Abstract
A vast diversity of microbes including macrofungi remain untapped for valuable bioactivities including antimicrobial activity. Searching wild sources may bring novel natural products with antimicrobial properties that can provide protection against infectious diseases. The present study was designed to identify the diverse forms of mushrooms being used as an ethnomycological source of food and medicine by the tribes of Meghalaya, India, and microscopically study the structures of mushrooms along with observing their antimicrobial effects on pathogens. Fruiting bodies of mushrooms were viewed morphologically and microscopically, and were identified using molecular markers. The dried aerial parts of the fruiting bodies were extracted with methanol and screened for their antimicrobial activity using 2,3,-triphenyl tetrazolium chloride against two Gram-negative and two Gram-positive bacteria. The average diameter of the inhibitory zone induced by fungal extracts ranged from 9 mm to 22 mm for Gram-negative and from 16 mm to 24 mm for Gram-positive bacteria, indicating that this dietary source is a good antimicrobial agent. Mushroom structures were examined using optical microscopy, while the deformities on the pathogens inflicted by mushroom extracts were visualized using scanning electron microscopy, which showed accumulation and formation of biofilm in Gram-positive and shrinkage with cavity formation in Gram-negative bacteria.
Keywords: ethnomycology, metabolites, microscopy, minimum inhibitory concentration, mushroom
1. Introduction
Infectious diseases remain one of the major concerns for humankind since the 20th century. Development of resistance by clinical pathogens against a wide range of antibiotics is a global issue. Infections by multidrug-resistant clinical isolates of Escherichia coli, Staphylococcus aureus, and Klebsiella pneumoniae are a major drive to stimulate the search of novel biological samples as antimicrobial agents [1].
Although a wide spectrum of antibiotics produced by fungi, including penicillin, streptomycin, cephalosporin, griseofulvin, erythromycin, and rifamycin, are used commercially, antibiotics from imperfect fungi are scarcely documented. Imperfect fungi or macrofungi have been used as a source of food and medicine since the Roman and Greek times [2,3]. The first ever report of basidiomycetes being used as a source of antimicrobial agent was first reported by Anchel et al. [4]. The mycelium and the fruiting body contain a wide range of bioactive compounds that can be used for their antimicrobial properties. Besides the cell wall glucans and secondary metabolites, basidiomycetes are also known to contain phenolic compounds, flavonoids, triterpenoids, lectin, dietary fibers, terpenes, and alkaloids, which have been proven to show significant bioactivities against microbial infections and anti-inflammatory, antiviral, anticancer, cardiovascular, and gastrointestinal disorders along with immunomodulatory properties [5,6,7].
The methanolic extract of polyporus mushrooms such as Ganoderma luciderm, Phellinus rimosus, and Navesporous floccose is known to show antibacterial activity against a wide spectrum of clinical isolates [8]. As reported by Belsare et al. [9], chloroform and methanolic extracts of Phellinus switeniae showed potent activity against Acinetobacter baumannii.
Since multidrug resistance of pathogenic microorganisms has been rampant due to the extensive use of commercial drugs for treatment of infectious diseases, there is a need for potent novel antimicrobial sources that can act against a wide range of clinical pathogens. The local tribal communities of Northeast India use mushrooms as a source of food rather than as a source of medicine. The present study aims to identify the diverse range of basidiomycetes that are predominant in the local subtropical forest of Northeast India and to screen them for their antimicrobial potential against the most common clinical pathogenic bacteria. This study also tried to correlate the electron micrographs of the subcellular ultrastructural deformities in pathogens induced by the mushroom extracts.
2. Materials and methods
2.1. Sampling location
Five mushroom species were collected from the forest of Meghalaya (25°26.7’N and 91°44.92’E) at an altitude of 1788 m with 75% humidity.
2.2. Sample collection and processing
Mushroom samples were collected in separate sterile bags and brought to the laboratory. These samples were washed gently with double-distilled water to remove the dirt, while maintaining complete safety by using sterile gloves and mask. Morphological identification of wild macrofungi was performed freshly according to both phenotypic and microscopic characteristics, following the standard parameters and online key [10]. A part of the collection was preserved in formaldehyde with the soft tissue in 2% formaldehyde and leathery in 4% formaldehyde solution in glass jars and stored in the in-house culture collection of the parent university. The remaining parts were shade dried for 3–4 days, wrapped with aluminum foil, and stored at −20 °C for further analysis.
2.3. Optical microscopic examination
Small pieces of the pileus were cut from the fruit body and were mounted on a cover slip over a filter paper. The plate was immediately covered with a lid. The spore print was observed after 24 hours on the surface of the filter paper. The spores were observed under microscope using two different dyes: lactophenol cotton blue and Melzer's reagent (0.5 g of iodine, 1.5 of potassium iodide, 22 g of chloral hydrate, and 20 g of water). Melzer's staining reaction was used for detecting amyloid, pseudoamyloid, or nonamyloid of the spores [11].
2.4. Molecular characterization of the macrofungi
Freeze-dried fungal tissues were considered for extraction of DNA. Genomic DNA was extracted using HiPurA fungal DNA isolation kit (Himedia, Mumbai, India) and characterized by two universal primers—lTS1F (5’-TCCGTAGGTGAACCTGCGG-3’) and 1TS4R (5’-TCCTCCGCTTATTGATATGC-3’) [12]. Amplification was carried out in a GeneAmp 9700 thermal cycler (Applied Biosystems, Foster City, California, USA) under the following conditions: initial denaturation at 95 °C for 5 minutes; 35 cycles of denaturation at 9 °C for 1 minute, annealing at 52 °C for 30 seconds, extension at 72 °C for 1 minute; and a final extension at 72 °C for 10 minutes. The amplified Polymerase Chain Reaction products were electrophoretically analyzed on 1.5% agarose gel along with a 100 bp marker ladder. Amplified ITS ribotypes were purified using the QIAquick gel extraction kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. All the purified amplicons were sequenced (Xcelris Lab, Ahmedabad, lndia).
2.5. Sequence and phylogenetic analysis
The sequences were searched for similarity with other deposited sequences in the National Center for Biotechnology Information (NCBI) database. The aligned sequences were computed using ClustalW software and sequence homologies were determined using a Basic Local Alignment Search Tool (BLAST) search to create an evolutionary distance matrix using MEGA 6 software. The sequences were submitted to NCBI and accession numbers were obtained.
2.6. Preparation of crude aqueous methanolic extract
The fruiting body was separated, washed with doubledistilled water to remove dirt, oven dried at 40°C, and powdered using a mortar and pestle. Ten grams of fine powder were mixed with 70% methanol at room temperature with stirring at 150 rpm for 5 days. Methanol was then evaporated to dryness under vacuum pressure at a temperature of 45 °C in a rotary evaporator, to obtain the crude extract of the macrofungi [13]. The dried biomaterial was finally dissolved in dimethylsulfoxide (DMSO) at a stock concentration of 1 mg/mL, which was further analyzed for antimicrobial activity.
2.7. Antimicrobial assay by disc diffusion method
In vitro antimicrobial assay was carried out using the disc diffusion method against two Gram-positive and two Gram-negative bacterial pathogens, following the protocol of Katoch et al. [14] with minor modifications. The bacterial strains Streptococcus pyogenes (MTCC1925), S. aureus (MTCC96), K. pneumoniae (MTCC109), and E. coli (MTCC730) were grown on brain–heart infusion at a temperature of 37 °C for18–24 hours until it reached a turbidity of 0.5 McFarland standards (106 colony forming units/mL). The antimicrobial assay was performed on Mueller Hinton Agar plates using a sterile susceptibility disc (Himedia), 6 mm in diameter, into which 40 μL of the crude methanolic extract and the pure extraction solvent (DMSO) as a negative control were loaded separately. The plates were preincubated at 4 ° C for 1 hour for uniform diffusion. After preincubation, all the plates were incubated at 37°C for 24 hours. Chloramphenicol (30 μg) was used as a positive control to determine the sensitivity of the strains. Antimicrobial activity of the crude extracts was determined by measuring the zone of inhibition around the sterile filter paper disc.
2.8. Minimum inhibitory concentration of the extracts using micro dilution technique
The minimum inhibitory concentration (MIC) of the macrofungal extract was determined in vitro using the microdilution technique in 96 multiwell microtiter plates, with minor modifications [15]. For susceptibility analysis, 50 μL of the bacterial broth was distributed from the second to the 12th well in the plate (A2–A12). Stock concentration (1 mg/mL) of the crude extract was further diluted with DMSO to reach a final concentration of 25 mg/mL. The crude extract of concentration 25 mg/mL (100 μL) was added into the first row of the plate. To the rest of the wells, serial dilution was performed using a micropipette (A2–A10). A single antimicrobial crude extract, in progressive dilutions, was added to each of the wells. Then, 50 μL of the bacterial suspension of around 106 colony forming units/mL was added to each well. Each plate has a set of both growth in A11 and sterility control in A12. Plates were covered with lids to avoid dehydration of the microbial cells and were placed in an incubator at a growth condition of 37 °C for 24 hours. After incubation, 10 μL of 0.2% 2,3,5-triphenyl tetrazoliumchloride was added in each well. The microtiter plates with 2,3,5-triphenyl tetrazoliumchloride were further incubated at the same temperature for 1 hour. A visible color change from purple to pink in 1 hour indicated the growth of microbes and was recorded as a negative control. The MIC of the macrofungal extract was recorded as the lowest concentration that showed no microbial growth.
2.9. Scanning electron microscopy
In order to analyze the morphological deformities of the test bacteria under scanning electron microscopy (SEM), the test organisms were treated with 4× concentration of the MIC of the macrofungal extract and were kept for overnight incubation at 37 ° C. At the end of the incubation, bacterial cultures were washed three times with distilled water and then fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.3) for 4 hours. Glutaraldehyde was drained off, and the bacterial pellets were subjected to three consecutive 1-hour washes with 0.1 M cacodylate buffer, followed by one wash with distilled water. The samples were further dehydrated with a series of acetone wash (30%, 50%, 70%, 80%, 90%, 95%, and 100%), and then the cells were immersed in trimethylsilane for twice at a time interval of 10–15 minutes after every wash at 4 °C and brought to room temperature for drying. Finally, the samples were sputter coated with a thin layer of gold-palladium and scanned under SEM (JSM-6360; Jeol, Peabody, Massachusetts, USA).
3. Results
3.1. Isolation and identification
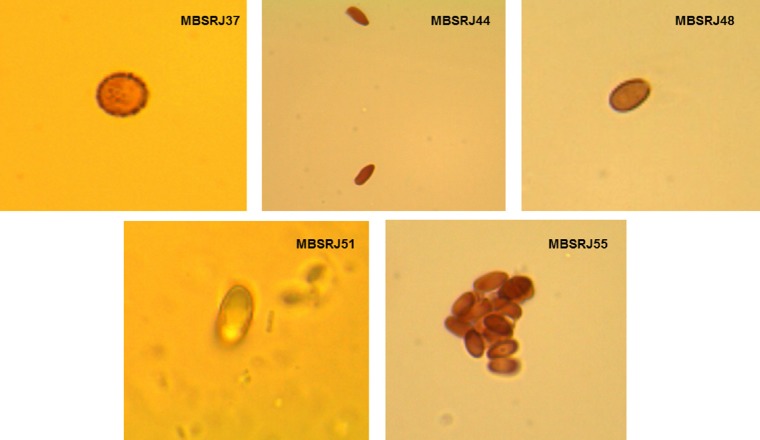
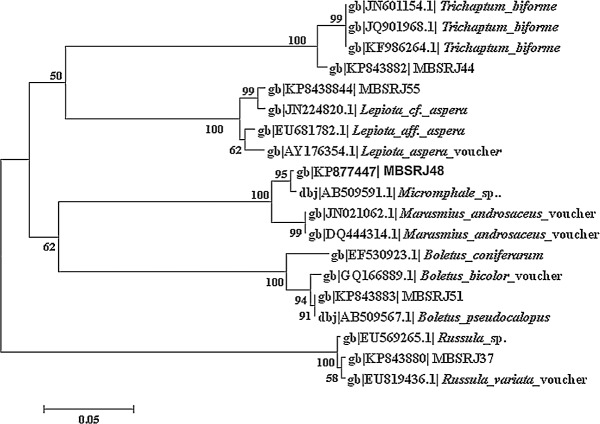
A total of five macrofungi with different morphological, sporological, and molecular traits were screened (Figure 1). The phenotypic trait such as the macrofungal color, shape, size of the pileus and stalk, color, texture of the spore (Table 1) was used for tentative identification of the macrofungi. Microscopic analysis of the spores was conducted to determine whether they are amyloid, dextrinoid, and inamyloid in nature (Figure 2). Molecular characterization of the conserve region of the macrofungi to amplify rDNA-ITS region was considered to be the confirmatory identification where the sequence data were aligned using BLAST, and the macrofungi were found to be the closest homolog of Russula variata, Trichaptum bio-forme, Baorangia pseudocalopus, Echinoderma aspera, and Micromphale foetidum (Figure 3). The ITS sequence for the macrofungi was deposited to NCBI, and the accession numbers were obtained (KP843880, KP843882-KP843884, and KP877447).
Fig. 1.
Morphological features of macrofungi selected in the study.
Table 1.
Macroscopic/microscopic features of the studied macrofungi.
| Isolates | Macroscopic/microscopic features |
|---|---|
| MBSRJ37 | Cap: 2.5–6.5 cm broad, convex in young, depressed shallow, green to olive green in the center, dry leathery smooth surface |
| Stalk: 6 cm long, 1.5–2 cm thick, whitish, brittle, smooth | |
| Gills: whitish color continuous running down the stem | |
| Spores: 8.01–9.23 μm × 4.01 μm, whitish spore print, elliptical, dextrinoid in Melzer | |
| MBSRJ44 | Cap: polyporus up to 3.3 cm, semicircular with wavy edges, leathery texture |
| Stalk: absent | |
| Gills: absent | |
| Pores: pale brownish | |
| Spores: 3.75–3.67 μm × 2.00 μm, whitish spore print, oval to slightly allantoids, dextrinoid | |
| MBSRJ48 | Cap: pale to dark brown, depressed in the center, convex, scales are distinct, up to 3.6 cap diameter, dry scaly surface |
| Stalk: 3.2 cm, dark brown to black toward the end | |
| Gills: whitish to pale brownish | |
| Spores: 5.2 μm × 2.88 μm, whitish spore print, amyloid | |
| MBSRJ51 | Cap: 6.2 cm broad, hemispherical, flat convex, soft velvety, reddish yellow |
| Stalk: 6 cm long, 1–3 cm thick, swollen at the base | |
| Gills: absent | |
| Pores: tubular, reddish yellow | |
| Spores: 4.63–5.83 μm, brownish spore print, inamyloid in Melzer | |
| MBSRJ55 | Cap: 9.6 cm broad, flat convex, covered with dusty fibers, soft velvety, pale brownish at the center, whitish at the edge |
| Stalk: 6–9.5 cm long, 1 cm thick, dark brown at the base | |
| Gills: present, whitish in color | |
| Spores: 3.58–3.67 μm, long elliptical whitish spore print, dextrinoid in Melzer |
Fig. 2.
Morphological nature of spores observed among mushrooms.
Fig. 3.
Evolutionary positions of macrofungi with their related species based on rRNA-ITS sequence similarity.
3.2. Antimicrobial assay
All the five voucher fungi showed antibacterial efficacy. The extract of the macrofungus T. bioforme (MBSRJ44) showed the highest antimicrobial activity against pathogenic microbes, followed by the extracts of R. variata (MBSRJ37) and B. pseudocalopus (MBSRJ51). The control disc with DMSO did not show any zone of inhibition (Table 2).
Table 2.
Antimicrobial activity of the macrofungal extract against pathogenic microbes (inhibition zone in mm).
| Isolates | K. pneumoniae | E. coli | S. aureus | S. pyogenes |
|---|---|---|---|---|
| MBSRJ37 | 10 | 25 | 14 | 13 |
| MBSRJ44 | 12 | 26 | 22 | 11 |
| MBSRJ48 | 17 | 10 | 15 | 9 |
| MBSRJ51 | 21 | 16 | 8 | 8 |
| MBSRJ55 | 17 | 16 | 10 | 10 |
| DMSO | – | – | – | – |
The symbol “—”indicates no zone of inhibition.
DMSO = dimethylsulfoxide.
3.3. MIC of crude extracts
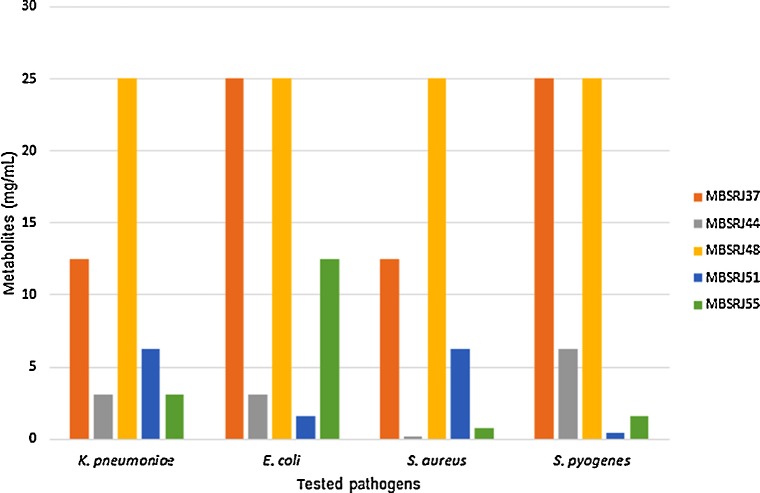
All the crude extracts with antimicrobial activity were further evaluated for their MIC by the 96-well microdilution technique. According to the primary antimicrobial screening, the MIC ranged from 0.195 mg/mL to 25 mg/mL against the tested strains. The MIC value of the macro-fungal extract of T. bioforme (MBSRJ44), which showed an antagonistic effect against Gram-positive S. aureus, was 0.195 mg/mL; this was followed by the MIC value (0.78mg/mL) of the macrofungal extract of B. pseudocalopus (MBSRJ51) against S. pyogenes (Figure 4). The findings of the MIC values endorse the finding of the primary screening using the disc diffusion technique. It is also evident from the results that the macrofungal extract showed a more synergistic effect against Gram-positive bacteria than against Gram-negative bacteria, which may be attributed to the difference in the cell wall composition. The experiments were repeated three times and the average MICs were obtained.
Fig. 4.
Inhibition of tested pathogens by the mushroom extracts, expressed as MIC.
MIC = minimum inhibitory concentration.
3.4. Scanning electron micrograph analysis
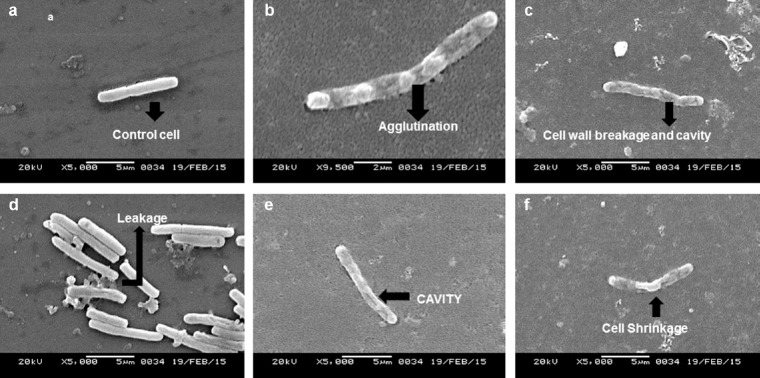
The extracts of the voucher fungi were found to possess potent antibacterial effects against the pathogenic strains and showed alteration of cell morphology, as revealed by scanning electron micrographs. Control strains showed a smooth, unruffled regular surface, whereas strains treated with the macrofungal extracts showed cavity formation, and shrunken and distorted cell walls. Compounds in the crude extracts were able to disrupt the intact cell membrane of the strains, allowing the formation of cavities and swelling, shrinkage, and leakage of intercellular components. The healthy cells of K. pneumoniae have a regular and smooth shape (Figure 5A), whereas the cells damaged by the effect of the extracts were found to contain bleb-like irregular protrusions with shrunken and agglutinated inner compositions (Figures 5B-F). However, E. coli did not reveal any physical deformities under the scanning electron microscope (Figures 6A and 6B). The macrofungal extracts may have certain irregularities in the metabolic activities of E. coli. The control strain, Gram-positive S. aureus, formed cell-to-cell smooth grape-like structures (Figure 7A), whereas the treated pathogens showed cellular deformities with sticky membrane and leakage of the internal compositions (Figures 7B-D). The micrograph of the treated S. pyogenes revealed the formation of biofilm, cell agglutination, and discrete clumps that were not seen in the case of control cells (Figures 8A and 8B). The SEM micrograph of both Gram-negative and Gram-positive bacteria on treatment with the macrofungal extract showed perforation in the cell wall membrane along with leakage of inner compositions.
Fig. 5.
(a—f) Scanning electron micrographs of untreated K. pneumoniae showing normal morphology (a) and those of the treated cells showing agglutination (b) of cellular components, cell wall breakage (c), leakage (d), cavity formation (e) and shrinkage of cellular components (f).
Fig. 6.
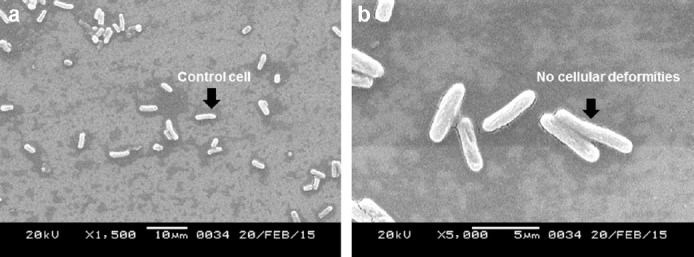
(a and b) Scanning electron micrographs of normal (a) and treated E. coli showing no physical damages in the cellular structures (b).
Fig. 7.
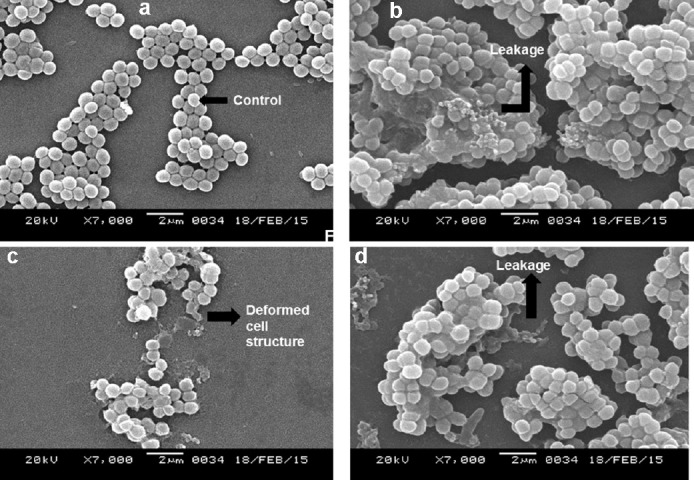
(a–d) Micrographs showing control (a) and treated cells of S. aureus with deformed cellular structure and leakage (b–d).
Fig. 8.
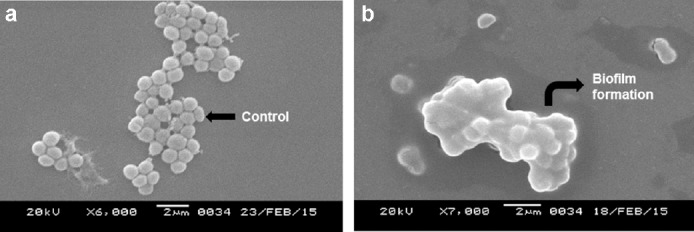
(a and b) Scanning electron micrographs of untreated (a) and biofilm forming treated S. pyogenes (b).
4. Discussion
Subcellular deformities were examined using electron microscopy, which supports the hypothesis that the extracts of higher fungi can be a potent source of antimicrobial agents causing cellular deformities in clinical pathogens. The findings of this study correlated with the findings of Ozturk et al. [16] as the MIC of the macrofungal extract is more potent against Gram-positive bacteria than against Gram-negative pathogens. However, the effect was much more prominent in the morphology of Gram-negative K. pneumoniae. Electron microscopy provided the ultrastructural details of deformities in the cell architecture on treating the cells with the extracts. SEM micrographs provided evidence of cell shrinkage and cell wall destruction. An increased number of antimicrobial agents from macrofungi are anticipated, and the traditional use of macrofungi by ethnic communities as a source of medicine holds importance in this regard. All the macrofungi considered in the present study exhibited an antagonistic effect against the tested clinical pathogens. The macrofungus E. aspera demonstrated the highest antimicrobial activity, which suggests that it can be explored as a potential source of an antimicrobial agent.
Identification of the macrofungi and their in vitro activity provides favorable clinical evidence of their use as antimicrobial agents. Visualization of the cellular deformities of treated clinical pathogens and their scanning electron micrographs give a platform to understand the nature of deformities and damages induced by the fungal extracts on the pathogens. With increasing resistance of clinical pathogens toward commercial antibiotics, methanolic extract of macrofungi holds promise as a novel drug candidate in the modern era. The present study brings out the first report on macrofungi consumed by ethnic tribes from Northeast India, which were surveyed as potential antimicrobial agents and provide promising observations.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
The authors acknowledge the financial support received from the Department of Science and Technology (SERB/SR/SO/PS/49/2012), Government of India, to carry out the present work. They also thank Sophisticated Analytical Instrument Facility, North Eastern Hill University, Shillong, for providing the SEM service.
References
- [1].Thomson KS, Moland ES. Version 2000: the new blactamases of Gram-negative bacteria at the dawn of the new millennium. Microb Infect. 2000;2:1225–35. doi: 10.1016/s1286-4579(00)01276-4. [DOI] [PubMed] [Google Scholar]
- [2].Sagakami H, Aohi T, Simpson A, Tanuma S. Induction of immunopotentiation activity by a protein-bound polysaccharide PSK. Anti-cancer Res. 1991;11:993–1000. [PubMed] [Google Scholar]
- [3].Wasser SP, Weis AL. Medicinal properties of substances occurring in higher Basidiomycetes mushrooms: current perspectives (review) Int J Med Mushrooms. 1999;1:31–62. [PubMed] [Google Scholar]
- [4].Anchel M, Hervey A, Kavanagh F, Polatnick J, Robbins WJ. Antibiotic substances from Basidiomycetes III. Caprius similis and Lentinus degener. Proc Nat Acad Sc. 1948;34:498–502. [PMC free article] [PubMed] [Google Scholar]
- [5].Kim S, Fung DYC. Antibacterial effect of water-soluble arrowroot (Puerariae radix) tea extracts on food borne pathogens in ground beef and mushroom soup. J Food Prot. 2004;67:1953–6. doi: 10.4315/0362-028x-67.9.1953. [DOI] [PubMed] [Google Scholar]
- [6].Akyuz M, Onganer AN, Erecevit P, Kirbag S. Antimicrobial activity of some edible mushrooms in the Eastern and Southeast Anatolia Region of Turkey. Gazi Univ J Sci. 2007;23:125–30. [Google Scholar]
- [7].Barros L, Calhelha RC, Vaz JA, Ferreira ICFR, Bapista P, Estevinho LM. Antimicrobial activity and bioactive compounds of Portuguese wild edible mushrooms methanolic extract. Eur Food Res Technol. 2007;225:151–6. [Google Scholar]
- [8].Sheena N, Ajith TA, Mathew A, Janardhanan KK. Antibacterial activity of three macrofungi, Ganoderma lucidum, Navesporus floccosa and Phellinus srimosus occurring in South India. Pharm Biol. 2013;41:54–6. [Google Scholar]
- [9].Belsare MH, Ranadive KR, Bapat GS, Garad S, Deokule SS, Vaidya JG. Screening of mushroom Phellinus switeniae (Murr) S. Herrera and Bondart against clinical isolates of Acinetobacter baumannii Bouvet & Grimont. Elixir Appl Botan. 2013;54:12398–9. [Google Scholar]
- [10].Peterson JH, Gaba A, Laessøe T. [accessed 12 August, 2015]. Available at: http://www.mycokey.com .
- [11].Kuo M. Using a microscope: equipment. Retrieved from the MushroomExpert.com. 2006. [accessed 18 July, 2013]. Web site: http://www.mushroomexpert.com/microscope equipment.html .
- [12].White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequenc-ing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, editor. PCR protocols, a guide to methods and applications. San Diego, CA: Academic Press Publishers; 1990. pp. 315–32. [Google Scholar]
- [13].Gbolagade JS, Fasidi IO. Antimicrobial activities of some selected Nigerian mushrooms. Afr J Biomed Res. 2005;8:83–7. [Google Scholar]
- [14].Katoch M, Singh G, Sharma S, Gupta N, Sangwan PL, Saxena AK. Cytotoxic and antimicrobial activities of endophytic fungi isolated from Bacoba monnieri (L.) Pennell (Scrophulariaceae) BMC Complement Altern Med. 2014;14:52. doi: 10.1186/1472-6882-14-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nath A, Joshi SR. Ultrastructural effect on mastitis pathogens by the extract of endophytic fungi associated with ethnoveterinary plant, Hibiscus sabdariffa L. J Microsc Ultrastruct. 2014;3:38–43. doi: 10.1016/j.jmau.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ozturk M, Emin Duru M, Kivrak S, Mercan-Dogan N, Turkoglu A, Ozler MA. In vitro antioxidant, anticholinesterase and antimicrobial activity studies on three Agaricus species with fatty acid compositions an iron content: a comparative study on the three most edible mushrooms. Food Chem Toxicol. 2011;49:1353–60. doi: 10.1016/j.fct.2011.03.019. [DOI] [PubMed] [Google Scholar]