In Tetrahymena, different condensins are required to segregate chromosomes in the germline versus the somatic nuclei. Germline condensins promote condensation and individualization of chromosomes. In the polyploid somatic nucleus, condensin acts as an insulator, preventing the multiple copies of each chromosome from clustering.
Abstract
Condensin is a protein complex with diverse functions in chromatin packaging and chromosome condensation and segregation. We studied condensin in the evolutionarily distant protist model Tetrahymena, which features noncanonical nuclear organization and divisions. In Tetrahymena, the germline and soma are partitioned into two different nuclei within a single cell. Consistent with their functional specializations in sexual reproduction and gene expression, condensins of the germline nucleus and the polyploid somatic nucleus are composed of different subunits. Mitosis and meiosis of the germline nucleus and amitotic division of the somatic nucleus are all dependent on condensins. In condensin-depleted cells, a chromosome condensation defect was most striking at meiotic metaphase, when Tetrahymena chromosomes are normally most densely packaged. Live imaging of meiotic divisions in condensin-depleted cells showed repeated nuclear stretching and contraction as the chromosomes failed to separate. Condensin depletion also fundamentally altered chromosome arrangement in the polyploid somatic nucleus: multiple copies of homologous chromosomes tended to cluster, consistent with a previous model of condensin suppressing default somatic pairing. We propose that failure to form discrete chromosome territories is the common cause of the defects observed in the absence of condensins.
INTRODUCTION
Condensin is a multi-subunit protein complex that was originally identified as a primary requirement for chromosome condensation in cell-free Xenopus oocyte extracts (Hirano and Mitchison, 1994). Subsequent studies in diverse organisms have shown it to be a highly conserved structural component of chromosomes that is important for proper chromosome segregation during both mitosis and meiosis (reviewed in Hirano, 2012, 2016). Condensin is proposed to act by encircling or extruding multiple DNA loops within the same chromatid, thus promoting or ensuring chromosome compaction (Nasmyth, 2001; Cuylen et al., 2011). In addition, condensin has been shown to function in dosage compensation (Caenorhabditis elegans), maintenance of rDNA repeats (Saccharomyces cerevisiae), tRNA gene clustering (S. cerevisiae and Schizosaccharomyces pombe), and DNA damage response and repair (Heale et al., 2006; Johzuka et al., 2006; Haeusler et al., 2008; Csankovszki et al., 2009; Iwasaki et al., 2010; Sakamoto et al., 2011). These combined functions indicate that condensin is a crucial factor in genomic stability and is therefore implicated in cancer biology, genetic disorders, and reproductive health (Trimborn et al., 2006; Ham et al., 2007; Davalos et al., 2012; Je et al., 2014).
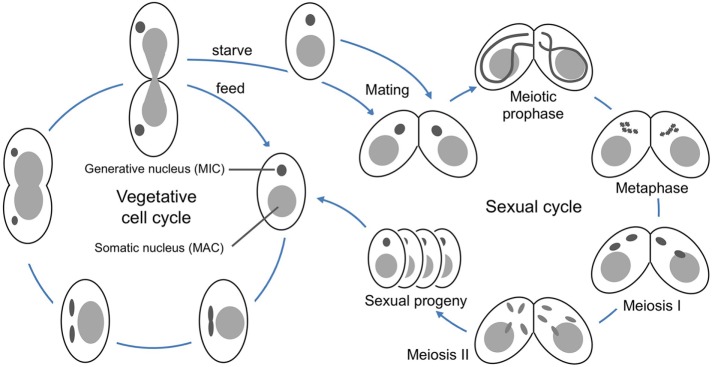
Tetrahymena thermophila is a free-living, freshwater ciliated protist. Like other ciliates, it exhibits nuclear dualism, that is, the germline and somatic genomes exist in separate nuclei within the same cell (Karrer, 2012). The germline nucleus has a diploid chromosome number of 10 and undergoes closed mitosis and meiosis (Figure 1). It is transcriptionally silent: its only role is to pass on genetic information during sexual reproduction. The somatic nucleus is derived from the germline nucleus during sexual reproduction in a process that eliminates ∼30% of the total DNA, cleaves the chromosomes at specific sites, adds telomeres, and, finally, amplifies the genome to produce ∼50 copies each of 181 chromosomes (Eisen et al., 2006; Karrer, 2012; Hamilton et al., 2016). These transcriptionally active somatic chromosomes lack centromeres, and the somatic nucleus divides via the poorly characterized process of amitosis, in which chromosomes are randomly segregated into two roughly equal portions prior to cell division (Fujiu and Numata, 2000; Cole and Sugai, 2012).
FIGURE 1:
Vegetative and sexual cycles of Tetrahymena thermophila. During vegetative growth (left), the two nuclei of Tetrahymena divide asynchronously. First, the germline nucleus divides mitotically, then the somatic nucleus elongates and pinches off to form daughter nuclei, and, finally, the cleavage furrow closes to divide the cell. To induce meiosis, starved strains of two different mating types are mixed (right). The cells pair and their germline nuclei undergo synchronous meiosis, with pronounced nuclear elongation during prophase.
Although condensin is evolutionarily conserved, research to date shows that the contribution of condensin to overall chromosome condensation is highly variable (Hirano, 2012). Its mechanism of action is not well understood, and it remains unclear whether the role of condensin is primarily structural or enzymatic. Studying condensin in an organism such as Tetrahymena, which is phylogenetically distant from more conventional plant, fungal, and animal models, provides a unique opportunity to investigate conservation of function and might lead to the discovery of new roles for condensin in chromosome dynamics.
A previous study showed that partial knockout of the core condensin gene SMC4 led to defects in somatic nuclear division (Cervantes et al., 2006). In this study, we explore the composition and function of condensin complexes in the somatic and germline nuclei of Tetrahymena.
RESULTS
Bioinformatic identification of condensin subunits
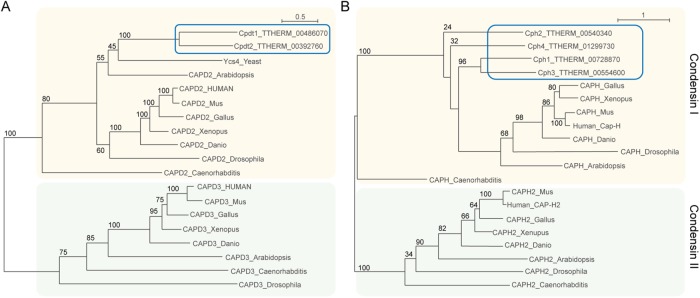
BLASTp searches were conducted using the protein sequences of budding yeast condensin subunits against predicted Tetrahymena open reading frames. We identified a single homologue of each of the core subunit genes SMC2 and SMC4 in the Tetrahymena genome (http://ciliate.org/) (Eisen et al., 2006) as previously reported (Cervantes et al., 2006). A search using the yeast HEAT IB subunit Ycg1 also yielded a single hit, TTHERM_00919690, which we designated CPG1. Searches using the budding yeast BrnI sequence identified four strong candidates for the kleisin (Brn/Cap-H) subunit: TTHERM_00728870 (1.7e-46), TTHERM_00540340 (2.6e-30), TTHERM_00554600 (1.5e-74), and TTHERM_01299730 (6.7e-33). These genes were designated CPH1, CPH2, CPH3, and CPH4. A BLASTp search using the yeast HEAT IA subunit Ycs4 yielded two significant hits: TTHERM_00486070 and TTHERM_00392760. We designated these genes CPDT1 and CPDT2 for Cap-D Two 1 and 2 (Table 1). Phylogenetic analysis (Guindon et al., 2010; Sievers et al., 2011) of kleisins and Cap-D homologues showed that all had greater similarity to condensin I than condensin II components in metazoans or plants, confirming a previous prediction that Tetrahymena condensins are derived from condensin I (Figure 2) (Hirano, 2012).
TABLE 1:
Localization and depletion phenotypes of Tetrahymena condensin subunits.
| Localization | ||||
|---|---|---|---|---|
| Subunit | Gene ID | Germline | Soma | RNAi phenotype |
| Common condensins | ||||
| Smc2 | TTHERM_00812950 | * | * | Failure to segregate germline chromosomes in mitosis and meiosis, failure to divide somatic nucleus, somatic chromosomes cluster |
| Smc4 | TTHERM_00446400 | + | + | Not done |
| Cpg1 | TTHERM_00919690 | + | + | Not done |
| Cpdt1 | TTHERM_00486070 | + | + | Failure to segregate germline chromosomes in mitosis and meiosis, failure to divide somatic nucleus, somatic chromosomes cluster |
| Germline condensins | ||||
| Cph1 | TTHERM_00728870 | + | Anaphase bridging in mitosis and meiosis, partially redundant to Cph2 | |
| Cph2 | TTHERM_00540340 | + | Anaphase bridging in mitosis and meiosis, partially redundant to Cph1 | |
| Somatic condensins | ||||
| Cph3 | TTHERM_00554600 | * | Failure to divide somatic nucleus, somatic chromosomes cluster | |
| Cph4 | TTHERM_01299730 | + | None detected | |
| Cpdt2 | TTHERM_00392760 | + | Failure to complete sexual reproduction |
*Localization was inferred from the RNAi phenotype.
FIGURE 2:
Phylogenetic tree reconstruction indicates that Tetrahymena condensins are more closely related to condensin I than condensin II. To determine relationships of (A) Cap-D and (B) Cap-H proteins, sequences were aligned with Clustal Omega, and, subsequently, maximum-likelihood trees were generated using PhyML v3.1 with the LG substitution model (Guindon et al., 2010; Sievers et al., 2011). Colored boxes highlight the condensin I (yellow) and condensin II (green) genes. Branch support is indicated by bootstrap values in percentages shown near each branch. The branch-length scale bar represents the estimated number of substitutions per amino acid site. Both Tetrahymena Cap-D homologues group within the Cap-D2 subfamily of HEAT repeat proteins and all four Tetrahymena Cap-H homologues group within the Cap-H kleisin subfamily with high bootstrap support, confirming that all Tetrahymena condensin complexes were derived from condensin I.
Condensin subunit localization
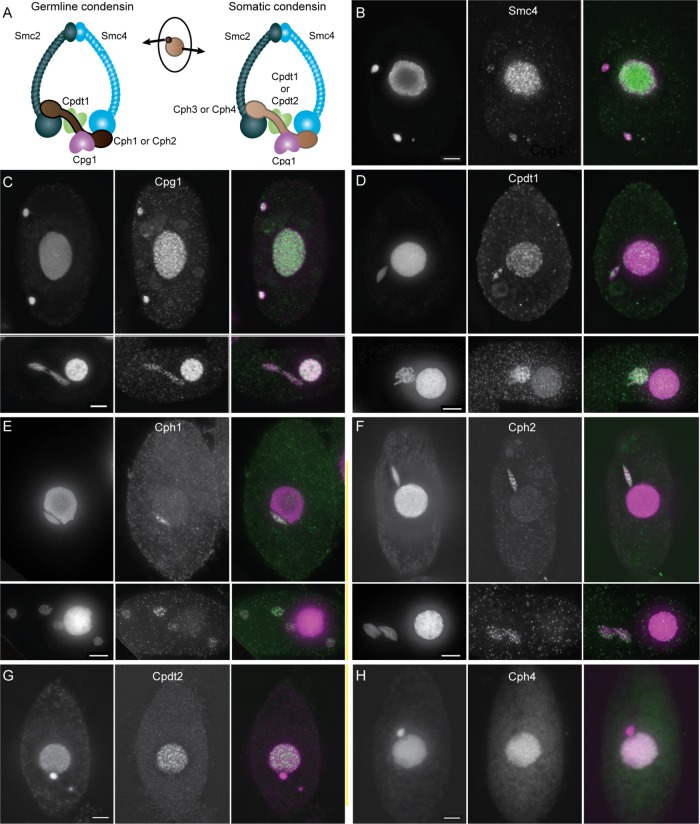
Tetrahymena is likely to use condensin complexes with specific compositions to fulfill the different functions necessary for performing mitosis and meiosis in the germline nucleus and amitosis in the somatic nucleus. To determine which subunits are active in which nucleus, we epitope tagged the protein subunits identified in our bioinformatic searches. With the exception of Cph1 and Cph4, all subunits were C-terminally tagged by knock-in of the tag epitope (see Materials and Methods). C-terminal tagging of Cph1 and Cph4 was unsuccessful, and therefore N-terminal HA tagging constructs were used to ectopically express these proteins. The ectopically expressed proteins could be visualized by immunofluorescence but caused aberrant nuclear divisions, indicating that either the proteins are not fully functional or that alteration of normal expression levels changed the dynamics of condensin function. Localization of tagged proteins is summarized in Table 1, and representative images are shown in Figure 3, B–H. In all cases, the condensin proteins were present throughout the cell cycle in both vegetative and mating cells. The localization of subunits indicates that the germline mitotic and meiotic functions of condensin are probably performed by complexes containing Smc2, Smc4, Cpdt1, Cpg1, and Cph1 or Cph2. Condensin in the vegetative somatic nucleus is composed of Smc2, Smc4, Cpg1, Cpdt1 or Cpdt2, and Cph3 or Cph4, as shown in Figure 3A.
FIGURE 3:
Composition and localization of condensin proteins in Tetrahymena. (A) Condensin is a multi-subunit protein complex composed of two “structural maintenance of chromosomes” proteins, Smc2 and Smc4, each consisting of a hinge domain joined to an ATPase head domain by a long coiled coil. The two Smcs associate via the hinge domains, and the head domains interact with a kleisin subunit, Cph1, 2, 3, or 4. The Cph subunit interacts with two additional subunits, Cpg1 and the Cpdt1 or Cpdt2 HEAT repeat protein. In Tetrahymena, germline and somatic nuclei contain condensin complexes with distinct subunit composition, as shown by epitope tagging experiments (B–H). Scale bars equal 5 μm. Merged images show DAPI staining in magenta and tagged proteins in green. (B) Smc4 localizes to both the somatic and germline nuclei, a mitotic cell is shown. Smc4 appears to be highly abundant and distributed throughout the somatic nucleus. The protein appears less abundant in the germline and forms some brighter foci on the dividing chromosomes. (C, D) Cpg1 and Cpdt1 localize to both somatic and germline nuclei in mitotic cells (top panels) and meiotic cells (bottom panels). Cpg1 (C) is shown in anaphase of meiosis I, and Cpdt1 (D) is shown at metaphase/diakinesis. (E, F) Cph1 and Cph2 are found only in the germline nucleus. Top panels show mitotic cells, bottom panels show cells in meiosis II. Cph1 is shown at telophase II (E), Cph2 at early anaphase II. Again, bright foci can be seen within the germline nuclei. (G, H) Cpdt2 and Cph4 localize only to somatic nuclei. Vegetative cells are shown.
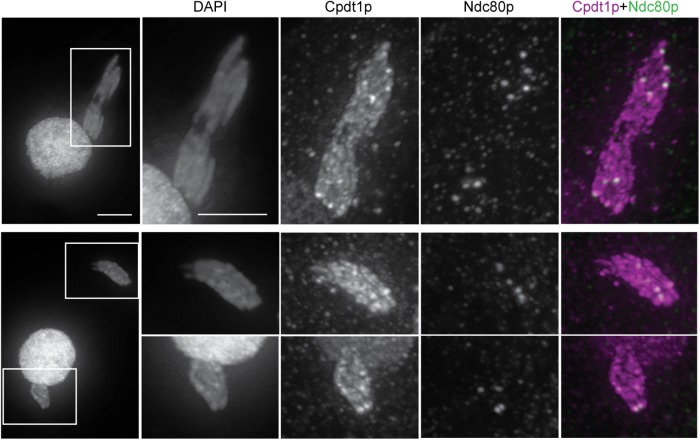
Condensin does not appear to be homogeneously distributed on germline chromosomes: areas of higher fluorescence intensity can be seen on meiotic chromosomes (Figure 4). Dual expression of HA-tagged Cpdt1 and GFP-tagged Ndc80 (a kinetochore protein) revealed that condensin is enriched at centromeres during meiotic divisions (Figure 4), consistent with observations made in other organisms (Ono et al., 2004; Savvidou et al., 2005; Shintomi and Hirano, 2011).
FIGURE 4:
Colocalization of condensin and centromeres. Cells expressing GFP-tagged Ndc80 were mated with cells expressing Cpdt1-HA. One cell of each pair is shown; scale bar represents 5 μm. Higher intensity Cpdt1-HA foci colocalizing with Ndc80-GFP are visible on chromosomes from metaphase onward but are most obvious at anaphase of meiosis I. Top panels, early anaphase; bottom panels, late anaphase.
Depletion of germline condensin causes defects in mitosis and meiosis
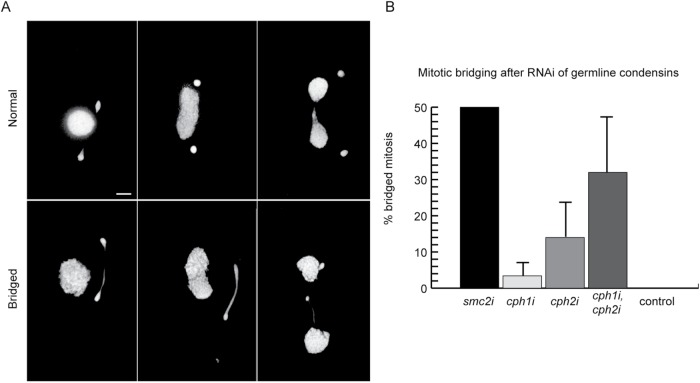
To learn about the function of condensin in Tetrahymena, we depleted condensin subunits using RNA interference (RNAi), which allows conditional knockdown of essential genes (Howard-Till and Yao, 2006). Knockdown was confirmed either by immunoblotting or quantitative reverse transcription PCR (RT-qPCR) (Supplemental Figure S1). The results are summarized in Table 1. RNAi depletion of the common condensins Smc2 and Cpdt1 (smc2i, cpdt1i) produced severe phenotypes during both vegetative growth and meiosis. Germline chromosomes did not segregate properly in mitosis, resulting in large DNA bridges at anaphase and telophase, unequal daughter nuclei, and, occasionally, complete loss of the germline nucleus (Figure 5A). In all, 50% of smc2i cells showed anaphase bridges, whereas all control cells showed normal mitotic anaphase (Figure 5B).
FIGURE 5:
RNAi-mediated knockdown of germline condensin genes during vegetative growth causes segregation defects in germline nuclei. (A) In wild-type cells, the germline nucleus divides prior to somatic amitosis and formation of the cleavage furrow. RNAi of germline condensin genes resulted in anaphase bridges during mitosis. Top panel, normal mitotic divisions; bottom panel, examples of bridged mitosis in cph1i/cph2i cells. Scale bar equals 5 μm. (B) Mitotic chromosome segregation was assayed by counting anaphase cells in which the two new nuclei were separated by at least the diameter of the somatic nucleus. If the two chromatin masses of the dividing germline were not completely separated, anaphase was classified as “bridged.” Quantitation of RNAi-treated cells versus controls shows that anaphase bridging is more frequent in cph1i/cph2i double RNAi cells than in single RNAi cells. Four strains each of control, cph1i, cph2i, and double RNAi cells were evaluated, with 50 anaphases counted under each condition. Two strains of smc2i cells were evaluated, with 100 anaphases counted for each.
Single knockdowns of germline-specific CPH1 and CPH2 showed milder defects such as occasional DNA bridging at mitotic anaphase. In cph1i strains, 3.4% of cells in anaphase had DNA bridges; in cph2i strains, 14% of anaphase cells had DNA bridges (Figure 5B). To test for functional redundancy between these two subunits, double RNAi knockdown strains were produced. These showed DNA bridging at anaphase in 31% of cells, suggesting at least partial redundancy (Figure 5B).
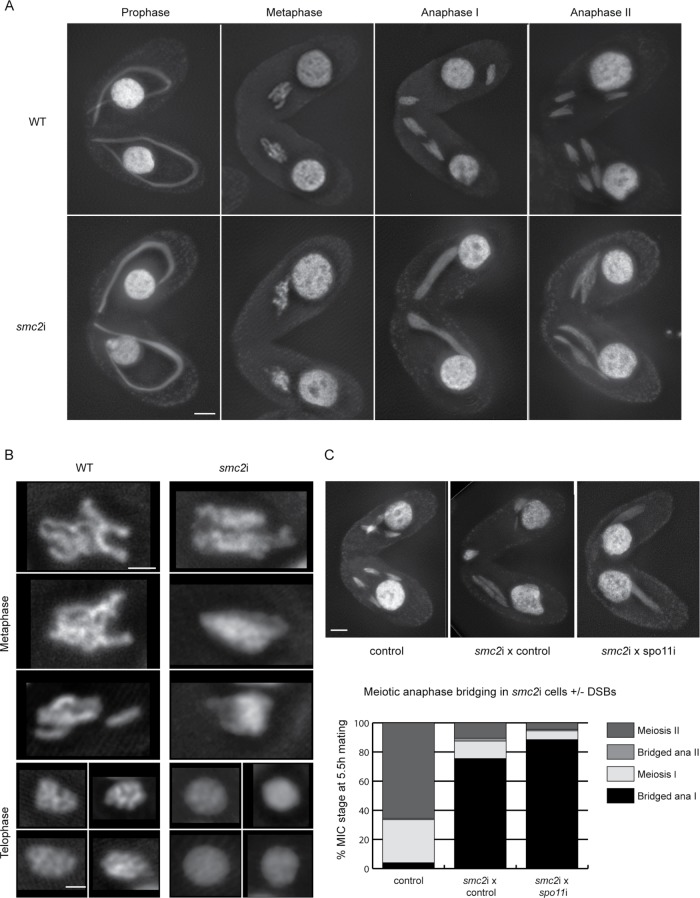
Next, RNAi-mediated depletion of germline condensin components was performed in meiotic cells. Germline chromosome behavior is more easily studied in meiosis than in mitosis because individual chromosomes are well separated, and meiotic divisions are more synchronous. RNAi-mediated SMC2 or CPDT1 knockdown in mating cells caused defects in meiotic DNA condensation and segregation (Figure 6, A and B). Chromosomes failed to condense into distinct bivalents, and in cells undergoing meiotic divisions, extensive DNA bridging was observed at anaphase. A time course of fixed smc2i or cpdt1i cells showed delayed progression of meiosis. At 5.5 h after induction of meiosis, only 20% of smc2i or 12% of cpdt1i cells had reached anaphase II, compared with 56% of SMC2 control or 28% of CPDT1 control matings. Meiotic divisions were also abnormal in cph1i, cph2i double-depleted cells: at 4.5 h after induction of meiosis, 77% of cells in meiosis I or II showed anaphase bridges, compared with 0% in wild-type (WT) cells (average of three matings, 100 cells counted).
FIGURE 6:
Condensin is required for DNA segregation in meiosis. (A) RNAi against SMC2 has no effect on nuclear stretching at meiotic prophase but results in reduced condensation of metaphase chromosomes and extensive DNA bridging at anaphase I and II compared with WT cells. Scale bar equals 5 μm. (B) Enlarged images of metaphase and telophase chromosomes show that chromosomes are less condensed in smc2i cells than in WT cells. Scale bar equals 2 μm. (C) Matings between smc2i and spo11i cells show DNA bridging at anaphase I, indicating that bridge formation is independent of double-strand breaks. Scale bar equals 5 μm. For each sample, 100 meiotic cells were counted and scored as showing either normal meiotic divisions (meiosis I and II categories) or bridged anaphase. Values plotted are the average of two experiments.
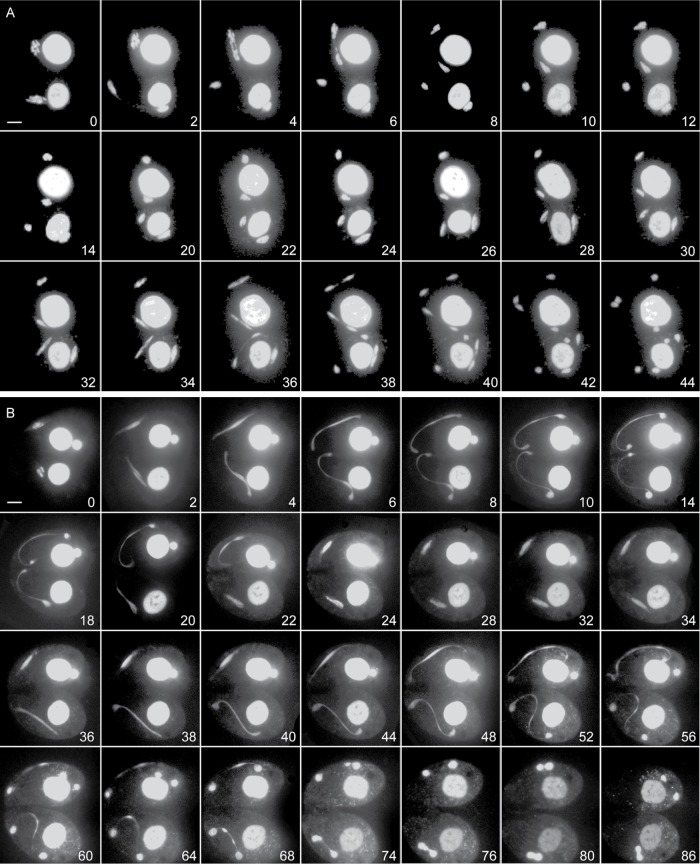
Live cell imaging of meiosis in smc2i cells was performed to observe germline chromosome segregation defects in real time (Figure 7 and Supplemental Movies S1 and S2). In vivo observation of the meiotic divisions revealed spectacular failures in segregating the chromosomes. One such example is shown in Figure 7B (Supplemental Movie S2). As cells attempted the first meiotic division, two masses of chromatin separated, with some DNA remaining between. This DNA bridge most likely represents entangled chromosomes that act like an elastic band, subsequently pulling the two masses of chromatin back toward each other after the failed attempt at division. However, the meiotic program progressed, and another attempt at division was made. The second attempt resulted in two masses of chromatin that were mostly separate but appeared to remain connected by chromatin links. In some cells, a large amount of DNA was unable to segregate to the poles before the spindle collapsed; this later appeared as a separate chromatin body in the cytoplasm. Therefore, at the end of meiosis, cells often contained three small bodies of germline chromatin (i.e., aberrant nuclei), which were eventually degraded, leaving only the somatic nuclei.
FIGURE 7:
Live imaging of meiosis. Images of cells expressing mCherry-tagged histone H3 (Hht2-mCherry) were captured throughout meiosis to visualize chromosome condensation and segregation. Numbers in each panel indicate the time (in minutes) elapsed since the first image. Scale bar equals 5 μm. (A) Control cells undergo normal meiotic divisions, starting with well-condensed chromosomes. (B) Cells expressing SMC2 RNAi start with less condensed chromosomes, and meiotic divisions show extensive DNA bridging. Ultimately, the chromosomes fail to segregate into four meiotic products.
Movie S1.
Timelapse video of live imaging of normal meiosis. Cells undergo normal meiotic divisions, starting with well-condensed chromosomes.
Movie S2.
Timelapse video of live imaging of meiosis in smc2i cells. Cells expressing SMC2 RNAi start with less condensed chromosomes, and meiotic divisions show extensive DNA bridging. Ultimately, the chromosomes fail to segregate into four meiotic products.
Segregation defects are not dependent on meiotic DNA double-strand breaks
Clearly, some type of DNA entanglement prevents segregation of mitotic and meiotic chromosomes in the absence of condensin. These DNA linkages could merely represent a failure of sister chromatid decatenation after replication; alternatively, in meiosis, they could be caused by incomplete repair of meiotic double-strand breaks (DSBs). In meiosis in budding yeast condensin mutants, anaphase bridges were shown to be recombination dependent; they were alleviated in the spo11 mutant background, in which DSBs are not formed (Yu and Koshland, 2003). To investigate whether anaphase bridges during meiosis in Tetrahymena smc2i cells are recombination dependent, we mated smc2i cells with spo11i cells (Figure 6C). As RNAi expressed in one cell can pass into the partner cell during mating, in this way we could achieve the double knockdown of SMC2 and SPO11. In control matings, 65.5% of cells completed meiosis II by 5.5 h after induction of meiosis. In matings between smc2i and control cells, 75.5% of cells had bridged anaphase I nuclei and only 10.5% of cells completed meiosis II by 5.5 h postinduction. In smc2i × spo11i matings, the number of bridged anaphase I cells increased to 88.5%, and only 4.5% of cells reached anaphase II. This result indicates that in the absence of Spo11, the delay in meiosis and the defects in segregation were slightly increased rather than reduced. Therefore, the linkages that prevent segregation are not dependent on persistent homologous connections but rather on nonhomologous linkages or entanglements that occur in the absence of proper condensation.
Depletion of condensin from the somatic nucleus prevents chromosome segregation and alters nuclear organization
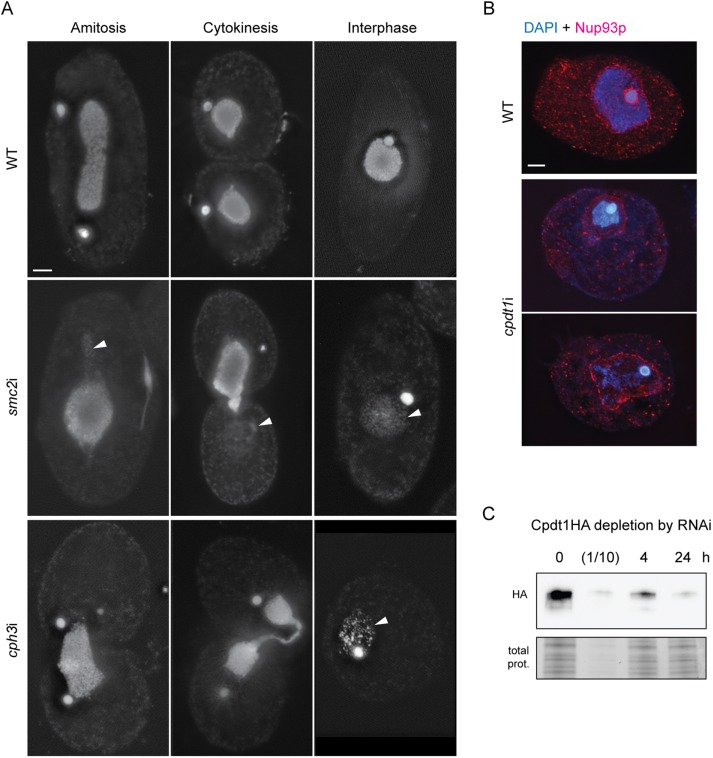
As described previously, after partial SMC4 knockdown (Cervantes et al., 2006), chromatin is unevenly distributed in the dividing somatic nucleus. Here, we similarly found that when the somatic nucleus divides in smc2i, cpdt1i, or cph3i strains, one half usually receives most of the DNA, while the other half receives almost none. In other cells, the cleavage furrow ends up cutting through the mass of chromatin, and much of the DNA in the middle is lost (Figure 8A). Interestingly, the size of the nearly DNA-free nucleus is almost normal; this was particularly evident when the nuclear membrane was stained using an antibody against the nuclear pore protein Nup93 (Figure 8B).
FIGURE 8:
RNAi-mediated knockdown of somatic condensin genes disrupts amitotic division. (A) In WT cells, the somatic nucleus elongates and splits into two new nuclei prior to cell division. In contrast, smc2i or cph3i cells divide with unequal distribution of DNA in somatic nuclei, or DNA remains in the cleavage furrow and is lost when the cells separate. Interphase cells often contain somatic nuclei with very little DNA. Arrowheads indicate areas of nuclear membranes lacking DAPI-stained chromatin. (B) cpdt1i cells show a similar phenotype to smc2i and cph3i cells, that is, loss of somatic DNA. Control cells stained with an anti-Nup93 antibody (red) show the nuclear membrane closely surrounding the DNA. cpdt1i nuclei have large, DAPI-free areas within the nuclear membrane, indicating lower DNA content. Scale bar equals 5 μm in A and B. (C). Western blotting of protein extracts from Cpdt1HA + cpdt1i cells before induction or 4 and 24 h after induction of RNAi. After 24 h, protein levels are comparable to 1/10 that of uninduced cells (lane 2). Total protein (Bio-Rad stain free visualization) is shown as a loading control.
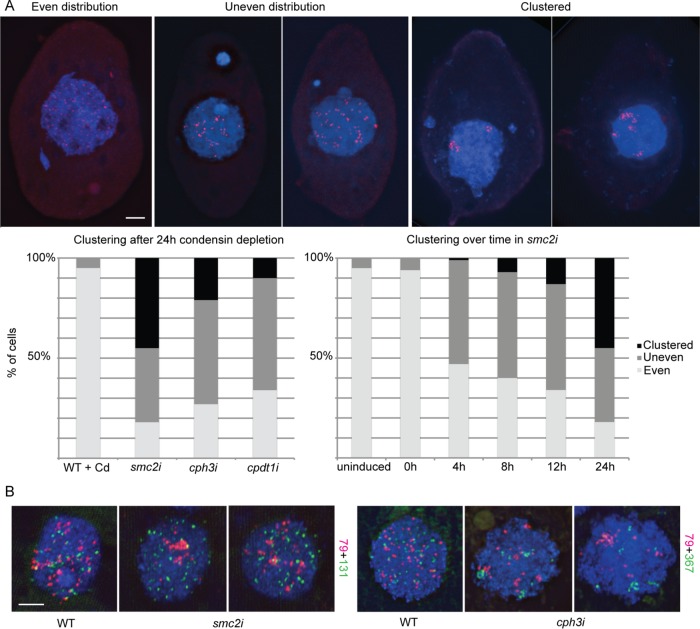
To investigate the possible causes of the somatic nuclear division defects in the absence of condensin, we used fluorescence in situ hybridization (FISH) of individual somatic chromosomes to assess alterations in chromosome condensation or arrangement. We first chose to label one of the smaller chromosomes (79 kb). In wild-type cells, FISH signals formed distinct spots that were distributed fairly evenly throughout the nucleus; the size and appearance of foci did not appear to change throughout the cell cycle (Figure 9). In cells in which Smc2, Cph3, or Cpdt1 had been depleted for 24 h, FISH signals in many nuclei showed a much more clustered distribution (Figure 9A). However, the signals remained distinct spots and did not appear substantially larger or more diffuse than those of WT nuclei, suggesting that condensin may have a more important role in organizing the somatic chromosomes in three-dimensional space than in condensing them.
FIGURE 9:
Condensin depletion causes chromosome clustering in somatic nuclei. (A) Representative images from FISH of a 79-kb chromosome in the somatic nucleus show examples of evenly distributed, unevenly distributed, or clustered chromosomes. Quantitation of chromosome clustering in smc2i, cph3i, and cpdt1i cells shows the various effects of depleting different condensin subunits. Temporal analysis after induction of RNAi in smc2i cells shows a gradual increase in chromosome clustering. RNAi was induced in starved cells, which were then refed to induce cell division and fixed at the indicated time points. (B) FISH of two different chromosomes shows nonoverlapping clusters, with various degrees of clustering for different probes. Scale bar equals 5 μm in A and B.
Chromosome clustering became progressively more severe over time after induction of SMC2 RNAi (Figure 9A). To determine whether clustering was due to general clumping of chromatin within one area of the nucleus, we labeled a second chromosome (131 kb) in a different color (Figure 9B). Double FISH showed that the two chromosome clusters did not colocalize; hence, FISH signal clustering was not due to general chromatin clumping. The 131-kb chromosome showed only moderate clustering compared with the 79-kb or 367-kb chromosomes, indicating that clustering is independent of chromosome size (Figure 9B).
Notably, chromosome clustering is not due to a failure to separate DNA copies after replication. In most cells at 4 h after RNAi induction, somatic chromosomes are not dividing; therefore, if replication continued without separation of newly replicated copies, we would expect the foci to become brighter but would not expect the overall number or distribution to change. Instead, our results support the somewhat controversial theory that daughter or homologous chromosomes have a tendency to interact unless actively prevented from doing so (Joyce et al., 2016) and suggest that condensin may act as an anti-pairing factor. This action is also consistent with the concept of chromosome territories, in which each chromosome maintains its own distinct space and is not comingled or intertwined with other chromosomes (Cremer and Cremer, 2010).
DISCUSSION
The evolutionary diversification of condensins
In multicellular eukaryotes, two forms of condensin (condensin I and II) have evolved from a common ancestor by the diversification of kleisins and HEAT repeat subunits, with the Smc subunits remaining unchanged. These two forms have different functions in axial versus lateral chromosome compaction and centromere organization (Hirano, 2012). Caenorhabditis elegans employs a third condensin that has a modified Smc4 subunit but is otherwise identical to condensin I, with a function in sex chromosome dosage compensation (Csankovszki et al., 2009). This degree of functional radiation is surpassed by the diversification of condensin components in Tetrahymena, in which we identified a total of seven condensin genes, one each for homologues of Smc2, Smc4, and Cap-G, two Cap-D2 homologues, and four kleisin Cap-H/Brn1 subunit homologues. Different combinations of these subunits could form at least five different condensin complexes with as many different functions. Such evolutionary radiation may be partly due to the separation of gene expression and sexual gene propagation into two functionally and morphologically distinct nuclei within this organism. Indeed, protein epitope tagging and RNAi against individual condensin genes showed that the somatic and germline nuclei of Tetrahymena each utilize different configurations of condensin to perform different functions. Whereas SMC2, SMC4, CPG1, and CPDT1 all contribute to chromosome segregation in both nuclei, the CPH genes seem to have specific functions in each nucleus. Cph1 and Cph2 localize to the germline nucleus and together are required for chromosome segregation at mitosis and meiosis. Cph3 and Cph4 localize to the somatic nucleus. Cph3 is required for chromosome segregation and nuclear organization, whereas the function of Cph4 is not clear: its function may either be redundant with Cph3 or the depletion phenotype may be subtle. Cpdt2 also localizes exclusively to the somatic nucleus and has an as-yet unclear function in nuclear differentiation in young sexual progeny. Altogether, our data indicate the great flexibility of condensins to diversify and adopt novel functions.
Questions remain as to the exact contribution of the Cph proteins to the condensin composition in each nucleus throughout the Tetrahymena life cycle. Unfortunately, tagged versions of these proteins appeared to have reduced or altered function and caused phenotypes associated with the loss of condensin. In the future, it will be interesting to study the Cph proteins using antibodies generated to specific subunits. It may then be possible to determine if condensins with specific Cph subunits are preferentially bound to discrete chromosomal locations or if differences in expression or condensin composition exist at different times in the cell cycle.
Condensin’s contribution to mitosis and meiosis
Tetrahymena has small chromosomes and performs closed mitosis and meiosis. Mitotic chromosomes are difficult to study because of their small size, and the only visible effect of condensin depletion in mitosis was difficulty in separating daughter nuclei (Figure 5). In contrast, meiosis is more amenable to cytological inspection, and the observed meiotic anomalies in condensin mutants may also pertain to mitosis. Tetrahymena has a somewhat unusual meiotic program. When starved cells of different mating types are mixed, they pair and initiate sexual reproduction (Figure 1) (reviewed in Cole and Sugai, 2012). The prophase germline nucleus elongates to about twice the length of the cell (Figure 6A). During this stage, the chromosomes are roughly aligned within the tubular nucleus with the centromeres at one end and the telomeres at the other (Loidl and Scherthan, 2004; Loidl et al., 2012). As the nucleus shortens, DSBs are repaired and chromatin condenses until bivalents become visible at diakinesis; then the two meiotic divisions occur (Lukaszewicz et al., 2010, 2015; Shodhan et al., 2014, 2017). In cells depleted of condensin proteins that function in the germline nucleus (Smc2, Cpdt1, Cph1, or Cph2), prophase appears normal and the elongated nucleus can still shorten, indicating that nuclear shortening is not driven by condensin-mediated chromosome condensation. However, complete condensation of chromosomes into distinct bivalents is inhibited by loss of condensin, confirming a conserved role for condensin in germline chromosome condensation.
Although condensin-depleted cells can begin meiotic divisions, extensive anaphase bridging occurs that prevents chromosome segregation. The anaphase bridging phenotype is commonly associated with loss of condensin (Yu and Koshland, 2003; Hartl et al., 2008b; Lee et al., 2011). In budding yeast, there is evidence that meiotic bridging is caused by physical linkage of chromosomes, because condensin is required in meiosis for cohesin removal and resolution of recombination-dependent linkages (Yu and Koshland, 2003). In Tetrahymena, however, inhibition of meiotic recombination by SPO11 depletion increased rather than abrogated the severity of DNA bridging in condensin-depleted cells. Without recombination, homologues do not become linked and should separate freely in meiosis I. Therefore, the linkages causing persistent bridging in anaphase I must occur between either homologous or heterologous chromosomes, as reported during male meiosis in Drosophila cap-H2 mutants (Hartl et al., 2008b). Without condensin, it is likely that proper chromosome territories are not formed, resulting in intertwines between heterologous chromosomes.
Condensin in the organization of the polyploid nucleus
In diploid nuclei, cohesion of sister chromatids prior to cell division is critical for achieving equal segregation during mitosis. However, amitosis of Tetrahymena’s polyploid somatic nucleus requires a different strategy to ensure nearly equal distribution of the ∼50 copies of each chromosome. Experiments demonstrating phenotypic assortment imply that chromosome copies are distributed randomly to daughter nuclei (Allen and Nanney, 1958; Orias and Flacks, 1975). If correct, then this would require a mechanism for maintaining the separation, rather than cohesion, of chromosome copies (Figure 10). Condensin appears to be performing such a function in the somatic nucleus of Tetrahymena. We found dramatic clustering of somatic chromosome copies in cells depleted of the core condensin gene SMC2, the HEAT repeat gene CPDT1, or the soma-specific kleisin CPH3. In addition to a role for condensin in promoting territory formation (Hartl et al., 2008b; Bauer et al., 2012; Ito and Narita, 2015; Iwasaki et al., 2016) and decatenation (Leonard et al., 2015; Sen et al., 2016), there are several reports that the complex has a specific function in separating homologous sequences. In mouse neuronal stem cells, condensin II was found to prevent hyperclustering of pericentromeric regions known as chromocenters (Nishide and Hirano, 2014). In Drosophila, condensin II was reported to promote dissolution of polytene chromosomes and antagonize transvection, a process in which homologous loci influence each other’s transcription through their physical interaction (Hartl et al., 2008a; Nguyen et al., 2015). The action of condensin in the Tetrahymena somatic nucleus seems to be yet another example of such anti-interaction functions of condensin in interphase nuclei.
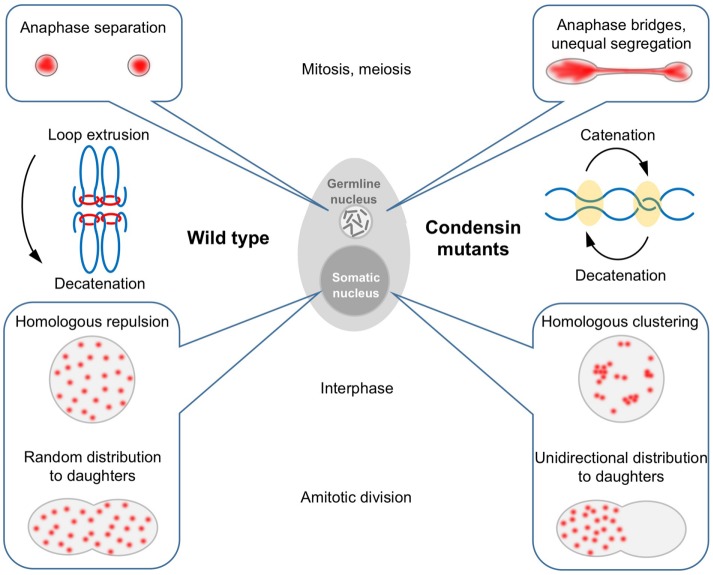
FIGURE 10:
Condensin has adapted to perform multiple functions in Tetrahymena’s two nuclei. In the germline nucleus, the conserved functions of condensin are required to condense and segregate chromosomes in meiosis and mitosis. In the presence of condensin, loop formation may compact chromosomes laterally and reduce the interface between sister chromatids. This could drive the action of Topo II toward decatenation, thus allowing chromatids to be separated easily by spindle forces. In the polyploid somatic nucleus, condensin acts as an anti-pairing factor that promotes the separation of newly replicated copies and maintains chromosome territories to prevent interaction between similar DNA molecules. These functions may help to maintain the even spacing of chromosome copies to aid the approximately equal segregation of copies during amitotic nuclear division.
The complete failure to segregate somatic chromatin in the absence of condensin cannot be entirely explained by clustering of chromosome copies. Massive interlinkage of all chromosomes would have to occur to prevent segregation; however, this is improbable due to the number and small size of somatic chromosomes. Therefore, it seems likely that condensin plays an additional, more active, role in somatic nuclear division. A previous study showed that partial knockdown of SMC4 disrupted microtubule formation within the dividing somatic nucleus (Cervantes et al., 2006). Exploration of the molecular or genetic interactions between condensin and microtubules in the dividing somatic nucleus may therefore lead to a better understanding of the unconventional amitotic segregation mechanism.
A universal model for condensin action
In this study, we show that condensin is required to segregate chromosomes in the two very different nuclei of Tetrahymena. Condensin mediates the condensation and resolution of germline chromosomes and promotes the spatial distribution and segregation of the smaller chromosomes of the polyploid somatic nucleus (Figure 10). One proposed mechanism for chromosome condensation is “loop extrusion” (Nasmyth, 2001; Goloborodko et al., 2016a,b). In this scenario, cohesin or condensin complexes bind a linear DNA molecule and, through the extrusion of loops, create a compact “noodle” closely resembling a condensed eukaryotic chromosome. By taking into account specific parameters for condensin occupancy, loop size, and the presence of a Topo II-like activity to release intertwines, computational modeling of loop extrusion can remarkably recapitulate the process of chromosome territory formation, sister chromatid resolution, and condensation (Goloborodko et al., 2016a). Condensin has been shown to mediate Topo II recruitment and resolution of DNA intertwines on chromosome arms (Leonard et al., 2015). A recent study showed that Topo II can both create and resolve DNA intertwines and that close physical proximity of sister chromatids promotes the catenation reaction in the absence of condensin-dependent supercoiling (Sen et al., 2016). Therefore, it is likely that the combined actions of cohesin removal and condensin supercoiling promote decatenation of sister chromatids (Figure 10). This model fits well with the action of condensin on germline Tetrahymena chromosomes and can be extended to account for condensin’s functions in the somatic nucleus. Chromosomes of Tetrahymena’s polyploid somatic nucleus are much smaller than most eukaryotic chromosomes, but their spacing nevertheless implies that they form distinct territories. Therefore, if a loop extrusion mechanism can produce chromosome territories within large chromosomes, then it should also be sufficient to separate replicated copies of small chromosomes, as well as untangle any unwanted interactions that occur between homologous sequences. In many organisms, it is now assumed that cohesin complexes are involved in loop extrusion (Schwarzer et al., 2017). However, cohesin is not present in Tetrahymena’s somatic nucleus, and therefore condensin may have been harnessed for this function.
Duplication and divergence of condensin subunits in Tetrahymena may explain the evolution of condensin complexes with various DNA-binding positions, on–off rates, or loop processivity, as well as the ability to interact with additional chromatin-bound proteins. Altering these properties, but not the basic action (i.e., DNA looping) of each condensin complex, may be sufficient to produce the range of condensin functions in both the germline and somatic nuclei.
MATERIALS AND METHODS
Strains and growth conditions
Tetrahymena strains B2086 and Cu428 obtained from the Tetrahymena stock center (https://tetrahymena.vet.cornell.edu/) were used as wild-type strains for transformations and as genomic DNA sources for amplification of regions used in tagging and RNAi constructs. Both strains were grown in modified Neff medium using standard methods (Orias et al., 2000).
Protein tagging and localization
Endogenous C-terminal protein tagging was performed by a knock-in strategy, as previously described (Howard-Till et al., 2013). In short, 500 base pairs of the C terminus of the gene of interest and 500 base pairs downstream of the gene were amplified by PCR and combined with the tagging epitope and Neo4 cassette by Gibson assembly using the NEBuilder HiFi DNA assembly master mix (New England BioLabs, Frankfurt, Germany). Primers used for amplification and assembly are listed in Supplemental Table S1. Plasmids pHA-Neo4, pGFP-Neo4, and pmCherry-Neo4 were used as the sources of tagging cassettes (gifts of K. Mochizuki). Transformation of strains by biolistic particle bombardment was performed as previously described (Cassidy-Hanley et al., 1997; Bruns and Cassidy-Hanley, 2000). Smc4-GFP, Cpg1-HA, Cpdt1-mCherry, Cpdt2-HA, and Cph2-mCherry strains were all constructed in this way, and protein localization was visualized in fixed cells by either direct detection or immunofluorescence (IF), as previously described (Loidl and Scherthan, 2004; Howard-Till et al., 2013). Primary antibodies used for IF were rabbit polyclonal anti-HA (1:100; Sigma, St. Louis, MO) and Living Colors mouse monoclonal JL-8 anti-GFP (1:50) and rabbit polyclonal anti-dsRed (1:100; Clontech Laboratories, Mountainview, CA). Endogenous C-terminal tagging of the other Cph subunits was unsuccessful; therefore, we ectopically expressed N-terminal HA-tagged proteins from the Mtt1 promoter. HA-Cph1 and HA-Cph4 expression constructs were created by amplifying the entire coding region of each gene and inserting it into pBNMB2-HA (a gift of K. Kataoka). Transformed strains were selected in 120 μg/ml of paromomycin and tagged protein expression was induced by the addition of 0.5 μg/ml CdCl2 to the growth medium.
RNAi
RNAi is performed by expressing a hairpin RNA molecule from an inducible promoter and can reduce RNA levels by up to 90% (Howard-Till and Yao, 2006; Howard-Till et al., 2013). RNAi constructs were created and introduced into cells as previously described for pREC8hpCYH (Howard-Till et al., 2013). Primers used to amplify gene fragments are listed in Supplemental Table S1. RNAi was induced by adding 0.3 μg/ml CdCl2 to cells growing in Neff medium or 0.05 μg/ml CdCl2 to cells in starvation medium. Starved cells were centrifuged and resuspended in starvation medium lacking cadmium prior to mixing to induce mating. Additional RNAi constructs for CPH1 and CPH2 were constructed to perform double RNAi experiments. These constructs were integrated into the BTU1 locus using the NEO5 selection cassette and expressed the RNAi hairpin from the MTT2 copper inducible promoter (Boldrin et al., 2008). RNAi from these constructs was induced by adding 100 μM CuSO4 to growing cells or 10 μM CuSO4 to starving or mating cells. Wild-type controls carried the relevant unexpressed RNAi construct. Confirmation of CPDT1 RNAi was performed by Western blotting analysis of tagged protein levels for a Cpdt1HA strain transformed with the CPDT1 hairpin construct (Figure 8C). Protein extracts were run on a Mini-PROTEAN TGX Stain-Free 4–20% gradient gel (Bio-Rad, Hercules, CA). The gel was blotted, and the membrane was probed with rabbit polyclonal anti-HA (1:1000; Sigma, St. Louis, MO). For the remaining constructs, RT-qPCR was used to assay RNA levels for the appropriate gene (Supplemental Figure S1). RNA was extracted from cells using TriFAST reagent (VWR, Radnor, PA), and RT-qPCR was performed using the LUNA Universal One-Step RT-qPCR kit (New England BioLabs, Frankfurt, Germany).
Live cell imaging
For live imaging, a Commodore Compressor device was used to flatten and immobilize the cells (Yan et al., 2014). Mating cells were concentrated by centrifugation and resuspended in 3% polyethylene oxide (Sigma, St. Louis, MO) to increase the viscosity of the medium, and 1 μl of the cell suspension was placed into the compressor. Cells were transformed with an HHT2-mCherry construct to visualize DNA (gift of K. Kataoka). Imaging was performed with a Zeiss Axioskop 2 wide-field fluorescence microscope, and images were aligned and processed using Adobe Photoshop and ImageJ software.
Fluorescence in situ hybridization
FISH was performed as previously described (Loidl and Scherthan, 2004), using probes generated by PCR amplification of 8–10 different regions (each ∼10 kb long) spanning the entire length of the somatic chromosome of interest. The total amount of DNA labeled was similar for each chromosome. Primers are listed in Supplemental Table S1. Somatic chromosome scaffolds of probed chromosomes were as follows: scf_8253887 (367,094 base pairs), scf_8254632 (79,368 base pairs), and scf_8254505 (131,308 base pairs). Sequences were retrieved using the genome browser of the Tetrahymena Genome Database (www.ciliate.org).
Supplementary Material
Acknowledgments
We are grateful to Kensuke Kataoka (National Institute of Basic Biology, Okazaki, Japan) for plasmid pBNMB2-HA and the HHT2-mCherry construct. Ece Sahi perfomed the live imaging experiments, and Emine Ali (Max F. Perutz Laboratories, Vienna, Austria) assisted with epitope tagging. Thanks also to Kazufumi Mochizuki (National Center for Scientific Research, Montpellier, France) for tagging and expression constructs. Finally, we thank Maria Novatchkova (Research Institute of Molecular Pathology, Vienna, Austria) for bioinformatics advice. This work was supported by grant P 28336-B28 from the Austrian Science Fund (FWF).
Abbreviations used:
- DSB
double-strand breaks
- FISH
fluorescence in situ hybridization
- RNAi
RNA interference
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-07-0451) on December 13, 2017.
REFERENCES
- Allen SL, Nanney DL. An analysis of nuclear differentiation in the selfers of tetrahymena. Am Nat. 1958:139–160. [Google Scholar]
- Bauer CR, Hartl TA, Bosco G. Condensin II promotes the formation of chromosome territories by inducing axial compaction of polyploid interphase chromosomes. PLoS Genet. 2012:e1002873. doi: 10.1371/journal.pgen.1002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldrin F, Santovito G, Formigari A, Bisharyan Y, Cassidy-Hanley D, Clark TG, Piccinni E. MTT2, a copper-inducible metallothionein gene from Tetrahymena thermophila. Comp Biochem Physiol C Toxicol Pharmacol. 2008:232–240. doi: 10.1016/j.cbpc.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Bruns PJ, Cassidy-Hanley D. Biolistic transformation of macro- and micronuclei. Methods Cell Biol. 2000:501–512. doi: 10.1016/s0091-679x(08)61553-8. [DOI] [PubMed] [Google Scholar]
- Cassidy-Hanley D, Bowen J, Lee JH, Cole E, VerPlank LA, Gaertig J, Gorovsky MA, Bruns PJ. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics. 1997:135–147. doi: 10.1093/genetics/146.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes MD, Coyne RS, Xi X, Yao MC. The condensin complex is essential for amitotic segregation of bulk chromosomes, but not nucleoli, in the ciliate Tetrahymena thermophila. Mol Cell Biol. 2006:4690–4700. doi: 10.1128/MCB.02315-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole E, Sugai T. Developmental progression of Tetrahymena through the cell cycle and conjugation. Methods Cell Biol. 2012:177–236. doi: 10.1016/B978-0-12-385967-9.00007-4. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Petty EL, Collette KS. The worm solution: a chromosome-full of condensin helps gene expression go down. Chromosome Res. 2009:621–635. doi: 10.1007/s10577-009-9061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuylen S, Metz J, Haering CH. Condensin structures chromosomal DNA through topological links. Nat Struct Mol Biol. 2011:894–901. doi: 10.1038/nsmb.2087. [DOI] [PubMed] [Google Scholar]
- Davalos V, Suarez-Lopez L, Castano J, Messent A, Abasolo I, Fernandez Y, Guerra-Moreno A, Espin E, Armengol M, Musulen E, et al. Human SMC2 protein, a core subunit of human condensin complex, is a novel transcriptional target of the WNT signaling pathway and a new therapeutic target J Biol Chem 2012. 43472 43481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote PLoS Biol 2006. e286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiu K, Numata O. Reorganization of microtubules in the amitotically dividing macronucleus of tetrahymena. Cell Motil Cytoskeleton. 2000:17–27. doi: 10.1002/(SICI)1097-0169(200005)46:1<17::AID-CM3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Goloborodko A, Imakaev MV, Marko JF, Mirny L. Compaction and segregation of sister chromatids via active loop extrusion. eLife. 2016a:e14864. doi: 10.7554/eLife.14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloborodko A, Marko JF, Mirny LA. Chromosome compaction by active loop extrusion. Biophys J. 2016b:2162–2168. doi: 10.1016/j.bpj.2016.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008:2204–2214. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham MF, Takakuwa T, Rahadiani N, Tresnasari K, Nakajima H, Aozasa K. Condensin mutations and abnormal chromosomal structures in pyothorax-associated lymphoma. Cancer Sci. 2007:1041–1047. doi: 10.1111/j.1349-7006.2007.00500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton EP, Kapusta A, Huvos PE, Bidwell SL, Zafar N, Tang H, Hadjithomas M, Krishnakumar V, Badger JH, Caler EV, et al. Structure of the germline genome of Tetrahymena thermophila and relationship to the massively rearranged somatic genome eLife 2016. e19090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl TA, Smith HF, Bosco G. Chromosome alignment and transvection are antagonized by condensin II. Science. 2008a:1384–1387. doi: 10.1126/science.1164216. [DOI] [PubMed] [Google Scholar]
- Hartl TA, Sweeney SJ, Knepler PJ, Bosco G. Condensin II resolves chromosomal associations to enable anaphase I segregation in Drosophila male meiosis. PLoS Genet. 2008b:e1000228. doi: 10.1371/journal.pgen.1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heale JT, Ball AR, Jr, Schmiesing JA, Kim JS, Kong X, Zhou S, Hudson DF, Earnshaw WC, Yokomori K. Condensin I interacts with the PARP-1-XRCC1 complex and functions in DNA single-strand break repair. Mol Cell. 2006:837–848. doi: 10.1016/j.molcel.2006.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 2012:1659–1678. doi: 10.1101/gad.194746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. Condensin-based chromosome organization from bacteria to vertebrates. Cell. 2016:847–857. doi: 10.1016/j.cell.2016.01.033. [DOI] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Howard-Till RA, Lukaszewicz A, Novatchkova M, Loidl J. A single cohesin complex performs mitotic and meiotic functions in the protist tetrahymena. PLoS Genet. 2013:e1003418. doi: 10.1371/journal.pgen.1003418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard-Till RA, Yao MC. Induction of gene silencing by hairpin RNA expression in Tetrahymena thermophila reveals a second small RNA pathway. Mol Cell Biol. 2006:8731–8742. doi: 10.1128/MCB.01430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Narita M. The expanding territories of condensin II. Cell Cycle. 2015:2723–2724. doi: 10.1080/15384101.2015.1063356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki O, Corcoran CJ, Noma K. Involvement of condensin-directed gene associations in the organization and regulation of chromosome territories during the cell cycle. Nucleic Acids Res. 2016:3618–3628. doi: 10.1093/nar/gkv1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki O, Tanaka A, Tanizawa H, Grewal SI, Noma K. Centromeric localization of dispersed Pol III genes in fission yeast. Mol Biol Cell. 2010:254–265. doi: 10.1091/mbc.E09-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Je EM, Yoo NJ, Lee SH. Mutational and expressional analysis of SMC2 gene in gastric and colorectal cancers with microsatellite instability. Acta Pathol Microbiol Immunol Scand. 2014:499–504. doi: 10.1111/apm.12193. [DOI] [PubMed] [Google Scholar]
- Johzuka K, Terasawa M, Ogawa H, Ogawa T, Horiuchi T. Condensin loaded onto the replication fork barrier site in the rRNA gene repeats during S phase in a FOB1-dependent fashion to prevent contraction of a long repetitive array in Saccharomyces cerevisiae. Mol Cell Biol. 2006:2226–2236. doi: 10.1128/MCB.26.6.2226-2236.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce EF, Erceg J, Wu CT. Pairing and anti-pairing: a balancing act in the diploid genome. Curr Opin Genet Dev. 2016:119–128. doi: 10.1016/j.gde.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer KM. Nuclear dualism. Methods Cell Biol. 2012:29–52. doi: 10.1016/B978-0-12-385967-9.00003-7. [DOI] [PubMed] [Google Scholar]
- Lee J, Ogushi S, Saitou M, Hirano T. Condensins I and II are essential for construction of bivalent chromosomes in mouse oocytes. Mol Biol Cell. 2011:3465–3477. doi: 10.1091/mbc.E11-05-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard J, Sen N, Torres R, Sutani T, Jarmuz A, Shirahige K, Aragon L. Condensin relocalization from centromeres to chromosome arms promotes Top2 recruitment during anaphase. Cell Rep. 2015:2336–2344. doi: 10.1016/j.celrep.2015.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J, Lukaszewicz A, Howard-Till RA, Koestler T. The Tetrahymena meiotic chromosome bouquet is organized by centromeres and promotes interhomolog recombination. J Cell Sci. 2012:5873–5880. doi: 10.1242/jcs.112664. [DOI] [PubMed] [Google Scholar]
- Loidl J, Scherthan H. Organization and pairing of meiotic chromosomes in the ciliate Tetrahymena thermophila. J Cell Sci. 2004:5791–5801. doi: 10.1242/jcs.01504. [DOI] [PubMed] [Google Scholar]
- Lukaszewicz A, Howard-Till RA, Loidl J. Mus81 nuclease and Sgs1 helicase are essential for meiotic recombination in a protist lacking a synaptonemal complex. Nucleic Acids Res. 2013:9296–9309. doi: 10.1093/nar/gkt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz A, Howard-Till RA, Novatchkova M, Mochizuki K, Loidl J. MRE11 and COM1/SAE2 are required for double-strand break repair and efficient chromosome pairing during meiosis of the protist Tetrahymena. Chromosoma. 2010:505–518. doi: 10.1007/s00412-010-0274-9. [DOI] [PubMed] [Google Scholar]
- Lukaszewicz A, Shodhan A, Loidl J. Exo1 and Mre11 execute meiotic DSB end resection in the protist Tetrahymena. DNA Repair. 2015:137–143. doi: 10.1016/j.dnarep.2015.08.005. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- Nguyen HQ, Nye J, Buster DW, Klebba JE, Rogers GC, Bosco G. Drosophila casein kinase I alpha regulates homolog pairing and genome organization by modulating condensin II subunit Cap-H2 levels. PLoS Genet. 2015:e1005014. doi: 10.1371/journal.pgen.1005014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishide K, Hirano T. Overlapping and non-overlapping functions of condensins I and II in neural stem cell divisions. PLoS Genet. 2014:e1004847. doi: 10.1371/journal.pgen.1004847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Fang Y, Spector DL, Hirano T. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol Biol Cell. 2004:3296–3308. doi: 10.1091/mbc.E04-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orias E, Flacks M. Macronuclear genetics of Tetrahymena. I. Random distribution of macronuclear genecopies in T. pyriformis, syngen 1. Genetics. 1975:187–206. doi: 10.1093/genetics/79.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orias E, Hamilton EP, Orias JD. Tetrahymena as a laboratory organism: useful strains, cell culture, and cell line maintenance. Methods Cell Biol. 2000:189–211. doi: 10.1016/s0091-679x(08)61530-7. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Inui YT, Uraguchi S, Yoshizumi T, Matsunaga S, Mastui M, Umeda M, Fukui K, Fujiwara T. Condensin II alleviates DNA damage and is essential for tolerance of boron overload stress in Arabidopsis. Plant Cell. 2011:3533–3546. doi: 10.1105/tpc.111.086314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvidou E, Cobbe N, Steffensen S, Cotterill S, Heck MM. Drosophila CAP-D2 is required for condensin complex stability and resolution of sister chromatids. J Cell Sci. 2005:2529–2543. doi: 10.1242/jcs.02392. [DOI] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber WCHH, Mirny L, Spitz F. Two independent modes of chromatin organization revealed by cohesin removal. Nature. 2017:51–56. doi: 10.1038/nature24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen N, Leonard J, Torres R, Garcia-Luis J, Palou-Marin G, Aragon L. Physical proximity of sister chromatids promotes Top2-dependent intertwining. Mol Cell. 2016:134–147. doi: 10.1016/j.molcel.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintomi K, Hirano T. The relative ratio of condensin I to II determines chromosome shapes. Genes Dev. 2011:1464–1469. doi: 10.1101/gad.2060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shodhan A, Kataoka K, Mochizuki K, Novatchkova M, Loidl J. A Zip3-like protein plays a role in crossover formation in the SC-less meiosis of the protist Tetrahymena. Mol Biol Cell. 2017:825–833. doi: 10.1091/mbc.E16-09-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shodhan A, Lukaszewicz A, Novatchkova M, Loidl J. Msh4 and Msh5 function in SC-independent chiasma formation during the streamlined meiosis of Tetrahymena. Genetics. 2014:983–993. doi: 10.1534/genetics.114.169698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega Mol Syst Biol 2011. 539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimborn M, Schindler D, Neitzel H, Hirano T. Misregulated chromosome condensation in MCPH1 primary microcephaly is mediated by condensin II. Cell Cycle. 2006:322–326. doi: 10.4161/cc.5.3.2412. [DOI] [PubMed] [Google Scholar]
- Yan Y, Jiang L, Aufderheide KJ, Wright GA, Terekhov A, Costa L, Qin K, McCleery WT, Fellenstein JJ, Ustione A, et al. A microfluidic-enabled mechanical microcompressor for the immobilization of live single- and multi-cellular specimens Microsc Microanal 2014. 141 151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HG, Koshland DE. Meiotic condensin is required for proper chromosome compaction, SC assembly, and resolution of recombination-dependent chromosome linkages. J Cell Biol. 2003:937–947. doi: 10.1083/jcb.200308027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.