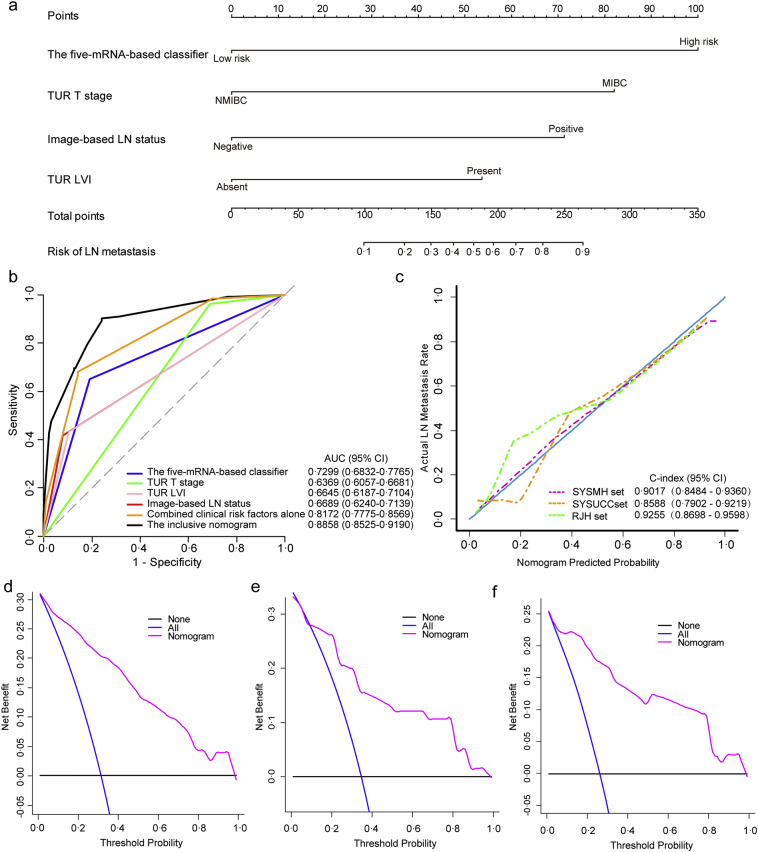
To construct a clinical nomogram, we first evaluated the clinicopathological risk factors and selected TUR T stage, image-based LN status and TUR LVI using a multivariable logistic regression model (Table 3). By combining the selected clinicopathological risk factors with the five-mRNA-based classifier, we constructed an inclusive nomogram that incorporated the genomic classifier and clinicopathologic factors, thus providing clinicians with an efficient tool to predict the individual risk of LN metastasis in a BCa patient preoperatively (Fig. 5a). To compare the discrimination ability of the inclusive nomogram with the five-mRNA-based classifier and clinicopathological risk factors, we conducted ROC analyses of all patients (n = 424) (Fig. 5b). The results suggested that the inclusive nomogram had higher prediction efficacy (AUC 0.8858, 95% CI 0.8525–0.9190) compared with the five-mRNA-based classifier or the model containing clinicopathological risk factors alone. We also defined an optimal risk score cutoff value of −1.779 in the training set based on the Youden index. The calibration curve of the nomogram is shown in Fig. 5c. The Hosmer-Lemeshow test suggested that there was no significant departure from excellent fit (P = .9640). The C-index value of 0.9017 (95% CI 0.8484–0.9360) for the bootstrapping validation in the SYSMH set also indicated good discrimination by the nomogram.
Table 3.
Multivariate logistic regression model for lymph node metastasis in bladder cancer.
| Variable and intercept | β | OR (95% CI) | P |
|---|---|---|---|
| The five-mRNAs-based classifier | 2·667 | 14·400 (5·562, 37·284) | <0·001 |
| TUR T stage | 2·188 | 8·915 (2·036, 39·033) | 0·004 |
| TUR LVI | 1·430 | 4·181 (1·551, 11·269) | 0·005 |
| Image-based LN status | 1·902 | 6·698 (2·389, 18·780) | <0·001 |
| Intercept | −9·694 | – | <0·001 |
Fig. 5.
The genomic-clinicopathologic nomogram for LN metastasis prediction in patients with bladder cancer.
(a) The genomic-clinicopathologic nomogram.
(b) ROC curve analyses of the models to compare the predictive performance in all patients (n = 424).
(c) Calibration curves and C-indexes (95% CI) of the genomic-clinicopathologic nomogram in SYSMH set, SYSUCC set and RJH set. Calibration curves depict the calibration of the nomogram in terms of agreement between predicted risk of LN metastasis and observed pN outcomes. The 45-degree blue line represents a perfect prediction, and the dotted lines represent the predictive performance of the nomogram. The closer the pink line fit to the ideal line, the better predictive accuracy the nomogram shows.
(d–f) The decision curve analyses (DCAs) for the genomic-clinicopathologic nomogram in the SYSMH set (d), the SYSUCC set (e) and the RJH set (f). The Y-axis represents the net benefit. The pink line represents the genomic-clinicopathologic nomogram. The blue line represents the hypothesis that all patients were involved in LN metastases. The black line represents the hypothesis that no patients are involved in LN metastases. The X-axis represents the threshold probability. The threshold probability is where the expected benefit of treatment is equal to the expected benefit of avoiding treatment. For example, if the possibility of LN metastasis involvement of a patient was over the threshold probability, treatment strategy for LN metastasis should be adopted. All the decision curves in three sets showed that if the threshold probability is between 10% and 100%, using the genomic-clinicopathologic nomogram to predict LN metastases adds more benefit than either all of patients were treated or none of them were treated.
In the validation stage, the nomogram was externally validated in the SYSUCC set (n = 142) and the RJH set (n = 104). Calibration curves were plotted to assess the calibration of the nomogram (Fig. 5c). The Hosmer-Lemeshow test indicated a satisfactory goodness-of-fit of the nomogram in the SYSUCC and RJH sets (P value 0.8501 and 0.8489, respectively). Notably, the C-indexes of the two validation sets were 0.8588 (95% CI 0.7902–0.9219) and 0.9255 (95% CI 0.8698–0.9598), respectively. We also obtained consistent results in all 424 patients from both of the training set and the validation sets, with a C-index of 0.8861 (95% CI 0.8492–0.9152) (see Supplementary Fig. S4B).
The DCA results for the nomogram are presented in Fig. 5d–f. The DCA showed that if the probability threshold for doctors or patients is 10% or more, LN status-related treatment decision-making based on the genomic-clinicopathologic nomogram adds more net benefit than treating either all patients or none in each independent dataset in this study. In other words, our nomogram was clinically useful for the study subjects.
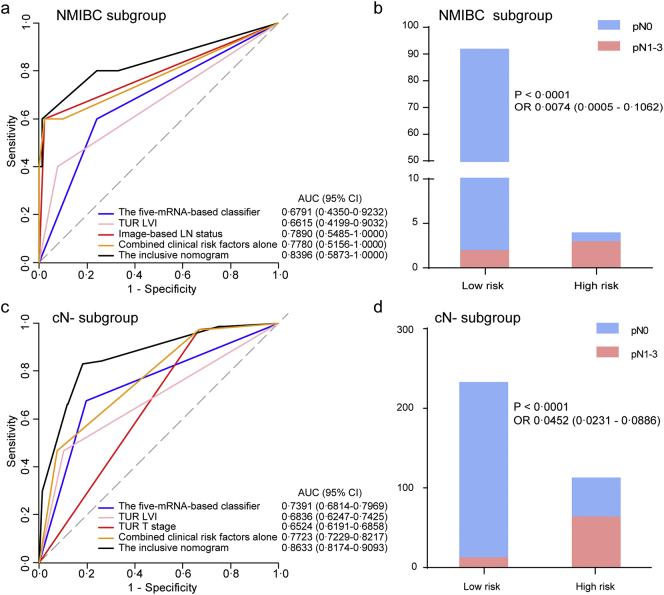
Furthermore, ROC analysis was performed to evaluate the discrimination ability of the inclusive nomogram in the clinically low-risk patient subgroups (NMIBC or cN-), and the results are shown in Fig. 6. Encouragingly, the inclusive nomogram also showed good discrimination ability in the NMIBC subgroup (AUC 0.8396, 95% CI 0.5873–1.0000; Fig. 6a) and the cN- subgroup (AUC 0.8633, 95% CI 0.8174–0.9093; Fig. 6c). Moreover, we defined optimal risk score cutoff values of −1.779 in the NMIBC subgroup and −1.636 in the cN- subgroup based on the Youden index. The patients in each subgroup were categorized into low- and high-risk groups according to the cutoff values. Notably, the high-risk group had a higher possibility of harboring LN metastasis in both subgroups (Fig. 6b and d).
Fig. 6.
The genomic-clinicopathologic nomogram for LN metastasis prediction in low-risk groups of patients with bladder cancer.
Left panels indicate the ROC curve analyses for the models in the low-risk subgroups of BCa patients, including NMIBC subgroup (n = 96) and cN- subgroup (n = 346). Right panels indicate the risk classification performance for the nomogram in the low-risk subgroups.
(a, b) NMIBC subgroup. (c, d) cN- subgroup. ROC = receiver operator characteristic. AUC = area under the curve. LN = lymph node.
Finally, we compared the inclusive nomogram with the clinicopathologic nomogram, which incorporated only clinicopathologic factors (including TUR T stage, image-based LN status and TUR LVI) (Supplementary Fig. S3). The results indicated that the calibration curve and DCA of the clinicopathologic nomogram were inferior to the inclusive genomic-clinicopathologic nomogram (Supplementary Figs. S4 and S5). Considering the significant relationship of pathologic node status and recurrence-free survival, we wondered whether risk stratification by the nomogram could also indicate recurrence-free survival post-cystectomy. We used Kaplan–Meier survival analysis to compare patients at low risk of LN metastasis with those at high risk according to the nomogram using the optimal cutoff mentioned above for the SYSMH set. The log-rank test showed that the high-risk subgroup had significantly shorter recurrence-free survival than the low-risk subgroup (P < .0001, Supplementary Fig. S6).
4. Discussion
The preoperative prediction of LN status is important for decision-making regarding BCa treatment regimens, specifically concerning the extent of PLND and the use of neoadjuvant chemotherapy. However, the accuracy of the current preoperative nodal staging is unsatisfactory; a considerable portion of patients are understaged, and some are overstaged. Therefore, accurate predictive tools for LN staging before surgery are urgently needed. In this study, we developed an inclusive nomogram that combined a five-mRNA-based classifier with three clinicopathological risk factors to predict LN metastasis in BCa patients. We validated the nomogram in two independent sample sets and achieved good performance in predicting LN metastasis preoperatively. Importantly, our model might benefit the subgroups of BCa patients who are clinically considered as having a low risk of nodal metastasis (NMIBC or cN-).
Current guidelines for BCa patients suggest that PLND should be considered as an essential part of RC, but the proximal extent of a PLND at the time of RC has always been an issue of controversy (Alfred Witjes et al., 2017). Shariat et al. developed a clinical nodal scoring model based on the preoperative T stage to help determining the minimum number of lymph nodes that should be removed at RC (Shariat et al., 2012). Several studies have also suggested that BCa patients benefit from extended PLND compared with standard PLND (Bruins et al., 2014). However, a recent randomized clinical trial presented a trend but no significant difference toward improved survival with an extended PLND, comparing with limited PLND (Gschwend et al., 2016). Nowadays, extended PLND has not been widely adopted in current clinical practice, in particular for clinically low-risk patients (NMIBC or cN-), because the population that will benefit and the potential harm caused by the enlargement of the PLND have not been determined. It is obvious that patients harboring LN metastasis will likely benefit from extended PLND. Thus, accurate preoperative prediction of LN status might help identify appropriate patients for extended PLND, in particular among those in the clinical low-risk subgroup (NMIBC or cN-).
In addition to identifying appropriate patients for PLND, accurate preoperative nodal staging might also help select appropriate patients for neoadjuvant chemotherapy and improve BCa patient survival. A previous phase III randomized trial indicated that patients with muscle-invasive bladder cancer (MIBC) benefited from neoadjuvant chemotherapy, with an increase in 10-year survival from 30% to 36% (Griffiths et al., 2011). In addition, the EAU guidelines recommend offering neoadjuvant chemotherapy to BCa patients with T2-T4a, cN0M0 stage disease. However, neoadjuvant chemotherapy has not been applied regularly in clinical practice and is rarely administered to T2 patients because a high percentage of patients will not benefit from it and it is difficult to predict who will have a therapeutic response (Advanced Bladder Cancer Overview Collaboration., 2005; Martini et al., 2016). The goal of neoadjuvant chemotherapy is to target micrometastatic lesions, including positive LNs (Mertens et al., 2014), indicating that pN1~3 patients are likely to benefit from neoadjuvant chemotherapy. Therefore, a preoperative tool for predicting LN status might help select appropriate patients for neoadjuvant chemotherapy.
It is important to construct a pre-cystectomy decision model to predict LN stage. Karakiewicz et al. developed a nomogram including “TUR T stage” and “TUR grade” to predict pN stage with a prediction accuracy of 63.1% (Karakiewicz et al., 2006). In addition to the clinicopathologic model, genomic models have emerged for disease prediction in recent years. To our knowledge, there are two studies involved genomic models that identify pN1–3 and pN0 BCa patients (Smith et al., 2011; Seiler et al., 2016). One study was conducted by Smith and colleagues, who constructed a 20-mRNA-based classifier based on the gene expression signature to evaluate the possibility of LN metastasis in cN- MIBC patients before RC. The model was validated externally with a modest AUC of 0.67 (Smith et al., 2011). Seiler et al. built a 51-RNA-based model and achieved an AUC of 0.82 (95% CI 0.71–0.93) (Seiler et al., 2016). Although the prediction power of the 51-RNA-based model was favorable, the model lacked external validation.
To achieve more efficient genomic classifiers for BCa patients with a distinct risk of LN metastasis, we developed a five-mRNA-based classifier and a genomic-clinicopathologic model in the training set and externally validated it in two independent sample sets. The five-mRNA-based classifier contains five mRNAs: ADRA1D, COL10A1, DKK2, HIST2H3D and MMP11. Among them, ADRA1D, COL10A1, DKK2 and MMP11 have been previously reported to be associated with cancer (Chapman et al., 2012; Colciago et al., 2016; Li et al., 2016; Xu et al., 2016). ADRA1D and DKK2 have been reported to promote metastasis in prostate cancer (Colciago et al., 2016; Xu et al., 2016). Notably, MMP11 overexpression has been demonstrated to be related to LN metastasis in BCa (Li et al., 2016). Consequently, further research on the biological function of these selected mRNAs is likely to offer new insights into BCa LN metastasis. The AUC of our five-mRNA-based classifier was 0.7867 (95% CI 0.7093–0.8642), which is comparable to that of the previously reported 51-RNA-based model (Seiler et al., 2016). When stratified by clinicopathologic factors, our five-mRNA-based classifier remained a favorable predictive model.
Furthermore, we postulated that the inclusive model combining the genomic signature and clinical risk factors might achieve a more favorable and reliable prediction efficacy for LN status prediction. Indeed, the genomic-clinicopathologic nomogram performed well (C-index 0.9017, 95% CI [0.8484–0.9360]) in LN prediction and was consistently validated in two independent sample sets. Hence, our genomic-clinicopathologic nomogram may represent a reliable and accurate tool for LN status prediction, which is helpful for BCa treatment decision-making.
Notably, a valuable feature of our inclusive nomogram might be the discrimination ability in clinically low-risk subgroups (NMIBC or cN-). BCa patients diagnosed with NMIBC or cN- disease are usually considered at low risk of LN metastasis and are therefore not subjected to neoadjuvant chemotherapy or extended PLND. However, some patients in the clinically low-risk subgroups have LN metastasis, and they are likely to benefit from neoadjuvant chemotherapy or extended PLND. It is a formidable challenge to precisely identify the patients who will experience LN metastasis. Encouragingly, our inclusive nomogram showed good discrimination ability in the NMIBC and cN- subgroups. Moreover, when categorized into low and high-risk groups according to the cutoff values of the risk score derived from the nomogram, the high-risk group had a significantly higher possibility of harboring LN metastasis in both clinically low-risk subgroups (NMIBC and cN-). Thus, our inclusive nomogram might benefit a large proportion of the patients who might be incorrectly considered at low risk of harboring LN metastasis.
Compared with previous predictive models, our model has several advantages. First, unlike the reported 20-mRNA-based model or 51-RNA-based model, we focused on cN0–3 patients rather than cN- patients only (Smith et al., 2011; Seiler et al., 2016). Some pN0 patients may be incorrectly diagnosed as LN positive (cN+) and thus may receive excessive treatment (Baltaci et al., 2008; Lodde et al., 2010; Goodfellow et al., 2014), such as unnecessary neoadjuvant chemotherapy or extended PLND, which come with the risks of side effects, delayed operation, or additional operative complications. Therefore, we included patients diagnosed at stage cN0–3 in the predictive model to enable us to potentially discriminate pN0 patients who were incorrectly diagnosed as cN+. Second, we used qRT-PCR, a simple, low-cost and reliable procedure, for the five-mRNA-based classifier, thus making our model more practical for clinical use. Third, we combined the genomic signature with clinicopathological risk factors to construct an inclusive nomogram for predicting LN metastasis, thereby taking advantage of both genomic and clinical information and perhaps improving accuracy. Such an inclusive model for BCa LN metastasis prediction has not yet been reported.
A limitation of our research is that our nomogram lacked prospective validation. Multicenter prospective clinical trials with a large sample size are needed to provide high-level evidence for clinical application. Nevertheless, randomized clinical trial examining tissues from transurethral resection of bladder tumor (TURBT) are warranted to validate our findings.
In conclusion, we developed a five-mRNA-based classifier for LN metastasis prediction in BCa patients. The inclusive nomogram incorporating the five-mRNA-based classifier and clinicopathologic factors might represent an efficient and valuable tool for individual LN metastasis prediction and BCa treatment decision-making, in particular for those patients diagnosed at stage NMIBC or cN-.
Funding Sources
This work was funded by National Natural Science Foundation of China (81572514, U1301221, 81472384, 81372729). These funding sources had no role in the data collection, analysis or interpretation; in the study design; in the writing of the manuscript; or in the decision to submit the paper for publication.
Conflicts of Interest
We declare that we have no conflicts of interest.
Author Contributions
T-XL, JH, Z-WL, H-GC and Q-QC designed the study. JH, Z-WL, H-GC and Q-QC provided samples or obtained clinical data or both. S-XW, PG, J-JZ and H-DQ analyzed and interpreted the data. S-XW, J-JZ, Z-SZ, XC, S-LC and R-YZ did the experiments. T-XL, S-XW and PG wrote the report, which was edited by all authors. T-XL and JH supervised the project. S-XW, JH, Z-WL, H-GC, PG and Q-QC contributed equally to this work.
Acknowledgments
We are thankful to the TCGA Research Network for its contribution. We thank UCSC Cancer Genomics Browser for the normalized gene expression data.
Appendix A. Supplementay data
Supplementary material
References
- Abol-Enein H., Tilki D., Mosbah A., El-Baz M., Shokeir A., Nabeeh A., Ghoneim M.A. Does the extent of lymphadenectomy in radical cystectomy for bladder cancer influence disease-free survival? A prospective single-center study. Eur. Urol. 2011;60(3):572–577. doi: 10.1016/j.eururo.2011.05.062. [DOI] [PubMed] [Google Scholar]
- Advanced Bladder Cancer Overview Collaboration Neoadjuvant chemotherapy for invasive bladder cancer. Cochrane Database Syst. Rev. 2005;(2) doi: 10.1002/14651858.CD005246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfred Witjes J., Lebret T., Comperat E.M., Cowan N.C., De Santis M., Bruins H.M., Hernandez V., Espinos E.L., Dunn J., Rouanne M., Neuzillet Y., Veskimae E., van der Heijden A.G., Gakis G., Ribal M.J. Updated 2016 EAU guidelines on muscle-invasive and metastatic bladder cancer. Eur. Urol. 2017;71(3):462–475. doi: 10.1016/j.eururo.2016.06.020. [DOI] [PubMed] [Google Scholar]
- Antoni S., Ferlay J., Soerjomataram I., Znaor A., Jemal A., Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur. Urol. 2017;71(1):96–108. doi: 10.1016/j.eururo.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Baltaci S., Resorlu B., Yagci C., Turkolmez K., Gogus C., Beduk Y. Computerized tomography for detecting perivesical infiltration and lymph node metastasis in invasive bladder carcinoma. Urol. Int. 2008;81(4):399–402. doi: 10.1159/000167836. [DOI] [PubMed] [Google Scholar]
- Baltaci S., Adsan O., Ugurlu O., Aslan G., Can C., Gunaydin G., Buyukalpelli R., Elhan A.H., Beduk Y. Reliability of frozen section examination of obturator lymph nodes and impact on lymph node dissection borders during radical cystectomy: results of a prospective multicentre study by the Turkish Society of Urooncology. BJU Int. 2011;107(4):547–553. doi: 10.1111/j.1464-410X.2010.09504.x. [DOI] [PubMed] [Google Scholar]
- Barentsz J.O., Engelbrecht M.R., Witjes J.A., de la Rosette J.J., van der Graaf M. MR imaging of the male pelvis. Eur. Radiol. 1999;9(9):1722–1736. doi: 10.1007/s003300050916. [DOI] [PubMed] [Google Scholar]
- Bassi P., Ferrante G.D., Piazza N., Spinadin R., Carando R., Pappagallo G., Pagano F. Prognostic factors of outcome after radical cystectomy for bladder cancer: a retrospective study of a homogeneous patient cohort. J. Urol. 1999;161(5):1494–1497. [PubMed] [Google Scholar]
- Bruins H.M., Veskimae E., Hernandez V., Imamura M., Neuberger M.M., Dahm P., Stewart F., Lam T.B., N'Dow J., van der Heijden A.G., Comperat E., Cowan N.C., De Santis M., Gakis G., Lebret T., Ribal M.J., Sherif A., Witjes J.A. The impact of the extent of lymphadenectomy on oncologic outcomes in patients undergoing radical cystectomy for bladder cancer: a systematic review. Eur. Urol. 2014;66(6):1065–1077. doi: 10.1016/j.eururo.2014.05.031. [DOI] [PubMed] [Google Scholar]
- Burger M., Catto J.W., Dalbagni G., Grossman H.B., Herr H., Karakiewicz P., Kassouf W., Kiemeney L.A., La Vecchia C., Shariat S., Lotan Y. Epidemiology and risk factors of urothelial bladder cancer. Eur. Urol. 2013;63(2):234–241. doi: 10.1016/j.eururo.2012.07.033. [DOI] [PubMed] [Google Scholar]
- Chapman K.B., Prendes M.J., Sternberg H., Kidd J.L., Funk W.D., Wagner J., West M.D. COL10A1 expression is elevated in diverse solid tumor types and is associated with tumor vasculature. Future Oncol. 2012;8(8):1031–1040. doi: 10.2217/fon.12.79. [DOI] [PubMed] [Google Scholar]
- Cho K.S., Seo H.K., Joung J.Y., Park W.S., Ro J.Y., Han K.S., Chung J., Lee K.H. Lymphovascular invasion in transurethral resection specimens as predictor of progression and metastasis in patients with newly diagnosed T1 bladder urothelial cancer. J. Urol. 2009;182(6):2625–2630. doi: 10.1016/j.juro.2009.08.083. [DOI] [PubMed] [Google Scholar]
- Colciago A., Mornati O., Ferri N., Castelnovo L.F., Fumagalli L., Bolchi C., Pallavicini M., Valoti E., Negri-Cesi P. A selective alpha1D-adrenoreceptor antagonist inhibits human prostate cancer cell proliferation and motility “in vitro”. Pharmacol. Res. 2016;103:215–226. doi: 10.1016/j.phrs.2015.11.017. [DOI] [PubMed] [Google Scholar]
- Culp S.H., Dickstein R.J., Grossman H.B., Pretzsch S.M., Porten S., Daneshmand S., Cai J., Groshen S., Siefker-Radtke A., Millikan R.E., Czerniak B., Navai N., Wszolek M.F., Kamat A.M., Dinney C.P. Refining patient selection for neoadjuvant chemotherapy before radical cystectomy. J. Urol. 2014;191(1):40–47. doi: 10.1016/j.juro.2013.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman R.E., Alpern M.B., Gross B.H., Sandler M.A. Upper abdominal lymph nodes: criteria for normal size determined with CT. Radiology. 1991;180(2):319–322. doi: 10.1148/radiology.180.2.2068292. [DOI] [PubMed] [Google Scholar]
- Epstein J., Eble J., Sauter G., Sesterhenn I. 2004. World Health Organization Classification of tumors: pathology and genetics of tumours of the urinary system and male genital organs. [Google Scholar]
- Goodfellow H., Viney Z., Hughes P., Rankin S., Rottenberg G., Hughes S., Evison F., Dasgupta P., O'Brien T., Khan M.S. Role of fluorodeoxyglucose positron emission tomography (FDG PET)-computed tomography (CT) in the staging of bladder cancer. BJU Int. 2014;114(3):389–395. doi: 10.1111/bju.12608. [DOI] [PubMed] [Google Scholar]
- Griffiths G., Hall R., Sylvester R., Raghavan D., Parmar M.K. International phase III trial assessing neoadjuvant cisplatin, methotrexate, and vinblastine chemotherapy for muscle-invasive bladder cancer: long-term results of the BA06 30894 trial. J. Clin. Oncol. 2011;29(16):2171–2177. doi: 10.1200/JCO.2010.32.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gschwend J.E., Heck M.M., Lehmann J., Ruebben H., Albers P., Heidenreich A., Geeter P.d., Wolff J.M., Frohneberg D., Schnoeller T., Kälble T., Stoeckle M., Stenzl A., Mueller M., Liehr U.-B., Truss M., Roth S., Leissner J., Retz M. Limited versus extended pelvic lymphadenectomy in patients with bladder cancer undergoing radical cystectomy: Survival results from a prospective, randomized trial (LEA AUO AB 25/02) J. Clin. Oncol. 2016;34(15_suppl):4503. [Google Scholar]
- Jensen J.B., Ulhoi B.P., Jensen K.M. Evaluation of different lymph node (LN) variables as prognostic markers in patients undergoing radical cystectomy and extended LN dissection to the level of the inferior mesenteric artery. BJU Int. 2012;109(3):388–393. doi: 10.1111/j.1464-410X.2011.10369.x. [DOI] [PubMed] [Google Scholar]
- Karakiewicz P.I., Shariat S.F., Palapattu G.S., Perrotte P., Lotan Y., Rogers C.G., Amiel G.E., Vazina A., Gupta A., Bastian P.J., Sagalowsky A.I., Schoenberg M., Lerner S.P. Precystectomy nomogram for prediction of advanced bladder cancer stage. Eur. Urol. 2006;50(6):1254–1260. doi: 10.1016/j.eururo.2006.06.010. (discussion 1261-2) [DOI] [PubMed] [Google Scholar]
- Karl A., Carroll P.R., Gschwend J.E., Knuchel R., Montorsi F., Stief C.G., Studer U.E. The impact of lymphadenectomy and lymph node metastasis on the outcomes of radical cystectomy for bladder cancer. Eur. Urol. 2009;55(4):826–835. doi: 10.1016/j.eururo.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Kluth L.A., Black P.C., Bochner B.H., Catto J., Lerner S.P., Stenzl A., Sylvester R., Vickers A.J., Xylinas E., Shariat S.F. Prognostic and prediction tools in bladder cancer: a comprehensive review of the literature. Eur. Urol. 2015;68(2):238–253. doi: 10.1016/j.eururo.2015.01.032. [DOI] [PubMed] [Google Scholar]
- Leissner J., Ghoneim M.A., Abol-Enein H., Thuroff J.W., Franzaring L., Fisch M., Schulze H., Managadze G., Allhoff E.P., el-Baz M.A., Kastendieck H., Buhtz P., Kropf S., Hohenfellner R., Wolf H.K. Extended radical lymphadenectomy in patients with urothelial bladder cancer: results of a prospective multicenter study. J. Urol. 2004;171(1):139–144. doi: 10.1097/01.ju.0000102302.26806.fb. [DOI] [PubMed] [Google Scholar]
- Li W.M., Wei Y.C., Huang C.N., Ke H.L., Li C.C., Yeh H.C., Chang L.L., Huang C.H., Li C.F., Wu W.J. Matrix metalloproteinase-11 as a marker of metastasis and predictor of poor survival in urothelial carcinomas. J. Surg. Oncol. 2016;113(6):700–707. doi: 10.1002/jso.24195. [DOI] [PubMed] [Google Scholar]
- Lodde M., Lacombe L., Friede J., Morin F., Saourine A., Fradet Y. Evaluation of fluorodeoxyglucose positron-emission tomography with computed tomography for staging of urothelial carcinoma. BJU Int. 2010;106(5):658–663. doi: 10.1111/j.1464-410X.2010.09212.x. [DOI] [PubMed] [Google Scholar]
- Martini T., Gilfrich C., Mayr R., Burger M., Pycha A., Aziz A., Gierth M., Stief C.G., Muller S.C., Wagenlehner F., Roigas J., Hakenberg O.W., Roghmann F., Nuhn P., Wirth M., Novotny V., Hadaschik B., Grimm M.O., Schramek P., Haferkamp A., Colleselli D., Kloss B., Herrmann E., Fisch M., May M., Bolenz C. The use of neoadjuvant chemotherapy in patients with urothelial carcinoma of the bladder: current practice among clinicians. Clin. Genitour. Cancer. 2016;15(3):356–362. doi: 10.1016/j.clgc.2016.09.003. [DOI] [PubMed] [Google Scholar]
- McKibben M.J., Woods M.E. Preoperative imaging for staging bladder cancer. Curr. Urol. Rep. 2015;16(4):22. doi: 10.1007/s11934-015-0496-8. [DOI] [PubMed] [Google Scholar]
- Mertens L.S., Meijer R.P., Meinhardt W., van der Poel H.G., Bex A., Kerst J.M., van der Heijden M.S., Bergman A.M., Horenblas S., van Rhijn B.W. Occult lymph node metastases in patients with carcinoma invading bladder muscle: incidence after neoadjuvant chemotherapy and cystectomy vs after cystectomy alone. BJU Int. 2014;114(1):67–74. doi: 10.1111/bju.12447. [DOI] [PubMed] [Google Scholar]
- Seiler R., Lam L.L., Erho N., Takhar M., Mitra A.P., Buerki C., Davicioni E., Skinner E.C., Daneshmand S., Black P.C. Prediction of lymph node metastasis in patients with bladder cancer using whole transcriptome gene expression signatures. J. Urol. 2016;196(4):1036–1041. doi: 10.1016/j.juro.2016.04.061. [DOI] [PubMed] [Google Scholar]
- Shariat S.F., Karakiewicz P.I., Palapattu G.S., Lotan Y., Rogers C.G., Amiel G.E., Vazina A., Gupta A., Bastian P.J., Sagalowsky A.I., Schoenberg M.P., Lerner S.P. Outcomes of radical cystectomy for transitional cell carcinoma of the bladder: a contemporary series from the Bladder Cancer Research Consortium. J. Urol. 2006;176(6 Pt 1):2414–2422. doi: 10.1016/j.juro.2006.08.004. (discussion 2422) [DOI] [PubMed] [Google Scholar]
- Shariat S.F., Ehdaie B., Rink M., Cha E.K., Svatek R.S., Chromecki T.F., Fajkovic H., Novara G., David S.G., Daneshmand S., Fradet Y., Lotan Y., Sagalowsky A.I., Clozel T., Bastian P.J., Kassouf W., Fritsche H.M., Burger M., Izawa J.I., Tilki D., Abdollah F., Chun F.K., Sonpavde G., Karakiewicz P.I., Scherr D.S., Gonen M. Clinical nodal staging scores for bladder cancer: a proposal for preoperative risk assessment. Eur. Urol. 2012;61(2):237–242. doi: 10.1016/j.eururo.2011.10.011. [DOI] [PubMed] [Google Scholar]
- Smith S.C., Baras A.S., Dancik G., Ru Y., Ding K.F., Moskaluk C.A., Fradet Y., Lehmann J., Stockle M., Hartmann A., Lee J.K., Theodorescu D. A 20-gene model for molecular nodal staging of bladder cancer: development and prospective assessment. Lancet Oncol. 2011;12(2):137–143. doi: 10.1016/S1470-2045(10)70296-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobin L.H., Gospodarowicz M., Wittekind C. Wiley-Blackwell; Hoboken, NJ: 2009. TNM Classification of Malignant Tumors. [Google Scholar]
- Stamatakis L., Godoy G., Lerner S.P. Innovations in radical cystectomy and pelvic lymph node dissection. Semin. Oncol. 2012;39(5):573–582. doi: 10.1053/j.seminoncol.2012.08.012. [DOI] [PubMed] [Google Scholar]
- Stein J.P., Lieskovsky G., Cote R., Groshen S., Feng A.C., Boyd S., Skinner E., Bochner B., Thangathurai D., Mikhail M., Raghavan D., Skinner D.G. Radical cystectomy in the treatment of invasive bladder cancer: long-term results in 1,054 patients. J. Clin. Oncol. 2001;19(3):666–675. doi: 10.1200/JCO.2001.19.3.666. [DOI] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B Methodol. 1996:267–288. [Google Scholar]
- Vazina A., Dugi D., Shariat S.F., Evans J., Link R., Lerner S.P. Stage specific lymph node metastasis mapping in radical cystectomy specimens. J. Urol. 2004;171(5):1830–1834. doi: 10.1097/01.ju.0000121604.58067.95. [DOI] [PubMed] [Google Scholar]
- Vickers A.J., Cronin A.M., Elkin E.B., Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med. Inform. Decis. Mak. 2008;8:53. doi: 10.1186/1472-6947-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers A.J., Van Calster B., Steyerberg E.W. Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. BMJ. 2016;352:i6. doi: 10.1136/bmj.i6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Pang K., Zhou Z.G., Chen Y.F., Mo T., Li M., Liu C.B. Dickkopf 2 promotes proliferation and invasion via Wnt signaling in prostate cancer. Mol. Med. Rep. 2016;14(3):2283–2288. doi: 10.3892/mmr.2016.5502. [DOI] [PubMed] [Google Scholar]
- Zargar-Shoshtari K., Zargar H., Lotan Y., Shah J.B., van Rhijn B.W., Daneshmand S., Spiess P.E., Black P.C. A multi-institutional analysis of outcomes of patients with clinically node positive urothelial bladder cancer treated with induction chemotherapy and radical cystectomy. J. Urol. 2016;195(1):53–59. doi: 10.1016/j.juro.2015.07.085. [DOI] [PubMed] [Google Scholar]
- Zehnder P., Studer U.E., Skinner E.C., Dorin R.P., Cai J., Roth B., Miranda G., Birkhauser F., Stein J., Burkhard F.C., Daneshmand S., Thalmann G.N., Gill I.S., Skinner D.G. Super extended versus extended pelvic lymph node dissection in patients undergoing radical cystectomy for bladder cancer: a comparative study. J. Urol. 2011;186(4):1261–1268. doi: 10.1016/j.juro.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Zehnder P., Studer U.E., Daneshmand S., Birkhauser F.D., Skinner E.C., Roth B., Miranda G., Burkhard F.C., Cai J., Skinner D.G., Thalmann G.N., Gill I.S. Outcomes of radical cystectomy with extended lymphadenectomy alone in patients with lymph node-positive bladder cancer who are unfit for or who decline adjuvant chemotherapy. BJU Int. 2014;113(4):554–560. doi: 10.1111/bju.12520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material