Abstract
Protein homeostasis in the secretory pathway is maintained by a hierarchy of quality control checkpoints, including endoplasmic reticulum–associated degradation (ERAD), which leads to the destruction of misfolded proteins in the ER, as well as post-ER proteolysis. Although most aberrant proteins are degraded by ERAD, some misfolded proteins escape the ER and are degraded instead by lysosomal/vacuolar proteases. To date, it remains unclear how misfolded membrane proteins are selected for these different fates. Here we designed a novel model substrate, SZ*, to investigate how substrate selection is mediated in yeast. We discovered that SZ* is degraded by both the proteasome and vacuolar proteases, the latter of which occurs after ER exit and requires the multivesicular body pathway. By interrogating how various conditions affect the fate of SZ*, we also discovered that heat-shock and substrate overexpression increase ERAD targeting. These conditions also increase substrate aggregation. We next found that aggregation of the membrane-free misfolded domain in SZ* is concentration dependent, and fusion of this misfolded domain to a post-ER quality control substrate instead targets the substrate for ERAD. Our data indicate that a misfolded membrane protein with a higher aggregation propensity is preferentially retained in the ER and targeted for ERAD.
INTRODUCTION
In eukaryotes, approximately one-third of all proteins enter the secretory pathway. Owing to genetic mutations, translational errors, or cellular stresses, protein misfolding can occur quite frequently (Chen et al., 2011; Amm et al., 2014). Accumulation of misfolded, aggregation-prone polypeptides is detrimental to cellular health, and in fact >70 human diseases arise from defects in secretory and membrane protein folding (Guerriero and Brodsky, 2012). To ensure the fidelity of the proteome and maintain cellular homeostasis, eukaryotes evolved a hierarchy of protein quality control mechanisms that operate along the secretory pathway. These include ER quality control (ERQC) via the endoplasmic reticulum–associated degradation (ERAD) pathway (McCracken and Brodsky, 1996) and ER-phagy (Lipatova and Segev, 2015), Golgi quality control (Reggiori and Pelham, 2002), and plasma membrane (PM) quality control (Lin et al., 2008). Therefore, proteins—especially membrane proteins with complex topologies and subunits of multimeric assemblies—will be tightly monitored by sequential quality control checkpoints before arriving at their final destinations (Pizzirusso and Chang, 2004; Liu et al., 2006).
Since all secreted and membrane proteins are synthesized and begin the process of maturation in the ER, the cell devotes considerable resources to the regulation of both their quality and quantity via ERQC (Hampton and Garza, 2009; Ruggiano et al., 2014). Most proteins that fail to pass this QC checkpoint are eliminated by ERAD. During ERAD, misfolded proteins are first recognized by molecular chaperones, residing both in the ER lumen (Braakman and Bulleid, 2011) and cytoplasm (Zhang et al., 2001; Han et al., 2007; Nakatsukasa et al., 2008; Stolz and Wolf, 2010), as well as by chaperone-like lectins (Bhamidipati et al., 2005; Kim et al., 2005; Szathmary et al., 2005; Hosokawa et al., 2006; Braakman and Hebert, 2013). Based on the location of a folding lesion, misfolded ER-resident proteins are then ubiquitinated by distinct E3 ubiquitin ligases that associate with a nonoverlapping group of ERAD-requiring partners in the ER (Vashist and Ng, 2004; Carvalho et al., 2006). Despite the multitude of ERAD pathways utilized by the diverse group of ERAD substrates identified, they must all be retrotranslocated (or dislocated). During retrotranslocation, the ubiquitinated substrate is extracted from the ER into the cytoplasm in a Cdc48/p97- and ATP-dependent manner (Ye et al., 2001; Jarosch et al., 2002; Rabinovich et al., 2002). Following retrotranslocation, the extracted ubiquitinated protein is then delivered to the 26S proteasome for degradation (Vembar and Brodsky, 2008).
If the ERAD pathway is compromised or overwhelmed by the accumulation of misfolded protein substrates, then the unfolded protein response (UPR) is induced, which alleviates ER stress by increasing the folding capacity of the ER and ERAD efficiency (Walter and Ron, 2011). UPR induction also triggers ER-phagy (Bernales et al., 2006a; Schuck et al., 2014), which may further reduce the unfolded protein load in the ER. In addition to the UPR, the cytoplasmic heat-shock response alleviates ER stress by altering multiple ER-associated activities, such as protein translocation, protein folding in the ER, and protein transport from the ER to the Golgi (Liu and Chang, 2008). Moreover, heat-shock directly affects ERAD activity (Kelly et al., 2007). Nevertheless, some misfolded proteins can still escape the ER and advance to the Golgi or even to the plasma membrane.
Like ERQC, post-ERQC—including Golgi QC (GQC) and plasma membrane QC (PMQC)—utilizes key components to target misfolded proteins for degradation by the vacuole/lysosome or in select cases by the proteasome. For example, some ERAD substrates are transiently exported to the cis-Golgi but are then retrieved for ERAD (Caldwell et al., 2001; Vashist et al., 2001; Taxis et al., 2002). In contrast, misfolded proteins that travel along the secretory pathway may be monitored by GQC and are recognized by the Tul1 or Rsp5 E3 ubiquitin ligases. Substrates for this pathway include Phm5, Cps1, and Pep12(D), which harbor polar residues in transmembrane domains. These substrates are recognized by Tul1 in the Golgi and sorted to the vacuole for degradation (as is the case for Pep12(D)) or maturation (as observed for vacuolar resident proteins like Phm5 and Cps1) (Reggiori et al., 2000; Reggiori and Pelham, 2002). Membrane proteins with lumenal folding lesions may also be targeted for vacuolar degradation, such as Wsc1* (Wang and Ng, 2010; Wang et al., 2011).
PMQC ensures the fidelity of the proteome at the plasma membrane. When cells are subject to stresses, such as heat-shock, a change in nutrient levels, or altered environmental conditions, conformational changes within the substrate are detected, which leads to selection for PMQC (Nikko and Pelham, 2009; Zhao et al., 2013). Key players during PMQC include the ART-Rsp5 network, which recognizes and ubiquitinates PM proteins and targets them for vacuolar/lysosomal degradation (MacGurn et al., 2012; O’Donnell, 2012). In mammalian cells, the PMQC of misfolded forms of the cystic fibrosis transmembrane conductance regulator (CFTR) utilizes the CHIP ubiquitin ligase and the Hsp70 chaperone (Okiyoneda et al., 2010; Bagdany et al., 2017). Ultimately, both the GQC and PMQC pathways converge at the multivesicular body (MVB) pathway. Here ubiquitinated membrane substrates are invaginated into the lumen of an endosome with the assistance of endosomal sorting complexes required for transport (ESCRT) (Henne et al., 2011). Concomitant with the formation of intralumenal vesicles, misfolded membrane substrates are then delivered to the vacuolar/lysosomal lumen for degradation (Okiyoneda et al., 2011; Babst, 2014).
Although many mechanistic details underlying protein QC, and especially ERAD, have been revealed, little is known about how these QC checkpoints are coordinated with one another to maintain cellular health. To date, five membrane proteins with folding lesions in either the lumenal or transmembrane domain (TMD) have been identified as post-ERQC substrates that are targeted for vacuole-dependent degradation in yeast (Reggiori et al., 2000; Reggiori and Pelham, 2002; Kincaid and Cooper, 2007; Wang and Ng, 2010; Wang et al., 2011). Each of these substrates contain alterations in the membrane spanning region or in the ER lumen. In this study, we sought instead to define how a substrate with a cytoplasmic misfolded domain might be selected for ERAD versus post-ERQC. To this end, we designed a novel misfolded membrane protein, SZ*. This substrate is a simplified version of a well-characterized ERAD substrate, Ste6-166 (also known as Ste6*) (Loayza et al., 1998; Huyer et al., 2004), but contains a single TMD appended to a truncated form of a cytosolic nucleotide binding domain 2 (NBD2) that functions as a “degron” (Varshavsky, 1991; Guerriero et al., 2013). SZ* is sorted for efficient degradation by both ERAD and post-ERQC, the latter of which requires the MVB pathway. We also discovered that both heat stress and SZ* overexpression increase ERAD targeting, which correlates with increased aggregation. Together, our results provide a new tool to investigate the mechanisms underlying degradation pathway selection and add new insights into the rules that govern the selection of ERAD substrates.
RESULTS
Characterization of a new ERAD substrate
To design a misfolded membrane protein that can be degraded by both ERAD and post-ERQC pathways, we used a truncated, cytosolic nucleotide binding domain (NBD2*) derived from Ste6-166. There are multiple advantages of using NBD2*: 1) under appropriate conditions, this truncated domain can to be targeted either to the cytoplasm (Guerriero et al., 2013) or to the ER (Prasad et al., 2012; Guerriero et al., 2017; Preston et al., 2018); 2) cytosolic folding lesions appear to be more easily detected than lumenal folding lesions. For example, the lumenal folding lesion in Wsc1* is not recognized by the ERQC machinery and is instead delivered for GQC (Wang and Ng, 2010; Wang et al., 2011), and some ERAD-L substrates exit the ER before being returned to this compartment for ERAD (Taxis et al., 2002; Vashist and Ng, 2004). In contrast, ERAD-C substrates are retained in the ER (Vashist and Ng, 2004); and 3) NBD2* possesses a putative diacidic ER-exit signal, TLEVENN and QDEILEIEMYD (Watanabe and Riezman, 2004). Because TMDs can contain sorting information, we chose a single-pass transmembrane domain from a yeast plasma membrane protein, Wsc1, which lacks an ER-retention signal. To further simplify the substrate, the lumenal domain only contains a triple human influenza hemagglutinin (HA) tag (Prasad et al., 2012). In parallel, we designed a “wild-type” version of our substrate, SZ, which instead deposits full length NBD2 in the cytoplasm (Figure 1A).
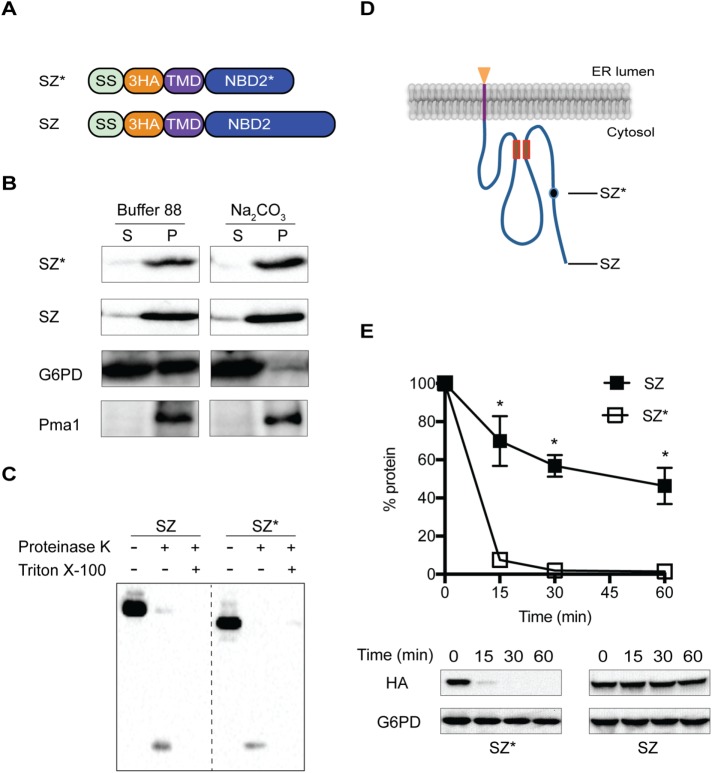
FIGURE 1:
Characterization of a novel ERAD substrate. (A) Domain schematics of SZ and SZ* (SS, signal sequence of Kar2; 3HA, triple HA tag; TMD, transmembrane domain; NBD2, full-length nucleotide binding domain 2 from Ste6p; NBD2*, truncated NBD2 from Ste6-166). (B) Microsomes from wild-type (BY4742) yeast containing SZ* or SZ were treated with or without 0.1 M sodium carbonate and protein residence in the supernatant (S) and pellet (P) fractions was analyzed after centrifugation (100,000 × g for 30 min) and immunoblotting (G6PD, soluble protein marker; Pma1, membrane protein marker). (C) Microsomes from wild-type yeast containing SZ* or SZ were treated with proteinase K in the presence or absence of Triton X-100 and protein remaining in the microsomes was analyzed by immunoblotting with anti-HA antibody. (D) Predicted topology of SZ and SZ*. Closed circle indicates the truncation position in SZ*. The Walker A and Walker B motifs are indicated as two small boxes, and the HA epitope is depicted as a triangle. (E) The stabilities of SZ and SZ* were measured by cycloheximide chase analysis. Data represent the means ± SE of three to six independent experiments; *p < 0.05.
To confirm that SZ and SZ* are integral membrane proteins, we conducted carbonate extraction analysis in yeast expressing each protein. After treatment, both species resided in the pellet along with the integral membrane protein, Pma1 (Figure 1B). In contrast, the cytosolic G6PD enzyme resided almost completely in the supernatant. Next, we established membrane topology using a protease accessibility assay (Vashist and Ng, 2004). Treatment of ER microsomes with limiting concentrations of proteinase K resulted in the generation of a ∼7-kDa peptide, which corresponds to the expected size of the protease inaccessible triple HA-tag and the appended TMD (Figure 1C). Also, both SZ and SZ* migrate at their expected molecular masses (39 and 35 kDa, respectively), indicating that the four cryptic glyosylation sites in NBD2 failed to enter the ER (Figure 1C). We conclude that both SZ and SZ* are type-I membrane proteins that deposit NBD2 in the cytoplasm and the HA tag in the lumen (Figure 1D). We then compared the stabilities of SZ and SZ* by a cycloheximide chase assay in a wild-type yeast strain. Similar to what was found for the completely soluble, cytoplasmic forms of NBD2 and NBD2* (Guerriero et al., 2013), SZ is stable and possesses a half-life of ∼44 min (Figure 1E), whereas SZ* is highly unstable with a half-life of ∼3.8 min (Figure 1E). These results indicate that the truncated NBD2 targets SZ* for rapid degradation.
SZ* is only partially selected by ERAD
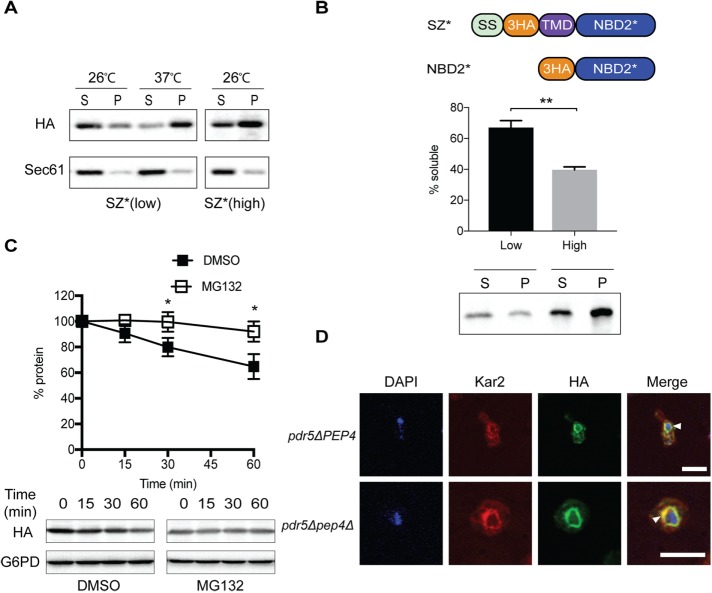
Because SZ* contains a prominent misfolded domain in the cytoplasm, we hypothesized that the substrate would utilize the ERAD-C pathway. Four lines of evidence support this conclusion: First, in yeast lacking the multidrug pump, Pdr5, which allows for effective treatment with proteasome inhibitors, SZ* was stabilized on treatment with MG132 (Figure 2A). In contrast, SZ was stable regardless of MG132 treatment (Supplementary Figure S1A). Second, when SZ* stability was assessed in a temperature-sensitive Cdc48 mutant, cdc48-2, SZ* degradation was slowed on incubation at the nonpermissive temperature (Figure 2B). Third, SZ* degradation was also slowed in yeast lacking the ERAD-C-specific E3 ligase, Doa10 (Figure 2C). And, fourth, mutations in both the cytosolic Hsp70, Ssa1, and Hsp40, Ydj1, reduced the SZ* degradation rate (Figure 2, D and E). In contrast, the ER lumenal Hsp70, Kar2, was dispensable for SZ* turnover (Supplemental Figure S1B). We next performed indirect immunofluorescence microscopy to examine the subcellular localization of SZ*. In the untreated pdr5Δ strain, SZ* colocalized with the ER-resident chaperone, Kar2 (Figure 2F), which is also consistent with the protein being targeted for ERAD. Interestingly, however, SZ* was stabilized to a similar extent in each of the ERAD defective strains, but was never completely stabilized. These data suggested that SZ* might be subject to another degradation pathway, such as post-ERQC, as well as ERAD.
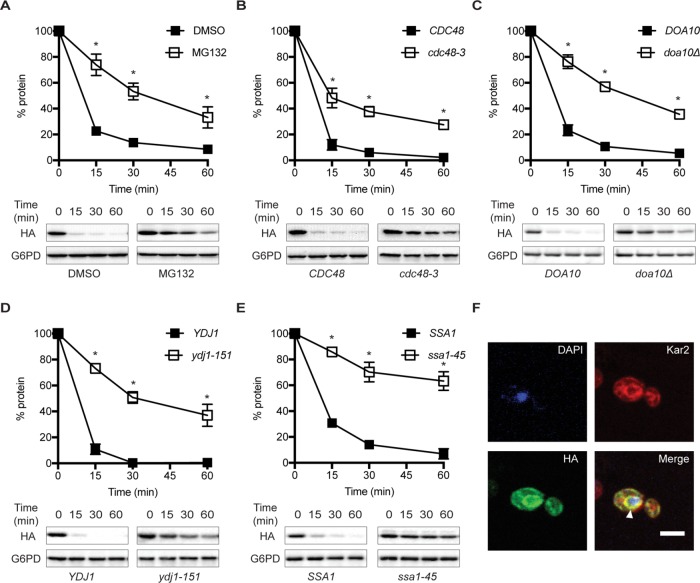
FIGURE 2:
SZ* is only partially degraded by the ERAD pathway. The stabilities of SZ* in the indicated wild-type and mutant strains were determined by cyloheximide chase analyses. (A) The yeast proteasome was inhibited with 100 μM MG132 in a strain lacking PDR5. (B) A strain containing a temperature-sensitive mutation in the AAA-ATPase Cdc48 (cdc48-3) was used. (C) Yeast lacking the ER-associated E3 ligase, Doa10, were examined. (D, E) Cells with a temperature-sensitive mutation in the cytosolic Hsp40, Ydj1 (ydj1-151), and the cytosolic Hsp70, Ssa1, (ssa1-45), were examined. Data represent means ± SE of three to six independent experiments; *p < 0.05. (F) The cellular localization of SZ* was determined by indirect immunofluorescence microscopy. SZ*, ER, and the nucleus were detected with anti-HA antibody, anti-Kar2 antiserum, and DAPI, respectively. Arrowhead denotes the ER. Scale bar: 5 μm.
SZ* is sorted to the vacuole for degradation after Golgi transit
Because SZ* is a single-pass membrane protein, like some other post-ERQC substrates (see Introduction), and is only partially stabilized in each of the ERAD defective mutants examined, we next assayed SZ* dependence on the vacuole for degradation. We therefore investigated SZ* stability in a pdr5Δ mutant that also lacks Pep4, which eliminates ∼90% of vacuolar protease activity (Jones, 1984). Even in the absence of MG132, most of the substrate was stable, with ∼70% of SZ* remaining after 60 min (Figure 3A). The residual degradation of SZ* in the pep4Δpdr5Δ strain was inhibited by MG132 treatment (Supplemental Figure S2A). Consistent with these data, SZ* also accumulated in the vacuole lumen in pep4Δpdr5Δ cells in the absence of MG132 treatment as shown by indirect immunofluorescence microscopy (Figure 3B). Therefore, the majority of SZ* is degraded in the vacuole.
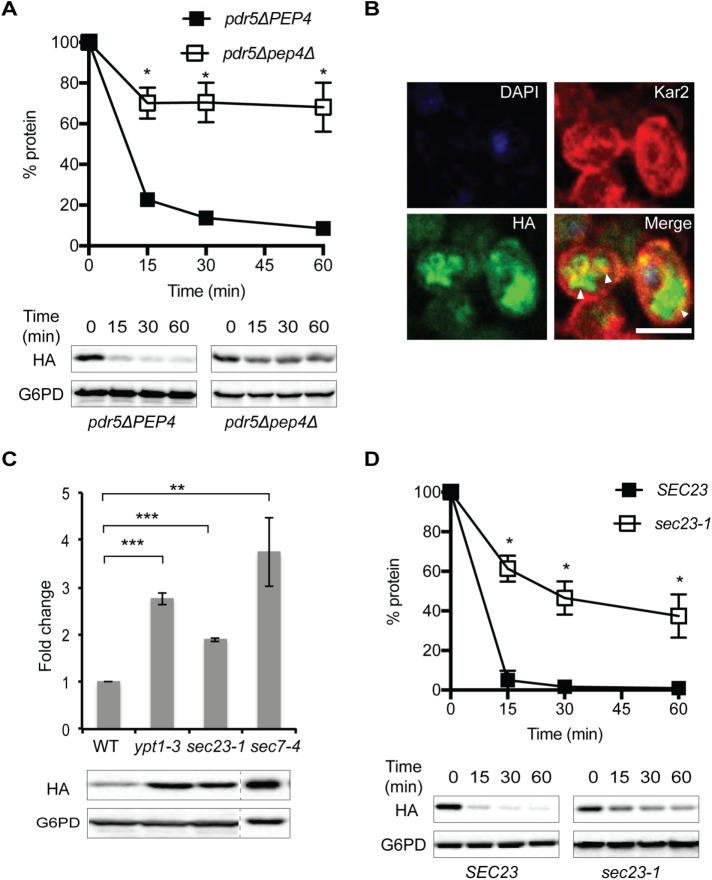
FIGURE 3:
SZ* is sorted into the vacuole for degradation after transit to the Golgi. (A) The stability of SZ* was determined by cyloheximide chase analysis in PEP4pdr5Δ and pep4Δpdr5Δ cells in the absence of proteasome inhibition. (B) The cellular localization of SZ*, the ER, and the nucleus was determined by indirect immunofluorescence microscopy in pep4Δpdr5Δ cells, as in Figure 2. Arrowheads denote the lobes of the vacuole and the whole vacuole. Scale bar: 5 μm. (C) The fold change in steady-state levels of SZ* in wild-type and mutant yeast defective in ER-Golgi transport or intra-Golgi trafficking was examined by immunoblotting. (D) Cycloheximide chase analyses in SEC23 and sec23-1 cells were performed, and data represent three independent experiments; *p < 0.05, **p < 0.005, ***p < 0.0005.
In general, misfolded membrane proteins are sorted into the vacuole/lysosome through either ER-phagy or post-ERQC pathways. To test if ER-phagy directs SZ* to the vacuole, we assessed SZ* stability in select autophagy-deficient yeast strains (Feng et al., 2014; Lipatova and Segev, 2015). As shown in Supplemental Figure S2B, deletion of the selected autophagy factors did not affect the turnover rate of SZ*. To test instead whether post-ERQC is required for the vacuolar degradation of SZ*, we measured the steady-state levels of SZ* in wild-type yeast and in strains defective for ER to Golgi transport (sec23-1 and ypt1-3) and intra-Golgi transport (sec7-4). In contrast to what was observed in the selected autophagy deficient yeast, the steady-state levels of SZ* at nonpermissive temperatures increased from two- to fourfold in these mutants when compared with the wild-type strain when cells were incubated at the nonpermissive temperature (Figure 2C). In a representative experiment, the degradation kinetics of SZ* were also measured in the sec23-1 background at the nonpermissive temperature, and the half-lives of the SZ* protein in the wild-type versus sec23-1 mutant strains were ∼3.3 min and ∼25 min, respectively (Figure 3D). Together, these results indicate that SZ* is sorted into the vacuole for degradation after Golgi transit and is therefore a substrate for both ERAD and post-ERQC.
SZ* transport to the vacuole requires the MVB pathway
The PMQC and GQC of membrane proteins require the multivesicular body (MVB) pathway, which is mediated by ESCRT (MacGurn et al., 2012). Therefore, we next tested whether SZ* sorting into the vacuole is compromised in strains mutated for distinct ESCRT components. However, due to the concern that the lumenal HA epitope in SZ* would be cleaved by vacuolar proteases, a cytosolic GFP-tagged SZ* chimera was also constructed in which the GFP moiety resides in the cytosol (Figure 4A). Our cycloheximide chase data showed that the GFP-tagged SZ* chimera behaved similarly to the HA-tagged version in that both substrates remained susceptible to ERAD (specifically, the Doa10 E3 ligase) and vacuolar protease-associated degradation (Supplemental Figure S3A). Therefore, we next assessed the steady-state level of SZ* in wild-type and several ESCRT mutants, including vps27Δ (ESCRT-0), vps23Δ (ESCRT-I), vps36Δ (ESCRT-II), snf7Δ(ESCRT-III), and vps4Δ (the AAA-ATPase required for ESCRT recycling). As shown in Figure 4B, SZ* cleavage was evident in the wild-type strain, but in the ESCRT mutants only the full-length SZ* chimera was observed, and the steady-state level of this species was higher than in the wild-type background. Moreover, in wild-type cells, the GFP tagged SZ* chimera resided in the vacuolar lumen, whereas in vps27Δ yeast the substrate was found in a prevacuolar compartment (Figure 4C). We conclude that SZ* is sorted into the vacuole through the MVB pathway.
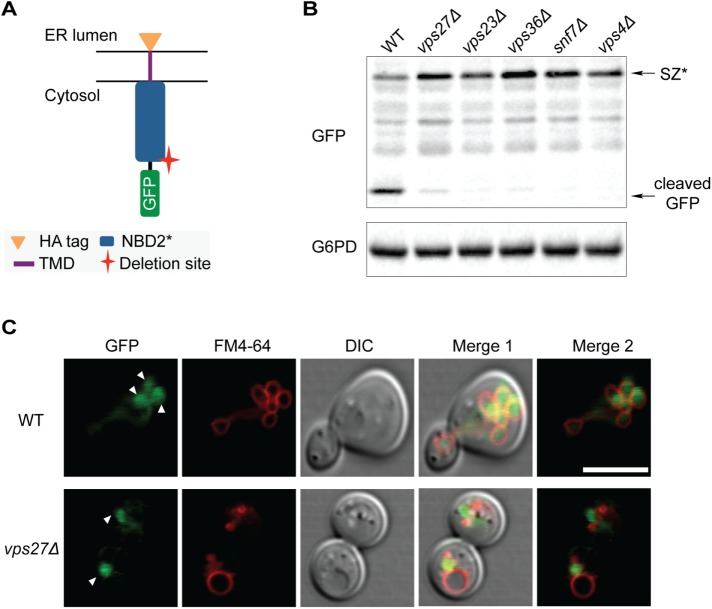
FIGURE 4:
SZ* is sorted into the vacuole through the MVB pathway. (A) Predicted topology of GFP-tagged SZ*. The cytosolic residence of GFP allows for analysis of MVB delivery to the vacuole. (B) GFP cleavage from GFP-SZ* in the wild-type and indicated mutant strains was determined by immonoblotting. G6PD serves as a loading control. (C) The cellular localization of SZ* in both wild-type and vps27Δ cells was determined by fluorescence microscopy. FM4-64 marks the vacuolar membrane. Arrowheads denote the lobes of the vacuole (Bohovych et al., 2016) and the prevacuolar compartment (bottom). Scale bar: 5 μm.
Since plasma membrane localization of SZ* was absent by either indirect immunofluorescence or live cell fluorescence microscopy (Figures 2–4), we hypothesized that SZ* is sorted into the vacuole directly from the Golgi but not after endocytic retrieval from the plasma membrane. To test this possibility, we assessed the degradation of SZ* in an endocytosis mutant strain, end3Δ, after a temperature shift to 37°C (Benedetti et al., 1994). End3 coordinates assembly of the cortical actin cytoskeleton, and therefore the absence of this protein leads to defects in actin-dependent endocytosis. However, we found that deletion of END3 had no effect on SZ* degradation (Supplemental Figure S3B), suggesting that the population of SZ* that escapes ERAD is sorted into the vacuole through an MVB intermediate.
Heat stress increases ERAD targeting of SZ* by inducing its aggregation
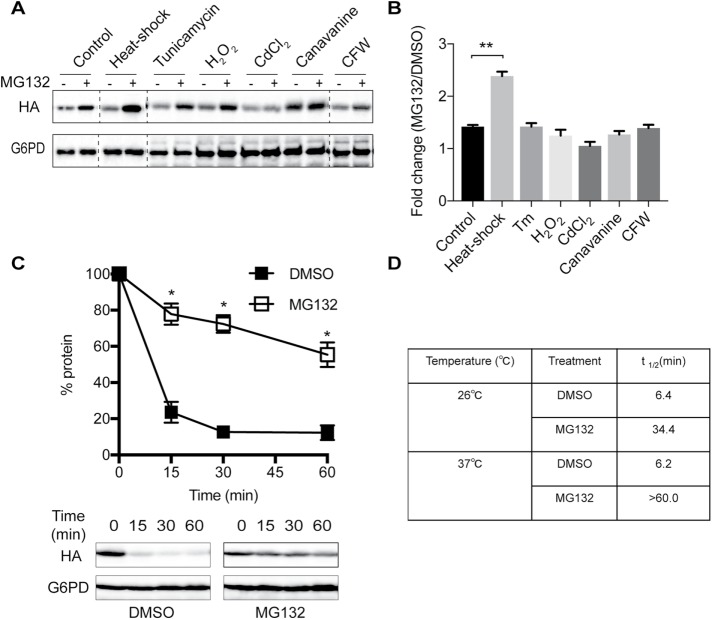
Various protein degradation pathways within the cell are differentially affected by stress (Bernales et al., 2006b; Liu and Chang, 2008; Bohovych et al., 2016). To examine whether SZ* sorting into either the proteasome or vacuolar degradation pathway is modified by exogenous stress, we first investigated whether a specific stress would lead to the preferential degradation of SZ* via ERAD. Cells expressing SZ* were incubated under a variety of stress conditions, including ER stress, heat stress, oxidative stress, heavy metal stress, protein misfolding stress, and cell wall stress. Next, ERAD dependence was assayed after treating pdr5Δ cells with either dimethyl sulfoxide (DMSO) (the vehicle) or MG132. Among these conditions, only heat stress significantly increased the relative steady-state level of SZ* compared with the control (Figure 5, A and B). We then confirmed the effect of heat stress on the ERAD-dependent degradation of SZ* by cycloheximide chase assay (Figure 5C). In the presence of MG132, the SZ* half-life was prolonged from ∼34 min at 26°C to >60 min at 37°C (Figure 5D).
FIGURE 5:
Heat stress increases the targeting of SZ* for ERAD. (A) A pdr5Δ mutant strain expressing SZ* was treated with the indicated stressors for 60 min prior to the addition of DMSO or 100 μM MG132. The level of SZ* was visualized by immunoblotting. (B) ERAD dependence was quantified by the relative fold change of SZ* levels after treatment with MG132 vs. DMSO as in A and normalized to the loading control G6PD. (C) ERAD dependence of SZ* was assayed by cycloheximide chase at 37°C after treatment with the proteasome inhibitor MG132 or the vehicle DMSO. (D) The half-life of SZ* was calculated from cycloheximide chase data. Data represent the means ± SE of three independent experiments. *p < 0.05 and **p < 0.005.
The effect of heat stress on SZ* degradation might have arisen from a general stress response, such as the heat-shock response (HSR), or from a temperature-dependent change in protein conformation. To examine whether HSR induction increased ERAD targeting of SZ*, we introduced a constitutively active Hsf1 mutant, Hsf1-R206S, into SZ*-expressing cells. The introduction of this mutant permits HSR activation in the absence of a temperature shift (Sewell et al., 1995), allowing us to differentiate whether the HSR versus a direct increase in temperature magnified SZ* targeting for ERAD. We first noted that the HSR was induced to an even greater extent than that produced by heat-shock in the Hsf1-R206S-expressing yeast (Supplemental Figure S4A). Nevertheless, Hsf1-R206S failed to increase the ERAD of SZ*, as established in either a steady-state (Supplemental Figure S4B) or cycloheximide chase assay (Supplemental Figure S4C). This finding suggests that the temperature increase instead altered the conformation of SZ*.
Based on an NBD2 homology model, the truncation site in NBD2* resides between two beta sheets, which may result in the generation of an aggregation-prone protein (Preston et al., 2018). Therefore, we surmised that heat stress induces SZ* aggregation, which in turn increases ERAD targeting. To test this hypothesis, we quantified SZ* detergent solubility under different stress conditions. First, ER-derived microsomes were prepared from SZ*-expressing yeast. The microsomes were then pretreated at either 26° or 37°C, incubated with dodecylmaltoside (DDM), and subjected to centrifugation. As shown in Figure 6A, the detergent solubility of SZ* was significantly reduced when microsomes were pretreated at 37°C compared with those pretreated at 26°C. As a control for these studies, we tested whether Wsc1* (Wang and Ng, 2010) is also subject to heat-induced aggregation in vitro. In contrast to SZ*, there was no change in Wsc1* detergent solubility. Moreover, Wsc1* was not targeted for ERAD but instead remained a post-ERQC substrate, even after heat stress (Supplemental Figure S5). Therefore, it appears that incubation at an elevated temperature increased ERAD targeting by decreasing SZ* detergent solubility.
FIGURE 6:
ERAD targeting of SZ* correlates with higher aggregation propensity. (A) Microsomes from wild-type yeast expressing SZ* under the control of the TEF promoter in either a CEN (low) or a 2μ (high) plasmid were treated at the indicated temperature and with 10% DDM. Protein residing in the supernatant (S) and pellet (P) fractions was analyzed after centrifugation at 18,000 × g for 30 min and immunolotting. Sec61 was used as a control. (B) Wild-type yeast lysate containing a soluble version of SZ* that lacks a TMD (“NBD2*”) was treated with 100 mM Na2CO3, followed by centrifugation as in A and immunoblotting. (C) ERAD dependence of SZ* at high expression was assayed by a cycloheximide chase at 26°C after treated with the proteasomal inhibitor MG132 or DMSO. Data represent the means ± SE of three independent experiments; *p < 0.05 and **p < 0.005. (D) The cellular localization of SZ* expressed at high level, and the position of the nucleus and ER was determined in both pdr5Δ and pep4Δ pdr5Δ cells by indirect immunofluorescence microscopy in the absence of proteasome inhibition as in Figure 2. Arrowheads denote the ER. Scale bar: 5 μm.
To better establish a correlation between detergent solubility and ERAD targeting, we devised a mechanism to induce the aggregation of SZ* in the absence of a temperature shift. Because protein aggregation is concentration dependent, we next tested the effect of SZ* overexpression on its aggregation propensity. A high-copy-number (2μ) plasmid under the control of a strong promoter was used to drive SZ* overexpression. As anticipated, SZ* overexpression resulted in a pronounced decrease in detergent solubility, even when the isolated microsomes were incubated at 26°C (Figure 6A, right panel). We then tested whether aggregation of the soluble cytosolic NBD2* was concentration dependent. A high- (p426TEF) and low- (p416TEF) copy-number NBD2* expression vector were introduced into yeast, and cell lysates were prepared and incubated with sodium carbonate to harvest insoluble aggregates. While ∼60% of NBD2* remained in solution in treated lysates from cells with the low-expression plasmid, overexpression of NBD2* was sufficient to reduce solubility by approximately twofold (Figure 6B). Moreover, overexpression of SZ*, which contains a single TMD, stabilized the protein, and the substrate was now exclusively targeted for ERAD in yeast (Figure 6C). We do note that the overall stability of the substrate was higher. This may arise from saturation of the ERAD pathway, even though the capacity of the ERAD pathway is known to be quite high, at least for some substrates (Spear and Ng, 2003; Vincenz-Donnelly et al., 2018). Substrate overexpression may also lead to the occlusion of regions that serve as ER exit motifs, which prohibits vacuole targeting but will also slow the overall degradation rate. In addition, compared with the significant accumulation of SZ* in the vacuole in the pep4Δ mutant when expressed at low levels (Figure 3B), overexpressed SZ* was retained in the ER in either the PEP4 or pep4Δ background (Figure 6D). Because SZ* degradation under low-expression conditions showed a partial ERAD dependence (Figure 2A), these data are consistent with a model that aggregation propensity is a trigger for ER retention and thus ERAD substrate selection.
We next asked whether the secretory pathway is saturated or compromised on SZ* overexpression. Therefore, the degradation of a post-ERQC substrate, Wsc1*, was assessed in conjunction with SZ* overexpression; Wsc1* utilizes the same transport pathway from the ER to the vacuole as SZ* when expressed at low levels (Wang and Ng, 2010). When Wsc1* was expressed in the presence or absence of overexpressed SZ*, both the half-life and the Wsc1* Golgi glycosylation pattern were similar, regardless of whether SZ* was present (Supplemental Figure S5C). These data suggest that the post-ERQC pathway does not become saturated on SZ* overexpression. Instead, we propose that SZ* aggregation induced by overexpression, even in the absence of a temperature shift, results in ER retention and increased ERAD targeting. This contrasts with the fate of an ERAD-L substrate, CPY*, whose overexpression still allows for ER exit and degradation in the vacuole (Spear and Ng, 2003).
Fusion of an aggregation-prone domain targets a post-ERQC substrate for ERAD
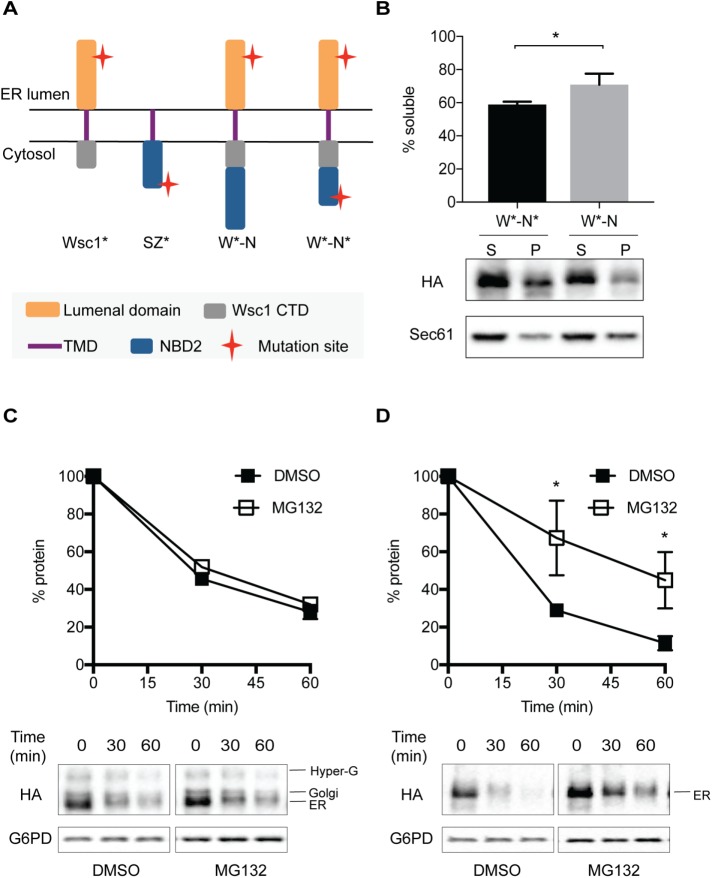
Can the aggregation-prone NBD2* degron act in a dominant manner to retain a post-ERQC substrate in the ER and induce degradation by the ERAD pathway? To answer this question, we fused both NBD2* as well as the full-length NBD2 domain to Wsc1*, thereby generating W*-N* and W*-N (Figure 7A). We first examined the detergent solubility of W*-N* and W*-N in isolated membranes. Because substrates containing NBD2*(but not NBD2) are targeted for proteasome-dependent degradation (Prasad et al., 2012; Guerriero et al., 2013), and because aggregation-propensity favors ERAD targeting (see above), we anticipated that W*-N* would be less soluble. As hypothesized, the detergent solubility of W*-N* was lower than that of W*-N (Figure 7B). Next, we examined the trafficking of these proteins in the cell. Previous work indicated that the Wsc1* lumenal domain is glycosylated in the Golgi (Wang and Ng, 2010), which serves as a read-out for ER exit as the protein migrates somewhat slower than the ER form in SDS–PAGE. As shown in Figure 7C, only W*-N exits the ER as shown by the presence of the slightly higher-molecular-weight species (Figure 7C; Western blot, bottom panels). However, W*-N* was unable to exit the ER since Golgi-associated glycosylation was absent (Figure 7D; Western blot, bottom panels). Additionally, we found that W*-N* is an ERAD substrate because degradation was proteasome- (i.e., MG132) dependent (Figure 7D). In contrast, the degradation of the Wsc1*-NBD2 protein (W*-N) was proteasome independent (Figure 7C). To confirm these results, we also performed pulse chase experiments (Supplemental Figure S6, A and B). In accordance with data from the cycloheximide chase experiments in Figure 7, C and D, only W*-N* exhibited partial proteasome-dependent degradation. In addition, when we investigated the E3 ligase requirements for the degradation of W*-N and W*-N*, W*-N* was degraded in a Doa10-dependent manner, whereas the turnover of W*-N was Doa10 independent (Supplemental Figure S7, A and B): in this experiment, we noted the conversion of the ER form of W*-N into the Golgi form, regardless of whether MG132 was present. However, MG132 stabilized the ER form of W*-N*. Notably, the hyperglycosylated species, which is present for both W*-N* as well as W*-N, does not represent a Golgi-resident form of the protein (Wang and Ng, 2010). Overall, these data demonstrate that a misfolded and aggregation-prone domain, NBD2*, is sufficient to target a post-ERQC substrate for ERAD. Moreover, this aggregation-prone ERAD-C determinant is dominant over the selection of a substrate for post-ERQC.
FIGURE 7:
Fusion of the aggregation-prone NBD2* domain targets a post-ER quality control substrate, Wsc1*, for ERAD. (A) The predicted topologies of the chimeric proteins. (B) The DDM solubility of two chimeric proteins, W*-N and W*-N*, in ER-derived microsomes was performed as described under Materials and Methods. ERAD dependence of (C) W*-N and (D) W*-N* was assayed by cycloheximide chase after treatment with the proteasomal inhibitor MG132 or the vehicle DMSO. Data represent means ± SE of three independent experiments; *p < 0.05.
DISCUSSION
Membrane proteins are subjected to multiple quality control checkpoints along the secretory pathway, including ERQC, GQC, and PMQC. During ERQC, the ERAD-C pathway specifically recognizes membrane proteins with cytosolic folding lesions (Vashist and Ng, 2004; Carvalho et al., 2006). However, the rules that define the selection of substrates for this pathway are still being developed. Thus, the first goal of this study was to characterize a new substrate and investigate how proteins are selected by ERAD-C. Therefore, we appended a cytosolic degron–-a truncated form of the second nucleotide binding domain from Ste6 (NBD2*)–-to the membrane-spanning domain from Wsc1. Previous work on a soluble form of this substrate (i.e., NBD2*) indicated that San1 targets NBD2* for Cyto-QC (Prasad et al., 2012; Guerriero et al., 2013). Additionally, a somewhat different construct (WS) created by Ng and colleagues indicated that the ER-resident E3, Doa10, targets this protein for ERAD (Prasad et al., 2012). Here, we similarly found that SZ* is sorted to the canonical ERAD-C pathway, as proteolysis required Doa10, the cytosolic Hsp70 Ssa1, and the ER-associated cytosolic Hsp40 Ydj1. Surprisingly, we also discovered that most of the SZ* is sorted to the vacuole for degradation and in fact undergoes selection by both ERQC and post-ERQC. We also found that aggregation induced by either heat stress or overexpression was sufficient for increased ERAD targeting. In contrast, a post-ERQC substrate, Wsc1*, was significantly less aggregation prone, even after heat-shock. These data are consistent with a recent report from our group in which more aggregated ERAD substrates have shorter protein half-lives than less aggregated species (Preston et al., 2018). The substrate used in this recent article similarly contained the NBD2* motif, but it was tethered to the ER via a helical hairpin, which trapped the protein in the ER. In contrast, the use of the Wsc1 single-spanning TMD provided us with a novel tool to dissect how aggregation propensity would dictate not only the “decision” to target a substrate for ERAD, but whether this property leads to ER retention. We were also able to establish that NBD2*-dependent aggregation is dominant; that is, that it was transferable and could deliver a post-ERQC substrate for ERAD. Thus, we have significantly extended prior findings to show unexpected features of how a substrate is selected for ERAD versus post-ERQC.
Previous work showed that the post-ERQC pathway in yeast selects membrane proteins that escape ERQC. These include Pep12(D), CPS1, Phm5, and Wsc1*(Reggiori et al., 2000; Reggiori and Pelham, 2002; Wang and Ng, 2010; Wang et al., 2011). Like SZ*, these misfolded membrane proteins possess a single TMD and harbor folding lesions in either the lumen or within the membrane domain. However, membrane proteins with cytosolic folding lesions, such as Ste6*, KSS, KWS, Pma1, and some mutant forms of CFTR, are thought to be retained in the ER and targeted exclusively for ERAD (Loayza et al., 1998; Wang and Chang, 1999; Vashist and Ng, 2004; Vij et al., 2006; Ahner et al., 2007). In contrast, we discovered that the majority of SZ* is routed for vacuolar degradation through the MVB pathway. This unexpected result could be explained by one of two features: first, two putative diacidic ER exit signals exist in the cytosolic domain of SZ* (TLEVENN and QDEILEIEMYD), which may allow a substantial fraction of the substrate to become captured in COPII vesicles. Second, the transmembrane segment, derived from a plasma membrane protein, Wsc1, may contain a sorting signal for post-ER trafficking (Roberts et al., 1992; Rayner and Pelham, 1997; Karsten et al., 2004; Singh and Mittal, 2016).
It remains elusive how ERAD cooperates with post-ERQC machinery to safeguard cellular homeostasis. One possibility is that ERAD retains and degrades substrates with ERAD-specific degrons, which reside in substrates like CPY* and Ste6-166 (Loayza et al., 1998; Wolf and Schafer, 2005), whereas substrates lacking the degron include post-ERQC substrates like Wsc1*, CPS1, and Phm5 (Reggiori et al., 2000; Reggiori and Pelham, 2002; Wang and Ng, 2010; Wang et al., 2011). The nature of the misfolded regions that lead to their designation for one pathway over another remains mysterious, but our results indicate that aggregation propensity may be one such feature. A correlation between ER retention and protein aggregation, and ER retention and proteasome-mediated degradation, is supported by previous studies in which the biogenesis of two aquaporin-2 mutants, T126M and E258K, was examined (Tamarappoo et al., 1999; Hirano et al., 2003). The data in these reports indicated that the T126M mutant was less soluble in 1% CHAPS than E258K, which correlated with their respective ER (T126M) and Golgi (E258K) localization.
Another view is that ERAD substrates are recognized efficiently by the major ER lumenal Hsp70, BiP, by ER-associated cytosolic chaperones, or by the chaperone-like lectins in the ER, whereas those that escape ERQC simply lack these recognition motifs. In fact, we previously reported that mutated forms of a small, model substrate (BPTI) that lack BiP binding sites are targeted exclusively for vacuolar degradation (Coughlan et al., 2004). Similarly, the post-ERQC substrate Wsc1*, which is unable to bind BiP, was efficiently degraded in the vacuole. However, when attached to a BiP binding ERAD determinant–-the C-terminal domain of CPY*–-the chimeric protein was targeted for ERAD (Wang and Ng, 2010). It is also important to note that the ERAD of some substrates appears to be chaperone independent but is instead metabolically regulated. For example, select lipids directly induce a conformational change in HMG-CoA reductase, which leads to its efficient degradation in yeast (Shearer and Hampton, 2005). Together, the many pathways followed by ERAD substrates reflects their diverse topologies, conformations, locations of folding lesions with respect to the ER membrane, and regulatory mechanisms.
Previous studies showed that when CPY* was fused to a functional ER exit signal, it traffics from the ER and is degraded in the vacuole (Kincaid and Cooper, 2007), and maximal degradation of CPY* by ERAD requires ER-Golgi trafficking (Taxis et al., 2002; Vashist and Ng, 2004). The retrieval mechanism for misfolded proteins targeted for ERAD-L most likely requires Kar2, the BiP homologue in yeast. Since Kar2 can exit the ER, misfolded proteins like CPY* bound by Kar2 at the Golgi may be retrieved together with the HDEL receptor, Rer1, or other receptors (Nishikawa and Nakano, 1993; Sato et al., 1995; Shibuya et al., 2015). In addition, while this article was in review, a report on the competition between ER exit and the ERAD of a membrane protein with a misfolded lumenal domain in plant cells was published. Specifically, a misfolded lumenal protein STRUBBELIG (SUBEX-C57Y) from Arabidopsis was fused to different TMDs (Shin et al., 2018). The authors of this study reported that a strong ER exit signal from a nine-pass Golgi resident protein together with its cytosolic tail could guide the chimeric protein—which is otherwise retained in the ER–to the vacuole. These results indicate that ER exit can outcompete chaperone-mediated ER retention. These results are also consistent with data from the Cooper lab. Here, ER export and the ERAD-L pathway were shown to compete for binding of misfolded substrates (Kincaid and Cooper, 2007). Of note, each of these reports utilized misfolded lumenal domains. In contrast, our report focused on the impact of misfolded cytoplasmic domains on protein quality control.
According to a homology model (Preston et al., 2018), NBD2* may be aggregation prone because the truncation site lies between two hydrophobic beta sheets that are otherwise buried in the full-length domain. We propose that the exposed hydrophobic patches on NBD2* drive amorphous aggregate formation after heat-shock and overexpression. Under those conditions, the ERAD machinery may outcompete the ER exit (i.e., COPII) machinery (Barlowe and Helenius, 2016). In this case, we propose that the more aggregated population of SZ* is selected for ERAD. Selection might be mediated by an active, chaperone-dependent process (e.g., by Hsp70s, Hsp40s, and Hsp104) (Preston et al., 2018) or may arise when the substrate simply fails to enter COPII vesicles and lingers in the ER. We note that prolonged residence is sufficient for the ERAD targeting of glycosylated proteins after mannose trimming (Helenius and Aebi, 2004; Xu and Ng, 2015). Because SZ* lacks glycosylation acceptor sites, there are two models in this scenario by which SZ* may be passively retained in the ER. First, aggregation may bury ER exit motifs so that the COPII machinery fails to recognize the substrate, thus facilitating ER retention (Gomez-Navarro and Miller, 2016). Second, the aggregate may simply be too large to enter COPII vesicles in yeast, which are ∼70–80 nm in diameter.
In summary, our work demonstrates for the first time that a single-spanning membrane protein with a cytosolic folding lesion can be targeted for degradation by both ERAD and post-ERQC. In addition, the aggregation state of SZ* determines this trafficking decision. In the future, it will be vital to identify the machinery that leads to the selection of the substrate for degradation in the vacuole via the MVB pathway and whether other substrates undergo a similar fate in the secretory pathway.
MATERIAL AND METHODS
Yeast growth conditions, strains, and plasmid construction
Yeast cells were grown at 26°C on rich or selective media (Alison Adams, 1997). The Saccharomyces cerevisiae strains employed in this study are listed in Supplemental Table S1. The primers and plasmids used in this study are listed in Supplemental Table S2.
To generate triple HA-tagged SZ and SZ* substrates, we first made SZ* through overlap PCR from plasmid pRP12 using primers OSZ01, OSZ02, OSZ03, and OSZ04. The SZ* fragment was cloned into the p416TEF plasmid (Mumberg et al., 1995) at the SpeI/XhoI sites, thus generating plasmid pSZ01. Next, SZ was PCR amplified from plasmid pSZ01 and pCG01 (see Supplemental Table S2) by overlapping PCR again using primers OSZ01, OSZ06, OSZ05, and OSZ07, thus generating plasmid pSZ02. To construct SZ*-GFP, SZ* with a 15-amino-acid ((GGGGS)3) flexible linker between the transmembrane domain and NBD2* was first constructed with primers OSZ08 and OSZ09 and inserted into the p416TEF plasmid, and superfolder GFP (Lee et al., 2013) was inserted next to the SZ*-linker at the BamHI/EcoRI sites, generating SZ*-GFP (pSZ03). W*-N was amplified by overlap PCR using primers OSZ07, OSZ10, OSZ11, and OSZ12 and plasmid pSW148 (see Supplemental Table S2) and pSZ02. Similarly, W*-N* was amplified by overlap PCR using primers OSZ04, OSZ10, OSZ11, and OSZ12 and plasmid pSW148 and pSZ01. Then, the fragments encoding W*-N and W*-N* were inserted into the p416TEF plasmid at the SpeI and XhoI sites, generating pSZ04, and pSZ05, respectively. 3HA-NBD2* was PCR amplified with primers OSZ13 and OSZ04 using pCG03 as a template. The 3HA-NBD2* fragment was then inserted into the p416TEF (low-expression) and p426TEF (high-expression) plasmids, generating pSZ06 and pSZ07, respectively. A high-expression vector for SZ* (pSZ08) was created by subcloning into p426TEF at the SpeI/XhoI sites. The DNA sequences of all constructs were confirmed by Genewiz.
Western blot analysis
After SDS–PAGE, proteins were transferred onto nitrocellulose (BioTrace NT; Pall Corp.) using the overnight wet transfer system (Amersham Biosciences), and Western blot analysis was performed as described previously (Guerriero et al., 2013) with the following primary antibodies: rabbit anti–glucose-6-phosphate dehydrogenase (G6PD) (A9521; Sigma-Aldrich) at 1:5000, polyclonal rabbit anti-Sec61p (Stirling et al., 1992) at 1:5000, rat anti–HA-peroxidase (Roche) at 1:10,000, monoclonal mouse anti-GFP antibody (Roche) at 1:5000, and mouse anti-Pma1, which was a gift from Amy Chang (University of Michigan, Ann Arbor, MI) at 1:1000. Secondary antibodies used in this study were as follows: HRP-conjugated goat anti-mouse or anti-rabbit secondary antibody (Cell Signaling Technology) at 1:5000. Proteins were visualized using SuperSignal Chemiluminescence (Thermo Scientific). Images of the blots were taken on a BIO-RAD ChemiDocXRS Image Station and quantified using ImageJ version 1.48v software (National Institutes of Health). Where indicated, protein half-life was calculated using PRISM GraphPad version 7.0C (GraphPad Software).
Carbonate extraction
A total of 30 ODs of wild-type yeast cells expressing the indicated substrates were lysed with glass beads in 400 μl of Buffer 88 (20 mM HEPES, pH 6.8, 5 mM MgOAc, 150 mM KOAc, 250 mM sorbitol) with protease inhibitors (3 mM phenylmethylsulfonyl fluoride [PMSF], 1.5 μg/ml pepstatin A, and 3 μg/ml leupeptin) and 1 mM dithiothreitol (DTT). Unbroken cells and cell debris were pelleted at 800 × g for 5 min at 4°C. Next, 300 μl of the crude lysate (supernatant) was mixed with 300 μl of 200 mM Na2CO3 (pH 11.5) and the mixture was incubated on ice for 30 min. The samples were then centrifuged at 100,000 × g for 30 min at 4°C. A trichloroacetic acid (TCA) precipitation (Zhang et al., 2001) was used to isolate proteins in the supernatant, and 80 μl of TCA sample buffer (80 mM Tris, pH 8, 15% glycerol, 8 mM EDTA, 0.08% Tris base, 3.5% SDS, 0.01% bromphenol blue supplemented with freshly added β-mercaptoethanol to a final concentration of 4%) was added to solubilize the proteins from supernantant and pellet fractions. Both the supernantant (S) and pellet (P) were then resolved by 10% SDS–PAGE, followed by Western blot analysis.
Proteinase accessibility assay
ER microsomes containing SZ or SZ* were prepared as previously described (Nakatsukasa et al., 2008). In brief, 100 ODs of the indicated cells expressing SZ or SZ* from the p416TEF plasmid were harvested and lysed with glass beads in Buffer 88 with protease inhibitors (3 mM PMSF, 1.5 μg/ml pepstatin A, and 3 μg/ml leupeptin). Unbroken cells and cell debris were pelleted at 800 × g for 5 min at 4°C, and the supernatant was removed and centrifuged at 18,000 × g for 30 min at 4°C to isolate a crude microsome fraction. After two washes with Buffer 88, the microsomes were again collected and resuspended, and equal amounts were treated in the presence or absence of proteinase K (Sigma-Aldrich; final concentration 5 μg/ml), or proteinase K in the presence of 1% Triton X-100. After 15 min on ice, PMSF (final concentration 10 mM) was added to terminate the reaction. Total protein was TCA precipitated, as above, and SDS–PAGE and Western blot analysis were performed.
Detection of steady-state protein levels
Cells expressing the indicated substrates were grown to log phase (A600 ∼ 1.0), and equal numbers of cells were harvested. Total protein was precipitated with TCA. When temperature-sensitive strains were examined, cells were shifted to 37°C for 30 min to induced the mutant phenotype before cells were harvested. The total TCA precipitated material was then subject to SDS–PAGE and Western blot analysis as described above.
Stress treatments and measurements of ERAD-dependent degradation
pdr5Δ cells expressing SZ* were grown to early log phase (A600 ∼ 0.8), and the following stressors were added to the culture media before the cells were incubated at 30°C for 1 h: Tunicamycin (4 μg/ml), H2O2 (1 mM), CdCl2 (50 μM), canavanine (1 mM), and calcofluor white (CFW; 100 μg/ml). For heat-shock treatments, cells were grown at 37°C for 1 h in the absence of any other treatment. After a 1-h treatment, MG132 (100 μM) or DMSO was added to the culture, and the cells were incubated with shaking at 26°C (or 37°C to induce heat stress) in a waterbath for 40 min. Finally, cells (1 ml) were taken from each culture and snap frozen in liquid nitrogen and total protein was obtained by TCA precipitation. The indicated proteins were resolved by 10% SDS–PAGE, followed by Western blot analysis as described above.
Assays to measure protein degradation
Cycloheximide chase assays were preformed to measure protein stability as described (Guerriero et al., 2013). In brief, the indicated yeast strains expressing the desired substrate were grown in synthetic complete (SC) medium lacking uracil (–ura) and containing glucose to mid–log phase (A600 ∼1.0), and cycloheximide was added to the culture to a final concentration of 150 μg/ml. A 1-ml aliquot was taken for the zero time point, and the culture was incubated at 26°C or 37°C, as indicated, in a shaking water bath. At each time point, 1-ml aliquots were taken and the cells were snap-frozen in liquid nitrogen. For total protein extraction, TCA precipitation was used and protein pellets were solubilized with TCA sample buffer. Before SDS–PAGE, insoluble material was removed by centrifugation at 10,000 rpm in a microcentrifuge at room temperature. Samples were resolved via 10% SDS–PAGE and subject to Western blot analysis, as described above.
Pulse chase assays were also performed to test the stabilities of distinct proteins, as described (Tansey, 2007). In brief, yeast expressing W*-N and W*-N* were grown in SC medium lacking uracil (–ura) and methionine (–met) but containing glucose to early log phase (A600 ∼ 0.5), and S-35 (Perkin Elmer-Cetus) was added to each culture to a final concentration of 100 μCi/ml for 10 min. After labeling, the cells were washed with prewarmed culture medium and then resuspended in culture medium supplemented with 13 mM methione, 40 mM cysteine, and 0.2 mg/ml cycloheximide. A 1-ml aliquot of the culture was immediately taken as the zero time point, which was then mixed with 400 μl of quench buffer (80 mM Cys, 80 mM Met, and 100 mM NaN3) and placed on ice. The rest of the culture was incubated at 26°C in a shaking water bath, and at each subsequent time point 1-ml aliquots were taken and similarly mixed with quench buffer. Cells were lysed by glass bead beating and W*-N and W*-N* were immunoprecipitated from these samples with protein A sepharose (GE healthcare) and anti-HA antibody (Roche). After the pellets were washed and proteins were subject to SDS–PAGE, gels were dried and exposed to a phosphorimager screen. The protein substrates were detected by phosphorimager analysis on a Typhoon FLA 7000 (GE Healthcare).
Detergent solubility assays
The solubility of the indicated proteins was measured in dodecylmaltoside (DDM) as described previously (Zhao et al., 2013). In brief, ER microsomes from wild-type yeast expressing the indicated substrates were prepared as described above and incubated on ice (Figure 7B) or at the indicated temperatures (26°C or 37°C indicated in Figure 6A and Supplemental Figure S5B) for 30 min in Buffer 88 with protease inhibitors (see above) in the presence or absence of 1% DDM (EMD Millipore). The mixture was then centrifuged at 18,000 × g for 30 min at 4°C, and the detergent soluble fraction (i.e., the supernatant) was precipitated with TCA (see above). Proteins from both the soluble and insoluble fractions were resuspended in TCA sample buffer and resolved by 10% SDS–PAGE, and Western blot analysis was performed as described above.
Assays to detect the heat-shock response
W303-A wild-type yeast were transformed with the HSE-LacZ reporter plasmid (Santoro et al., 1998) along with the Hsf1-R206S plasmid (Sewell et al., 1995), where indicated, and were grown to log phase (A600 ∼ 1.0). A total of 5 ODs of cells were harvested and lysed in Buffer 88 with protease inhibitors (3 mM PMSF, 1.5 μg/ml pepstatin A, and 3 μg/ml leupeptin) by bead beating. Unbroken cells and cell debris were removed as described above. The supernatant was then collected and cleared by centrifugation at 18,000 × g for 10 min (4°C). The overall protein concentration of the cleared lysate was determined by bicinchoninic acid (BCA) protein assay kit (ThermoFisher). A total of 100 μl of cleared lysate (25–80 μg) was added to 0.9 ml of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, pH 7.0). Next, the mixture was incubated at 30°C for 5 min and the reaction was initiated by adding 200 μl ortho-nitrophenyl-β-galactoside (ONPG) (4 mg/ml in Z buffer). The reaction was incubated at 30°C until the mixture acquired a pale yellow color and then terminated with 500 μl 1 M Na2CO3. The optical density was measured at A420 nm and β-galactosidase activity was calculated by using the following formula: OD420*1.7/(0.0045*total protein*time).
Indirect immunofluorescence microscopy
Yeast expressing HA-tagged SZ* from the p416TEF plasmid were grown in SC-ura medium with glucose to an A600 of 0.5–0.8, fixed for 1 h in 4% formaldehyde at 30°C, and the cell walls were digested by incubation with Zymolyase 20T (MP Biomedicals) at 37°C for 30 min. The cells were applied to polylysine pretreated slides and were then permeabilized with methanol/acetone. The slides were blocked for 1 h at 37°C with blocking buffer (0.5% BSA, 0.5% ovalbumin, 0.6% fish skin gelatin in phosphate-buffered saline buffer) and incubated with the following primary antibodies at 4°C: anti-HA at a 1:250 and anti-Kar2 at a 1:500. After washing, the primary antibodies were conjugated with Alexa Fluor 488 goat anti-mouse and Alexa Fluor 568 goat anti-rabbit (ThermoFisher), both at 1:500, and nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI). The slides were mounted with Prolong Antifade Gold (Invitrogen) and imaged with an Olympus FV1000-100 confocal microscope.
Live-cell fluorescence microscopy
The indicated yeast cells expressing GFP-tagged SZ* were grown to log phase (A600 ∼ 0.8), and cells were incubated at 30°C in the presence of FM4-64 (ThermoFisher) for 30 min. Unbound FM4-64 was removed by washing the cells with SC media. The treated cells were then harvested by centrifugation and imaged by fluorescence microscopy on an Olympus FV1000-100 confocal microscope.
Supplementary Material
Acknowledgments
This work was supported by grants GM075061 and DK079307 from the National Institutes of Health to J.L.B. We thank Charles Barlowe, Allyson O’Donnell, Davis Ng, Dennis Thiele, Dennis Winge, and Mike Preston for reagents and valuable discussions.
Abbreviations used:
- CFTR
cystic fibrosis transmembrane conductance regulator
- DAPI
4’,6-diamidino-2-phenylindole, dihydrochloride
- DMSO
dimethyl sulfoxide
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum–associated degradation
- ERAD-C
ERAD cytosolic
- ERAD-L
ERAD lumenal
- ERQC
endoplasmic reticulum quality control
- ESCRT
endosomal sorting complexes required for transport
- GQC
Golgi quality control
- HA
human influenza hemagglutinin
- MVB
multivesicular body
- PM
plasma membrane
- PMQC
plasma membrane quality control
- QC
quality control
- TMD
transmembrane domain
- UPR
unfolded protein response
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-02-0117) on April 24, 2018.
REFERENCES
- Ahner A, Nakatsukasa K, Zhang H, Frizzell RA, Brodsky JL. (2007). Small heat-shock proteins select deltaF508-CFTR for endoplasmic reticulum-associated degradation. Mol Biol Cell , 806–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alison Adams DG, Kaiser C, Stearns T. (1997). Media and stock preservation. In: Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Pres; s, 145–160. [Google Scholar]
- Amm I, Sommer T, Wolf DH. (2014). Protein quality control and elimination of protein waste: the role of the ubiquitin-proteasome system. Biochim Biophys Acta , 182–196. [DOI] [PubMed] [Google Scholar]
- Babst M. (2014). Quality control: quality control at the plasma membrane: one mechanism does not fit all. J Cell Biol , 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdany M, Veit G, Fukuda R, Avramescu RG, Okiyoneda T, Baaklini I, Singh J, Sovak G, Xu H, Apaja PM, et al. (2017). Chaperones rescue the energetic landscape of mutant CFTR at single molecule and in cell. Nat Commun , 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C, Helenius A. (2016). Cargo capture and bulk flow in the early secretory pathway. Annu Rev Cell Dev Biol , 197–222. [DOI] [PubMed] [Google Scholar]
- Benedetti H, Raths S, Crausaz F, Riezman H. (1994). The END3 gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol Biol Cell , 1023–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P. (2006a). Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol , e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, Papa FR, Walter P. (2006b). Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol , 487–508. [DOI] [PubMed] [Google Scholar]
- Bhamidipati A, Denic V, Quan EM, Weissman JS. (2005). Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell , 741–751. [DOI] [PubMed] [Google Scholar]
- Bohovych I, Kastora S, Christianson S, Topil D, Kim H, Fangman T, Zhou YJ, Barrientos A, Lee J, Brown AJ, Khalimonchuk O. (2016). Oma1 links mitochondrial protein quality control and TOR signaling to modulate physiological plasticity and cellular stress responses. Mol Cell Biol , 2300–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I, Bulleid NJ. (2011). Protein folding and modification in the mammalian endoplasmic reticulum. Annu Rev Biochem , 71–99. [DOI] [PubMed] [Google Scholar]
- Braakman I, Hebert DN. (2013). Protein folding in the endoplasmic reticulum. Cold Spring Harb Perspect Biol , a013201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell SR, Hill KJ, Cooper AA. (2001). Degradation of endoplasmic reticulum (ER) quality control substrates requires transport between the ER and Golgi. J Biol Chem , 23296–23303. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA. (2006). Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell , 361–373. [DOI] [PubMed] [Google Scholar]
- Chen B, Retzlaff M, Roos T, Frydman J. (2011). Cellular strategies of protein quality control. Cold Spring Harb Perspect Biol , a004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlan CM, Walker JL, Cochran JC, Wittrup KD, Brodsky JL. (2004). Degradation of mutated bovine pancreatic trypsin inhibitor in the yeast vacuole suggests post-endoplasmic reticulum protein quality control. J Biol Chem , 15289–15297. [DOI] [PubMed] [Google Scholar]
- Feng Y, He D, Yao Z, Klionsky DJ. (2014). The machinery of macroautophagy. Cell Res , 24–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Navarro N, Miller E. (2016). Protein sorting at the ER-Golgi interface. J Cell Biol , 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero CJ, Brodsky JL. (2012). The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol Rev , 537–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero CJ, Reutter KR, Augustine AA, Preston GM, Weiberth KF, Mackie TD, Cleveland-Rubeor HC, Bethel NP, Callenberg KM, Nakatsukasa K, et al (2017). Transmembrane helix hydrophobicity is an energetic barrier during the retrotranslocation of integral membrane ERAD substrates. Mol Biol Cell , 2076–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero CJ, Weiberth KF, Brodsky JL. (2013). Hsp70 targets a cytoplasmic quality control substrate to the San1p ubiquitin ligase. J Biol Chem , 18506–18520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Garza RM. (2009). Protein quality control as a strategy for cellular regulation: lessons from ubiquitin-mediated regulation of the sterol pathway. Chem Rev , 1561–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Liu Y, Chang A. (2007). Cytoplasmic Hsp70 promotes ubiquitination for endoplasmic reticulum-associated degradation of a misfolded mutant of the yeast plasma membrane ATPase, PMA1. J Biol Chem , 26140–26149. [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M. (2004). Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem , 1019–1049. [DOI] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. (2011). The ESCRT pathway. Dev Cell , 77–91. [DOI] [PubMed] [Google Scholar]
- Hirano K, Zuber C, Roth J, Ziak M. (2003). The proteasome is involved in the degradation of different aquaporin-2 mutants causing nephrogenic diabetes insipidus. Am J Pathol , 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Wada I, Natsuka Y, Nagata K. (2006). EDEM accelerates ERAD by preventing aberrant dimer formation of misfolded alpha1-antitrypsin. Genes Cells , 465–476. [DOI] [PubMed] [Google Scholar]
- Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, Michaelis S. (2004). Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J Biol Chem , 38369–38378. [DOI] [PubMed] [Google Scholar]
- Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, Wolf DH, Sommer T. (2002). Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol , 134–139. [DOI] [PubMed] [Google Scholar]
- Jones EW. (1984). The synthesis and function of proteases in Saccharomyces: genetic approaches. Annu Rev Genet , 233–270. [DOI] [PubMed] [Google Scholar]
- Karsten V, Hegde RS, Sinai AP, Yang M, Joiner KA. (2004). Transmembrane domain modulates sorting of membrane proteins in Toxoplasma gondii. J Biol Chem , 26052–26057. [DOI] [PubMed] [Google Scholar]
- Kelly SM, Vanslyke JK, Musil LS. (2007). Regulation of ubiquitin-proteasome system mediated degradation by cytosolic stress. Mol Biol Cell , 4279–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Spear ED, Ng DT. (2005). Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol Cell , 753–764. [DOI] [PubMed] [Google Scholar]
- Kincaid MM, Cooper AA. (2007). Misfolded proteins traffic from the endoplasmic reticulum (ER) due to ER export signals. Mol Biol Cell , 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lim WA, Thorn KS. (2013). Improved blue, green, and red fluorescent protein tagging vectors for S. cerevisiae. PLoS One , e67902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. (2008). Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell , 714–725. [DOI] [PubMed] [Google Scholar]
- Lipatova Z, Segev N. (2015). A role for macro-ER-phagy in ER quality control. PLoS Genet , e1005390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chang A. (2008). Heat shock response relieves ER stress. EMBO J , 1049–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Sitaraman S, Chang A. (2006). Multiple degradation pathways for misfolded mutants of the yeast plasma membrane ATPase, Pma1. J Biol Chem , 31457–31466. [DOI] [PubMed] [Google Scholar]
- Loayza D, Tam A, Schmidt WK, Michaelis S. (1998). Ste6p mutants defective in exit from the endoplasmic reticulum (ER) reveal aspects of an ER quality control pathway in Saccharomyces cerevisiae. Mol Biol Cell , 2767–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGurn JA, Hsu PC, Emr SD. (2012). Ubiquitin and membrane protein turnover: from cradle to grave. Annu Rev Biochem , 231–259. [DOI] [PubMed] [Google Scholar]
- McCracken AA, Brodsky JL. (1996). Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J Cell Biol , 291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. (1995). Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene , 119–122. [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. (2008). Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell , 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko E, Pelham HR. (2009). Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic , 1856–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S, Nakano A. (1993). Identification of a gene required for membrane protein retention in the early secretory pathway. Proc Natl Acad Sci USA , 8179–8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell AF. (2012). The running of the Buls: control of permease trafficking by alpha-arrestins Bul1 and Bul2. Mol Cell Biol , 4506–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiyoneda T, Apaja PM, Lukacs GL. (2011). Protein quality control at the plasma membrane. Curr Opin Cell Biol , 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiyoneda T, Barriere H, Bagdany M, Rabeh WM, Du K, Hohfeld J, Young JC, Lukacs GL. (2010). Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science , 805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzirusso M, Chang A. (2004). Ubiquitin-mediated targeting of a mutant plasma membrane ATPase, Pma1-7, to the endosomal/vacuolar system in yeast. Mol Biol Cell , 2401–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Kawaguchi S, Ng DT. (2012). Biosynthetic mode can determine the mechanism of protein quality control. Biochem Biophys Res Commun , 689–695. [DOI] [PubMed] [Google Scholar]
- Preston GM, Guerriero CJ, Metzger MB, Michaelis S, Brodsky JL. (2018) Substrate insolubility dictates Hsp104-dependent endoplasmic reticulum associated degradation. Mol Cell , 242–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S. (2002). AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol , 626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner JC, Pelham HR. (1997). Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO J , 1832–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Black MW, Pelham HR. (2000). Polar transmembrane domains target proteins to the interior of the yeast vacuole. Mol Biol Cell , 3737–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Pelham HR. (2002). A transmembrane ubiquitin ligase required to sort membrane proteins into multivesicular bodies. Nat Cell Biol , 117–123. [DOI] [PubMed] [Google Scholar]
- Roberts CJ, Nothwehr SF, Stevens TH. (1992). Membrane protein sorting in the yeast secretory pathway: evidence that the vacuole may be the default compartment. J Cell Biol , 69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiano A, Foresti O, Carvalho P. (2014). Quality control: ER-associated degradation: protein quality control and beyond. J Cell Biol , 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro N, Johansson N, Thiele DJ. (1998). Heat shock element architecture is an important determinant in the temperature and transactivation domain requirements for heat shock transcription factor. Mol Cell Biol , 6340–6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Nishikawa S, Nakano A. (1995). Membrane protein retrieval from the Golgi apparatus to the endoplasmic reticulum (ER): characterization of the RER1 gene product as a component involved in ER localization of Sec12p. Mol Biol Cell , 1459–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S, Gallagher CM, Walter P. (2014). ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J Cell Sci , 4078–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewell AK, Yokoya F, Yu W, Miyagawa T, Murayama T, Winge DR. (1995). Mutated yeast heat shock transcription factor exhibits elevated basal transcriptional activation and confers metal resistance. J Biol Chem , 25079–25086. [DOI] [PubMed] [Google Scholar]
- Shearer AG, Hampton RY. (2005). Lipid-mediated, reversible misfolding of a sterol-sensing domain protein. EMBO J , 149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya A, Margulis N, Christiano R, Walther TC, Barlowe C. (2015). The Erv41-Erv46 complex serves as a retrograde receptor to retrieve escaped ER proteins. J Cell Biol , 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin YJ, Vavra U, Veit C, Strasser R. (2018). The glycan-dependent ERAD machinery degrades topologically diverse misfolded proteins. Plant J , 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Mittal A. (2016). Transmembrane domain lengths serve as signatures of organismal complexity and viral transport mechanisms. Sci Rep , 22352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear ED, Ng DT. (2003). Stress tolerance of misfolded carboxypeptidase Y requires maintenance of protein trafficking and degradative pathways. Mol Biol Cell , 2756–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling CJ, Rothblatt J, Hosobuchi M, Deshaies R, Schekman R. (1992). Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol Biol Cell , 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A, Wolf DH. (2010). Endoplasmic reticulum associated protein degradation: a chaperone assisted journey to hell. Biochim Biophys Acta , 694–705. [DOI] [PubMed] [Google Scholar]
- Szathmary R, Bielmann R, Nita-Lazar M, Burda P, Jakob CA. (2005). Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol Cell , 765–775. [DOI] [PubMed] [Google Scholar]
- Tamarappoo BK, Yang B, Verkman AS. (1999). Misfolding of mutant aquaporin-2 water channels in nephrogenic diabetes insipidus. J Biol Chem , 34825–34831. [DOI] [PubMed] [Google Scholar]
- Tansey W.P. (2007). Pulse-chase assay for measuring protein stability in yeast. CSH Protoc , pdb.prot4641. [DOI] [PubMed] [Google Scholar]
- Taxis C, Vogel F, Wolf DH. (2002). ER-golgi traffic is a prerequisite for efficient ER degradation. Mol Biol Cell , 1806–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. (1991). Naming a targeting signal. Cell , 13–15. [DOI] [PubMed] [Google Scholar]
- Vashist S, Kim W, Belden WJ, Spear ED, Barlowe C, Ng DT. (2001). Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J Cell Biol , 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S, Ng DT. (2004). Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol , 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. (2008). One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol , 944–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vij N, Fang S, Zeitlin PL. (2006). Selective inhibition of endoplasmic reticulum-associated degradation rescues DeltaF508-cystic fibrosis transmembrane regulator and suppresses interleukin-8 levels: therapeutic implications. J Biol Chem , 17369–17378. [DOI] [PubMed] [Google Scholar]
- Vincenz-Donnelly L, Holthusen H, Korner R, Hansen EC, Presto J, Johansson J, Sawarkar R, Hartl FU, Hipp MS. (2018). High capacity of the endoplasmic reticulum to prevent secretion and aggregation of amyloidogenic proteins. EMBO J , 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science , 1081–1086. [DOI] [PubMed] [Google Scholar]
- Wang Q, Chang A. (1999). Eps1, a novel PDI-related protein involved in ER quality control in yeast. EMBO J , 5972–5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Ng DT. (2010). Evasion of endoplasmic reticulum surveillance makes Wsc1p an obligate substrate of Golgi quality control. Mol Biol Cell , 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Thibault G, Ng DT. (2011). Routing misfolded proteins through the multivesicular body (MVB) pathway protects against proteotoxicity. J Biol Chem , 29376–29387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe R, Riezman H. (2004). Differential ER exit in yeast and mammalian cells. Curr Opin Cell Biol , 350–355. [DOI] [PubMed] [Google Scholar]
- Wolf DH, Schafer A. (2005). CPY* and the power of yeast genetics in the elucidation of quality control and associated protein degradation of the endoplasmic reticulum. Curr Top Microbiol Immunol , 41–56. [DOI] [PubMed] [Google Scholar]
- Xu C, Ng DT. (2015). Glycosylation-directed quality control of protein folding. Nat Rev Mol Cell Biol , 742–752. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. (2001). The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature , 652–656. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Nijbroek G, Sullivan ML, McCracken AA, Watkins SC, Michaelis S, Brodsky JL. (2001). Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol Biol Cell , 1303–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Macgurn JA, Liu M, Emr S. (2013). The ART-Rsp5 ubiquitin ligase network comprises a plasma membrane quality control system that protects yeast cells from proteotoxic stress. Elife , e00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.