Abstract
Background
The aim of this study was to investigate the correlations between ADAMTSs expression and breast invasive ductal carcinoma (IDC), and to offer a theoretical basis for novel treatment methods for IDC patients.
Material/Methods
Non-proliferative catheter of breast fibroadenoma (FA) and IDC were used as the normal control and experimental group, respectively. Immunohistochemical (IHC) staining and Western blot (WB) analysis was used to assess protein expression levels of ADAMTS8, ADAMTS18, and ADAMTS20 in both FA and IDC tissues. The results of IHC, the relationship between the protein expression and the tumor molecular classification, and clinical pathological parameters were all evaluated.
Results
IHC and WB results showed that the expression of ADAMTS8/18 in IDC samples was higher than in FA samples, while the expression of ADAMTS20 in IDC samples was lower than that in FA samples. According to the results of WB, the level of ADAMTS8 was higher in the HER2+ group than in the HER2− group and FA group. The expression of ADAMTS18 in the HR+ (including ER+ and PR+) group was significantly higher than in the HR− group and FA group. The expression of ADAMTS18 protein was also higher in the Ki67+ group than in the Ki67− group. ADAMTS20 was higher in HER2+ IDC compared with the basal subtype of IDC.
Conclusions
ADAMTS8/18/20 levels were not significantly correlated to the molecular subtype of IDC. ADAMTS18/20 was significantly associated with histological grade of IDC. ADAMTS8 may predict poor prognosis results of IDC patients.
MeSH Keywords: Matrix Metalloproteinase 8; Carcinoma, Ductal, Breast; Matrix Metalloproteinase 20; Matrix Metalloproteinases
Background
Breast cancer is the most common cancer affecting women. Cancer statistics show that the lifetime risk of breast cancer is as high as 12% in the USA. It was estimated that the number of new cases diagnosed in the USA and Europe in 2015 alone was 230 000 and 470 000, respectively [1]. However, in China the estimated number of females who died of breast cancer reached 60 473 [2]. Due to its invasive characteristics, breast invasive ductal carcinoma (IDC) has become a leading cause of cancer-related deaths among women worldwide [3]. Therefore, the discovery of new markers, especially molecularly detectable ones, is noteworthy [4].
Biomarkers such as human epidermal growth factor estrogen receptors (ER), progesterone receptors (PR), receptor 2 (HER2), and Ki67 were detected in our study. HER2 is a key mediator in cell growth, differentiation, and survival [5]. HER2 is also a representative marker; approximately 20% of patients with IDC show increased HER2 levels [6–9]. In general, HER2-positive tumors had higher histological grade. Furthermore, compared to other tumor subtypes, HER2-positive tumors are more likely to invade lymph nodes [10,11]. Ki67 is a common proliferation parameter which can be used to predict the rate of multiplication of tumor cells. The expression of Ki67 is often upregulated in proliferating cells. Physicians can determine the growth fraction in clonal cell populations by detecting Ki67 expression [12]. In addition, IDC of Luminal types A and type B can be distinguished by Ki67 levels [13]. Patients with high Ki67 expression levels are usually recommended to accept clinical treatments [14].
ADAMTS, also known as ADAM with thrombospondin motifs, is a family of secreted extracellular protease enzymes. Evidence has suggested that ADAMTS expression is dysregulated in diverse types of cancers, including breast, gastric, lung, and colorectal cancer [15–18]. In the human genome, 19 ADAMTS members have been identified. Based on known substrates, ADAMTSs can be sub-grouped according to their characteristics. For instance, ADAMTS8 and ADAMTS20 are categorized in the group of aggrecanases or proteoglycanases, while ADAMTS18 is classified in the orphan enzymes group [19,20]. ADAMTS8, also known as METH-2, is thought to be an antiangiogenic factor [21]. There is a high frequency of promoter methylation in some cancers in which there is downregulation of ADAMTS8 [22,23]. In a very wide range of tumor tissue types, ADAMTS18 has been observed to be frequently hypermethylated, and a deficiency of it enhances tumorigenesis [24]. Relevant research has suggested that ADAMTS20 is correlated with migration of melanoblasts [25]. ADAMTS8, 18, and 20 have been shown to be associated with cancer development, yet little is known about the mechanism in tumorigenesis, especially in IDC development.
We conducted this study to investigate the association between ADAMTSs and IDC, and to offer a theoretical basis for new treatment methods for IDC patients.
Material and Methods
Tissue samples
A total of 278 breast IDC tissue samples were collected from IDC patients who had complete clinical data archived at the Affiliated Cancer Hospital of Zhengzhou University from January 2012 to June 2016, and non-proliferative catheter of breast fibroadenoma (FA) was used as the normal control. Patients had no previous diagnosis of carcinoma, no distant metastases at time of diagnosis, and no evidence of disease within 1 month after primary surgery. Furthermore, patients receiving neo-adjuvant therapy or with carcinoma in situ only were excluded. After primary surgery, a representative part of the tumor was macroscopically selected by a pathologist, formalin-fixed, and paraffin-embedded for further study. This study was approved by the Institute Review Ethics committee. Written informed consent was obtained from all participants.
Immunohistochemical (IHC) staining
We deparaffinized 3-μm–thick continuous sections using xylene I and II for 20 min and then dehydrated them with gradient anhydrous ethanol. The slides were soaked in 50 μl 3% H2O2 for 20 min, and then were placed into a 1-mM Tris-EDTA (pH=9) water bath (100°C) for 20 min, and cooled to room temperature. After washing with PBS, 50 μl of primary antibody solution (#PA5-64274, rabbit anti-ADAMTS8/18/20, 1: 100, Thermo Fisher Scientific, Waltham, MA, USA) was added and incubated with the slice at 4°C overnight. The UltraView Universal DAB Detection Kit (#760-500, Ventana Medical Systems, Tucson, Arizona, USA) was used to detect the rabbit primary antibodies used earlier, then the slides were visualized using hydrogen peroxidase substrate and DAB chromogen, producing a brown precipitate.
Western blot (WB)
Total proteins from tissues were extracted using radio immunoprecipitation assay (RIPA) buffer (1% NP-40 or Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 50 mM Tris-HCl, pH 7.8, 1 mM EDTA). Protein concentrations were examined with BCA (Thermo Scientific, Rockford, IL, USA), and the proteins were separated using SDS-PAGE. The proteins were then transferred onto PVDF membranes under a constant current of 200 mA. After blocking for 2 h with 5% non-fat dry milk, membranes were incubated for 2 h with the primary antibodies (mouse anti-ADAMTS8/18/20, 1: 900, Abcam, Cambridge, MA, USA), and incubated with HRP-conjugated secondary antibodies (1: 2000, Abcam, Cambridge, MA, USA) for another 2 h. The membranes were washed with TBST (1: 1000 in 2.5% BSA in TBST, Abcam, USA), and all blots were visualized using ECL (Bio-Rad, Hercules, CA, USA).
Prognostic analysis
The prognostic data of participants were collected at follow-up visits of 125 breast IDC patients. The date of the operation was set as the beginning date of follow-up, and time of death after the operation was set as the end of follow-up. By analyzing the relationship between clinical pathological parameters, such as the number of lymphatic metastases, ER, PR, HER2, Ki67, ADAMTS8/18/20, and disease-free survival (DFS) rate and mean survival time, the correlation of clinical pathological parameters and the disease-free survival (DFS) time or overall survival (OS) time of IDC patients could be evaluated.
Statistical analysis
SPSS 21.0 software was used to perform statistical analyses. P<0.05 was considered to indicate statistical significance. The chi-squared test was used to compare results between IDC groups, while the Spearman correlation analysis was used for the correlation analysis between ADAMTS 8/18/20 expression and clinical pathological parameters. The relative average densities of proteins are presented as the mean ±SD, and their differences were analyzed by the t test. The Kaplan-Meier method was assessed with the log-rank test. Cox’s proportional hazards model was used for univariate and multivariate analyses of prognostic values. In DFS analysis, the discovery of recurrence or distant metastatic lesions by imagological examination was considered as the end time. Patient death was considered as the end time of the survival event.
Results
Clinical pathology data of IDC patients
The average age in the 278 included cases of breast IDC patients was 49 years. About half of the patients had a ≤2 cm diameter tumor size. Diagnosis of the specimens was made according to the WHO classification of tumors. The number of patients with histological grade level II or III was significantly larger than in level I. Lymphatic metastasis was found in about 62.6% of patients, while 97.5% of patients did not have metastasis in any other areas. In follow-up visits of 125 cases, 9 (7.2%) patients had passed away, and 116 (92.8%) were still alive. Details are listed in Table 1.
Table 1.
Clinicopathologic data of 278 cases of IDC patients.
| The clinical pathological characteristics | Case number (n=278) | |
|---|---|---|
| Age (year) | <50 | 136 (48.9%) |
| ≥50 | 142 (51.1%) | |
| Diameter of tumor (cm) | ≤2 | 154 (55.4%) |
| >2 | 124 (44.6%) | |
| Histological grade | I | 17 (6.1%) |
| II–III | 261 (93.3%) | |
| Lymphatic metastasis | No | 105 (37.8%) |
| Yes | 173 (62.2%) | |
| Other parts metastasis | No | 271 (97.5%) |
| Yes | 7 (2.5%) | |
| Survival status | Survival | 116 (92.8%) |
| Dead | 9 (7.2%) | |
| Loss to follow-up | 153 (55.0%) |
Positive expressions of ADAMTS 8, ADAMTS 18, and ADAMTS 20
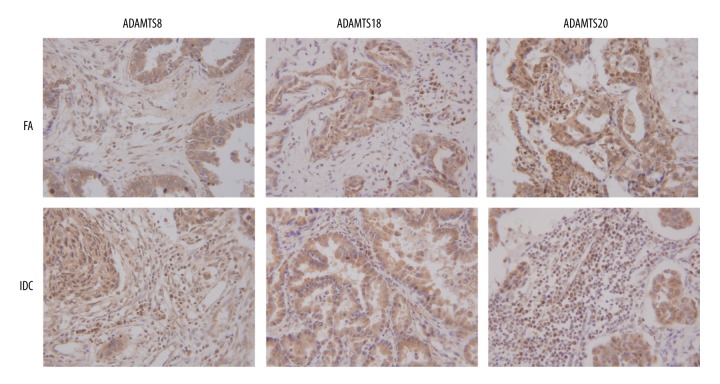
In 278 cases of breast IDC, the positive expressions of ADAMTS 8, ADAMTS 18, and ADAMTS 20 were 84.5%, 88.8%, and 72.3%, respectively. In 20 cases of FA, the positive expressions of these 3 proteins were 45%, 50%, and 90%, respectively. IHC staining results of breast IDC and non-proliferative catheter of FA are shown in Figure 1. Results are generally scored as 0, 1, 2, or 3. Positive cells <5% scored 0 point, 5–25% scored 1 point, 25–50% scored 2 points, 50–75% scored 3 points, and > 75% scored 4 points. Immunostaining intensity was scored as follows: 0, negative; 1, weak; 2, moderate; and 3, strong. When the staining was heterogeneous, the score was calculated, in which the percent positive rating was multiplied by the intensity rating. Those cells scored 0–1 were considered as negative. Therefore, ADAMTS 8, ADAMTS 18, and ADAMTS 20 were all positively expressed in breast IDC.
Figure 1.
IHC staining result of ADAMTS8, ADAMTS18, and ADAMTS20 in IDC and non-proliferative character of FA. In IDC, the positive expressions of ADAMTS8 and ADAMTS18 were significantly higher, while ADAMTS20 was lower, compared with those in FA. The sepia sections represent the stained proteins. The results showed that proteins were distributed in both cytoplasm and cytoplasm interstitial, mainly in cytoplasm.
Relationship between expressions of ADAMTS8/18/20 and IDC molecular classification
According to the consensus reached at the 2011 St. Gellen Conference, breast IDC is divided into 4 groups – Luminal A, Luminal B, HER-2, and basal-like groups – based on expression levels of ER, PR, HER2, and Ki67, respectively. In 278 breast IDC patients, there were 7 (2.5%) cases of Luminal A type, 63 (22.7%) cases of Luminal B type (HER2-), 113 (40.6%) cases of Luminal B type (HER2+), 66 (23.7%) cases of HER2+ type, and 29 (10.4%) cases of basal subtype. Significant ADAMTS20 differences existed between HER2 enriched and basal subtypes (P<0.05, Table 2).
Table 2.
Positive rate of ADAMTS8, ADAMTS18 and ADAMTS20 protein expressions in each breast IDC molecular subtyping.
| Subtype | Cases | ADAMTS8 | ADAMTS18 | ADAMTS20 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| + | − | P | + | − | P | + | − | P | ||
| Luminal A | 7 | 7 | 0 | 7 | 0 | 7 | 0 | |||
| Luminal B HER2(−) | 63 | 53 | 10 | 0.979@ | 56 | 7 | 0.471@ | 46 | 17 | 0.331@ |
| Luminal B HER2(+) | 113 | 93 | 20 | 0.757* | 100 | 13 | 0.937* | 79 | 34 | 0.663* |
| HER2 enriched | 66 | 58 | 8 | 0.348& | 57 | 9 | 0.628& | 51 | 15 | 0.331& |
| Basal subtype | 29 | 24 | 5 | 0.315# | 27 | 2 | 0.106# | 18 | 11 | 0.021# |
Compared with Luminal A;
compared with Luminal B HER2(−);
compared with Luminal B HER2(+);
compared with HER2 enriched.
P<0.05 indicated statistical significance.
Relationship between expressions of ADAMTS8/18/20 and clinical pathological parameters of IDC patients
The expression of ADAMTS18 was significantly higher, while ADAMTS20 was lower, in II phase patients than in I phase patients (P<0.01, Table 3). ADAMTS8 had no significant relation to the histological grading of tumors. Moreover, the positive expressions of ADAMTS8, ADAMTS18, and ADAMTS20 showed no association with patient age, tumor size, or lymphatic metastasis (P>0.05, Table 3).
Table 3.
The relationship between ADAMTS8, ADAMTS18 and ADAMTS20 protein expressions and clinical pathology parameters of IDC.
| Parameter | Cases | ADAMTS8 | ADAMTS18 | ADAMTS20 | |||
|---|---|---|---|---|---|---|---|
| Positive | P | Positive | P | Positive | P | ||
| Histological grade | |||||||
| I | 17 | 82.4% (14/17) | 76.5% (13/17) | 88.2% (15/17) | |||
| II–III | 261 | 84.7% (221/261) | 0.568 | 95.0% (248/261) | 0.002* | 54.0% (141/261) | 0.006* |
| Tumor size (cm) | |||||||
| ≤2 | 154 | 83.1% (128/154) | 87.7% (135/154) | 70.8% (109/154) | |||
| >2 | 124 | 86.3% (107/124) | 0.558 | 90.3% (112/124) | 0.651 | 74.2% (92/124) | 0.635 |
| Age (year) | |||||||
| <50 | 136 | 83.1% (113/136) | 87.5% (119/136) | 70.6% (96/136) | |||
| ≥50 | 142 | 86.0% (122/142) | 0.558 | 90.1% (128/142) | 0.651 | 74.0% (105/142) | 0.635 |
| Lymphatic metastasis | |||||||
| No | 105 | 88.6% (93/105) | 87.6% (92/105) | 70.5% (74/105) | |||
| Yes | 173 | 82.1% (142/173) | 0.16 | 89.0% (154/173) | 0.825 | 73.4% (127/173) | 0.753 |
P<0.05 indicated statistical significance.
Relationship between expressions of ADAMTS8/18/20 and HR, HER2, and Ki67 in IDC
In IDC, ADAMTS8 was closely correlated with HER2 expression and displayed a positive relationship (r=0.139, P=0.037). ADAMTS18 was closely related with ER, PR, and Ki67, and displayed a positive relationship (r=0.172, P=0.004; r=0.135, P=0.026; r=0.179, P=0.005). However, ADAMTS20 showed no correlation with HR (including ER and PR), HER2, or Ki67 (Table 4).
Table 4.
The correlation between ADAMTS8, ADAMTS18, ADAMTS20 protein expressions and expressions of HR, HER2, Ki67.
| ADAMTS8 | ADAMTS18 | ADAMTS20 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| + | − | r | P | + | − | r | P | + | − | r | P | |
| ER | ||||||||||||
| + | 146 | 30 | 157 | 19 | 111 | 65 | ||||||
| − | 89 | 13 | 0.087 | 0.153 | 90 | 12 | 0.172 | 0.004* | 90 | 12 | −0.078 | 0.2 |
| PR | ||||||||||||
| + | 131 | 27 | 141 | 17 | 118 | 40 | ||||||
| − | 104 | 16 | 0.035 | 0.562 | 106 | 14 | 0.135 | 0.026* | 83 | 37 | −0.069 | 0.193 |
| HER2 | ||||||||||||
| + | 151 | 28 | 156 | 23 | 131 | 50 | ||||||
| − | 84 | 15 | 0.139 | 0.037* | 91 | 8 | 0.036 | 0.576 | 70 | 27 | 0.027 | 0.712 |
| Ki67 | ||||||||||||
| + | 176 | 28 | 188 | 16 | 143 | 61 | ||||||
| − | 59 | 15 | 0.177 | 0.259 | 59 | 15 | 0.179 | 0.005* | 58 | 16 | 0.026 | 0.72 |
P<0.05 indicated statistical significance.
Expressions of ADAMTS8, ADAMTS18, and ADAMTS20 proteins in IDC
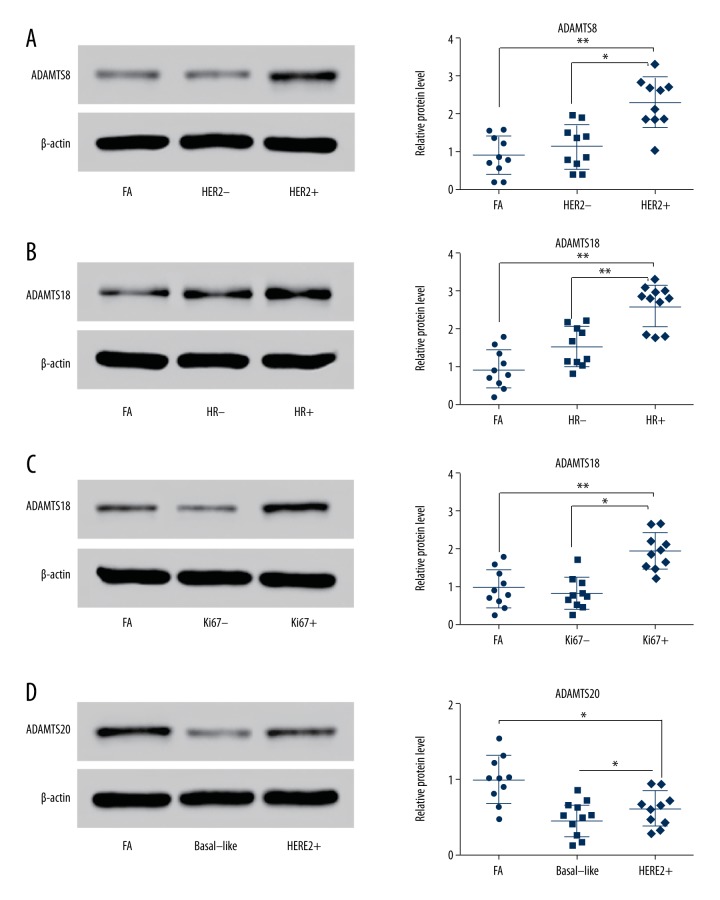
Western blot analysis was used to detect the variation in ADAMTS8, ADAMTS18, and ADAMTS20 protein expressions. For ADAMTS8, specific protein bands were observed in the FA group, HER2− group, and HER2+ group (each group contained 10 samples). The relative average grey values were significantly different (P<0.01, P<0.05, Figure 2A). For ADAMTS18, specific protein bands were observed in the FA group, HR− group, and HR+ group. The relative average grey value in the HR+ group was significantly higher than in the FA and HR− groups (P<0.01, P<0.01, Figure 2B). In addition, protein bands were observed in the FA group, Ki67+ group, and Ki67− group. The relative average grey values had statistically significant differences (P<0.01, P<0.05, Figure 2C). Finally, ADAMTS20 protein expression in the HER2+ group was significantly higher than in the basal subtype of IDC, but lower than in the FA group (P<0.05, P<0.05, Figure 2D).
Figure 2.
Western blot results of overexpression of ADAMTS8, ADAMTS18, and ADAMTS20 proteins in IDC tissue samples. (A) Specific ADAMTS8 protein bands were observed in the FA group, HER2− group, and HER2+ group. Overexpression of ADAMTS8 was positively correlated with the expression level of HER2. (B) Specific ADAMTS18 protein bands were observed in the FA group, HR− group, and HR+ group. Overexpression of ADAMTS18 was positively correlated with the expression level of HR. (C) Specific ADAMTS18 protein bands were observed in the FA group, Ki67− group, and Ki67+ group. Overexpression of ADAMTS18 was positively correlated with the expression level of Ki67. (D) Specific ADAMTS20 protein bands were observed in the FA group, basal-like group, and HER2+ group. The results indicated that ADAMTS20 in the HER2+ group was higher than in the Basal-like group, but lower than in the FA group.
Relationship between clinical pathological parameters and prognosis of IDC patients
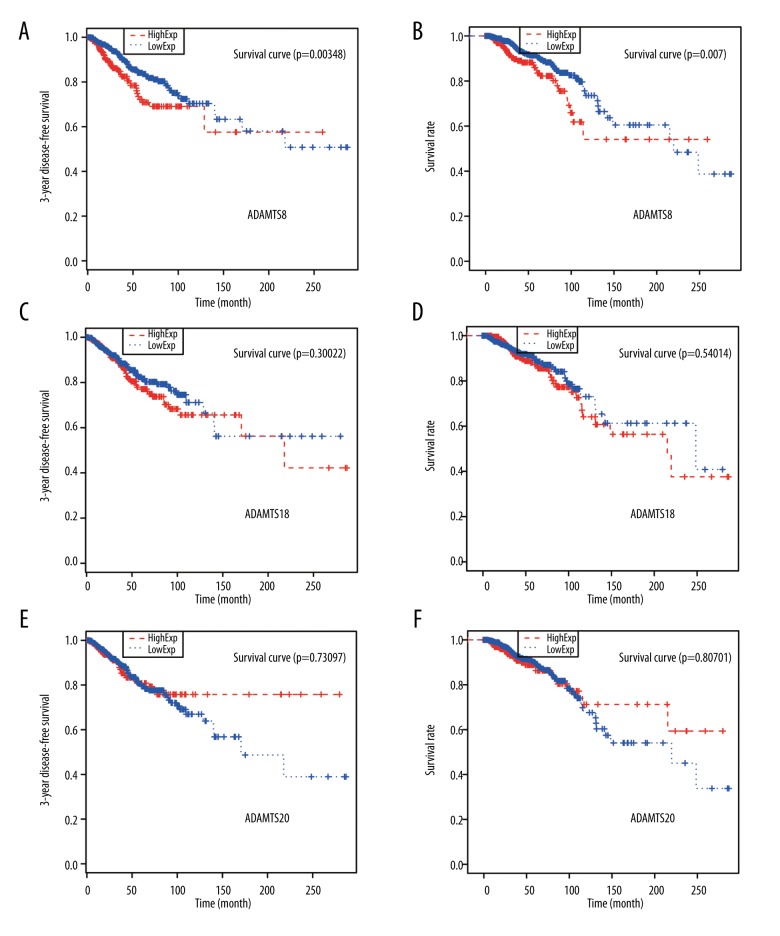
A higher ADAMTS8 level indicated worse DFS rate and OS rate (all P<0.01, Figure 3A, 3B). However, the levels of ADAMTS18 and ADAMTS20 were not significantly related to the postoperative DFS rate and OS rate (all P>0.05, Figure 3C–3F). In all clinical pathological parameters, lymphatic metastasis was the only factor that was significantly related to the postoperative DFS rate and the OS time (P<0.05, Table 5). The number of lymphatic metastases was inversely proportional to the total postoperative survival time of IDC patients. Lymph node metastasis was also correlated with OS rate (P<0.05, Table 6).
Figure 3.
Relationship between expressions of ADAMTS8, ADAMTS18, and ADAMTS20 and DFS rate and OS rate. (A, B) A high level of ADAMTS8 predicted poor DFS and OS rates. (C, D) The expression of ADAMTS18 was not significantly related to DFS or OS rate. (E, F) The expression of ADAMTS20 was not significantly related to DFS or OS rate. * P<0.05 indicates a significant difference.
Table 5.
The relationship between pathological parameters and disease-free survival rate and total survival time.
| Parameter | Case | Ratio (%) | 3-year disease-free survival rate | χ2 | P | Mean survival time (Month) | SE | P |
|---|---|---|---|---|---|---|---|---|
| Age (year) | ||||||||
| <50 | 63 | 42.9 | 90.5 | 29 | 1.3 | |||
| ≥50 | 62 | 57.1 | 87.1 | 0.408 | 0.523 | 29 | 1.3 | 0.813 |
| Tumor size (cm) | ||||||||
| ≤2 | 29 | 14.3 | 93.1 | 25 | 0.8 | |||
| >2 | 96 | 85.7 | 87.5 | 0.345 | 0.557 | 31 | 1.0 | 0.55 |
| Histological grade | ||||||||
| I | 12 | 7.1 | 91.7 | 34 | 1.7 | |||
| II–III | 113 | 92.9 | 88.5 | 0.304 | 0.581 | 28 | 20 | 0.257 |
| Number of lymph node metastasis | ||||||||
| 0 | 50 | 14.3 | 96.0 | 34 | 1.2 | |||
| 1≤L<4 | 43 | 21.4 | 93.0 | 30 | 0.7 | |||
| >4 | 32 | 64.3 | 71.9 | 14.643 | 0.001* | 26 | 2.8 | 0.004* |
P<0.05 indicated statistical significance.
Table 6.
The result of multivariate Cox regression analysis on clinicopathologic factors.
| Parameter | Overall survival (95%CI) | P |
|---|---|---|
| Age (year) | ||
| <50 | 1 | |
| ≥50 | 1.860 (0.348–9.943) | 0.468 |
| Tumor size (cm) | ||
| ≤2 | 1 | |
| >2 | 2.236 (0.554–10.942) | 0.236 |
| Histological grade | ||
| I | 1 | |
| II–III | 4.210 (0.075–2.369) | 0.327 |
| Number of lymph node metastasis | ||
| 1≤L<4 | 1 | |
| >4 | 16.231 (1.171–224.96) | 0.038* |
P<0.05 indicated statistical significance.
Discussion
This study elucidated the relationship between ADAMTS8/18/20 and breast IDC by studying the molecular subtypes and clinical pathological parameters of IDC patients. ADAMTS20 was distinctly different between HER2-enriched and basal subtype IDC. ADAMTS8 was associated with HER2 expression and ADAMTS18 was associated with HR (ER and PR) expression and Ki67 expression. ADAMTS18 indicated more a severe histological grade, and ADAMTS20 expression indicated a lower histological grade, but none of the 3 ADAMTS were correlated with lymph node metastasis. Lastly, a higher level of ADAMTS8 was correlated with worse prognosis, while ADAMTS 18 and ADAMTS 20 were not related to IDC prognosis.
Molecular subtypes are based on the expression of biological markers. Since breast cancer has many different patterns of metastasis, biological markers are used to distinguish the relevant variable biological features and clinical outcomes. The main subtypes taken into consideration were Luminal A, Luminal B+, Luminal B−, Basal, and HER2-enriched. Aure et al. [25] posited that Luminal A was the most frequent subtype found in breast cancer patients (approximately 40%). Phipps et al. [26] suggested that high hormone receptor expression is an important characteristic of Luminal A. Goldhirsch et al. [27] and Ejlertsen et al. [28] also found that both Ki67 and HER2 expressions in Luminal A were significantly reduced. Compared with other subtypes, Luminal A subtype was the most heterogeneous group with the best prognosis and lowest mutation rate [29,30]. Our study attempted to identify a correlation between ADAMTS and molecular subtypes, finding that ADAMTS8, ADAMTS18, and ADAMTS20 all had significantly higher rates of positive expression in Luminal A subtype (100%) than in other subtypes, indicating that the 3 ADAMTS family members chosen are correlated with these subtypes.
In the ADAMTS family, the members assessed were proven to have different levels of expression and various effects in breast cancer. In a study examining the expressions of all ADAMTS members in breast cancer [31], 7 (ADAMTS1, 3, 5, 8, 9, 10, and 18) were observed to be decreased in breast cancer. Reports on ADAMTS family members with roles in breast cancer have mainly concentrated on ADAMTS1 [32–36]. Lu et al. [36] found an overexpression of ADAMTS1 in nearly 40% of breast tumors, and also reported that ADAMTS1 had a positive correlation with the risk of bone metastasis. ADAMTS18 is usually regarded as a suppressor of breast cancer [37,38]. However, few researchers have undertaken studies on how ADAMTSs members affect IDC. Thus, our research focused on ADAMTS18 and 2 other rarely-studied ADAMTS members, analyzing the relationship between them and each clinical pathological parameter of IDC patients. However, expressions of both ADAMTS8 and ADAMTS18 were higher in IDC in the present study. In IDC, we observed that HER2 demonstrated a positive correlation with ADAMTS8, and HR and Ki67 were both positively correlated with ADAMTS18. Moreover, clinical pathological parameters were of great help in the prognostic analysis. In our findings, ADAMTS8 and lymphatic metastasis were both significantly correlated with the postoperative 3-year DFS rate and OS time.
However, the present study has certain limitations. Firstly, the small size of patients, especially the number of normal control patients (patients with FA), was not large enough to reveal other unseen differences. A small sample size may fail to clearly reflect the statistical significance of the relationships suggested. Our research also lacked cell experiments, and thus was unable to observe the direct functions of these 3 proteins involved in the development of IDC. We plan to address these issues in future research. In addition, the expression of ADAMTS8, ADAMTS18, and ADAMTS20 in lymph metastases areas should also be studied further to verify their lack of relationship with IDC tumor molecular classification and clinical pathological parameters. Nevertheless, in the present study, several previously unknown characteristics of ADAMTS family members were explored. We took ADAMTS20 into consideration and gained valuable insights into this.
Conclusions
In conclusion, all ADAMTSs examined in our research had high positive rates in Luminal A subtype. According to our findings, ADAMTS8 was associated with HER2 expression and ADAMTS18 was associated with HR (ER and PR) expression and Ki67 expression. ADAMTS18 indicated a more severe histological grade, and ADAMTS20 expression indicated lower histological grade, but none of these 3 ADAMTS were correlated with lymph node metastasis. In IDC patients, a high ADAMTS8 level predicted worse prognosis of IDC patients. These results may inspire more researchers to study IDC.
Footnotes
Conflict of interest
None.
Source of support: Departmental sources
References
- 1.DeSantis CE, Fedewa SA, Goding Sauer A, et al. Breast cancer statistics, 2015: Convergence of incidence rates between black and white women. Cancer J Clin. 2016;66:31–42. doi: 10.3322/caac.21320. [DOI] [PubMed] [Google Scholar]
- 2.Jia M, Zheng R, Zhang S, et al. Female breast cancer incidence and mortality in 2011, China. J Thorac Dis. 2015;7:1221–26. doi: 10.3978/j.issn.2072-1439.2015.05.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, et al. Global cancer statistics. Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Paryan M, Tavakoli R, Rad S, et al. Over-expression of NOTCH1 as a biomarker for invasive breast ductal carcinoma. 3 Biotech. 2016;6:58. doi: 10.1007/s13205-016-0373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moasser MM. The oncogene HER2: Its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene. 2007;26:6469–87. doi: 10.1038/sj.onc.1210477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stenehjem DD, Yoo M, Unni SK, et al. Assessment of HER2 testing patterns, HER2+ disease, and the utilization of HER2-directed therapy in early breast cancer. Breast Cancer. 2014;6:169–77. doi: 10.2147/BCTT.S69416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parise CA, Caggiano V. Breast cancer survival defined by the ER/PR/HER2 subtypes and a surrogate classification according to tumor grade and immunohistochemical biomarkers. J Cancer Epidemiol. 2014;2014:469251. doi: 10.1155/2014/469251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varga Z, Noske A, Ramach C, et al. Assessment of HER2 status in breast cancer: Overall positivity rate and accuracy by fluorescence in situ hybridization and immunohistochemistry in a single institution over 12 years: A quality control study. BMC Cancer. 2013;13:615. doi: 10.1186/1471-2407-13-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blows FM, Driver KE, Schmidt MK, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: A collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7:e1000279. doi: 10.1371/journal.pmed.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makki J. Diversity of breast carcinoma: histological subtypes and clinical relevance. Clin Med Insights Pathol. 2015;8:23–31. doi: 10.4137/CPath.S31563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai X, Li T, Bai Z, et al. Breast cancer intrinsic subtype classification, clinical use and future trends. Am J Cancer Res. 2015;5:2929–43. [PMC free article] [PubMed] [Google Scholar]
- 12.Hafeez F, Neboori HJ, Harigopal M, et al. Is Ki-67 expression prognostic for local relapse in early-stage breast cancer patients treated with breast conservation therapy (BCT)? Int J Radiat Oncol Biol Phys. 2013;87:344–48. doi: 10.1016/j.ijrobp.2013.05.052. [DOI] [PubMed] [Google Scholar]
- 13.Nishimura R, Osako T, Nishiyama Y, et al. Prognostic significance of Ki-67 index value at the primary breast tumor in recurrent breast cancer. Mol Clin Oncol. 2014;2:1062–68. doi: 10.3892/mco.2014.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldhirsch A, Ingle JN, Gelber RD, et al. Thresholds for therapies: highlights of the St Gallen International Expert Consensus on the primary therapy of early breast cancer 2009. Ann Oncol. 2009;20:1319–29. doi: 10.1093/annonc/mdp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filou S, Korpetinou A, Kyriakopoulou D, et al. ADAMTS expression in colorectal cancer. PLoS One. 2015;10:e0121209. doi: 10.1371/journal.pone.0121209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yi JM, Guzzetta AA, Bailey VJ, et al. Novel methylation biomarker panel for the early detection of pancreatic cancer. Clin Cancer Res. 2013;19:6544–55. doi: 10.1158/1078-0432.CCR-12-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J, Zhi Y, Chang X, et al. Expression of ADAMTS1 and its correlation with angiogenesis in primary gastric cancer and lymph node metastasis. Dig Dis Sci. 2013;58:405–13. doi: 10.1007/s10620-012-2379-x. [DOI] [PubMed] [Google Scholar]
- 18.Jin H, Wang X, Ying J, et al. Epigenetic identification of ADAMTS18 as a novel 16q23.1 tumor suppressor frequently silenced in esophageal, nasopharyngeal and multiple other carcinomas. Oncogene. 2007;26:7490–98. doi: 10.1038/sj.onc.1210559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelwick R, Desanlis I, Wheeler GN, Edwards DR. The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biol. 2015;16:113. doi: 10.1186/s13059-015-0676-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Apte SS. A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: Functions and mechanisms. J Biol Chem. 2009;284:31493–97. doi: 10.1074/jbc.R109.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunn JR, Reed JE, du Plessis DG, et al. Expression of ADAMTS-8, a secreted protease with antiangiogenic properties, is downregulated in brain tumours. Br J Cancer. 2006;94:1186–93. doi: 10.1038/sj.bjc.6603006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heighway J, Knapp T, Boyce L, et al. Expression profiling of primary non-small cell lung cancer for target identification. Oncogene. 2002;21:7749–63. doi: 10.1038/sj.onc.1205979. [DOI] [PubMed] [Google Scholar]
- 23.Masui T, Hosotani R, Tsuji S, et al. Expression of METH-1 and METH-2 in pancreatic cancer. Clin Cancer Res. 2001;7:3437–43. [PubMed] [Google Scholar]
- 24.Lu T, Dang S, Zhu R, et al. Adamts18 deficiency promotes colon carcinogenesis by enhancing beta-catenin and p38MAPK/ERK1/2 signaling in the mouse model of AOM/DSS-induced colitis-associated colorectal cancer. Oncotarget. 2017;8:18979–90. doi: 10.18632/oncotarget.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silver DL, Hou L, Somerville R, et al. The secreted metalloprotease ADAMTS20 is required for melanoblast survival. PLoS Genet. 2008;4:e1000003. doi: 10.1371/journal.pgen.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phipps AI, Buist DS, Malone KE, et al. Reproductive history and risk of three breast cancer subtypes defined by three biomarkers. Cancer Causes Control. 2011;22:399–405. doi: 10.1007/s10552-010-9709-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldhirsch A, Wood WC, Coates AS, et al. Strategies for subtypes – dealing with the diversity of breast cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22:1736–47. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ejlertsen B, Mouridsen HT, Jensen MB, et al. Cyclophosphamide, methotrexate, and fluorouracil; oral cyclophosphamide; levamisole; or no adjuvant therapy for patients with high-risk, premenopausal breast cancer. Cancer. 2010;116:2081–89. doi: 10.1002/cncr.24969. [DOI] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haque R, Ahmed SA, Inzhakova G, et al. Impact of breast cancer subtypes and treatment on survival: an analysis spanning two decades. Cancer Epidemiol Biomarkers Prev. 2012;21:1848–55. doi: 10.1158/1055-9965.EPI-12-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porter S, Scott SD, Sassoon EM, et al. Dysregulated expression of adamalysin-thrombospondin genes in human breast carcinoma. Clin Cancer Res. 2004;10:2429–40. doi: 10.1158/1078-0432.ccr-0398-3. [DOI] [PubMed] [Google Scholar]
- 32.Martino-Echarri E, Fernandez-Rodriguez R, Rodriguez-Baena FJ, et al. Contribution of ADAMTS1 as a tumor suppressor gene in human breast carcinoma. Linking its tumor inhibitory properties to its proteolytic activity on nidogen-1 and nidogen-2. Int J Cancer. 2013;133:2315–24. doi: 10.1002/ijc.28271. [DOI] [PubMed] [Google Scholar]
- 33.Freitas VM, do Amaral JB, Silva TA, et al. Decreased expression of ADAMTS-1 in human breast tumors stimulates migration and invasion. Mol Cancer. 2013;12:2. doi: 10.1186/1476-4598-12-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tyan SW, Hsu CH, Peng KL, et al. Breast cancer cells induce stromal fibroblasts to secrete ADAMTS1 for cancer invasion through an epigenetic change. PLoS One. 2012;7:e35128. doi: 10.1371/journal.pone.0035128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ricciardelli C, Frewin KM, Tan Ide A, et al. The ADAMTS1 protease gene is required for mammary tumor growth and metastasis. Am J Pathol. 2011;179:3075–85. doi: 10.1016/j.ajpath.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu X, Wang Q, Hu G, et al. ADAMTS1 and MMP1 proteolytically engage EGF-like ligands in an osteolytic signaling cascade for bone metastasis. Genes Dev. 2009;23:1882–94. doi: 10.1101/gad.1824809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu M, Lu T, Jing F, et al. ADAMTS-18 in the host tissues exerts little effect on breast tumor progress in a murine 4T1 breast cancer model. J Negat Results Biomed. 2016;15:2. doi: 10.1186/s12952-016-0045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nordgard SH, Johansen FE, Alnaes GI, et al. Genome-wide analysis identifies 16q deletion associated with survival, molecular subtypes, mRNA expression, and germline haplotypes in breast cancer patients. Genes Chromosomes Cancer. 2008;47:680–96. doi: 10.1002/gcc.20569. [DOI] [PubMed] [Google Scholar]