Abstract
The limited resolving power of conventional diffraction-limited microscopy hinders analysis of small, densely packed structural elements in cells. Expansion microscopy (ExM) provides an elegant solution to this problem, allowing for increased resolution with standard microscopes via physical expansion of the specimen in a swellable polymer hydrogel. Here, we apply, validate, and optimize ExM protocols that enable the study of Drosophila embryos, larval brains, and larval and adult body walls. We achieve a lateral resolution of ∼70 nm in Drosophila tissues using a standard confocal microscope, and we use ExM to analyze fine intracellular structures and intercellular interactions. First, we find that ExM reveals features of presynaptic active zone (AZ) structure that are observable with other superresolution imaging techniques but not with standard confocal microscopy. We further show that synapses known to exhibit age-dependent changes in activity also exhibit age-dependent changes in AZ structure. Finally, we use the significantly improved axial resolution of ExM to show that dendrites of somatosensory neurons are inserted into epithelial cells at a higher frequency than previously reported in confocal microscopy studies. Altogether, our study provides a foundation for the application of ExM to Drosophila tissues and underscores the importance of tissue-specific optimization of ExM procedures.
INTRODUCTION
Analysis of intercellular interactions and intracellular structures often requires optical resolution below the diffraction limit of light (∼250 nm). While several methods have been developed for superresolution imaging of biological samples using specialized microscopes (Huang et al., 2010), expansion microscopy (ExM) is compatible with standard optical microscopes that are already widely available, requires no specialized computational processing, can be used with a wide range of fluorophores, and is suitable for multicolor imaging at substantial depth. In ExM, a swellable hydrogel polymer is grown within a fixed specimen that is treated with proteolytic enzymes to soften the tissue and then expanded through dialysis in distilled water, yielding an approximately fourfold increase in lateral resolution (Chen et al., 2015; Chozinski et al., 2016; Ku et al., 2016; Tillberg et al., 2016). ExM is therefore dependent on the ability of the specimen to be successfully embedded in the swellable polymer hydrogel and to be rendered compliant to expansion through proteolytic digestion. However, many invertebrates, fungi, and plants are covered with a rigid exoskeleton important for maintaining organismal integrity. In some of these organisms, including Drosophila, this exoskeleton is composed of a lipid- and polysaccharide-rich cuticle that resists conventional proteolytic digestion, presenting a major impediment to implementation of ExM.
Here, we set out to extend the utility of ExM by adapting it for use in Drosophila. We find that some Drosophila tissues lacking a rigid cuticle are compatible with established ExM protocols, as has been also shown in two recent reports (Cahoon et al., 2017; Mosca et al., 2017). We also find that addition of a single step to established ExM protocols, treatment with cuticle-digesting enzymes, renders larval and adult body-wall specimens compatible with ExM. Using our optimized ExM protocol, we achieve a lateral resolution of ∼70 nm in various Drosophila tissues, facilitating analysis of structural elements that cannot be accurately studied with conventional optical microscopy, and we demonstrate the utility of this approach in three experimental contexts. First, we show that ExM allows for high-resolution analysis of presynaptic active zone (AZ) structure at the larval neuromuscular junction (NMJ) and that analysis of these structures with conventional confocal microscopy leads to systematic sampling errors. Second, we identify age-dependent changes in adult AZ structure using ExM. Third, we analyzed cell–cell interactions in the larval peripheral nervous system using ExM and found that epithelial ensheathment of somatosensory dendrites is more prevalent than previously reported, underscoring the likely importance of this intercellular interaction. Altogether, these studies establish ExM as an accessible superresolution imaging platform amenable to analysis of diverse Drosophila tissues.
RESULTS AND DISCUSSION
Expansion of Drosophila tissues with minimal distortion
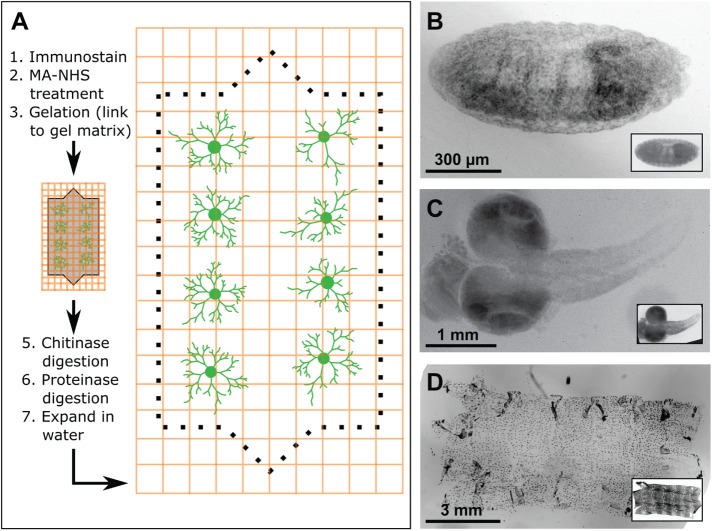
Prior studies demonstrated several specimens that are amenable to ExM (Chen et al., 2015; Chozinski et al., 2016; Ku et al., 2016), including various cultured cells and brain tissue. Given the prevalence of whole-mount imaging in analysis of Drosophila development, we first examined whether ExM could be applied to intact Drosophila embryos. To this end, we fixed embryos using a heptane/formaldehyde fixative and processed them for ExM, which includes gelation, digestion, and expansion steps (Figure 1A). Using this approach, Drosophila embryos were readily expanded ∼4× without obvious tearing or distortion (Figure 1B). To assess the fidelity of expansion, we recorded images of embryos before and after expansion. We then quantified distortion in the x, y, and z dimensions by comparing postexpansion images to digitally expanded preexpansion images with a nonrigid deformation algorithm (Supplemental Figures S1 and S2). We found that lateral distortion was generally below 1–2% over a range of length scales.
FIGURE 1:
Isotropic expansion of Drosophila tissues for fluorescence microscopy. (A) ExM workflow. (B–D) Correlative pre- and postexpansion imaging of Drosophila tissue. Drosophila embryos (B), larval brains (C), and larval body walls (D) were stained with 4′,6-diamidino-2-phenylindole (DAPI) and imaged both before and after expansion. Preexpansion (inset) and postexpansion images are shown at the same magnification, such that postexpansion images are approximately four times larger than the corresponding preexpansion images.
Next, we examined the compatibility of isolated Drosophila tissue with ExM. Similar to embryos, formaldehyde-fixed larval brains were readily expanded without gross distortion (Figure 1C). Staining larval brains with anti-Fas II antibody (Hummel et al., 2000) provided feature-rich labeling in x, y, and z dimensions (Supplemental Figure S1), allowing us to generate robust measures of lateral and axial distortion. As in embryo preps, distortion in expanded larval brains was generally low (<3%) over length scales of up to 30 μm (Supplemental Figure S1), comparable to results reported for ExM of other tissue samples (Chen et al., 2015; Chozinski et al., 2016; Ku et al., 2016; Tillberg et al., 2016).
Unlike embryonic and larval brain samples, larval body-wall samples failed to expand, instead detaching from the swellable hydrogel and becoming somewhat torn and distorted (Supplemental Figure S1), likely the result of incomplete digestion of the polysaccharide-rich cuticle by the standard proteinase K digestion in ExM. Indeed, treatment with chitinase enzymes after gelation, but before proteolytic digestion with proteinase K, allowed for isotropic body-wall expansion in larval (Figure 1D) and adult body-wall samples (see Figure 4, presented later), suggesting that this approach may be broadly useful for ExM on samples with chitin-rich exoskeletons, including other arthropods and fungi. Altogether, these results demonstrate that ExM is applicable to different types of Drosophila tissues and that fine structural elements are preserved during expansion of complex cellular assemblies, including whole-embryo preparations.
FIGURE 4:

ExM analysis of age-related changes in AZ structure. Brp staining at the CM9 NMJ in unexpanded (A) and expanded (B, C) tissue of 10- or 65-d adult flies. In unexpanded samples, Brp immunoreactivity appeared as regularly shaped spherical structures (A′) as well as larger structures possibly representing clusters of AZs (A″). Arrowheads mark elongated puncta that likely represent clusters of joined AZs that cannot be resolved in unexpanded tissue (A″) and clusters of joined AZs resolved by ExM (C). (D) Box plots depicting AZ area measurements using ExM. In this and subsequent panels, boxes mark 1st and 3rd quartiles, bands mark medians, whiskers mark 1.5 × interquartile range, and outliers are shown as open circles. **, p < 0.01 compared with 10-d adults, one-way ANOVA with a post hoc Dunnett’s test. (E) The proportion of AZs with individual (singlets) or multiple Brp rings is shown for the indicated time points. Error bars correspond to SD, *, p < 0.05 compared with 10-d adults (Fisher’s exact test). Measurements in D and E were performed blind to specimen age. (F) Box plots depicting area measurements for single, double, and multiple Brp ring containing AZs in 65-d adults. *, p < 0.05, **, p < 0.01 compared with singlets, one-way ANOVA with a post hoc Dunnett’s test. The number of NMJs analyzed for each time point is indicated. Scale bars: 1 µm. All areas and scale bars refer to preexpansion dimensions.
Superresolution imaging of subcellular structures with ExM
We next examined whether ExM would facilitate identification of fine structural details in Drosophila tissue that were not observable with standard confocal microscopy. We first focused on the imaging of mitochondria because mitochondrial morphology is dynamic during development, is altered in a broad range of disease states (Pernas and Scorrano, 2016), and is commonly studied with conventional fluorescence microscopy methods. We expressed a form of green fluorescent protein (GFP) that is targeted to the mitochondrial matrix (UAS-mitoGFP) in Drosophila class IV dendrite arborization (C4da) neurons, which innervate the body wall; fixed and immunostained larval body-wall fillets with anti-GFP antibodies; and processed the specimens for ExM. Confocal imaging of mitochondria in a cell before and after expansion (Figure 2, A–D) revealed much greater detail in the postexpansion state, with many loops, branches, and swellings that were visible after expansion but were difficult or impossible to detect before expansion. The preexpansion images correspond excellently with postexpansion images of the same region (2–6% distortion over a range of length scales, ∼4.08× expansion factor; Supplemental Figure S2), Thus, ExM allows for visualization of fine structural elements in organelles and should prove to be a valuable tool in studying organelle morphogenesis.
FIGURE 2:
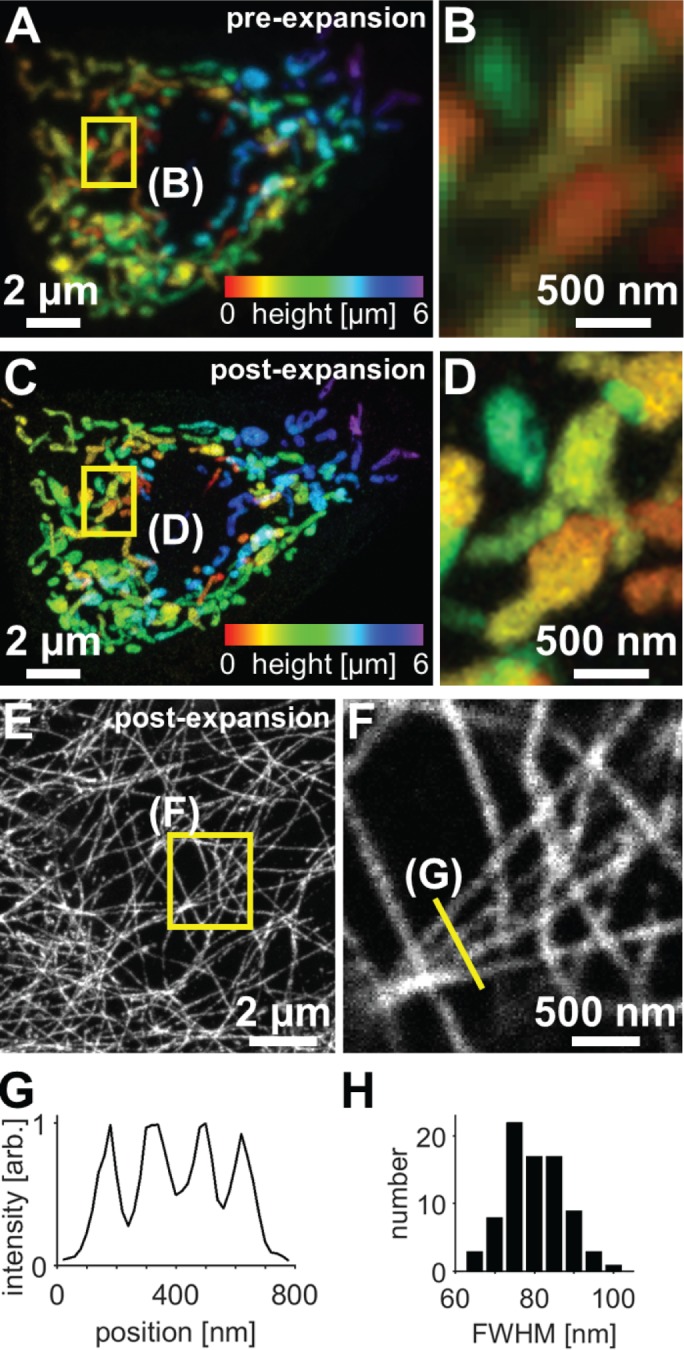
Confocal imaging of subcellular structures in expanded Drosophila tissue samples. (A–D) Maximum-intensity projections of C4da neurons expressing mito-GFP (ppk-Gal4, UAS-mito-GFP) and immunostained for GFP, where color indicates height within the specimen. The same field of view is shown before expansion (A, B) and after expansion (C, D). (E) Maximum-intensity projections of epithelial cells immunostained for acetylated tubulin in expanded third instar larval body walls. (F) Zoomed-in views of single focal plane image corresponding to boxed regions in E. Images of epithelial acetylated tubulin in unexpanded body walls are shown in Supplemental Figure S3. (G) Line profile from F (arb., arbitrary units). (H) Analysis of microtubule widths yielded average Gaussian-fitted full-width at half-maximum (FWHM) of 83 ± 9 nm (mean ± SD, 80 microtubule profiles). Here and in all subsequent figures containing ExM data, distances and scale bars have been divided by their respective measured expansion factors of approximately four times and therefore correspond to pre-expansion dimensions.
Second, we used ExM to visualize the microtubule cytoskeleton by immunostaining for acetylated tubulin. Before expansion, individual microtubules in epithelial cells were difficult to detect due to their high density (Supplemental Figure S3); however, expansion of specimens clearly revealed an intricate network of well-resolved microtubules (Figure 2, E and F). The cross-sectional profile of expanded microtubules had an average Gaussian-fitted full-width at half-maximum of 83 ± 9 nm (mean ± SD); given that indirect immunofluorescence leads to specimen broadening due to the nonnegligible size of primary and secondary antibodies, we estimate a spatial resolution of ∼70 nm, which is consistent with earlier work on other specimens (Chen et al., 2015; Chozinski et al., 2016).
Superresolution imaging of presynaptic AZs with ExM
The high density and small size of Drosophila synapses prevent accurate assessment of synapse number and structure with conventional light microscopy. In particular, features of AZs, which serve as sites of neurotransmitter release, are obscured with conventional light microscopy. With superresolution microscopy, AZs appear as hollow ring-like structures ∼200–400 nm in diameter (Kittel et al., 2006; Wagh et al., 2006; Ehmann et al., 2014). We reasoned that the increased lateral resolution provided by ExM, compared with conventional microscopy, should facilitate analysis of NMJ synapse number and structure while providing an opportunity to benchmark ExM against other superresolution imaging platforms such as SIM, STORM, and stimulated emission depletion (STED) that have been successfully applied to NMJ analysis (Kittel et al., 2006; Fouquet et al., 2009; Matkovic et al., 2013; Ehmann et al., 2014; Jepson et al., 2014; Lepicard et al., 2014).
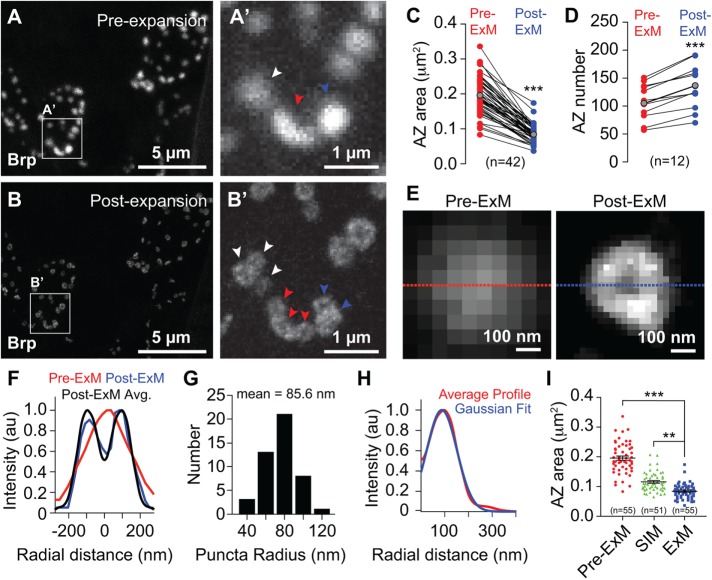
To examine the utility of ExM in analysis of AZ structure, we conducted correlative pre- and postexpansion analysis of the third instar NMJ immunostained for the structural component Bruchpilot (Brp). Before expansion, a Brp immunostain revealed many solid structures ∼300–500 nm in diameter and ∼0.1–0.3 μm2 in area (Figure 3, A and C). These structures exhibited either uniform signal intensity or a central maximum. Expanded specimens showed excellent correspondence with preexpansion images of the same region (< 2% distortion; Supplemental Figure S2), indicating a smooth and overall isotropic expansion, and we were able to observe fine details of AZs that were not evident in unexpanded specimens, as described later. First, AZs were less regularly shaped; whereas pre-expansion AZs were largely spherical, many postexpansion AZs were elliptical, elongated, and/or contained multiple lobes (Figure 3B). Second, AZs appeared uniformly smaller (Figure 3C). Third, many sites that appeared as individual structures before expansion were resolved into multiple independent structures (Figure 3, A′ and B′), and ratiometric analysis of pre- and postexpansion AZ counts revealed that ExM consistently facilitated identification of larger numbers of AZs (Figure 3D). Fourth, many Brp puncta in expanded samples exhibited hollow ring-like structures (Figure 3, B and E–H) with a maximum intensity in the periphery. While we were able to resolve these hollow ring-like structures with structured illumination microscopy (SIM) analysis (Supplemental Figure S4), we found that SIM images were intermediate in resolution between our pre- and post-ExM confocal images as assessed by apparent AZ area (Figure 3I). In contrast, prior reports using STED imaging yielded measured sizes of AZs comparable to our findings using ExM (Fouquet et al., 2009; Weyhersmüller et al., 2011; Ehmann et al., 2014).
FIGURE 3:
Superresolution imaging of synapses with ExM. Correlative confocal microscopy imaging of Brp staining (A) preexpansion and (B) postexpansion in third instar larvae. (A′, B′) Zoomed view of boxed regions in A and B showing AZ features resolved by ExM but not conventional confocal microscopy. Arrowheads mark individual AZs that can be resolved in correlated pre- and postexpansion samples. (C) AZ area and (D) AZ number in correlated pre- and postexpansion images. Red and blue points represent individual measurements from pre- and postexpansion samples, respectively, and lines connect paired measurements. Gray points mark mean values. ***, p < 0.001, unpaired t test with Welch’s correction. (E) Pre- and postexpansion images of a single AZ. (F) Cross-sectional profile of pre- and postexpansion images of puncta in E and average cross-sectional profile of 46 individual postexpansion AZs. (G) Distribution of puncta radii and (H) average radial Brp intensity profile of 46 individual AZs aligned according to their centers of mass. (I) AZ area measurements from Brp-stained third instar larvae imaged using a conventional confocal microscope (preexpansion and postexpansion) or using SIM. Note that SIM imaging was performed on unexpanded tissue (see Supplemental Figure S4). Number of AZs measured for each imaging modality is shown. **, p < 0.01, ***, p < 0.001 compared with ExM, one-way analysis of variance (ANOVA) with a post hoc Dunnett’s test. All distances, areas, and scale bars correspond to preexpansion dimensions.
Thus, ExM allowed for visualization of AZ substructure at the larval NMJ, including the hollow ring-like structures previously revealed by superresolution imaging using the same anti-Brp antibody (Fouquet et al., 2009; Ehmann et al., 2014), and more accurate scoring of AZ number than conventional fluorescence microscopy.
Age-dependent changes in AZ structure
In adult flies, the CM9 NMJs experience an increase in neurotransmitter release during aging, but the cellular mechanisms underlying this potentiation of release are unclear (Mahoney et al., 2014). The size of vesicle release events scale with presynaptic AZ size at many synapses, including small glutamatergic synapses of the CNS and large synapses like the NMJ (Propst and Ko, 1987; Weyhersmüller et al., 2011; Holderith et al., 2012; Südhof, 2012; Gupta et al., 2016). This includes changes in Brp levels at larval AZs during homeostatic plasticity (Weyhersmüller et al., 2011). We therefore used ExM to investigate whether changes in Brp staining patterns at the AZ could explain the age-dependent potentiation of neurotransmission at the CM9 NMJ.
In unexpanded tissue, Brp immunoreactivity at the CM9 NMJ appeared as regularly shaped spherical structures (Figure 4, A and A′) and as larger structures possibly representing clusters of AZs (Figure 4A″). As in unexpanded larval samples, Brp immunoreactivity in unexpanded CM9 NMJs appeared as solid structures with uniform signal intensity or a central maximum, though the average apparent size of the adult AZs was increased ∼20% (mean AZ area, 0.240 ± 0.099 μm2, n = 100). In contrast, imaging these tissues with ExM revealed Brp puncta with hollow ring-like structures that were less regularly shaped and smaller than unexpanded samples (Figure 4, B and C). We therefore used ExM to examine AZ morphology in aging flies and found a significant increase in the two-dimensional (2D) area of AZs across age (Figure 4D) as well as a significant increase with age in the presence of AZs containing multiple Brp rings (Figure 4E and Supplemental Figure S4). Notably, the AZs containing multiple Brp rings were larger than AZs harboring only a single Brp ring (Figure 4F), although the size of individual rings in multiple Brp ring–containing AZs was significantly smaller than Brp rings in singlet AZs (Supplemental Figure S4). Thus, it is possible that the AZs containing multiple Brp rings arise from fusion of singlet AZs. If this is indeed the case, these AZs could have enhanced release, given the likely increased abundance of voltage-gated calcium channels and larger pools of synaptic vesicles (Cooper et al., 1996; Holderith et al., 2012). Altogether, these results document an age-dependent change in AZ structure consistent with the fusion of neighboring AZs during aging that could not be resolved using conventional confocal microscopy.
Improved axial resolution with ExM
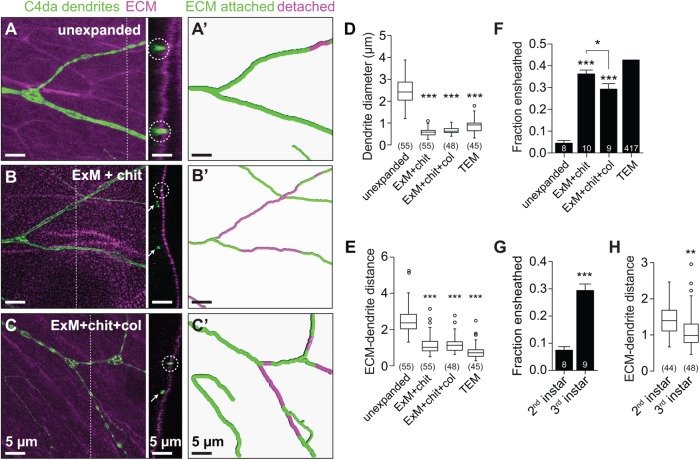
We next explored the utility of ExM for studying features of cell–cell interactions that are not observable with conventional confocal microscopy. Portions of Drosophila larval class IV da neuron (C4da) dendrite arbors, like peripheral arbors of other nociceptive sensory neurons, are ensheathed by the epidermis (Chalfie and Sulston, 1981; Han et al., 2012; Kim et al., 2012; O’Brien et al., 2012). The extent of epidermal dendrite ensheathment has been difficult to ascertain, in part because the size and spacing of sensory dendrites and epithelial membranes cannot be accurately measured using diffraction-limited microscopy. Although transmission electron microscopy (TEM) analysis suggests that ∼30% of C4da dendrites are ensheathed by the epidermis (Jiang et al., 2014), the difficulty in identifying terminal dendrites that are largely devoid of electron-dense material and the small cross-sectional area sampled by TEM limit the utility of TEM in a systematic analysis of epidermal ensheathment. We reasoned that the increased resolution afforded by ExM would allow for the most accurate analysis of C4da dendrite embedding to date. We therefore double-labeled the extracellular matrix (ECM) and C4da dendritic membranes and used confocal microscopy of unexpanded and expanded tissue to monitor apical displacement of dendrites from the ECM as a proxy for epithelial dendrite ensheathment (Han et al., 2012; Jiang et al., 2014).
In unexpanded tissues, dendrites appeared elongated in cross-section, with splayed features and axial lengths ranging from 1 to 4 μm, substantially higher than dendrite diameters observed by TEM (Figure 5, A and D). The vast majority of these dendrites appeared to be in contact with the ECM (Figure 5, A and F). However, the diameter of higher-order dendrite branches (∼150 nm) and the ECM thickness are smaller than the axial resolution of the confocal (Han et al., 2012; Jiang et al., 2014), precluding accurate assessment of the relative position of these structures.
FIGURE 5:
Improved axial resolution facilitates analysis of cell–cell interactions. (A–C) Axial position of dendrites and ECM. Maximum-intensity projections are shown for the following fillet preparations of larvae expressing the C4da-specific marker ppk-CD4-tdTomato: (A) unexpanded body walls, (B) expanded body walls of larvae treated with chitinase (ExM + chit), and (C) expanded body walls of larvae treated with chitinase and collagenase (ExM+chit+col), each stained with antibodies to perlecan to label the ECM and to DsRed to label C4da dendrites. Dashed lines mark y,z cross-sectional positions shown to the right of each image. Circles mark ECM-contacting dendrites, and arrows mark ECM-detached dendrites. (A′–C′) Traces depict ECM-contacting dendrites in green and ECM-detached dendrites in magenta. Plots depict (D) dendrite diameter, (E) the distance between detached dendrites and the ECM, and (F) the fraction of dendrites detached from the ECM measured using the indicated imaging approaches. (G) The fraction of dendrites detached from the ECM and (H) distance between detached dendrites and the ECM are shown for the indicated stages. Lines depict mean and SD in F and G. *, p < 0.05, ***, p < 0.005 compared with unexpanded samples, unless otherwise indicated, one-way ANOVA with a post hoc Dunnett’s test in D–F. TEM samples were excluded from statistical analysis in E because the number of neurons sampled by TEM is not known. **, p < 0.01, ***, p < 0.005 compared with second instar samples, unpaired t test with Welch’s correction in G and H. n values represent the number of dendrites scored (D, F, and H; TEM values in E) or neurons analyzed (F and G). Scale bars: 5 µm. All distances and scale bars refer to preexpansion dimensions.
In expanded tissue, we identified several differences in the apparent size and orientation of dendrites and ECM. First, dendrites appeared spherical with defined margins (Figure 5B). Second, dendrite diameter was significantly reduced compared with pre-expansion images and comparable to values from TEM (Figure 5D). Third, expansion allowed for identification of ECM detachment of dendrites over shorter length scales (Figure 5E). Finally, ExM revealed that a significantly larger proportion of dendrites was detached from the ECM and ensheathed by epithelial cells in third instar larvae (Figure 5F), comparable to estimates from TEM analysis (Jiang et al., 2014). By contrast, we observed significantly less dendrite detachment in second instar larvae, when epithelial ensheathment is initiated (Jiang et al., 2014). Taken together, these results strongly suggest that ExM sample processing does not induce significant levels of spurious ECM detachment of dendrites (Figure 5G).
We noted a “dry lakebed” effect in ECM staining following expansion with antibodies to perlecan (Figure 5B) or collagen (unpublished data), likely the result of incomplete ECM proteolysis in conventional ExM. To ascertain whether the lakebed effect caused distortion in measurements of dendrite–ECM interactions, we conducted ExM that included both chitinase treatment and a collagenase-mediated enzymatic ECM digestion step (ExM+chit+col). This collagenase treatment remedied the lakebed effect (Figure 5C), and although signal intensity for ECM staining was attenuated by this treatment, dendrite–ECM interactions with ExM+chit+col mirrored results from ExM with chitinase.
Epithelial dendrite ensheathment is thought to modulate somatosensation (Han et al., 2012; Kim et al., 2012; O’Brien et al., 2012) by reducing the distance between nerve endings and the skin surface or by some other means. If ensheathment acts primarily to increase dendrite proximity to the skin surface, we reasoned that ensheathed dendrites might be progressively displaced toward the body-wall surface. This is not what we found. Using ExM we found that apical dendrite displacement was not increased between the second larval instar, when ensheathment is initiated, and the end of the third larval instar, when >30% of the dendrite arbor is ensheathed (Figure 5, G and H).
In this work, we have optimized, validated, and applied ExM methods to interrogate Drosophila tissues at a spatial resolution of ∼70 nm using a standard confocal microscope. Using ExM, we demonstrated that optical diffraction-limited microscopy, which is frequently used to analyze NMJ structure and function, leads to systematic errors in AZ quantification; ExM provides a solution to this problem. We also documented changes in AZ structure that provide a plausible cellular mechanism to account for age-dependent changes in neurotransmitter release at CM9 NMJs. Finally, our analysis of sensory neuron–epidermis interactions revealed that peripheral arbors of nociceptive neurons are extensively embedded in epithelial cells. Taken together with findings that Caenorhabditis elegans and Danio rario nociceptive neurons and some mammalian unmyelinated intraepidermal nerve fibers are similarly ensheathed in the epidermis (Cauna, 1973; Chalfie and Sulston, 1981; O’Brien et al., 2012), our finding that large portions of nociceptive dendrite arbors are ensheathed by epithelial cells underscores the likelihood that epithelial ensheathment may contribute to nociceptor function. Intriguingly, some forms of peripheral neuropathy exhibit loss of unmyelinated intraepidermal nerves (Weis et al., 2011; Üçeyler et al., 2013); whether epithelial ensheathment plays a role in these neuropathies remains to be determined.
Two recent studies pointed toward the utility of ExM in superresolution imaging of Drosophila. One study used a version of ExM together with the optical superresolution microscopy method SIM to study organization of synaptonemal complexes in Drosophila ovaries at relatively high resolution (Cahoon et al., 2017), and another demonstrated the compatibility of ExM for imaging of Drosophila brain dissections (Mosca et al., 2017). While these studies demonstrated applications and potential advantages of ExM, both lacked validation through correlative imaging of specimens before/after expansion and applied ExM to only a single tissue. By contrast, we provide a thorough analysis of ExM applied to a variety of tissues.
We found that some Drosophila preparations, including whole-mount embryos and larval brains, expanded robustly with low distortion (<4%) when we used previously developed procedures based on digestion with the broad-specificity protease proteinase K to soften the specimen (Chen et al., 2015). Drosophila larvae, on the other hand, required an additional enzymatic treatment step with chitinase to soften the tough, polysaccharide-based, chitin-rich cuticle, and we found that robust expansion of the larval ECM required further treatment with collagenase. Our optimized procedures are detailed in a step-by-step protocol to aid others in evaluating or adopting the methodology (see the Supplemental Material).
In its current form, ExM will allow interrogation of many cellular structures that cannot be resolved with conventional diffraction-limited microscopy. We have demonstrated the utility of ExM in analysis of synapse structure and cell–cell interactions, and a natural extension of our results is the use of ExM to study neural circuits in Drosophila. We have shown that resolution limits lead to systematic undersampling of synapse number at the NMJ using conventional diffraction-limited microscopy, and the problem is likely worse in the neuropil, where synapses are both more tightly packed and distributed over a larger volume. One elegant solution to this problem has been to use GFP reconstitution as a proximity sensor (Feinberg et al., 2008). Splitting the GFP beta barrel yields two fragments that reassemble when stably juxtaposed within 7–10 nm of one another (Morell et al., 2008). Fusing these GFP fragments to inert membrane tethers, such as human CD4 lacking interaction domains, produces a sensor that yields GFP fluorescence when cells approach inside of ∼30 nm. While this tool has been useful in identifying synaptic partners, the technique is limited by spurious reassembly and the irreversibility of the reaction. ExM provides a complementary tool and should allow researchers to visualize juxtaposition of pre- and postsynaptic partners at sufficient resolution to map and count synapses, especially when used in combination with the recently completed EM connectivity maps (Eichler et al., 2017).
The vast number of epitope-tagged transgenes available for use in Drosophila, together with emerging techniques for tagging endogenous loci, provide many potential applications of ExM in superresolution imaging of these transgenes. For example, we have demonstrated the utility of ExM in visualization of fine structural elements of GFP-labeled mitochondria. In another application, we anticipate that ExM will facilitate mapping of protein subcellular localizations, for example, the asymmetric distribution of polarity determinants. The potential future applications of ExM to Drosophila and other model organisms will grow with further development of the methodology. For instance, recent technical advances by other researchers include expansion of large intact specimens (Ku et al., 2016), interrogation of mRNA species (Chen et al., 2016), and the development of iterative expansion microscopy to achieve higher spatial resolution by means of larger (multistep) hydrogel expansion (Chang et al., 2017). While ExM is a particularly exciting methodology, owing to its accessibility and strong performance with multicolor and volumetric imaging, the potential for artifacts is high, and we believe that it is important to carefully validate and optimize ExM for individual specimens as we have done here.
MATERIALS AND METHODS
Fly stocks
Flies were maintained on standard cornmeal–molasses–agar media and reared at 25°C under 12 h alternating light–dark cycles. The following fly lines were used in this study: w1118 (BDSC6326); w1118; ppk-Gal4 (BDSC32079); w1118; UAS-mito-GFP (BDSC8443); w1118, ppk-CD4-tdTomato (BDSC35845).
Antibody staining
A detailed list of antibodies and dilutions is presented in Supplemental Table S1. Details on fixation/staining are outlined in the following sections.
Larval ventral nerve cord staining
Third instar larvae were pinned dorsal side up on a Sylgard plate (Dow Corning) and filleted along the dorsal midline. Brains were carefully removed using forceps, fixed in EM-grade paraformaldehyde (PFA; Electron Microscopy Sciences), freshly diluted to 4% final concentration in phosphate-buffered saline (PBS) for 30 min at room temperature, and washed 5× for 5 min each time in PBSTx (PBS with 0.2% Triton X-100) before being blocked in blocking/permeabilization buffer (PBS with 5% bovine serum albumin and 0.2% Triton X-100) for 30 min. Samples were incubated with primary antibodies in blocking/permeabilization buffer overnight at 4°C, washed 3× for 15 min each time in PBSTx, and incubated for 6 h with secondary antibodies in blocking/ permeabilization buffer. After three 20-min washes in PBSTx, samples were treated with the amine-reactive small molecule MA-NHS (methacrylic acid N-hydroxy succinimidyl ester) to preserve fluorescence signal (Chozinski et al., 2016) at room temperature for 1 h; this was followed by three 20-min washes with PBS.
Whole-mount embryo staining
Embryos were collected on yeasted grape juice agar plates for 4 h and aged at 25°C in a moist chamber for 10–14 h. Embryos were dechorionated in 50% bleach for 4 min and rinsed in water. Embryos were then fixed in a 1:1 suspension of heptane and 4% formaldehyde (in PBS) for 15 min with vigorous shaking. The formaldehyde was removed, 1 volume of methanol was added, and the heptane:methanol suspension was vigorously shaken to devitellinize the embryos. After fixation, embryos were recovered and processed as described above for the nerve cord samples.
Larval body-wall fillets
Third instar larvae were pinned on a Sylgard plate, filleted along the ventral midline, and pinned open. After removal of the intestines, fat bodies, imaginal disks, and ventral nerve cord, fillets were fixed in PBS with 4% PFA for 15 min (for anti-brp staining) or 30 min (for anti-GFP staining). For microtubule staining, we extracted the tissue with PEM (0.1M PIPES, pH 7, 1 mM EDTA, 1 mM MgCl2) containing 0.5% Triton-X-100 for 30 s immediately before fixation. Samples were then fixed for 15 min in a solution containing 3.2% PFA and 0.1% glutaraldehyde or 3.2% PFA alone in PEM, followed by a 30-s wash with extraction buffer. After fixation, samples were processed as above for staining.
Gelation, digestion, and expansion of nerve cord and embryo samples
Samples were incubated in monomer solution (2 M NaCl, 8.625% sodium acrylate, 2.5% acrylamide, 0.15% bisacrylamide in PBS) for 1 h at 4°C before gelation. A stock of 4-hydroxy-2,2,6,6-tetramenthylpiperidin-1-oxyl (4-hydroxy-TEMPO) at 1% (wt/wt) in water was added to the incubation solution and diluted to a concentration of 0.01%. Concentrated stocks of tetramethylethylenediamine and ammonium persulfate at 10% (wt/wt) in water were added sequentially to the incubation solution and diluted to concentrations of 0.2% (wt/wt). The tissues were then incubated at 37°C for 2–2.5 h. After the samples were gelled, the gels were cut and placed in a small 12-well chamber and were digested in 8 U/ml proteinase K solution in digestion buffer (40 mM Tris, pH 8, 1 mM EDTA, 0.5% Triton, 0.8 M guanidine HCl) for 1 h at 37°C. Subsequently, samples were removed from the digestion solution and were allowed to expand in excess water overnight.
Gelation, digestion, and expansion of Drosophila body-wall samples
Samples were incubated in monomer solution for 1 h at 4°C before gelation. Larval fillets were gelled with the same solution as above, but were incubated at 37°C for 3–4 h. After gelation, the gels were cut and placed in a small 12-well chamber and 1 mg/ml chitinase in PBS (pH 6.0) was used to digest the cuticles for ∼4 d at 37°C. Samples were then rinsed 2× with PBS for 5 min each, digested with proteinase K solution, and expanded as described above.
Before expansion, chitinase-treated samples were incubated with 1000 U/ml collagenase solution (prepared with buffer 1× HBSS lacking calcium, magnesium, and phenol red) with 0.01 M CaCl2 and 0.01 M MgCl2 overnight in a 37°C shaking incubation chamber. Samples were then rinsed with PBS twice for 5 min and digested in 8 U/ml proteinase K solution in digestion buffer for 1 h at 37°C.
Mounting and imaging
Before expansion, samples were mounted on lysine-coated #1.5 coverglasses in wells made of polydimethylsiloxane. Samples were transferred onto coverglass in a drop of PBS, moved to the desired position/orientation with forceps, and excess liquid was wicked away to ensure that samples adhered to the coverglass. Next, PBS was added to the well, submerging the tissue to prevent it from drying out. After expansion, the expanded gel was trimmed to fit onto the coverglass, excess water was removed, and the gel was mounted on a lysine-coated coverglass for imaging. Confocal microscopy was performed on a Leica SP5 inverted confocal scanning microscope using a 63×/1.2 NA water lens (Figures 1–5) or a 20×/0.7 NA air lens (Supplemental Figure S1).
TEM
TEM was as previously described (Jiang et al., 2014). Briefly, larvae were perforated with insect pins, fixed in 2.5% glutaraldehyde/0.1 M sodium cacodylate buffer, washed in PBS, and postfixed in 2% osmium tetroxide. Samples were embedded in epon-araldite, and 70-nm sections were stained with lead citrate and viewed on a JEOL-1230 microscope with an AMT XR80 camera.
Measurements
Distortion analysis of expanded specimens was performed as previously described (Chozinski et al., 2016). Briefly, postexpansion images were aligned with corresponding preexpansion images using only linear transformations (rotation, scaling, and translation). The linearly transformed postexpansion images were then nonlinearly deformed to match the preexpansion images. Quantitative comparison of the linearly and nonlinearly transformed postexpansion images produced values for root-mean-square (RMS) deviation over a range of length scales. All analyses were performed in three dimensions, and each RMS plot was generated with one set of corresponding (pre/post) images.
Active zones.
AZ numbers were scored as the number of discrete Brp-positive structures in 2D projections of confocal stacks from corresponding areas of pre- and postexpansion tissue using the cell-counting function of ImageJ. AZ area was measured by computer-assisted tracing of the perimeter of Brp-positive structures in 2D projections of confocal stacks, and density measurements represent the number of Brp-positive structures per unit area (in 2D projections of confocal stacks). AZ area and density measurements were taken from at least five independent samples. AZ architecture (singlets, doublets, multiples) represents the proportion of Brp-positive structures with the respective number of distinct ring-like structures.
Dendrite–epithelia interactions.
Epithelial enclosure of somatosensory dendrites was measured in confocal image stacks as previously described (Han et al., 2012). Briefly, image stacks were captured at a z-depth of 0.15 microns, and where indicated, image stacks were deconvolved using the Leica LAS deconvolution plug-in set to adaptive point-spread function for 10 iterations. Three-dimensional reconstruction was performed with Imaris, and colocalization was measured between fluorescent signals labeling dendrites (GFP immunoreactivity) and ECM (perlecan immunoreactivity) using the Imaris Coloc module. The dendrites were traced in Imaris, portions of the arbor that failed to colocalize with the ECM (apically detached dendrites) were pseudocolored in traces, and the proportion of the arbor that was detached from the ECM was measured in these traces. Dendrites were identified in TEM images as processes near the basal epithelial surface containing arrays of parallel microtubules. A total of 417 dendrites were scored (in sections of four larvae) as ECM attached (in direct contact with the basement membrane) or detached (internalized in the epidermis).
Supplementary Material
Acknowledgments
This work was supported by a National Institutes of Health (NIH) grant (NINDS R01 NS076614), a UW Research Innovation award, and startup funds from the University of Washington (J.Z.P.); by an NIH grant (NIMH R01 MH115767), a Burroughs-Wellcome Career Award at the Scientific Interface, and startup funds from the University of Washington (J.C.V.); and by National Science Foundation Graduate Research Fellowship DGE-1256082 (T.J.C.). We thank the Center on Human Development and Disability for assistance with TEM sample preparation. Anti-perlecan antibodies were provided by Stefan Baumgartner, and the Brp antibody was obtained from the Developmental Studies Hybridoma Bank, created by the National Institute of Child Health and Human Development (NICHD) of the NIH and maintained at the University of Iowa. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
Abbreviations used:
- 2D
two-dimensional
- AZ
active zone
- ECM
extracellular matrix
- ExM
expansion microscopy
- GFP
green fluorescent protein
- NMJ
neuromuscular junction
- PBS
phosphate-buffered saline
- PFA
paraformaldehyde
- RMS
root-mean-square
- SIM
structured illumination microscopy
- STED
stimulated emission depletion microscopy
- TEM
transmission electron microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-10-0583) on April 24, 2018.
REFERENCES
- Cahoon CK, Yu Z, Wang Y, Guo F, Unruh JR, Slaughter BD, Hawley RS. (2017). Superresolution expansion microscopy reveals the three-dimensional organization of the Drosophila synaptonemal complex. Proc Natl Acad Sci USA , E6857–E6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauna N. (1973). The free penicillate nerve endings of the human hairy skin. J Anat , 277–288. [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Sulston J. (1981). Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev Biol , 358–370. [DOI] [PubMed] [Google Scholar]
- Chang J-B, Chen F, Yoon Y-G, Jung EE, Babcock H, Kang JS, Asano S, Suk H-J, Pak N, Tillberg PW, et al (2017). Iterative expansion microscopy. Nat Methods , 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tillberg PW, Boyden ES. (2015). Expansion microscopy. Science , 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Wassie AT, Cote AJ, Sinha A, Alon S, Asano S, Daugharthy ER, Chang JB, Marblestone A, Churg GM, et al (2016). Nanoscale imaging of RNA with expansion microscopy. Nat Methods , 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chozinski TJ, Halpern AR, Okawa H, Kim H-J, Tremel GJ, Wong ROL, Vaughan JC. (2016). Expansion microscopy with conventional antibodies and fluorescent proteins. Nat Methods , 485–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper RL, Winslow JL, Govind CK, Atwood HL. (1996). Synaptic structural complexity as a factor enhancing probability of calcium-mediated transmitter release. J Neurophysiol , 2451–2466. [DOI] [PubMed] [Google Scholar]
- Ehmann N, van de Linde S, Alon A, Ljaschenko D, Keung XZ, Holm T, Rings A, DiAntonio A, Hallermann S, Ashery U, et al (2014). Quantitative super-resolution imaging of Bruchpilot distinguishes active zone states. Nat Commun , 4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler K, et al. (2017). The complete connectome of a learning and memory centre in an insect brain. Nature , 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, Bargmann CI. (2008). GFP reconstitution across synaptic partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron , 353–363. [DOI] [PubMed] [Google Scholar]
- Fouquet W, Owald D, Wichmann C, Mertel S, Depner H, Dyba M, Hallermann S, Kittel RJ, Eimer S, Sigrist SJ. (2009). Maturation of active zone assembly by Drosophila Bruchpilot. J Cell Biol , 129–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta VK, Pech U, Bhukel A, Fulterer A, Ender A, Mauermann SF, Andlauer TF, Antwi-Adjei E, Beuschel C, Thriene K, et al (2016). Spermidine suppresses age-associated memory impairment by preventing adverse increase of presynaptic active zone size and release. PLoS Biol , e1002563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Wang D, Soba P, Zhu S, Lin X, Jan LY, Jan Y-N. (2012). Integrins regulate repulsion-mediated dendritic patterning of Drosophila sensory neurons by restricting dendrites in a 2D space. Neuron , 64–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holderith N, Lorincz A, Katona G, Rózsa B, Kulik A, Watanabe M, Nusser Z. (2012). Release probability of hippocampal glutamatergic terminals scales with the size of the active zone. Nat Neurosci , 988–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Babcock H, Zhuang X. (2010). Breaking the diffraction barrier: super-resolution imaging of cells. Cell , 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel T, Krukkert K, Roos J, Davis G, Klämbt C. (2000). Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron , 357–370. [DOI] [PubMed] [Google Scholar]
- Jepson JEC, Shahidullah M, Liu D, le Marchand SJ, Liu S, Wu MN, Levitan IB, Dalva MB, Koh K. (2014). Regulation of synaptic development and function by the Drosophila PDZ protein Dyschronic. Development , 4548–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Soba P, Parker E, Kim CC, Parrish JZ. (2014). The microRNA bantam regulates a developmental transition in epithelial cells that restricts sensory dendrite growth. Development , 2657–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ME, Shrestha BR, Blazeski R, Mason CA, Grueber WB. (2012). Integrins establish dendrite-substrate relationships that promote dendritic self-avoidance and patterning in Drosophila sensory neurons. Neuron , 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittel RJ, Wichmann C, Rasse TM, Fouquet W, Schmidt M, Schmid A, Wagh DA, Pawlu C, Kellner RR, Willig KI, et al (2006). Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science , 1051–1054. [DOI] [PubMed] [Google Scholar]
- Ku T, Swaney J, Park J-Y, Albanese A, Murray E, Cho JH, Park Y-G, Mangena V, Chen J, Chung K. (2016). Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat Biotechnol , 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepicard S, Franco B, de Bock F, Parmentier M-L. (2014). A presynaptic role of microtubule-associated protein 1/Futsch in Drosophila: regulation of active zone number and neurotransmitter release. J Neurosci , 6759–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney RE, Rawson JM, Eaton BA. (2014). An age-dependent change in the set point of synaptic homeostasis. J Neurosci , 2111–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matkovic T, Siebert M, Knoche E, Depner H, Mertel S, Owald D, Schmidt M, Thomas U, Sickmann A, Kamin D, et al (2013). The Bruchpilot cytomatrix determines the size of the readily releasable pool of synaptic vesicles. J Cell Biol , 667–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell M, Czihal P, Hoffmann R, Otvos L, Avilés FX, Ventura S. (2008). Monitoring the interference of protein-protein interactions in vivo by bimolecular fluorescence complementation: the DnaK case. Proteomics , 3433–3442. [DOI] [PubMed] [Google Scholar]
- Mosca TJ, Luginbuhl DJ, Wang IE, Luo L. (2017). Presynaptic LRP4 promotes synapse number and function of excitatory CNS neurons. eLife , e27347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien GS, Rieger S, Wang F, Smolen GA, Gonzalez RE, Buchanan J, Sagasti A. (2012). Coordinate development of skin cells and cutaneous sensory axons in zebrafish. J Comp Neurol , 816–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernas L, Scorrano L. (2016). Mito-morphosis: mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu Rev Physiol , 505–531. [DOI] [PubMed] [Google Scholar]
- Propst JW, Ko CP. (1987). Correlations between active zone ultrastructure and synaptic function studied with freeze-fracture of physiologically identified neuromuscular junctions. J Neurosci , 3654–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC. (2012). The presynaptic active zone. Neuron , 11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillberg PW, Chen F, Piatkevich KD, Zhao Y, Yu CC, English BP, Gao L, Martorell A, Suk HJ, Yoshida F, et al (2016). Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat Biotechnol , 987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Üçeyler N, Zeller D, Kahn A-K, Kewenig S, Kittel-Schneider S, Schmid A, Casanova-Molla J, Reiners K, Sommer C. (2013). Small fibre pathology in patients with fibromyalgia syndrome. Brain J Neurol , 1857–1867. [DOI] [PubMed] [Google Scholar]
- Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Dürrbeck H, Buchner S, Dabauvalle MC, Schmidt M, Qin G, et al (2006). Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron , 833–844. [DOI] [PubMed] [Google Scholar]
- Weis J, Katona I, Müller-Newen G, Sommer C, Necula G, Hendrich C, Ludolph AC, Sperfeld A-D. (2011). Small-fiber neuropathy in patients with ALS. Neurology , 2024–2029. [DOI] [PubMed] [Google Scholar]
- Weyhersmüller A, Hallermann S, Wagner N, Eilers J. (2011). Rapid active zone remodeling during synaptic plasticity. J Neurosci , 6041–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.