Hemidesmosomes are epithelial-specific cell-matrix adhesions stably anchoring the intracellular keratin network to the extracellular matrix and providing mechanical resilience to epithelia. The small GTPase Arf6 and caveolae are essential for their remodeling, notably in response to external mechanical cues.
Abstract
Hemidesmosomes (HDs) are epithelial-specific cell–matrix adhesions that stably anchor the intracellular keratin network to the extracellular matrix. Although their main role is to protect the epithelial sheet from external mechanical strain, how HDs respond to mechanical stress remains poorly understood. Here we identify a pathway essential for HD remodeling and outline its role with respect to α6β4 integrin recycling. We find that α6β4 integrin chains localize to the plasma membrane, caveolae, and ADP-ribosylation factor-6+ (Arf6+) endocytic compartments. Based on fluorescence recovery after photobleaching and endocytosis assays, integrin recycling between both sites requires the small GTPase Arf6 but neither caveolin1 (Cav1) nor Cavin1. Strikingly, when keratinocytes are stretched or hypo-osmotically shocked, α6β4 integrin accumulates at cell edges, whereas Cav1 disappears from it. This process, which is isotropic relative to the orientation of stretch, depends on Arf6, Cav1, and Cavin1. We propose that mechanically induced HD growth involves the isotropic flattening of caveolae (known for their mechanical buffering role) associated with integrin diffusion and turnover.
INTRODUCTION
The physical interaction of cells with their environment is essential for their function and the maintenance of tissue architecture and homeostasis. Cell adhesion to the extracellular matrix (ECM) provides a link and allows the cell to probe the physical and chemical properties of its microenvironment (Balaban et al., 2001; Riveline et al., 2001; Galbraith et al., 2002). Two classes of cell–ECM adhesion complexes, with their distinctive integrins and adaptor proteins, enable cell attachment and migration: focal adhesions (FAs) and epithelial-specific hemidesmosomes (HDs) (Kim et al., 2011; Hopkinson et al., 2014). While numerous reports have described how the polarized maturation of FAs depends on mechanical cues that promote the recruitment of further adaptor proteins (Moore et al., 2010; Parsons et al., 2010), our knowledge of the mechanisms controlling HD remodeling remains incomplete. In particular, the input of mechanical forces on their reorganization and maturation has not been investigated in vertebrates.
The HD-specific integrin α6β4 links components of the extracellular basement membrane, in particular the epithelial-specific laminin-332, to the intracellular plectin and BPAG1e adaptors, which in turn anchor the keratin cytoskeleton (Rezniczek et al., 1998; Litjens et al., 2005; Walko et al., 2011). Hemidesmosome-mediated adhesion to the extracellular basement membrane, coupled with the high deformability of the keratin cytoskeleton, protect the epithelial layer from mechanical strain (Herrmann et al., 2007; Osmani and Labouesse, 2015). Indeed, patients with defective HDs due to inherited (epidermolysis bullosa) or autoimmune skin diseases develop severe blistering syndromes (Sawamura et al., 2010; Iwata and Kitajima, 2013).
Vertebrate HDs undergo dynamic remodeling during keratinocyte migration and wound healing (Osmani and Labouesse, 2015). Biochemical and cellular analyses have highlighted the contribution of β4 integrin phosphorylation in regulating HD disassembly (Rabinovitz et al., 2004; Wilhelmsen et al., 2007; Germain et al., 2009; Frijns et al., 2010) (for reviews, see Litjens et al., 2006; Hopkinson et al., 2014; Walko et al., 2014; Osmani and Labouesse, 2015). In Caenorhabditis elegans, HD-like structures (CeHDs), which attach the epidermis to the cuticle apically and to an ECM shared with the underlying muscles basally, must also be remodeled during embryonic elongation (Zhang and Labouesse, 2010). We recently established that muscle contractions contribute to CeHD remodeling by inducing an epidermal mechanotransduction pathway that stimulates intermediate filament phosphorylation and reorganization (Zhang et al., 2011).
Vesicle trafficking has been recognized to play an important role in cell–cell junction remodeling (Le et al., 1999; Green et al., 2010), as well as in the turnover of cell–ECM adhesion complexes (Donaldson and Jackson, 2011; Margadant et al., 2011; Paul et al., 2015; Osmani and Labouesse, 2015). In particular, integrin endocytosis and recycling play an important role in cell migration and invasion, with a prominent role for Rab11+ or Arf6+ endosomes in integrin recycling (Donaldson and Jackson, 2011; Margadant et al., 2011; Paul et al., 2015), including for integrin α6β4 in breast carcinoma (Yoon et al., 2005). Other processes known to influence HD remodeling include the phosphorylation of the cytoplasmic moiety of the integrin β4 chain to regulate its binding to plectin (Litjens et al., 2006; Margadant et al., 2011; Osmani and Labouesse, 2015).
To identify the mechanisms controlling HD remodeling in vertebrates and assess how tension might impact on the process, we searched for trafficking regulators that colocalize with integrin α6β4 in vertebrate keratinocytes. We subsequently examined whether these regulators affect HD turnover in migrating cells and in cells submitted to mechanical tension. Their characterization led us to demonstrate that external mechanical cues can drive HD remodeling.
RESULTS
HD integrins colocalize with Arf6 and caveolin1
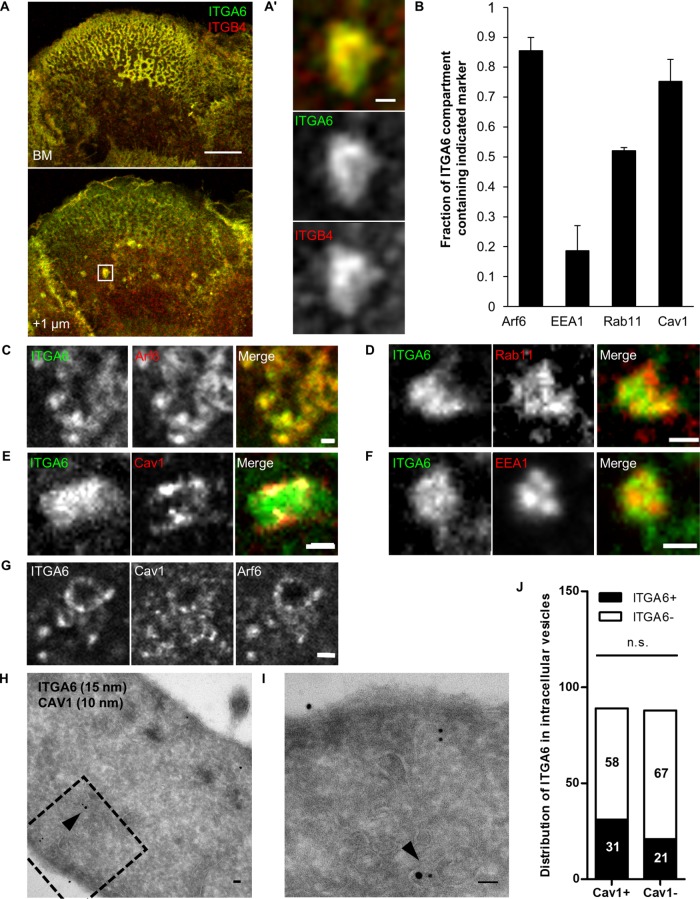
In primary keratinocytes or keratinocyte-derived cell lines such as the HaCaT cell line we used, α6β4 integrins are highly enriched at the basal plasma membrane in dense HD structures with a typical rosette-like morphology (Figure 1A). Hemidesmosomes integrins colocalize with the spectraplakin Plectin at the basal membrane, which is required to anchor keratin intermediate filaments (Supplemental Figure S1). We carefully examined α6β4 integrin localization in HaCaT keratinocytes and found that integrin α6 (ITGA6) and integrin β4 (ITGB4) were enriched in intracellular compartments (ICs) located roughly 1 µm above HDs (Figure 1, A and A′). Their size and distribution was consistent with the possibility they were endosomal compartments. To define their identity, we costained HaCaT cells with antibodies against ITGA6 and various endosomal markers or transfected cells with tagged Arf6 prior to staining (Figure 1B). We found that 85% ITGA6+ ICs were positive for the small GTPase Arf6, less than 20% of them contained the early endosome marker early endosome antigen-1 (EEA1), and more than 50% had the recycling endosome (RE) marker Rab11 (Figure 1, B–D and F, and Supplemental Figure S2). It suggests that they correspond, at least for a part of them, to this compartment, consistent with the known role of Arf6 in recycling (Aikawa and Martin, 2003; Powelka et al., 2004). Integrin-positive ICs also contained Cav1 (75%), a marker for caveolae (Ω-shaped membrane invagination of ∼100 nm in diameter) (Parton and del Pozo, 2013), previously associated with α6β4 in lipid rafts (Gagnoux-Palacios et al., 2003) (Figure 1, B and E). Significantly, ITGA6+/Arf6+ ICs were also always positive for Cav1 (Figure 1G). This result was further confirmed using immunogold labeling coupled to transmitted electron microscopy. We observed that ITGA6 and Cav1 were localized in the same ICs (Figure 1, H and I). We quantified the localization of ITGA6 and Cav1 in 177 internal vesicles from 30 different cells. We observed that 60% of ITGA6+ vesicles were also Cav1+ (31/52), whereas 46% of ITGA6- vesicles were Cav1+ (58/125; Figure 1J).
FIGURE 1:
Hemidesmosome integrins are located in Arf6-intracellular compartments containing Cav1. (A) Confocal plane of HaCaT cells at the level of the basal membrane (BM) or +1 µm above stained for ITGA6 and ITGB4. Scale bar 10 µm. (A′) Magnification of the area boxed in A. Scale bar = 1 µm. (B) Quantification of the various markers found at ITGA6-containing intracellular compartments. Number of ICs Arf6 = 46, EEA1 = 16, Rab11 = 22, Cav1 = 43 cells from three independent experiments. (C–G) Close-up on ICs in a confocal section +1 µm above the basal plasma membrane of cells coimmunostained for ITGA6 and (C) expressing EGFP-Arf6(wt); (D) for endogenous Rab11; (E) expressing RFP-Cav1(wt); (F) for endogenous EEA1; and (G) for endogenous Cav1 and expressing EGFP-Arf6(wt). Scale bar = 1 µm. (H, I) Immunoelectron micrographs of HaCaT cells after staining with antibodies against ITGA6 (15-nm gold beads) and Cav1 (10-nm gold beads) showing colocalization of ITGA6 and Cav1 in vesicles 500 nm above the plasma membrane (arrowhead). The dotted square in H shows the magnified region in I. Scale bar = 50 nm. (J) Quantification of the localization of ITGA6 and Cav1 in intracellular vesicles from EM micrographs (177 vesicles from 30 cells) from three independent experiments. Fischer test: ITGA6+ vs. ITGA6– in Cav1+ or Cav1– ICs.
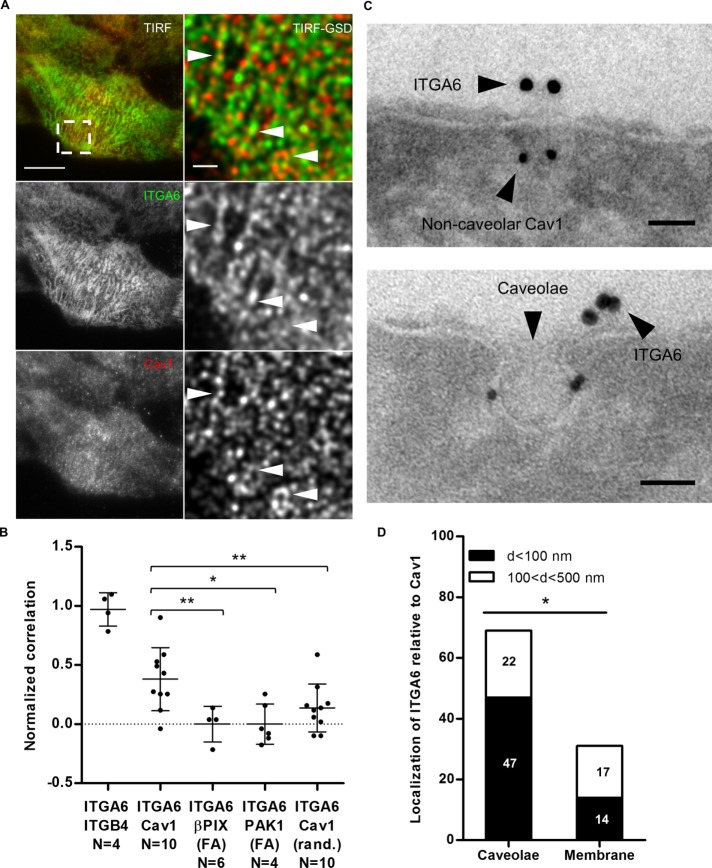
We used two parallel approaches to examine ITGA6 and Cav1 distribution at the plasma membrane (the most common localization of caveolae). Using total internal reflection fluorescence–ground-state depletion (TIRF-GSD) nanoscopy, we observed endogenous ITGA6 and Cav1 colocalization at the basal plasma membrane with a correlation coefficient of 0.48 (n = 10 cells). We normalized the correlation by comparing ITGA6/Cav1 colocalization frequency with that of ITGA6 and ITGB4, ITGA6 partner in HDs, as a positive control, and with that of βPIX or PAK1, two FA markers (Delorme-Walker et al., 2011; Kuo et al., 2011) taken as a negative control (Supplemental Figure S3). We found it to be significantly higher than the negatives controls (Figure 2, A and B). To further confirm, we randomized the GSD images of cells stained for endogenous ITGA6 and Cav1 and performed a similar analysis. We observed a ∼60% drop in the correlation between ITGA6 and Cav1, suggesting that these proteins colocalize at the basal plasma membrane (Figure 2, A and B). These observations were further confirmed by immunogold labeling coupled to electron microscopy showing that ITGA6 clusters were localized either close to or just at the edge of Cav1+ caveolae (Figure 2, C and D). To a lesser extent, ITGA6 was also localized close to noncaveolar Cav1 (Figure 2D). Finally, we observed that non-HD ITGA6 and Arf6 colocalized at the plasma membrane (Supplemental Figure S2). Collectively, our data strongly suggest that Arf6 and Cav1 colocalize with ITGA6 in internal compartments and that ITGA6 is localized in the same subdomains as Cav1 and Arf6 at the basal plasma membrane of keratinocytes.
FIGURE 2:
ITGA6 localizes close to caveolar and noncaveolar Cav1 at the basal membrane. (A) HaCaT cell immunostained for ITGA6 and Cav1. Left panel: TIRF image, scale bar = 10 µm; right panel: GSD-TIRF image (pixel size = 20 nm), scale bar = 1 µm. Arrowheads highlight colocalization. (B) Mean correlation of both channels measured in single GSD images for each pair of markers. Values are normalized to the mean correlation coefficient of ITGA6/ITGB4 GSD images used as positive control and compared with the correlation of ITGA6 at focal adhesions marked by βPIX or PAK1 taken as a negative control from a least four independent experiments. Mann-Whitney tests: R(ITGA6/Cav1) vs. R(ITGA6/βPIX(FA)), R(ITGA6/PAK1(FA)), or R (ITGA6/Cav1 randomized). (C) Immunoelectron micrographs of the plasma membrane of HaCaT cells after staining with antibodies against ITGA6 (15-nm gold beads) and Cav1 (10-nm gold beads). Scale bar = 50 nm. (D) Quantification of the localization of ITGA6 relative to caveolar and noncaveolar Cav1 (noted caveolae and membrane, respectively) from EM micrographs (N = 30) from three independent experiments. Fischer test: d < 100 nm vs. 100 < d < 500 nm in caveolae or at the membrane.
Arf6 and caveolin1 control HD biogenesis
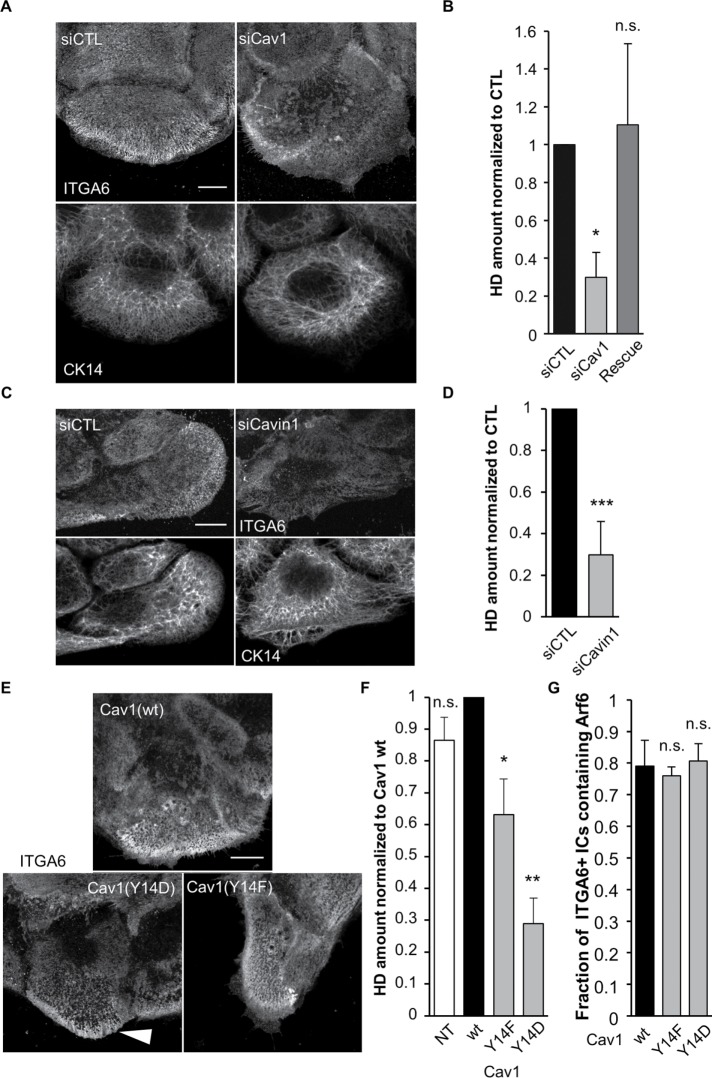
To assess whether Arf6 and Cav1 control HD biogenesis, we affected their function using small interfering RNAs (siRNAs) or mutant forms. Arf6 depletion induced a strong loss of HDs at the basal plasma membrane, associated with a thin and less dense IF network, which is indicative of partially defective HDs (Figure 3, A and B). Likewise, dominant-negative Arf6(T27N)-expressing cells reduced the HD area to the same extent as Arf6-depleted cells (Figure 3, C and D) and reduced the fraction of ITGA6/Arf6 double positive ICs (Figure 3E). The constitutively active Arf6(Q67L) form induced milder HD defects characterized by ITGA6 and Arf6 clustering centrally at the basal plasma membrane in 57/63 cells (Figure 3, D–E′), without any associated plectin or keratins (unpublished data). Moreover, these cells had the same fraction of ITGA6/Arf6Q67L as Arf6(wt)-expressing cells (Figure 3F), suggesting that HD integrin internalization was not affected but rather that the recycling step was overactivated, leading to HD disorganization. Importantly, nontransfected cells and Arf6-expressing cells had the same amount of HDs at the basal plasma membrane, suggesting that Arf6 overexpression did not affect HD organization (Figure 3D). These results thus suggest that Arf6 depletion and expression of Arf6 mutants affect HD integrin recycling.
FIGURE 3:
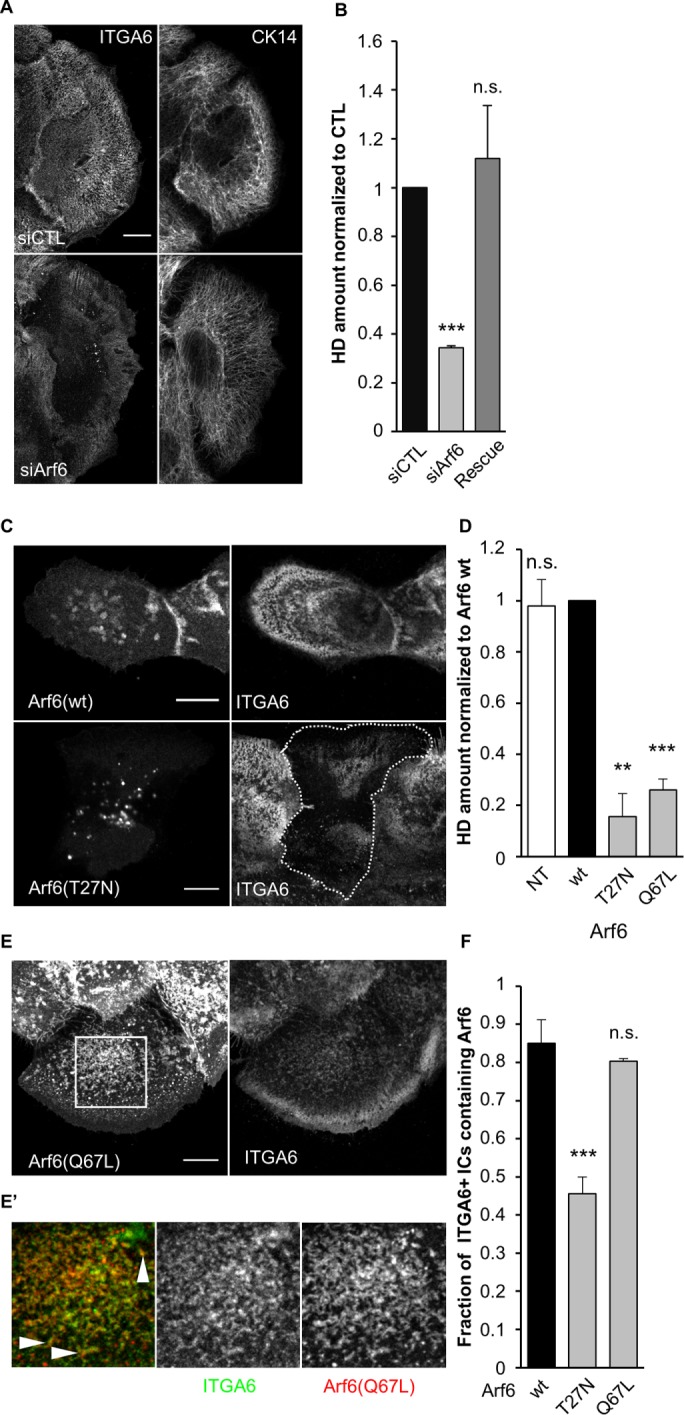
Arf6 is essential for proper HD organization. (A, B) Confocal images of the basal membrane and associated quantification of the amount of HDs (from ITGA6 staining) at the basal membrane of cells transfected with control or (A, B) or Arf6 siRNAs and then immunostained for ITGA6 and CK14 (A). The Rescue column (B) corresponds to cells transfected with Arf6 siRNAs plus a plasmid encoding wild-type RFP-Arf6 plasmid mutated to be Arf6 siRNA-resistant (Rescue). Scale bar = 10 µm. Data are from three independent experiments; number of cells = 40, 46, and 45. Student’s t tests: siCTL vs. siArf6 or Rescue. (C) Confocal images of the basal membrane of cells expressing EGFP-Arf6(wt) or (T27N) immunostained for ITGA6. Scale bar = 10 µm. (D) Quantification of the amount of HDs (from ITGA6 staining) at the basal membrane of cells expressing mutant forms of Arf6 from three independent experiments; number of cells = 37, 42, 61, and 63. Student’s t tests: Arf6(wt) vs. NT, Arf6(T27N). or Arf6(Q67L). (E) Confocal images of the basal membrane of cells expressing EGFP-Arf6(Q67L) immunostained for ITGA6 and CK14. Scale bar = 10 µm. (E′) Magnification of the boxed region in E (magnification ×3). Arrowheads highlight the colocalization of Arf6 (red) and ITGA6 (green) at the basal plasma membrane. (F) Quantification of the presence of Arf6 in ITGA6-enriched intracellular compartments in cells expressing mutant forms of Arf6 from three independent experiments; number of cells = 45, 44, and 46. Student’s t tests: Arf6(wt) vs. Arf6(T27N) or Arf6(Q67L).
Cav1 depletion also reduced HDs at the basal plasma membrane but did not affect keratins (Figure 4, A and B). Cav1 is a major caveolae component but also has a caveolae-independent membrane scaffolding function (Lajoie et al., 2009). To distinguish between both functions, we specifically affected caveolae by depleting Cavin1, another key caveolae component (Gambin et al., 2014). Cavin1 depletion also reduced HDs at the basal plasma membrane without affecting keratin organization (Figure 4, C and D). We also confirmed by transmission electron microscopy that Cav1- and Cavin1-depleted cells had significantly fewer caveolae at the basal plasma membrane (Supplemental Figure S4, A and B). It suggests that HD formation depends on caveolae rather than on the trafficking and signaling functions of Cav1. In other cell types, Cav1 function is regulated by Src-mediated Y14 phosphorylation (Parton and del Pozo, 2013). To test whether Cav1 phosphorylation alters its function in HD biogenesis, we expressed a phosphomimetic Cav1(Y14D) or a nonphosphorylatable Cav1(Y14F) mutant form (there was no significant difference between Cav1 wt-expressing cells and nontransfected cells; Figure 4F). Both forms significantly decreased HDs at the basal plasma membrane, with Cav1(Y14D) having stronger effects (Figure 4, E and F), without affecting ITGA+ Arf6+ ICs (Figure 4G). Conversely, we found that affecting Arf6 activity reduced Cav1 levels at the basal plasma membrane (Supplemental Figure S4, C–E) to an extent almost comparable to Cavin1 depletion, suggesting that Arf6 is also required to recycle Cav1 to the plasma membrane. This was further supported by the observation that Cav1 is transported in Arf6+ vesicles (Supplemental Video 1).
FIGURE 4:
Caveolae proteins are essential for proper HD organization. (A–D) Confocal images of the basal membrane and associated quantification of the amount of HDs (from ITGA6 staining) at the basal membrane of cells transfected with control (A, B) Cav1 siRNAs or (C, D) Cavin1 siRNAs then immunostained for ITGA6 and CK14 (A, C). The Rescue column (B) corresponds to cells transfected with Cav1 siRNAs plus a plasmid encoding wild-type RFP-Cav1 mutated to be Cav1 siRNA resistant. Scale bar = 10 µm. Data are from three independent experiments with (B) number of cells = 50, 65, and 46 and (D) N = 31 and 31. Student’s t tests: siCTL vs. (B) siCav1 or Rescue and (D) vs. siCavin1. (E) Confocal images of the basal membrane of cells expressing wt or mutant forms of RFP-Cav1 immunostained for ITGA6 and CK14. Scale bar = 10 µm. The white arrowhead highlights integrin accumulation at the edge of a Cav1(Y14D) expressing cell. (F) Quantification of the amount of HDs (from ITGA6 staining) at the basal membrane of cells expressing Cav1 mutant forms from three independent experiments; number of cells = 29, 59, 58, and 59. Student’s t tests: Cav1(wt) vs. NT, Cav1(Y14F), or Cav1(Y14D). (G) Quantification of the presence of Arf6 in ITGA6-enriched intracellular compartments in cells expressing mutant forms of Cav1 from three independent experiments; number of cells = 27, 38, 33. Student’s t tests: Cav1(wt) vs. Cav1(Y14F) or Cav1(Y14D).
Video S1.
Arf6/Cav1 vesicles are transported between intracellular organelles. Spinning disk confocal time lapse of a section +1 μm above the plasma membrane of a cell expressing EGFP-Arf6(wt) (green) and RFP-Cav1(wt) (red). Images were acquired every 250 ms. The movie is played at 20 frame/s.
Since Arf6 and Cav1 are also required for FA dynamics (Margadant et al., 2011; Paul et al., 2015), we examined whether HD defects would not indirectly result from FA defects. We found that affecting FA formation by inhibiting focal adhesion kinase (FAK) activity with PF-228 (Schaller, 2010) did not affect HDs (Supplemental Figure S5, A and B). Likewise, RNA interference–mediated knock-down of β1 integrin (ITGB1), one of the major adhesion receptor in keratinocyte FAs (Litjens et al., 2006; Hopkinson et al., 2014; Walko et al., 2014; Osmani and Labouesse, 2015), did not affect HDs (Supplemental Figure S5C). Altogether, we conclude that HD formation requires Arf6 and caveolae and that Arf6 acts upstream of Cav1.
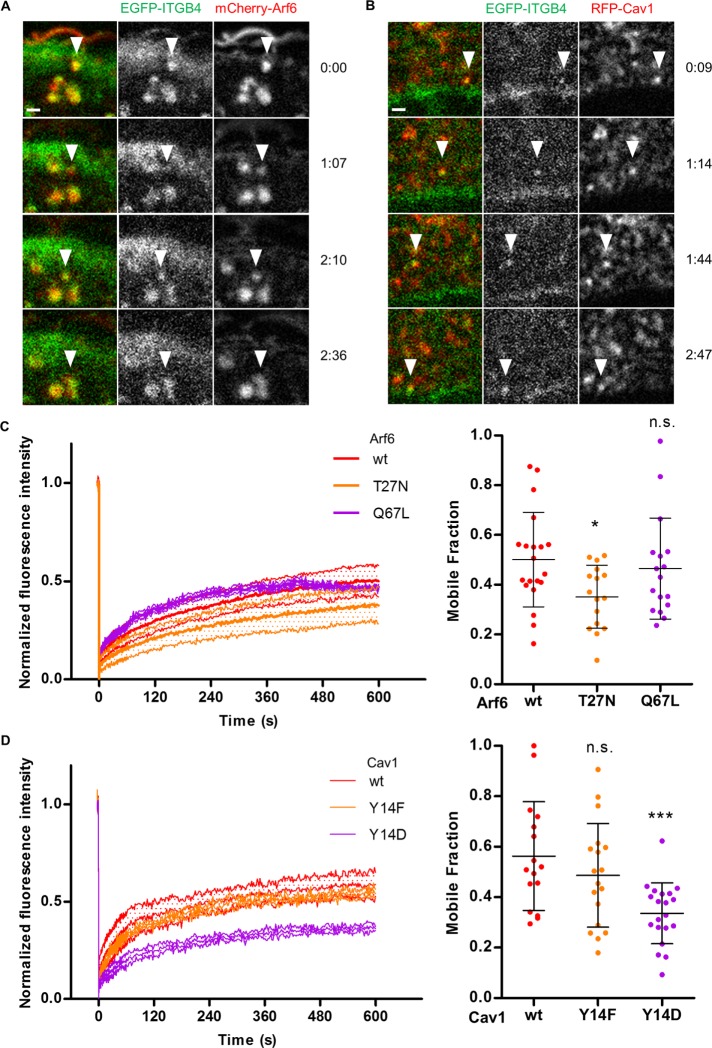
Arf6 and caveolae are essential for HD integrin dynamics
To further define the role of Arf6 and caveolae, we examined their function in integrin turnover and in integrin endocytosis assays. First, we used confocal video microscopy on cells expressing EGFP-β4 integrin (ITGB4) and mCherry-Arf6(wt) or RFP-Cav1(wt) to track Arf6-positive or Cav1-positive vesicles containing ITGB4. We saw that HD integrins were cotransported with Arf6 and Cav1 in such vesicles (Figure 5, A and B; Supplemental Videos 2 and 3). Second, we measured integrin dynamics using fluorescence recovery after photobleaching (FRAP) in HDs. In wild-type keratinocytes expressing EGFP-ITGB4, the integrin mobile fraction was ∼50% (Figure 5, C and D, and Supplemental Figure S6A). In keratinocytes expressing the dominant-negative Arf6(T27N) or the phosphomimetic Cav1(Y14D) mutant, the mobile fraction dropped to 36±7% or 34±2%, respectively (Figure 5, C and D). The constitutively active Arf6(Q67L) and the nonphosphorylatable Cav1(Y14F) mutants did not affect ITGB4 dynamics (Figure 5, C and D). Hence, affecting Arf6 activity or mimicking Cav1 phosphorylation compromises the effective turnover of HD integrins.
FIGURE 5:
Arf6-dependent traffic and Cav1 regulate HD integrin dynamics. Time-lapse acquisition of EGFP-ITGB4 (green) and (A) mCherry-Arf6(wt) (red) extracted from Supplemental Video 2 or (B) RFP-Cav1(wt) (red) extracted from Supplemental Video 3 (see arrowheads). Time is shown in seconds. Scale bar = 1 µm. (C, D) FRAP analysis of EGFP-ITGB4 in HDs of cells expressing (C) m-Cherry-Arf6 or (D) RFP-Cav1 constructs (MF: mobile fraction) from three independent experiments, number of cells (C) N = 24, 16, and 17 (D) and 16, 18, and 20. Mann-Whitney tests: (C) Arf6(wt) vs. Arf6(T27N) or Arf6(Q67L); (D) Cav1(wt) vs. Cav1(Y14F) or Cav1(Y14D). See Supplemental Figure S5.
Video S2.
ITGB4/Arf6 vesicles are transported between intracellular organelles. Spinning disk confocal time lapse of a section +1 μm above the plasma membrane of a cell expressing EGFP-ITGB4 (green) and mCherry-Arf6(wt) (red). Images were acquired every 250 ms. The movie is played at 20 frame/s.
Video S3.
ITGB4/Cav1 vesicles are transported between intracellular organelles. Spinning disk confocal time lapse of a section +1 μm above the plasma membrane of a cell expressing EGFP-ITGB4 (green) and RFP-Cav1(wt) (red). Images were acquired every 250 ms. The movie is played at 20 frame/s.
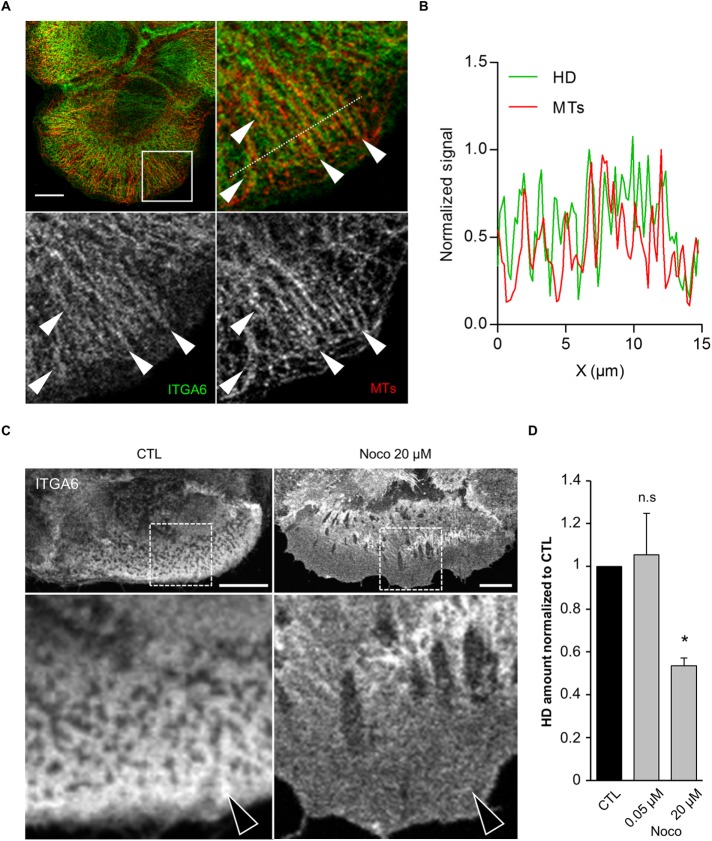
The directional motions of Arf6+/ITGB4+ and Cav1+/ITGB4+ vesicles suggest that they might move along microtubule (MT) tracks (Soldati and Schliwa, 2006). Consistent with this notion, ITGA6 often formed filamentous-like structures running parallel to MTs at the cell edge, presumably where MT tips are located (Figure 6, A and B). Furthermore, MT depolymerization with high doses of nocodazole (20 µM) severely affected HDs at the basal plasma membrane with a diffuse localization of ITGA6; doses that inhibit MT dynamics without compromising their structure (0.05 µM) had no effect (Figure 6, C and D) (Liao et al., 1995; Vasquez et al., 1997). This is also supported by our recent data showing that MTs are required to address the adhesion receptor LET-805 to CeHDs in C. elegans epidermal cells (Quintin et al., 2016).
FIGURE 6:
Microtubule-dependent traffic is required for HD maintenance. (A) Confocal plane of the basal membrane of a cell stained for ITGA6 (green) and microtubules (MTs, red) and close-ups of the boxed region (arrowheads highlight colocalization. Scale bar = 10 µm. (B) Normalized signal along the dashed line in the HD channel (ITGA6 staining, green) and MT channel (α-tubulin, red). (C) Cells were incubated with vehicle (CTL) or nocodazole (Noco) before fixation and immunostained for ITGA6. Scale bar = 10 µm. Arrows highlight the differences observed. (D) Quantification of the amount of HDs (from ITGA6 staining) at the basal membrane of cells treated with vehicle or the MT depolymerizing drug nocodazole from three independent experiments; number of cells N = 15, 16, and 11. Student’s t tests: CTL vs. Noco 0.05 µM or Noco 20 µM.
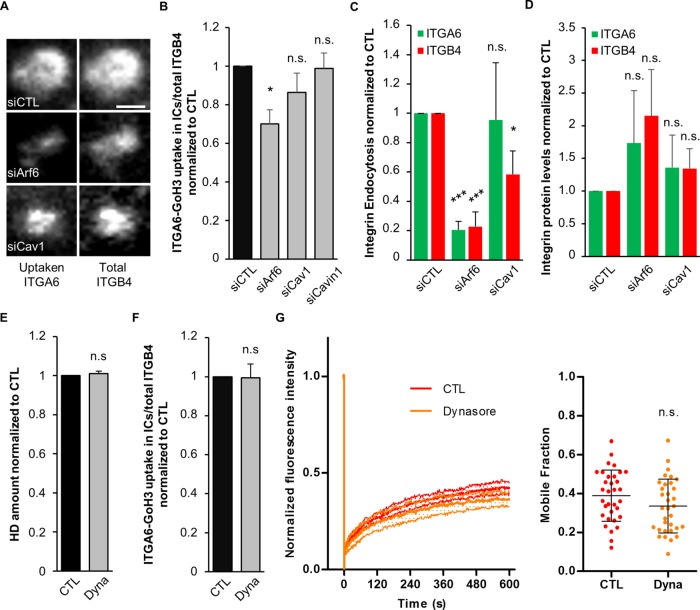
To define whether integrin turnover involves its trafficking between recycling vesicles and the plasma membrane, we next monitored the uptake of GoH3 antibodies, which recognize the extracellular domain of ITGA6. In control cells, we observed that ITGA6-bound GoH3 antibodies were localized in ITGB4+ ICs. In Arf6-depleted cells, ITGA6-GoH3 uptake was significantly reduced (Figure 7, A and B). By contrast, Cav1 or Cavin1 depletion had no effect on ITGA6 endocytosis (Figure 7B). We further confirmed these data using a biochemical quantification of the internalization of ITGA6 and ITGB4 based on the biotinylation of plasma membrane proteins coupled to a pulse chase experiment revealed by enzyme-linked immunosorbent assay (ELISA). Arf6 depletion significantly decreased the endocytosis of both ITGA6 and ITGB4, suggesting that Arf6 is a key regulator of HD integrin turnover. Once again, Cav1 depletion did not significantly affect ITGA6 endocytosis and only mildly reduced that of ITGB4, suggesting it may only affect ITGB4 subunits that are not incorporated within HDs (Figure 7C), as previously reported (Seltmann et al., 2015). We also quantified the amount of ITGA6 and ITGB4 at the protein level but did not observe statistically significant difference in Arf6- and Cav1-depleted cells, suggesting that their expression or degradation are not affected (Figure 7D). As outlined above, the mutation Cav1(Y14D) also strongly affected HD formation. Depending on the cell type, Cav1 Y14 phosphorylation can either drive dynamin-dependent caveolae internalization (del Pozo et al., 2005) or promote caveolar coat reorganization and caveolae swelling (Zimnicka et al., 2016). We examined integrin localization in Cav1(Y14D)-expressing cells and found that in 37/59 cells the HD remnants were present at the cell edge (Figure 4E), potentially coinciding with MT tips. In parallel, we found that dynamin inhibition using Dynasore did not affect either HD (Figure 7E), ITGA6 uptake (Figure 7F), or ITGB4 turnover, as measured by FRAP (Figure 7G), although it prevented the uptake of epidermal growth factor–bound (EGF-bound) epidermal growth factor receptor (EGFR) (Supplemental Figure S7), showing that HaCaT cells are sensitive to it. Thus, caveolin most likely does not directly modulate ITGA6 or ITGB4 trafficking, in contrast to what has been found for FA integrin recycling (Chao and Kunz, 2009; Ezratty et al., 2009) or caveolae internalization (Upla et al., 2004; Bass et al., 2011). One interpretation of these results accounting for both the decrease in ITGB4 mobile fraction observed in Cav1(Y14D) cells and the previously described effect of Cav1-Y14 phosphorylation could be that Cav1(Y14D) slows down HD integrin turnover to induce their accumulation at the cell edge.
FIGURE 7:
Hemidesmosome integrin endocytosis requires Arf6. (A, B) Antibody uptake assay after incubating HaCaT cells with the monoclonal antibody (mAb) GoH3 (anti-ITGA6) on ice and shifting cells to 37°C for 1 h. (A) Representative confocal images of ICs +1 µm above HDs in siRNA-transfected cells showing uptaken anti-ITGA6 antibody bound to the extracellular domain of ITGA6 (GoH3 – left panel) and total ITGB4 revealed by immunostaining of the endogenous protein after fixation (right panel). Scale bar = 1 µm. (B) Ratio of internalized GoH3-ITGA6/total ITGB4 measured in several ICs of individual cells for each condition following siRNA transfection from three independent experiments; number of ICs siCTL = 30/262, siArf6 = 27/185, siCav1 = 30/218, and siCavin1 = 25/196 cells/ICs. Student’s t tests: siCTL vs. siArf6, siCav1, or siCavin1. (C) Amount of endocytosed ITGA6 and ITGB4 integrins, following cell surface biotinylation at 4°C followed by internalization at 37°C for 30 min. The amount of biotinylated α6 and β4 integrins was measured by capture ELISA with peroxidase-conjugated streptavidin, corrected for the total amount of ITGA6 or ITGB4 measured by Western blotting; ratios are expressed relative to control cells. (D) ITGA6 and ITGB4 protein levels assessed by Western blotting in HaCaT cells transfected with control or anti-Arf6 or Cav1 siRNAs. (C, D) All data from four independent experiments. Student’s t tests: siCTL vs. siArf6 or siCav1. (E) Quantification of the area covered by HDs at the basal membrane (from ITGA6 staining) of cells treated with vehicle or the dynamin inhibitor Dynasore (Dyna) from three independent experiments; number of cells = 31 and 36. Studen’ts t test: CTL vs. Dyna. (F) Ratio of internalized GoH3-ITGA6/total ITGB4 measured in several ICs of individual cells for each condition following treatment with vehicle or Dynasore from three independent experiments; number of ICs CTL = 18/105, Dyna = 18/131 cells/ICs. Student’s t test: CTL vs. Dyna. (G) FRAP analysis of EGFP-ITGB4 in HDs of cells treated with vehicle or Dynasore from four independent experiments; number of cells = 33 and 33. Mann-Whitney test: CTL vs. Dyna.
We conclude that α6β4 integrin recycling is regulated though the Arf6-dependent dynamin-independent pathway and that functional caveolae are required for α6β4 integrin stabilization at the basal plasma membrane. These two steps are critical for HD formation and maintenance.
HD remodeling in response to mechanical stress depends on Cav1 and Arf6
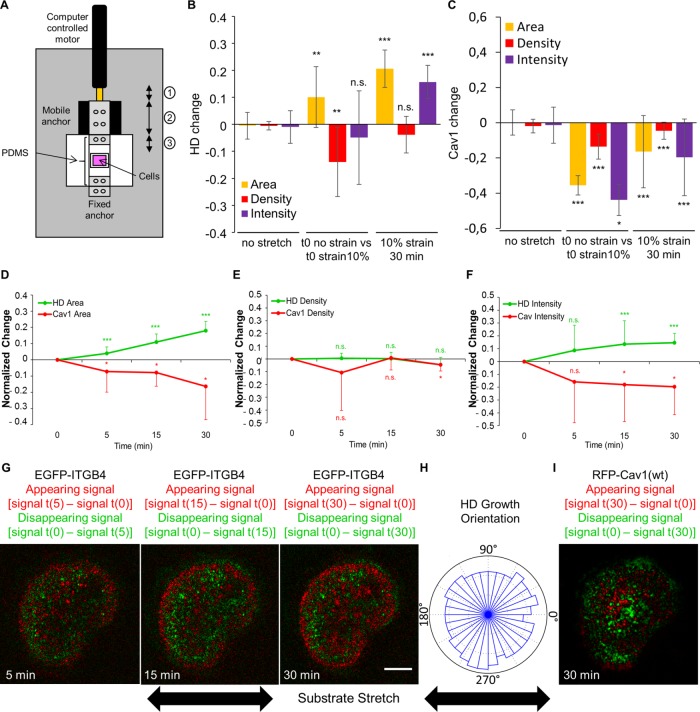
The results described so far identify a pathway promoting HD biogenesis. It might prove particularly necessary when tissues experience higher mechanical load such as during growth or wound healing. To directly assess whether HDs respond to mechanical cues, as in C. elegans (Zhang et al., 2011), we used a home-built stretching device compatible with confocal live imaging to apply a constant uniaxial 10% stretch on the cell substrate (Figure 8A). Immediately after stretch, HDs were passively stretched: the cell area increased by 10%, while HD density (HD integrin concentration at the membrane) decreased by 10% (Figure 8B and Supplemental Figure S8). In parallel, immediately after stretching, we observed a ∼40% decrease of Cav1 at the basal plasma membrane. Following this initial response, the HD amount increased by 20% over 30 min and maintained a constant density, suggesting a mechanically induced recruitment of HD integrins (Figure 8, A, B, and D–G, and Supplemental Video 4). Under strain, the increased HD amount correlated with a decrease of Cav1(wt) at the basal plasma membrane where HDs appeared (Figure 8, C–F and I, and Supplemental Video 4). Importantly, as outlined above, the mechanical challenge on HaCaT cells did not result in the complete disappearance of Cav1 (Figure 8C), whereas the effect of siCav1 results in a nearly complete absence of Cav1 (Supplemental Figure S9E). We take this observation to mean that a significant fraction of Cav1 (as well as caveolae) must be locally removed to make room for new HDs.
FIGURE 8:
Hemidesmosomes respond to external mechanical stimuli. (A) Scheme of the custom-built stretcher used. The motor motion (1) induces the displacement of the mobile anchor (2) driving the stretching of the PDMS membrane (3). (B, C) Quantification at the basal plasma membrane of the area (yellow), density (red), and total intensity (purple) changes of (B) EGFP-ITGB4 or (C) RFP-Cav1(wt) without strain (left), before and immediately after a 10% uniaxial strain (middle), or after 30 min of a 10% uniaxial strain (right), from 3–5 independent experiments; number of cells = 20, 12, and 12. Mann-Whitney tests: (no stretch) vs. (t0 no strain vs. t0 strain 10%) or (10% strain 30 min). (D–F) Quantification of the area (D) and total intensity (E) and density (F) of HDs (green) and Cav1 (red) in cells expressing EGFP-ITGB4 and RFP-Cav1(wt) after 5, 15, and 30 min of stretch with a 10% uniaxial strain. All values are normalized to time 0. Data are from three to five independent experiments; number of cells = 12. Mann-Whitney tests: 0 vs. 5 min, 15 min, or 30 min. (G, I) Confocal images of a cell expressing EGFP-ITGB4 (G) and RFP-Cav1 (I) under a 10% uniaxial strain after 5, 15, and 30 min. The green channel shows what disappears by subtracting from the signal in the first frame t(0) the signal at time t (5, 15, or 30 min): signal t(0) – signal t(x); the red channel shows what appears by subtracting from the signal at time t (5, 15, or 30 min) the signal in the first frame t(0): signal t(x) – signal t(0). Scale bar = 10 µm. (H) Quantification of the orientation of HD growth after 30 min of strain. Data are from three to five independent experiments; number of cells = 12. Images are related to Supplemental Video 4.
Video S4.
HD grow under mechanical strain. Spinning disk confocal time lapse of the basal plasma membrane plane of a cell expressing EGFP-ITGB4 (upper left/green) and RFP-Cav1 wt (upper right/red). Images were acquired every minute for 30 minutes. Bottom left shows HD appearing over time (see methods for details).
The analysis of the HD mechanoresponse over time showed a linear increase in HD amount, indicating an active process. We measured the speed of HD growth from all the movies (N = 12) acquired over a course of 30 min (1 image per min) and extracted a median speed of 1.5 ± 0.6 µm2/min. We also measured from our movies the density of integrin enrichment in HDs compared with integrins freely diffusing in the plasma membrane outside of HDs in the basal plasma membrane (i.e., close to the ECM) over the course of the 30-min 10% stretch by comparing between the initial frame immediately after the initiation of the substrate stretch and the frame after 30 min of stretch. We found that there is 2.6 ± 0.7-fold more integrins in the HD-enriched area of the basal plasma membrane compared with integrins diffusing in the membrane close to the ECM. Taking our FRAP data to estimate the diffusion coefficient (D) of integrins in HDs (Kang et al., 2012), we estimated a diffusion speed value of 1.4 ± 0.1 µm2/min (mean ± SEM, from four independent experiments, number of cells: N = 33). This suggests that the values of HD growth under external mechanical strain are consistent with a diffusion mechanism for integrin recruitment when the membrane is flattened. Interestingly, these values are also consistent with the reported measures of ∼1 µm2/min for β1 integrin diffusion and a twofold density enrichment in FAs compared with diffusing integrins close to the surface (Rossier et al., 2012). Finally, this is also consistent with the growth mechanism through membrane flattening and diffusion, first reported for cadherin-based adhesions (Delanoë-Ayari et al., 2004). In parallel, we measured the isotropy of HD growth: each cell was subdivided into quadrants of π/16 (i.e., ∼11.25°) and HD growth was measured in each quadrant. This revealed that, in contrast to FAs (Riveline et al., 2001) and adherens junctions (AJs), which grow along the direction of stretching (Brevier et al., 2008), the HD amount increased in all directions (Figure 8H), which suggests that HD increase depends on an isotropic structure such as caveolae modifying its organization (see Discussion).
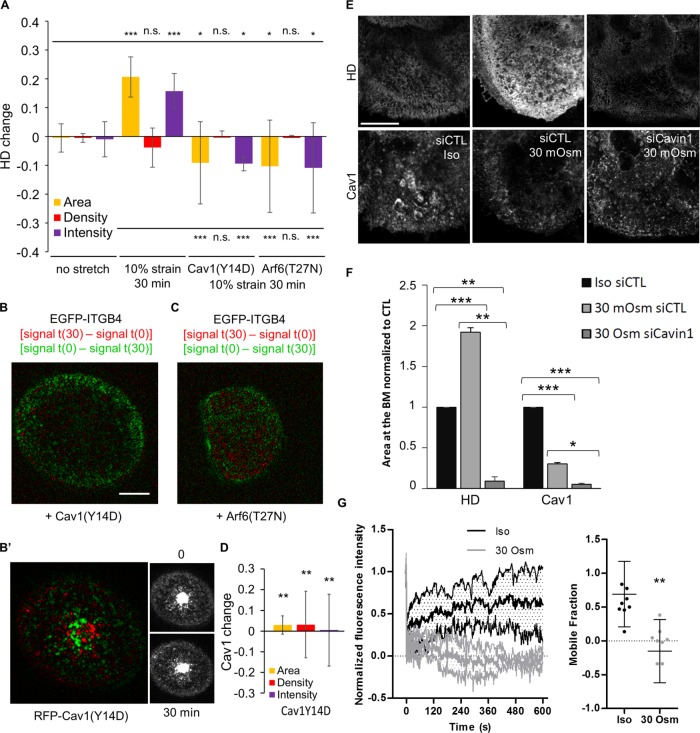
Since Arf6 and caveolae components were required for HD formation and integrin turnover, we tested their role in the mechanoresponse of HDs. In Cav1(Y14D)- or Arf6(T27N)-expressing cells, HDs almost disappeared, while Cav1(Y14D) was not removed (Figure 9, A–D), suggesting that the response of such cells to a mechanical challenge was defective. Recent results have suggested that caveolae can act as a mechanical buffer by flattening in response to the change in membrane tension, whether induced by cell stretching or a hypotonic shock (Sinha et al., 2011). To test whether HD growth could be connected to a similar mechanically induced caveolar flattening, we also used osmotic swelling as an alternate mechanical cue. We observed that a 10-fold decrease in osmolarity led to a twofold increase in HD area correlated with a twofold decrease in caveolae at the basal plasma membrane (Figure 9, E and F). This effect was dependent on caveolae integrity, since HDs did not respond to mechanical input in Cavin1-depleted cells (Figure 9, E and F). Finally, to assess whether mechanically induced HD growth relied on the Arf6-dependent recycling of HD integrins from the ICs, we measured the mobile fraction of EGFP-ITGB4 in either isotonic or hypotonic conditions (Supplemental Figure S6B). Focusing specifically on ICs, we observed a 69% recovery of EGFP-ITGB4 in cells grown in isotonic medium. By contrast, we failed to observe any recovery after a 10-min osmotic shock (Figure 9G). Although these results do not directly prove that Arf6 acts to recycle ITGB4 from ICs to the plasma membrane, a parsimonious interpretation is that ICs could not be replenished during a hypo-osmotic shock. Combined with the observation that Cav1 presence at the basal membrane depended on Arf6, this supports the notion that ICs serve as a reservoir for HD integrins used to shuttle a pool of free HD integrins from ICs towards the plasma membrane when cells are mechanically challenged. Altogether, our data strongly suggest that we have identified a novel trafficking-dependent pathway involved in mechanoresponse at HDs relying on Arf6 and Cav1.
FIGURE 9:
Cav1 and Arf6 are required for HD mechanoresponse to external forces. (A) Quantification of the area (yellow), density (red), and total intensity (purple) changes of HDs after 30 min without stretch in cells expressing EGFP-ITGB4 or after 30 min of a 10% uniaxial stretch in cells expressing EGFP-ITGB4 and RFP-Cav1(wt), RFP-Cav1(Y14D), or mCherry-Arf6(T27N) from three to five independent experiments; number of cells = 20, 12, 14, and 11. Mann-Whitney tests: upper bar (no stretch) vs. (10% strain 30 min), Cav1(Y14D) or Arf6(T27N); lower bar (10% strain 30 min) vs. Cav1(Y14D) or Arf6(T27N). (B, C) Representative confocal images of the basal membrane of a cell submitted to a 10% strain during 30 min expressing EGFP-ITGB4 and (B–B′) RFP-Cav1(Y14D) or (C) Arf6(T27N); image representation as in Figure 8G. (D) Quantification of the area (yellow), density (red), and total intensity (purple) changes of Cav1(Y14D) after 30 min of a 10% uniaxial stretch in RFP-Cav1(Y14D) from three independent experiments; number of cells = 14. Mann-Whitney tests: (10% strain 30 min) vs. Cav1(Y14D) (related to B′). (E) Confocal images of the basal membrane of cells transfected with control or anti-Cavin1 siRNAs and stained for ITGA6 and Cav1 in isotonic medium (300 mOsm) or after a 10-min hypotonic shock (30 mOsm). Brightness and contrasts are unmodified. Scale bar = 10 µm. (F) Quantification of the amount of HDs (from ITGA6 staining) and Cav1 at the basal membrane of cells in isotonic medium (Iso: 300 mOsm) or after a 10-min hypotonic shock (30 mOsm) in control cells or cells transfected with siCavin1 siRNAs to remove caveolae from three independent experiments; number of cells = 41, 32, and 30, respectively. Student’s t test Iso siCTL vs. 30 Osm siCTL or 30 Osm siCavin1 and 30 Osm siCTL vs. 30 Osm siCavin1. (G) FRAP analysis of EGFP-ITGB4 in the ICs of cells incubated with isotonic medium (300 mOsm) or after a 10-min hypotonic shock (30 mOsm) from three independent experiments; number of cells = 15 and 15. Mann-Whitney test: Iso vs. 30 Osm.
DISCUSSION
In this study, we characterize a pathway-mediating HD remodeling in keratinocytes during cell spreading and in response to mechanical tension. From a molecular standpoint, this pathway features a central role for Arf6 in shuttling HD integrins between presumptive recycling endosomes and the basal plasma membrane. From a mechanical standpoint, caveolae play a key role by allowing HDs to remodel and α6β4 integrin to bind the ECM, probably as further discussed below by flattening under stress (Figure 10). Our analysis of HD mechanoresponse reveals both similarities and differences with FA mechanoresponse to mechanical stress.
FIGURE 10:
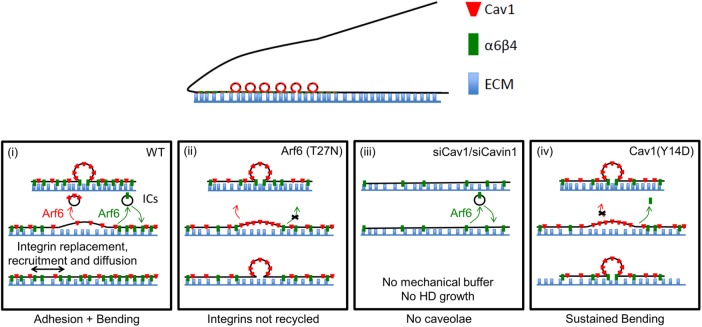
Proposed mechanism for the role of Arf6 and caveolae components in HD remodeling and mechanoresponse. (i) In wt cells, on transient mechanical challenge (external stretch or intracellular forces generated by actomyosin), caveolae are flattened, acting as mechanical buffers by providing extra plasma membrane that is then further colonized by HD integrins either from the plasma membrane by diffusion or through Arf6-dependent trafficking. When the mechanical input disappears, HD integrins may be recycled in an Arf6-dependent manner as part of the dynamic maintenance of HDs at the basal membrane. (ii) If Arf6 activity is compromised, then HD integrins are less recycled, and therefore HDs are less responsive to mechanical signals and unable to remodel, leading to compromised HD maintenance. (iii) In the absence of caveolae, the cells are unable to respond to mechanical challenges and HDs are not remodeled. This leads to poorly organized HDs with compromised maintenance. (iv) Prolonged Cav1 phosphorylation alters HD integrin dynamics and prevents proper HD remodeling in response to mechanical cues due to larger caveolae (as suggested by Zimnicka et al., 2016), which do not flatten as effectively. See Discussion for details.
Two lines of evidence support the notion that Arf6, a regulator of membrane trafficking (Donaldson and Jackson, 2011), acts upstream in HD formation. First, we observed HD integrins in large compartments containing Arf6 and the basal plasma, and we observed the movement of ITGB4+ Arf6+ vesicles. Second, we demonstrated that altering Arf6 activity affects ITGB4 turnover and HD formation. Third, Arf6 overactivation induced an abnormal accumulation of integrins at the basal plasma membrane. Fourth, although we did not directly observe ITGB4 trafficking from large compartments to the plasma membrane, we found that EGFP-ITGB4 fluorescence did not recover when ICs were photobleached after a hypotonic shock, suggesting that these compartments are a source of ITGB4 during HD spreading. Hence, our data together with the well-established role of Arf6 in integrin recycling (Donaldson and Jackson, 2011; Margadant et al., 2011; Paul et al., 2015) suggest that Arf6 regulates the endocytosis and the recycling of HD integrins and that HD formation requires a dynamic shuttling of α6β4 integrins.
We argue that Cav1, and presumably also Cavin1, act downstream of Arf6, since the latter was needed for Cav1 basal plasma membrane localization. We further argue that Cav1 and Cavin1 are required for HD integrin turnover through a mechanism that does not involve Cav1-mediated endocytosis. Indeed, depleting Cavin1, which has not been associated with caveolae-mediated endocytosis, affects integrin turnover in mechanical assays (see below) but not its uptake by endocytosis. Moreover, dynamin inhibition had no effect on HDs, nor on integrin uptake, suggesting that HD integrin endocytosis occurs through an Arf6-dependent recycling pathway (Doherty and McMahon, 2009) rather than through dynamin-dependent caveolae internalization as reported in some cell types (del Pozo et al., 2005). Last, expressing the phosphomimetic mutant Cav1(Y14D) reduced HD integrin mobility and induced their abnormal accumulation at the cell edge where MT tips are located. This abnormal accumulation could indicate that caveolae are indirectly required to enable HD integrin trafficking through an Arf6-dependent pathway. Interestingly, increasing the expression of Cav1 by a factor 2 in wt HaCaT cells (Supplemental Figure S9F) improved the recovery speed by almost twofold (half-time recovery: wt cells = ∼60 s and RFP-Cav1(wt) expressing cells = ∼35 s), which further supports our idea that caveolae are essential for HD turnover. Below, we propose a model to reconcile these data and account for the role of caveolae and Arf6 in HD remodeling.
One of our key findings is that HDs are mechanoresponsive, since they grow under external forces. We propose that the mechanism involved in this response relies on the mechanical stress buffer function of caveolae (Sinha et al., 2011) and on Arf6-dependent integrin recycling. As previously reported for FAs (Balaban et al., 2001; Riveline et al., 2001; Galbraith et al., 2002; Moore et al., 2010; Parsons et al., 2010) and adherens junctions (Delanoë-Ayari et al., 2004; Brevier et al., 2007, 2008), we found that HD intensity and area, but not density, increased under external forces, strongly suggesting that integrin recruitment depends on mechanical tension. However, whereas FA/AJ growth directions are set by actomyosin force orientation, HD integrin recruitment was not directional. Two arguments suggest that the nondirectionality of growth results from the isotropic flattening of caveolae. First, stretching was associated with an immediate decrease in integrin density (integrin concentration in HDs), suggesting that a novel area of plasma membrane devoid of integrin appears in close vicinity to the ECM and with Cav1 disappearance, indicating its unbinding from caveolae during flattening. Second, caveolae have a mostly “spherical” symmetry, and hence their flattening should be isotropic.
Biophysical studies have suggested that the size specificity of caveolae results from a competition between the bending energy induced by Cav1 and the large surface tension of the plasma membrane (Sens and Turner, 2004, 2006). We propose that, on stretching, caveolae are flattened and the new membrane facing the ECM can either return to its caveolar Ω shape, because the bending energy is favored, or remain flat if α6β4 integrins invade the newly available membrane by diffusion from basal plasma membrane or transport from RE and form new bonds thereby stabilizing new HD adhesions (Figure 10i). This mechanism is encoded in a competition between caveolar flattening and integrin/ECM adhesion energies. We further propose that HDs serve as pinning points in the vicinity of caveolae that must be removed by Arf6-dependent HD recycling i) to release adhesions and allow spreading and ii) to promote the formation of HD complexes at new adhesion sites (Figure 10i). Compromising Arf6 activity prevents such a recycling, thereby yielding to membrane bending and caveolae reformation (Figure 10ii). Likewise, we suggest that compromising with Cav1 or Cavin1 function (Figures 4, A–D, and 9E) denies cells from any mechanical buffer and prevents basal plasma membrane reorganization (Figure 10iii). Finally, expressing the phosphomimetic Cav1(Y14D) form may prevent Cav1 unbinding from caveolae and energetically favor their return to a caveolar Ω shape (Figure 10iv). This is further supported by recent findings suggesting that Cav1 phosphorylation, and hence the phosphomimetic mutant Cav1(Y14D), favors the transition of Cav1 oligomers toward monomers and induces caveolae swelling (Zimnicka et al., 2016). This would indeed account for a larger energy cost in flattening the caveolar Ω shape.
A recent study recently showed that microtubules associate with growing FAs in migrating HaCaT cells through MT-associated CLASPs that bind to the membrane-associated adaptor protein LL5β to FAs turnover (Stehbens et al., 2014). It was also documented that LL5β colocalizes with ITGA6 in mammary epithelial cells and that ITGA6 is required for the localization of LL5β at the basal plasma membrane (Hotta et al., 2010). Thus, it is tempting to speculate that MTs precede intermediate filaments anchoring in growing HDs and favor their growth by shuttling integrin from internal compartments. Keratins would then bind to immature HDs and stabilize them into more mature compact HDs.
Experimental considerations can also account for why growth is restricted to the cell edge over a region of ∼1 µm, where microtubule tips are also found, highlighting a transport-mediated local regulation of adhesion (Figure 6A). First, it has been proposed that the local competition between spreading of a spherical cell and adhesion triggers an increase in membrane line tension at the very edge (Bar-Ziv et al., 1999). Moreover, we previously showed that the cell cortex locally senses the force over an approximately 1-µm distance (Delanoë-Ayari et al., 2004), which is consistent with the HD growth from the cell border. Cav1(Y14D)-induced accumulation of HD integrins at the very edge (Figure 4E), suggesting the lack of caveolae mechanoresponse, prevents HD integrin reorganization and, consequently, HD growth. Finally, a recent superresolution mapping of HD components suggests that HDs forming at the edge of the cell are lacking BP180 and BP230 (so-called type II HDs) and might thus be easier to reorganize than type I HDs including BP180 (so-called type I HDs) (Nahidiazar et al., 2015). Altogether, the HD mechanical response appears to be mediated by the cortex in response to the local force.
Interestingly, HD remodeling in C. elegans also depends on a mechanical input (Zahreddine et al., 2010; Zhang et al., 2011), although it does not seem to feature a prominent role for ECM receptor trafficking as observed in vertebrates. Two important differences are that C. elegans neither forms caveolae (Kirkham et al., 2008), most likely due to the absence of a Cavin homologue in its genome, nor organizes its CeHDs with integrins (Osmani and Labouesse, 2015).
The phosphorylation of the ITGB4 cytoplasmic tail had been shown to be a major driver of HD disassembly (Rabinovitz et al., 2004; Wilhelmsen et al., 2007; Germain et al., 2009; Frijns et al., 2010), but the pathways regulating their reassembly had remained elusive until the present work. Collectively, our data support a mechanism whereby, in response to external mechanical cues, caveolae flattening promotes integrin mobilization to reinforce HDs in an isotropic mode.
MATERIALS AND METHODS
Plasmids and siRNAs
pcDNA3-ITGB4-EGFP was a gift from Arnoud Sonnenberg (NKI, Amsterdam, The Netherlands). pmCherry-Arf6 constructs were generated by subcloning pEGFP-Arf6 constructs into a pmCherry-N1 vector. All siRNAs are ON-TARGETplus SMARTpools and were purchased from Dharmacon. We confirmed that the siRNAs specifically depleted at least 75% of each targeted mRNA (Supplemental Figure S9). SMART pool siRNA-resistant human Arf6 and Cav1 cDNAs with silent mutations on siRNA-targeted regions were generated and cloned in pD673-CRc containing mCayenneRFP in C-terminal position by DNA2.0.
Antibodies
Antibodies against ITGA6 (rat monoclonal GoH3), ITGB4 (mouse monoclonal 7/CD104), Cav1 (mouse monoclonal 2297/Caveolin1 and polyclonal rabbit), Rab11 (mouse monoclonal 47/Rab11), and EEA1 (mouse monoclonal 14/EEA1) were purchased from BD Transduction Laboratories; against CK14 (rabbit polyclonal) from Convance; against Plectin (polyclonal guinea pig) from Progen; against Arf6 (rabbit polyclonal) and Cavin1 (rabbit polyclonal) from AbCam; against pY397-FAK (polyclonal rabbit) from Invitrogen; and against α-tubulin (monoclonal mouse DM1A). EGF and all secondary antibodies used in immunofluorescence staining are coupled to Alexa Fluor 488, 594, or 647 dyes and were purchased from Molecular Probes.
Drugs
FAK inhibitor PF-228 was purchased from Millipore and incubated with cells at 1 μM during 2 h before fixation. Nocodazole and Dynasore were purchased from Sigma and incubated with cells at 0.05 µM (low concentration), 20 µM (high concentration), and 80 µM, respectively, during 1 h prior to fixation.
Cell culture
HaCaT cells were obtained from the DKFZ (Heidelberg, Germany) and stocks were maintained at the IGBMC cell culture facility. Cells were mycoplasma tested every 4 mo. Cells were cultured in DMEM 1 g/l glucose (Invitrogen) supplemented with 10% fetal calf serum (PAN Biotech) and 10 μg/ml gentamicine (Kalys). For live imaging experiments, the medium was changed just before mounting cells on microscope slides with DMEM 1 g/l glucose supplemented with 10% fetal calf serum (FCS) and 10 μg/ml gentamicine without phenol red and supplemented with 20 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid).
Transfection
DNA plasmids were nucleofected in cells using Nucleofector device and Cell Line Nucleofector Kit V (Lonza) following the manufacturer’s instructions, and experiments were performed 24 h later. siRNAs were transfected into cells with Lipofectamine RNAiMAX (Invitrogen) following the manufacturer’s instructions, and experiments were performed 72 h later.
Immunofluorescence
Cells were fixed with paraformaldehyde (PFA) 4%, 10 min, at room temperature and permeabilized with phosphate-buffered saline– (PBS) 0.2% Triton for 10 min at room temperature. Aldehydes were reduced with PBS-NaBH4 for 10 min at room temperature. Primary antibodies diluted in PBS were incubated for 1 h at room temperature and corresponding secondary antibodies diluted in PBS were incubated 30 min at room temperature.
Confocal microscopy
All images from fixed samples were acquired with a TCS SP5 confocal microscope equipped with a HCX PL APO 63× NA 1.4 oil objective (Leica). All data sets were acquired in a row with the same settings.
HD and Cav1 image analysis
Single plane images corresponding to the plasma membrane were analyzed using the ImageJ. To quantify the area occupied by HDs, briefly, a region of interest (ROI) was manually drawn around each individual cell to isolate them. Control cells (control plasmids or siRNA) were used to determine a detection threshold. The same threshold was applied on the rest of the sample (mutant constructs or siRNAs). For each cell, the areas of the whole cell and the thresholded signal at the basal membrane were measured to determine a ratio (thresholded signal area)/(cell area). Values were then normalized to values for control cells. Each experiment was reproduced at least three times independently with a total sample number of at least 30 cells.
Anti-ITGA6 uptake
Cells were washed twice with ice-cold medium and incubated with anti-ITGA6 antibody (GoH3) at 1 µg/ml on ice for 30 min before two additional washes with ice-cold medium. The internalization of ITGA6-bound antibodies was induced by adding prewarmed medium and incubating cells at 37°C for 1 h. After fixation, permeabilization, and PFA reduction, ITGA6-bound antibodies were revealed with an Alexa Fluor 488-conjugated anti-rat antibody, and cells were immunostained for total ITGB4 with mouse monoclonal anti-ITGB4 and Alexa Fluor 647-conjugated anti-mouse antibody. Single confocal images at 1 µm above the basal membrane were acquired. ITGA6 internalization was measured using a custom-made macro in ImageJ software by drawing ROIs around individual ICs in each cell. The total intensities in the ITGA6 and ITGB4 channels were measured in each IC of individual cells for each condition. Internalized ITGA6 was normalized to total ITGB4 as the ratio [ITGA6 total intensity]/[ITGB4 total intensity] in each IC.
Biochemical measure of α6 and β4 integrin internalization
HD integrin endocytosis was measured biochemically by adapting the protocol described in Roberts et al. (2001). Cells were washed twice in cold PBS and their surface was labeled with 0.2 mg/ml NHS-SS-biotin in PBS for 30 min at 4°C. Cells were washed twice in cold PBS, and surface protein internalization was induced by adding prewarmed medium and incubating cells at 37°C for 30 min. After the medium was removed, cells were washed twice in cold PBS. Biotin was removed from proteins remaining at the cell surface by incubating the cells in 50 mM Tris, pH 8.6, 100 mM NaCl, and 20 mM MesNa for 15 min at 4°C. MesNa was quenched by adding 20 mM of iodoacetamide for 10 min at 4°C. Cells were lysed in 75 mM Tris, pH 8.6, 0.2 M NaCl, 7.5 mM EDTA, 7.5 mM EGTA (ethylene glycol-bis(β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid), Igepal CA-630 0.75%, Triton X-100 1.5%, and Complete Protease inhibitor cocktail (Roche). Lysates were fractioned with a 27G needle and centrifuged at 10,000 × g 4°C for 10 min. The amount of biotinylated integrins α6 and β4 were measured by capture ELISA. Briefly, COSTAR 96-well High Bind EIA/RIA plates (Corning) were coated with each anti-integrin antibodies separately at 5 µg/ml in 50 mM Na2CO3, pH 9.6, overnight at 4°C. The plate was blocked with PBS–0.05% Tween–5% bovine serum albumin (BSA) for 90 min at room temperature. Integrins α6 and β4 were captured separately in duplicates by adding 50 µl lysates in each well for 2 h at room temperature under gentle agitation. Plates were extensively washed with PBS–0.05% Tween. Biotin was revealed by incubating wells with horseradish peroxidase-conjugated streptavidin (Pierce) in PBS–0.05% Tween–1% BSA for 1 h at room temperature. After washing, integrin biotinylation was measured by a chromogenic reaction with 3,3′,5,5′-tetramethylbenzidine (TMB) ELISA substrate (Interchim). The amount of α6 and β4 integrins in each sample was measured by Western blotting to then normalize ELISAs.
EGF uptake
Cells were starved overnight with serum-free medium. Cells were washed twice with ice-cold medium and incubated with EGF coupled to Alexa Fluor 488 (Thermo Fischer) at 2 µg/ml in serum-free medium containing the vehicle or Dynasore (± Dynasore) on ice for 1 h min. Cells were then either washed two times with ice-cold serum-free medium ± Dynasore and fixed as a time 0 (t = 0) or washed two times with serum-free medium ± Dynasore preheated at 37°C. The internalization of EGF was induced by incubating cells at 37°C for 1 h. Cells were then fixed (t = 10 min). Cells were then processed for immunostaining for endogenous EEA1.
Superresolution microscopy and image analysis
Samples were immunostained as mentioned above using Alexa Fluor 488- and 647-conjugated secondary antibodies in glass-bottom dishes (μDish, 35 mm high; ibidi) and set in a 10 mM MEA, 25 mM HEPES in PBS. All images were acquired with a Leica SR GSD TIRF microscope equipped with a HCX PL APO 100× NA 1.47 oil objective and a 1.6× lens and a iXON ULTRA DU897V electron-multiplying charge-coupled device (EM-CCD) camera (Andor). Superresolution images were reconstructed from GSD raw images using a nonparametric kernel smoothing custom-made method with ImageJ software. Colocalization was measured using MATLAB (Mathworks) with the Pearson’s correlation coefficient defined as
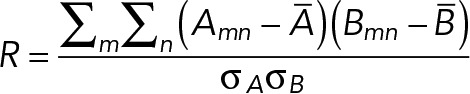 |
where ·– is the average and σ. the SD. For βPIX and PAK1, the correlation was measured specifically in FAs and isolated with a specific ROI around them and in the rest of the cell. The results were normalized to the correlation value of the positive control ITGA6/ITGB4. To randomize images, we used the TransformJ plug-in (https://imagescience.org/meijering/software/transformj/). In brief, numbers were randomly chosen between 10 and 50 pixels to translate images along the x and y axes using the interpolation cubic B-spline. Images were further processed as previously described.
Live imaging and FRAP experiments
Cells were seeded on glass-bottom dishes (μDish, 35 mm high; ibidi). Time-lapse acquisitions were performed on cells expressing EGFP-ITGB4(wt) and RFP-Arf6 or Cav1(wt or mutants) with an inverted Zeiss Axio Observer Z1 microscope equipped with a PLAN APO 100× NA 1.4 oil objective (Zeiss), a Yokogawa CSU-X1 confocal head, an Evolve 512 EM-CCD camera (Photometrics), an iLAS FRAP illumination module (Roper Scientific), and a 37°C incubation chamber (Pecon). The hardware was controlled with the MetaMorph software (Molecular Devices). For classical time-lapse acquisitions, 300–600 sequential dual color images of a single z-plane 1 μm above the basal membrane were acquired every 250 ms with 491- and 561-nm lasers. For FRAP experiments, single color acquisition with a 491-nm laser was performed on the basal membrane plane. Twenty-five prebleach frames were acquired at a rate of 5 frames per second (fps). A 30-pixel circle region in the basal membrane was bleached by a double laser pulse without acquisition. Fluorescence recovery was followed by a stepwise acquisition of 2 s at a rate of 10 fps, then 1 min at 2 fps, and, finally, 9 min at 0.5 fps. The nonspecific decay of the signal intensity in the bleached region due to the imaging process was corrected by the fluorescence decrease of the whole cell at each time point. The curves obtained were normalized so that the first postbleach frame was set to zero and were then subject to curve fitting.
FRAP quantifications were made using a standard modeling with chemical dominant interactions (Phair et al., 2004). First, raw intensity measurements were normalized using a double normalization to take into account the photobleaching and the background intensity. After the normalization, the intensities before and at the bleach time should reach, respectively, 1 and 0.
The recovery was estimated using the single exponential modeling
The parameters of this equation have been estimated using a nonlinear fitting within MATLAB (Mathworks) with the constraint I(0) = 0.
Once the parameters are estimated, the mobile fraction and the half-time recovery are given by
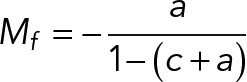 |
Each experiment was reproduced at least three times independently with a total sample number of at least 15 cells. Dynasore and hypotonic medium were added 2 h and 10 min before the experiment, respectively.
Stretching experiments
Briefly, membranes and stencils were fabricated using polydimethylsiloxane (PDMS). PDMS (Sylgard 184 Silicon Elastomer; Dow Corning) polymer was prepared by mixing the base prepolymer with the cross-linking agent in a 10:1 ratio, and vacuum atmosphere was used for 1 h to remove gas bubbles from the mixture. The polymer was then poured over a plastic Petri dish and spin coated (Laurell WS-650-23B, Laurell Technologies) at 1000 rpm to achieve a 100-μm-thick layer. Membranes were then cured for 12 h at 65°C. An analogous procedure was used to prepare the stencils: prepolymer was prepared at 10:1 ratio, degassed, poured over a Petri dish with a thickness of 1 mm, and cured at 65°C for 12 h. Membranes (5 × 2.5 cm) and stencils (2 × 2 cm) were cut off. Membranes were placed on a glass slide with parafilm. Stencils with vacuum grease (Dow Corning) on the bottom were placed on top of membranes. The setup was UV sterilized under the cell culture hood, and membranes were coated with poly-d-lysine, mol wt 70,000–150,000 10 μg/ml, in PBS for 1 h at 37°C. Transfected cells were seeded on membrane and left to adhere for 24 h. Imaging was performed on cells expressing EGFP-ITGB4(wt) and RFP-Arf6 or Cav1(wt or mutants) using an inverted Nikon Eclipse Ti equipped with HCX PL APO 63× NA 1.2 water objective (Leica), a Yokogawa CSU-X1 confocal head, an Evolve 512 EM-CCD camera (Photometrics), and a custom-made 37°C incubation chamber driven by MetaMorph software (Molecular Devices). Membranes were mounted on a custom-built cell-stretcher so that the membrane remained perfectly flat at the beginning of the experiment. An additional 10% uniaxial strain was applied on the cell stretcher by a computer controlled motor (Thorlabs). An image of each individual cell was taken at resting state before applying the strain, and a time-lapse was run at a rate of one image per minute during 30 min under step stretch conditions.
HD mechanoresponse analysis
Single plane images corresponding to the plasma membrane were analyzed using ImageJ software. To quantify the area occupied by HDs, an ROI was manually drawn around each individual cell. For each cell, the first frame was used to determine the detection threshold. The same threshold was applied to the other frames.
HD growth and Cav1 disappearance between frames x and y was calculated as follows:
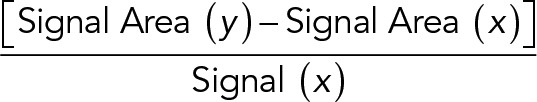 |
HD growth speed over 30 min was calculated as follows:
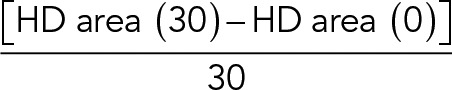 |
The final HD growth speed was computed as the median value of all individual cell values.
The orientation of HD growth was quantified using MATLAB (Mathworks). The appearing HD signal was segmented for each individual cell. Each cell was then centered on a polar coordinate system. Cells were subdivided in angular quadrants of π/16 radians. Angular histograms were computed as the proportion of overall intensity located in this angular portion. The final angular histogram is computed as an average of all cell’s angular histograms.
Osmotic shock
Cells were incubated for 10 min at 37°C with normal medium or with medium diluted in deionized water (1:9 for a 30 mOsm hypotonic shock). Cells were then fixed and stained with antibodies specific for ITGA6 (HDs) and for Cav1 (caveolae), and their respective amount at the basal membrane was either quantified from confocal images or used for live cell FRAP experiments as previously described.
Transmission electron microscopy
Cells transfected in six-well plates were fixed for 1 h at room temperature with 2.5% glutaraldehyde and washed in 0.1 M Na Cacodylate buffer. Cells were then postfixed in 1% OsO4 for 30 min at 4°C and washed with 0.1 M Na Cacodylate buffer and, finally, with deionized water. Samples were secondary postfixed in 4% water solution of uranyl acetate for 30 min at room temperature. The samples were stepwise dehydrated in ethanol (70% 20 min, 3 × 95%, 3 × 100% each 15 min) and infiltrated in diluted Epon (ethanol/Epon = 1/1) for 1 h. Samples were left in absolute Epon (EmBed812) overnight. The following day, the samples were placed in a fresh absolute Epon for 1 h and left to polymerize at 60°C for 24–48 h. Once polymerized, samples were reoriented and reembedded in empty epon blocks. Thin sections (100 nm) were sectioned using ultramicrotome (Leica Ultracut UCT), collected on formvar-coated slot grids and contrasted with 4% water solution of uranyl acetate for 10 min and Reynolds lead citrate for 3 min. Thin sections (100 nm) were imaged with a CM120 transmission electron microscope (Philips Bio Tween) operating at 120 kV. Images were recorded with a Veleta 2 k × 2 k (Olympus-SIS) camera using the iTEM software. Caveolae were manually counted on at least 20 pictures of a region of 3–4 µm of plasma membrane in 20 different cells.
Immunoelectron microscopy
Cells were grown to confluence in 75-cm2 flasks and chemically fixed with 2% paraformaldehyde + 0.2% glutaraldehyde in 0.1 M Na Cacodylate buffer for 30 min at room temperature and then fixed again with fresh fixative for 1 h at room temperature. Cells were collected over five steps of centrifugation at 2500 rpm and resuspended in 1% glycin in PBS. The supernatant was replaced by liquid gelatin, and cells were centrifugated for 3 min at 14,000 rpm and incubated for 30 min on ice. Samples of size 1 mm2 were incubated overnight in 2.3 M saccharose at 4°C under gentle agitation. Samples were cryo-fixed in liquid nitrogen, and ultrathin sections (∼60 nm) were cut with an ultramicrotome (Leica EM FCS). Samples were washed with PBS and then 1% glycin in PBS, saturated in 1% BSA in PBS for 15 min at room temperature, and incubated with Cav1 antibodies (BD rabbit polyclonal) at dilution 1:50 in 1% BSA in PBS for 30 min. Samples were washed in 1% BSA in PBS, incubated with protein A coupled to 10 nm gold beads for 15 min at room temperature, and washed with 1% BSA in PBS and then with PBS. Samples were fixed in 1% glutaraldehyde in PBS for 5 min at room temperature and washed in PBS and then 1% glycin in PBS. Samples were saturated in 1% BSA in PBS for 15 min and incubated with ITGA6 antibodies (BD rat monoclonal) at dilution 1:50 in 1% BSA in PBS for 30 min, washed in 1% BSA in PBS, incubated with anti-rat antibodies at dilution 1:200 in 1% BSA in PBS for 15 min at room temperature, washed with 1% BSA in PBS, incubated with protein A coupled to 15 nm gold beads for 15 min at room temperature, and washed with 1% BSA in PBS and then PBS. Samples were fixed in 1% glutaraldehyde in PBS for 5 min at room temperature, washed in PBS and then deionized water, and contrasted with 4% water solution of uranyl acetate and methyl cellulose on ice for 10 min. Thin sections (60 nm) were imaged with CM120 transmission electron microscope (Philips Bio Tween) operating at 120 kV. Images were recorded with a Veleta 2 k × 2 k (Olympus-SIS) camera using the iTEM software.
Statistics
Student’s t test was used for all the statistical analysis, except for GSD image colocalization, FRAP and stretch analysis, and transmission electron microscopy caveolae quantification where a Mann-Whitney test was used and immuno-EM image analysis where a Fischer test was used. All the statistical analysis was performed using Prism 5.0 software (GraphPad). *p < 0.05; **p < 0.01; ***p < 0.001; n.s.: not significant.
Supplementary Material
Acknowledgments
We thank Arnoud Sonnenberg for the gift of the ITGB4-EGFP construct, Nicolas Vitale for constructs and helpful discussions, the IGBMC Imaging Center for help with microscopy, and Anita Eckly-Michel and Fabienne Proamer for their help with electron microscopy. D.R. acknowledges Pierre Sens for discussions on the physical mechanism. We thank Bruno Klaholz and Leonid Andronov for granting access to the GSD microscope, which is supported by the French Infrastructure for Integrated Structural Biology (FRISBI) ANR-10-INSB-05-01 and Instruct as part of the European Strategy Forum on Research Infrastructures (ESFRI). We also thank Romeo Ricci, Nicolas Vitale, Patricia Simon-Assman, Sophie Quintin, and Adèle de Arcangelis for critical reading of the manuscript. This work was supported by Agence Nationale de la Recherche and European Research Council grants to M.L.; by grants from ISIS, the ci-FRC of Strasbourg, and Fondation Simone et Cino del Duca to D.R.; by institutional funds from INSERM and the University of Strasbourg to J.G.G.; by grant ANR-10-LABX-0030-INRT, a French State fund managed by the Agence Nationale de la Recherche under the frame programme Investissements d’Avenir labeled ANR-10-IDEX-0002-02 to the IGBMC; and by institutional funds from the CNRS, INSERM, University of Strasbourg. This work is dedicated to the memory of Elisabeth Georges-Labouesse, who passed away in 2012.
Abbreviations used:
- Arf
ADP-ribosylation factor
- Cav1
caveolin 1
- CeHD
Caenorhabditis elegans hemidesmosome
- ECM
extracellular matrix
- FA
focal adhesion
- FAK
focal adhesion kinase
- FRAP
fluorescence recovery after photobleaching
- GSD
ground-state depletion
- HD
hemidesmosome
- ITGA6
integrin α6
- ITGB4
integrin β4
- MT
microtubule
- NT
nontransfected cell
- ROI
region of interest
- TIRF
total internal reflection fluorescence
- wt
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-06-0356) on December 13, 2017.
REFERENCES
- Aikawa Y, Martin TFJ. ARF6 regulates a plasma membrane pool of phosphatidylinositol(4,5)bisphosphate required for regulated exocytosis. J Cell Biol. 2003:647–659. doi: 10.1083/jcb.200212142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, Mahalu D, Safran S, Bershadsky A, Addadi L, Geiger B. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001:466–472. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- Bar-Ziv R, Tlusty T, Moses E, Safran SA, Bershadsky A. Pearling in cells: a clue to understanding cell shape. Proc Natl Acad Sci USA. 1999:10140–10145. doi: 10.1073/pnas.96.18.10140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass MD, Williamson RC, Nunan RD, Humphries JD, Byron A, Morgan MR, Martin P, Humphries MJ. A syndecan-4 hair trigger initiates wound healing through Caveolin- and RhoG-regulated integrin endocytosis. Dev Cell. 2011:681–693. doi: 10.1016/j.devcel.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brevier J, Montero D, Svitkina T, Riveline D. The asymmetric self-assembly mechanism of adherens junctions: a cellular push–pull unit. Phys Biol. 2008:016005. doi: 10.1088/1478-3975/5/1/016005. [DOI] [PubMed] [Google Scholar]
- Brevier J, Vallade M, Riveline D. Force-extension relationship of cell-cell contacts. Phys Rev Lett. 2007:268101. doi: 10.1103/PhysRevLett.98.268101. [DOI] [PubMed] [Google Scholar]
- Chao W-T, Kunz J. Focal adhesion disassembly requires clathrin-dependent endocytosis of integrins. FEBS Lett. 2009:1337–1343. doi: 10.1016/j.febslet.2009.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo MA, Balasubramanian N, Alderson NB, Kiosses WB, Grande-Garcia A, Anderson RGW, Schwartz MA. Phospho-caveolin-1 mediates integrin-regulated membrane domain internalization. Nat Cell Biol. 2005:901–908. doi: 10.1038/ncb1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanoë-Ayari H, Al Kurdi R, Vallade M, Gulino-Debrac D, Riveline D. Membrane and acto-myosin tension promote clustering of adhesion proteins. Proc Natl Acad Sci USA. 2004:2229–2234. doi: 10.1073/pnas.0304297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme-Walker VD, Peterson JR, Chernoff J, Waterman CM, Danuser G, DerMardirossian C, Bokoch GM. Pak1 regulates focal adhesion strength, myosin IIA distribution, and actin dynamics to optimize cell migration. J Cell Biol. 2011:1289–1303. doi: 10.1083/jcb.201010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezratty EJ, Bertaux C, Marcantonio EE, Gundersen GG. Clathrin mediates integrin endocytosis for focal adhesion disassembly in migrating cells. J Cell Biol. 2009:733–747. doi: 10.1083/jcb.200904054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frijns E, Kuikman I, Litjens S, Raspe M, Jalink K, Ports M, Wilhelmsen K, Sonnenberg A. Phosphorylation of threonine 1736 in the C-terminal tail of integrin β4 contributes to hemidesmosome disassembly. Mol Biol Cell. 2012:1475–1485. doi: 10.1091/mbc.E11-11-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frijns E, Sachs N, Kreft M, Wilhelmsen K, Sonnenberg A. EGF-induced MAPK signaling inhibits hemidesmosome formation through phosphorylation of the integrin β4. J Biol Chem. 2010:37650–37662. doi: 10.1074/jbc.M110.138818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnoux-Palacios L, Dans M, van’t Hof W, Mariotti A, Pepe A, Meneguzzi G, Resh MD, Giancotti FG. Compartmentalization of integrin α6β4 signaling in lipid rafts. J Cell Biol. 2003:1189–1196. doi: 10.1083/jcb.200305006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. J Cell Biol. 2002:695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambin Y, Ariotti N, McMahon K-A, Bastiani M, Sierecki E, Kovtun O, Polinkovsky ME, Magenau A, Jung W, Okano S, et al. Single-molecule analysis reveals self assembly and nanoscale segregation of two distinct cavin subcomplexes on caveolae eLife 2014. e01434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain EC, Santos TM, Rabinovitz I. Phosphorylation of a novel site on the {beta}4 integrin at the trailing edge of migrating cells promotes hemidesmosome disassembly. Mol Biol Cell. 2009:56–67. doi: 10.1091/mbc.E08-06-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KJ, Getsios S, Troyanovsky S, Godsel LM. Intercellular junction assembly, dynamics, and homeostasis. Cold Spring Harb Perspect Biol. 2010:a000125. doi: 10.1101/cshperspect.a000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H, Bar H, Kreplak L, Strelkov SV, Aebi U. Intermediate filaments: from cell architecture to nanomechanics. Nat Rev Mol Cell Biol. 2007:562–573. doi: 10.1038/nrm2197. [DOI] [PubMed] [Google Scholar]
- Hopkinson SB, Hamill KJ, Wu Y, Eisenberg JL, Hiroyasu S, Jones JC. Focal contact and hemidesmosomal proteins in keratinocyte migration and wound repair. Adv Wound Care (New Rochelle) 2014:247–263. doi: 10.1089/wound.2013.0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta A, Kawakatsu T, Nakatani T, Sato T, Matsui C, Sukezane T, Akagi T, Hamaji T, Grigoriev I, Akhmanova A, et al. Laminin-based cell adhesion anchors microtubule plus ends to the epithelial cell basal cortex through LL5α/β J Cell Biol 2010. 901 917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata H, Kitajima Y. Bullous pemphigoid: role of complement and mechanisms for blister formation within the lamina lucida. Exp Dermatol. 2013:381–385. doi: 10.1111/exd.12146. [DOI] [PubMed] [Google Scholar]
- Kang M, Day CA, Kenworthy AK, DiBenedetto E. Simplified equation to extract diffusion coefficients from confocal FRAP data. Traffic. 2012:1589–1600. doi: 10.1111/tra.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Ye F, Ginsberg MH. Regulation of integrin activation. Annu Rev Cell Dev Biol. 2011:321–345. doi: 10.1146/annurev-cellbio-100109-104104. [DOI] [PubMed] [Google Scholar]
- Kirkham M, Nixon SJ, Howes MT, Abi-Rached L, Wakeham DE, Hanzal-Bayer M, Ferguson C, Hill MM, Fernandez-Rojo M, Brown DA, et al. Evolutionary analysis and molecular dissection of caveola biogenesis J Cell Sci 2008. 2075 2086 [DOI] [PubMed] [Google Scholar]
- Kuo J-C, Han X, Hsiao C-T, Yates Iii JR, Waterman CM. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for [beta]-Pix in negative regulation of focal adhesion maturation. Nat Cell Biol. 2011:383–393. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie P, Goetz JG, Dennis JW, Nabi IR. Lattices, rafts, and scaffolds: domain regulation of receptor signaling at the plasma membrane. J Cell Biol. 2009:381–385. doi: 10.1083/jcb.200811059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TL, Yap AS, Stow JL. Recycling of E-cadherin: a potential mechanism for regulating cadherin dynamics. J Cell Biol. 1999:219–232. [PMC free article] [PubMed] [Google Scholar]
- Liao G, Nagasaki T, Gundersen GG. Low concentrations of nocodazole interfere with fibroblast locomotion without significantly affecting microtubule level: implications for the role of dynamic microtubules in cell locomotion. J Cell Sci. 1995:3473–3483. doi: 10.1242/jcs.108.11.3473. [DOI] [PubMed] [Google Scholar]
- Litjens SHM, de Pereda JM, Sonnenberg A. Current insights into the formation and breakdown of hemidesmosomes. Trends Cell Biol. 2006:376–383. doi: 10.1016/j.tcb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Litjens SHM, Wilhelmsen K, de Pereda JM, Perrakis A, Sonnenberg A. Modeling and experimental validation of the binary complex of the plectin actin-binding domain and the first pair of fibronectin type III (FNIII) domains of the β4 integrin. J Biol Chem. 2005:22270–22277. doi: 10.1074/jbc.M411818200. [DOI] [PubMed] [Google Scholar]
- Margadant C, Monsuur HN, Norman JC, Sonnenberg A. Mechanisms of integrin activation and trafficking. Curr Opin Cell Biol. 2011:607–614. doi: 10.1016/j.ceb.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Dev Cell. 2010:194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahidiazar L, Kreft M, Broek B, van den Secades P, Manders EMM, Sonnenberg A, Jalink K. The molecular architecture of hemidesmosomes, as revealed with super-resolution microscopy. J Cell Sci. 2015:3714–3719. doi: 10.1242/jcs.171892. [DOI] [PubMed] [Google Scholar]
- Osmani N, Labouesse M. Remodeling of keratin-coupled cell adhesion complexes. Curr Opin Cell Biol. 2015:30–38. doi: 10.1016/j.ceb.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol. 2010:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton RG, del Pozo MA. Caveolae as plasma membrane sensors, protectors and organizers. Nat Rev Mol Cell Biol. 2013:98–112. doi: 10.1038/nrm3512. [DOI] [PubMed] [Google Scholar]
- Paul NR, Jacquemet G, Caswell PT. Endocytic trafficking of integrins in cell migration. Curr Biol. 2015:R1092–R1105. doi: 10.1016/j.cub.2015.09.049. [DOI] [PubMed] [Google Scholar]
- Phair RD, Gorski SA, Misteli T. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 2004:393–414. doi: 10.1016/s0076-6879(03)75025-3. [DOI] [PubMed] [Google Scholar]
- Powelka AM, Sun J, Li J, Gao M, Shaw LM, Sonnenberg A, Hsu VW. Stimulation-dependent recycling of integrin β1 regulated by ARF6 and Rab11. Traffic. 2004:20–36. doi: 10.1111/j.1600-0854.2004.00150.x. [DOI] [PubMed] [Google Scholar]
- Quintin S, Wang S, Pontabry J, Bender A, Robin F, Hyenne V, Landmann F, Gally C, Oegema K, Labouesse M. Non-centrosomal epidermal microtubules act in parallel to LET-502/ROCK to promote C. elegans elongation. Development. 2016:160–173. doi: 10.1242/dev.126615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovitz I, Tsomo L, Mercurio AM. Protein kinase C-a phosphorylation of specific serines in the connecting segment of the b4 integrin regulates the dynamics of type II hemidesmosomes. Mol Cell Biol. 2004:4351–4360. doi: 10.1128/MCB.24.10.4351-4360.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezniczek GA, de Pereda JM, Reipert S, Wiche G. Linking integrin α6β4-based cell adhesion to the intermediate filament cytoskeleton: direct interaction between the β4 subunit and plectin at multiple molecular sites. J Cell Biol. 1998:209–225. doi: 10.1083/jcb.141.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveline D, Zamir E, Balaban NQ, Schwarz US, Ishizaki T, Narumiya S, Kam Z, Geiger B, Bershadsky AD. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J Cell Biol. 2001:1175–1186. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M, Barry S, Woods A, van der Sluijs P, Norman J. PDGF-regulated rab4-dependent recycling of αvβ3 integrin from early endosomes is necessary for cell adhesion and spreading. Curr Biol. 2001:1392–1402. doi: 10.1016/s0960-9822(01)00442-0. [DOI] [PubMed] [Google Scholar]
- Rossier O, Octeau V, Sibarita J-B, Leduc C, Tessier B, Nair D, Gatterdam V, Destaing O, Albigès-Rizo C, Tampé R, et al. Integrins β1 and β3 exhibit distinct dynamic nanoscale organizations inside focal adhesions Nat Cell Biol 2012. 1057 1067 [DOI] [PubMed] [Google Scholar]
- Sawamura D, Nakano H, Matsuzaki Y. Overview of epidermolysis bullosa. J Dermatol. 2010:214–219. doi: 10.1111/j.1346-8138.2009.00800.x. [DOI] [PubMed] [Google Scholar]
- Schaller MD. Cellular functions of FAK kinases: insight into molecular mechanisms and novel functions. J Cell Sci. 2010:1007–1013. doi: 10.1242/jcs.045112. [DOI] [PubMed] [Google Scholar]
- Seltmann K, Cheng F, Wiche G, Eriksson JE, Magin TM. Keratins stabilize hemidesmosomes through regulation of [beta]4-integrin turnover. J Invest Dermatol. 2015:1609–1620. doi: 10.1038/jid.2015.46. [DOI] [PubMed] [Google Scholar]
- Sens P, Turner MS. Theoretical model for the formation of Caveolae and similar membrane invaginations. Biophys J. 2004:2049–2057. doi: 10.1016/S0006-3495(04)74266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sens P, Turner MS. The forces that shape Caveolae. In: Lipid Rafts and Caveolae, New York, Wiley-VCH Verlag GmbH & Co. 2006. pp. 25–44.
- Sinha B, Köster D, Ruez R, Gonnord P, Bastiani M, Abankwa D, Stan RV, Butler-Browne G, Vedie B, Johannes L, et al. Cells respond to mechanical stress by rapid disassembly of Caveolae Cell 2011. 402 413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati T, Schliwa M. Powering membrane traffic in endocytosis and recycling. Nat Rev Mol Cell Biol. 2006:897–908. doi: 10.1038/nrm2060. [DOI] [PubMed] [Google Scholar]
- Stehbens SJ, Paszek M, Pemble H, Ettinger A, Gierke S, Wittmann T. CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nat Cell Biol. 2014:561–573. doi: 10.1038/ncb2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upla P, Marjomaki V, Kankaanpaa P, Ivaska J, Hyypia T, van der Goot FG, Heino J. Clustering induces a lateral redistribution of a2b1 integrin from membrane rafts to Caveolae and subsequent protein kinase C-dependent internalization. Mol Biol Cell. 2004:625–636. doi: 10.1091/mbc.E03-08-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez RJ, Howell B, Yvon AM, Wadsworth P, Cassimeris L. Nanomolar concentrations of nocodazole alter microtubule dynamic instability in vivo and in vitro. Mol Biol Cell. 1997:973–985. doi: 10.1091/mbc.8.6.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walko G, Castañón M, Wiche G. Molecular architecture and function of the hemidesmosome. Cell Tissue Res. 2014:363–378. doi: 10.1007/s00441-014-2061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walko G, Vukasinovic N, Gross K, Fischer I, Sibitz S, Fuchs P, Reipert S, Jungwirth U, Berger W, Salzer U, et al. Targeted proteolysis of plectin ioform 1a accounts for hemidesmosome dysfunction in mice mimicking the dominant skin blistering disease EBS-Ogna PLoS Genet 2011. e1002396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsen K, Litjens SHM, Kuikman I, Margadant C, van Rheenen J, Sonnenberg A. Serine phosphorylation of the integrin beta4 subunit is necessary for epidermal growth factor receptor induced hemidesmosome disruption. Mol Biol Cell. 2007:3512–3522. doi: 10.1091/mbc.E07-04-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S-O, Shin S, Mercurio AM. Hypoxia stimulates carcinoma invasion by stabilizing microtubules and promoting the Rab11 trafficking of the α6β4 integrin. Cancer Res. 2005:2761–2769. doi: 10.1158/0008-5472.CAN-04-4122. [DOI] [PubMed] [Google Scholar]
- Zahreddine H, Zhang H, Diogon M, Nagamatsu Y, Labouesse M. CRT-1/Calreticulin and the E3 ligase EEL-1/HUWE1 control hemidesmosome maturation in C. elegans development. Curr Biol. 2010:322–327. doi: 10.1016/j.cub.2009.12.061. [DOI] [PubMed] [Google Scholar]
- Zhang H, Labouesse M. The making of hemidesmosome structures in vivo. Dev Dyn. 2010:1465–1476. doi: 10.1002/dvdy.22255. [DOI] [PubMed] [Google Scholar]
- Zhang H, Landmann F, Zahreddine H, Rodriguez D, Koch M, Labouesse M. A tension-induced mechanotransduction pathway promotes epithelial morphogenesis. Nature. 2011:99–103. doi: 10.1038/nature09765. [DOI] [PubMed] [Google Scholar]
- Zimnicka AM, Husain YS, Shajahan AN, Sverdlov M, Chaga O, Chen Z, Toth PT, Klomp J, Karginov AV, Tiruppathi C, et al. Src-dependent phosphorylation of caveolin-1 Tyr-14 promotes swelling and release of caveolae Mol Biol Cell 2016. 2090 2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.