Abstract
There is an evident difference in the intensity of morbidity caused by Schistosoma haematobium in North-African zones compared to Sub-Saharan ones. Clinical outcome dichotomy corresponds to two geographically distinct intermediate host snail species that are only infected by the related strain of the parasite. In concert, there is a manifest hybridization of the parasite with other Schistosoma species confined to certain regions of Africa. This raises a reasonable suggestion that S. haematobium has no less than two phylogenetic clusters that have different virulence. The aim of the study was to examine the possible diversity among S. haematobium using simultaneous amplification of genomic DNA of selected isolates. Random amplified polymorphic DNA-polymerase chain reaction markers were used to study the genetic diversity among S. haematobium natural isolates from selected regions of Africa (Egypt, Zimbabwe, and South Africa) that represent different ecological conditions, different species of intermediate host, and different possibilities of field hybridization with other schistosomes. A moderate to high level of genetic diversity was evident among the three isolates. More bands were shared by the isolates from Zimbabwe and South Africa (similarity index = 0.721) than those shared by each with the Egyptian isolate (similarity index = 0.551 and 0.566, respectively), suggesting that at least two phylogenetic groups of S. haematobium do exist in distinct geographic regions of Africa. The elucidation of the possible genetic diversity among S. haematobium parasites may explain many ambiguous aspects of the biology of the parasite-like virulence, immune evasion and drug resistance.
Keywords: clinical outcome, genetic diversity, polymerase chain reaction, random amplified polymorphic DNA, Schistosoma haematobium, virulence
1. Introduction
Human schistosomiasis is a serious chronic disease caused by trematode parasites of the genus Schistosoma, which is more prevalent in developing countries. Schistosomiasis is second after malaria among the parasitic diseases that cause human morbidity and mortality. Schistosoma haematobium is one of the most widespread schistosome species and is responsible for human urogenital schistosomiasis [1]. Although it has sporadic worldwide distribution, S. haematobium is probably truly African in origin. The parasite tends to be less focal, more prevalent, and more widely distributed throughout the continent than other human schistosomes [2].
The distribution of S. haematobium is restricted by its molluscan intermediate hosts. In Northern Africa, they are represented by bulinid snails of the truncatus group, while in Southern Africa, the hosts are members of the africanus group [3]. The northern strains of the parasite are not infective for the southern host snails and vice versa [4].
Schistosomiasis has a significant variation in morbidity and pathology among human hosts. In endemic regions, morbidity is related to the age of the human host, with an increase in early childhood, reaching a peak during adolescence and declining thereafter [5]. More interestingly, significant disparities in clinical outcome of S. haematobium infection have been linked to different geographical regions, ranging from trivial symptoms to serious pathology of the urinary tract. While the majority of infections in Sub-Saharan Africa result in mild pathology [6], the degree of pathology caused by S. haematobium in Egypt is severe and is considered a significant threat to public health [7]. The degree of pathology inflicted by S. haematobium in countries of Sub-Saharan Africa like Mali [8], Zimbabwe, Malawi [9], Ghana [10], Nigeria [11], and South Africa [12] is less severe compared to that observed in Egypt. Morbidity, due to urinary schistosomiasis in these countries, is mild and does not represent a serious public health problem, in spite of high prevalence rates as in Malawi, where half of the population is believed to be infected [9]. By contrast, urinary schistosomiasis in Egypt is usually manifested by obstructive uropathy that predisposes to pyelonephritis, lower urinary tract infections and urinary calculi. It also predisposes to metaplastic urothelial changes, leading to a high percentage of squamous cell carcinoma and adenocarcinoma [7].
The reasons behind these discrepancies in clinical outcome remain ambiguous. However, the genetic structure of the parasite is believed to play a role in this phenomenon. Genetic diversity is believed to have a major influence on many parasite-related characteristics, including the dynamics of transmission, host-parasite interaction, infectivity and virulence [13]. The genetic variability within S. haematobium species is largely understudied compared to that of its more serious counterpart Schistosoma mansoni [14], primarily due to many constraints in the process of laboratory passage of the parasite and absence of sufficient specific markers [15]. Studying the population genetics of S. haematobium is crucial for better understanding of the epidemiology of infection and for advancing novel treatment and vaccination strategies [16].
Random amplified polymorphic DNA (RAPD) primers represent sensitive markers for exploring genetic variability and degree of gene flow among parasitic populations. RAPD-polymerase chain reaction (PCR) has been developed in an attempt to discriminate between species, strains, and individuals [17]. With such technology, it became feasible to investigate population genetics of schistosomes with few available sequence data [18]. RAPD primers perform a whole genome scan, identifying a large number of loci and have emerged as a powerful tool in distinguishing both inter-as well as intraspecific variations [19,20]. Several studies have utilized RAPD primers to explore the genetic variability within schistosome populations in different hosts [21,22]. The small amount of DNA required for such a technique make it feasible to use cercarial or miracidial DNA, rather than DNA derived from adult schistosomes, to investigate genetic background, thus reducing the expected selection exerted by atypical hosts [23,24].
A few studies have investigated the relation between morbidity caused by S. haematobium infection and genetic diversity of the parasite, with conflicting results [25,26]. The aim of the current study was to explore genetic diversity within natural populations of S. haematobium selected from different geographic areas across Africa, using parasite isolates of Schistosoma eggs sampled directly from their natural human hosts, thereby avoiding the biological biases encountered on using laboratory-passed adult worms. This study provides the opportunity to investigate an important and understudied issue, which is the contribution of parasite genetics to discrepancies in disease severity and clinical outcome.
2. Materials and methods
2.1. Collection of samples
S. haematobium eggs were a generous gift from Dr. Clive Shiff, Johns Hopkins University, Baltimore, MD, USA. The eggs were retrieved by a filtration technique [27] from three noon urine samples (10 mL each) from heavily infected patients from Egypt, Zimbabwe, and South Africa. The process of egg washing and filtration was repeated until absence of any host cells was assured.
2.2. DNA preparation
Genomic DNA extraction from all collected samples was performed utilizing QIAamp DNA Mini kit (QIAGEN, Hilden, Germany). Genomic DNA pellet was dissolved in Tris-EDTA (TE) buffer to a working concentration of 5 ng/μL.
2.3. RAPD-PCR
Four arbitrarily selected 10-bp oligonucleotide primers (Operon Technologies, Alameda, CA, USA) were used; Primer number (P#)2: TGCCGAGCTG, P#7: GAAACGGGTG, P #9: GGGTAACGCC, and P # 10: GTGATCGCAG. Genomic DNA of S. haematobium (10 ng) was used in a PCR [23] with a total reaction volume of 20 μL in a DNA thermal cycler (Perkin–Elmer, Waltham, MA, USA) for 40 cycles, each of 10 seconds denaturation at 94 °C, 1 minute annealing at 36 °C, and extension for 2 minutes at 72°C with transition time 1 °C/s between the different temperature phases.
2.4. Gel electrophoresis
DNA electrophoresis was done on a 1.2% agarose gel in TE buffer stained with ethidium bromide. Samples were run with two molecular weight standards: high-molecular weight and a low-molecular weight 100-bp ladder (Gibco, Waltham, Massachusetts, USA). Gels were visualized under UV light and photographed using Polaroid 667 film (Polaroid corporation, Waltham, MA).
2.5. Data analysis
DNA bands from different isolates produced by RAPD-PCR were compared to a DNA molecular weight standard to guarantee consistent scoring between different gels and to determine the molecular weight of the band. Faint, unclear bands and high-molecular weight (> 4 kb) bands were not scored and disregarded. All bands generated by each isolate were pooled together. Comparison between isolates was done by scoring the total bands of each isolate, the bands shared between two different isolates, and the bands that are absent from one isolate and present in the other one. The similarity between different isolates was estimated by two different formulas. The first [28] depended only on shared bands between the two compared isolates:
Similarity index(F) = 2nxy/(nX + ny) × 100 (1)
where nx and ny represented all bands generated by Isolates x and y, respectively, and nxy represented the bands shared by them. In the second method, Jacard similarity coefficient [29] depended on both shared and absent bands:
(J 1,2) = 100a/(a + b + c) × 100 (2)
where a was the number of bands shared between Isolates 1 and 2, b was the number of bands present in Isolate 1 but absent in Isolate 2, and c was the number of bands present in Isolate 2 but absent in Isolate 1. With Jacard's coefficient values, a dendrogram was constructed using the simple agglomerative (bottom-up) hierarchical clustering method UPGMA (unweighted pair group method with arithmetic mean) performed on SPSS version 16 (SPSS Inc., Chicago, IL, USA).
3. Results
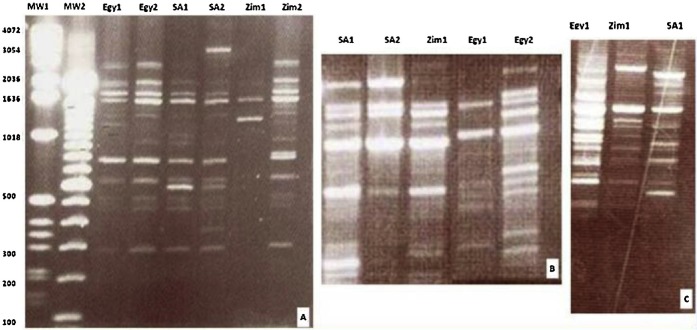
All primers used showed a consistent pattern of evident similarity between Zimbabwean and South African S. haematobium isolates, and a clear variability of either of them compared with the Egyptian isolate. P #2 showed the band pattern of two isolates from each country with interspecific variations between isolates of different countries and even some intraspecific individual variations between isolates of the same country most evident within the two Zimbabwean isolates (Figure 1A). P #7 showed a marked similarity of Zimbabwean and South African isolates while it failed to identify any loci in the genome of the two Egyptian isolates, revealing the great distance of the Egyptian isolate from the other two isolates in the loci screened by this primer. The fragments pattern generated by P #9 showed of two Egyptian, two South African, and one Zimbabwean isolates a moderate heterogeneous pattern between isolates of the different localities (Figure 1B). Magnification pattern of P #10 (Figure 1C) demonstrated a great difference between the Egyptian and the other two isolates.
Fig. 1.
Three photographs of 1.2% agarose gel showing random amplified polymorphic DNA-polymerase chain reaction patterns generated by P #2 (A), P #9 (B) and P #10 (C) of Schistosoma haematobium isolates. The first two lanes show high- and low-molecular weight standards.
Egy = Egyptian isolate; P # = Primer number; SA = South African isolate; Zim Zimbabwean isolate.
However as RAPD-PCR primers scan the whole genome and create a lot of bands, visual comparison between band patterns is not enough to obtain conclusive results. Thus, numerical transformation of the results, of each isolate, was done by scoring the total number of bands generated by each primer as well as the shared bands between different isolates (Table 1).
Table 1.
Total and shared amplified DNA fragments between Schistosoma haematobium isolates of the three countries revealed by each primer.
| Primer No. | Egypt(Egy) | Zimbabwe(Zim) | SouthAfrica (SA) | Shared fragments | ||
|---|---|---|---|---|---|---|
| Eg/SA | Eg/Zim | Zim/SA | ||||
|
#2 |
12 |
15 |
14 |
11 |
12 |
12 |
|
#7 |
Nil |
21 |
16 |
Nil |
Nil |
12 |
|
#9 |
13 |
16 |
14 |
9 |
10 |
11 |
|
#10 |
16 |
13 |
13 |
7 |
8 |
9 |
The sum of all shared bands between the different isolates is scored in Table 2, showing a great band sharing between Zimbabwean and South African isolates (44 bands), while there is moderate sharing between each of them and the Egyptian isolate (30 and 27 bands, respectively). This results in a similarity index (F) of 0.551 for Egyptian/South African, 0.566 for Egyptian/Zimbabwean, and a high similarity index of 0.721 for Zimbabwean/South African isolates (Table 2).
Table 2.
Shared amplified DNA fragments and similarity index (F) between isolates of three countries.
| Isolate | Egypt | Zimbabwe | South Africa | |
|---|---|---|---|---|
| Total No. of generated fragments |
41 | 65 | 57 | |
| Shared fragments |
Similarity index (F) | |||
| Egypt |
All | 30 | 27 | Egypt/Zimbabwe 0.566 |
| Zimbabwe |
30 | All | 44 | Egypt/South Africa 0.551 |
| South Africa |
27 | 44 | All | Zimbabwe/South Africa 0.721 |
Another comparison of the band pattern was done on the basis of band sharing and on bands lost in one isolate and present in the other (Table 3). Similarly, Jacard's coefficient (J) resulting from this concept showed 0.38 for Egyptian/South African, 0.395 for Egyptian/Zimbabwean, and a relatively high similarity coefficient of 0.564 for Zimbabwean/South African isolates (Table 3).
Table 3.
DNA fragments absent/present and Jacard's similarity coefficient between Schistosoma haematobium isolates.
| Isolate | Egypt | Zimbabwe | South Africa | |
|---|---|---|---|---|
| Total No. of generated fragments | 41 | 65 | 57 | |
| Absent/present fragments | Jacard's similarity coefficient (J) | |||
| Egypt |
Nil | 11 | 14 | Egypt/Zimbabwe 0.395 |
| Zimbabwe |
35 | Nil | 21 | Egypt/South Africa 0.38 |
| South Africa |
30 | 13 | Nil | Zimbabwe/South Africa 0.564 |
A constructed dendrogram (Figure 2) based on Jacard's similarity coefficients of RAPD data showed a clear split of the three S. haematobium isolates into two phylogenetic clusters, with the Egyptian isolate far from both Zimbabwean and South African isolates, which were close together and converged to represent a single cluster.
Fig. 2.
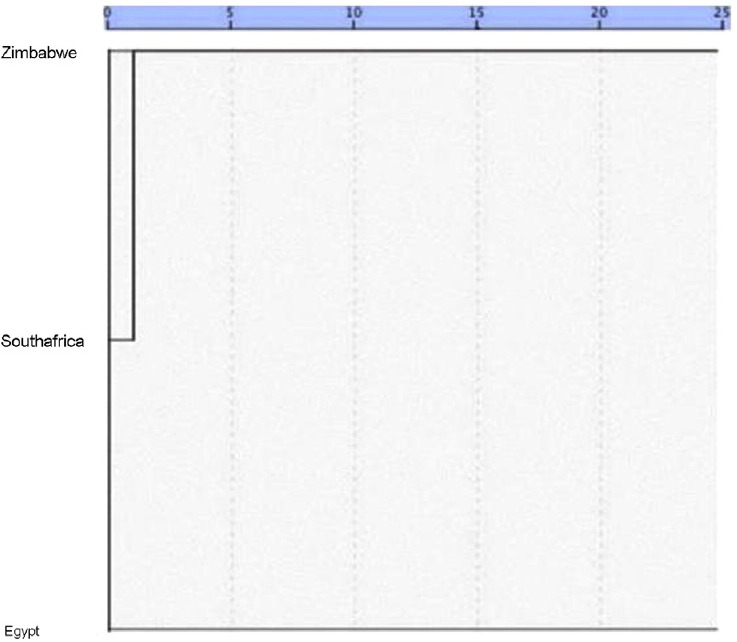
Dendrogram of Schistosoma haematobium genotypes constructed using UPGMA (unweighted pair group method with arithmetic mean) based on Jaccard's similarity coefficients shows a clear split of the three isolates into two phylogenetic clusters.
4. Discussion
Variations in intensity of the urinary tract pathology inflicted by S. haematobium infection have been noticed between different regions of Africa and even within communities [25]. Whether these discrepancies in disease severity are related to or caused by a variance in parasite genetic background is still an unsettled issue.
Many factors influence patterns of genetic variability of different parasites, like the mating system, the intermediate hosts and vehicle of parasite transmission [30]. S. haematobium has a cycle with two alternating hosts. The asexual reproduction occurs in the Bullinus snail, while the sexual reproduction among adult worms takes place in the human definitive host. The genetic variability of schistosomes can be influenced by a variety of factors, mainly the bionomics of their two hosts [31].
Our results indicate moderate to high genetic diversity among schistosome populations. This result agrees with a previous study [32,33] that reported a high level of genetic variability within selected populations of S. haematobium. Our results also agree with a recent study [1] reporting that S. haematobium genotypes across Africa split into two distinct groups; one that is predominately from mainland Africa and the other from the islands of the Indian Ocean and their neighboring coastal regions, with a considerable net divergence between the two groups.
The genetic differences between S. haematobium populations in Egypt and Sub-Saharan Zimbabwe and South Africa might be linked to morbidity that is more severe in Egypt [7]. Genetic variability may alter some of the virulence-related parasitic features like fecundity and immunogenicity. Emergence of parasitic strains that are more immunogenic or fecund than others might shape the clinical outcome of such infection [25,26].
The different local ecological phenomena in Egypt versus Zimbabwe and South Africa may explain our observation of moderate genetic diversity among the corresponding S. haematobium isolates. In Egypt, there is a unique occupational pattern of repeated exposure to the parasite with consequent prolonged acquired concomitant immunity of human hosts. There is also an isolated one river system along with almost no snail host migration. All these phenomena, together with limited movement of human populations, may lead to isolated populations of the parasite favoring higher susceptibility to mutation and genetic drift that could lead to the unique lineage of the parasite in Egypt [36].
By contrast, Zimbabwe and South Africa have an overlap of contact sites, with open river systems allowing free snail movements [34]. Maintenance of genetic diversity among adult infrapopulations is one of two main consequences of infection of snails with multiple schistosome genotypes [35]. These unique phenomena, together with an evident hybridization of S. haematobium with other human and animal schistosomes in South-East Africa, including Zimbabwe and South Africa [23], may lead to coexistence of multiple genotypes of the parasite in one locality. Genetic exchange and hybridization between native genotypes and imported ones could lead to the emergence of new parasitic strains [36].
Major movements of human populations during famines and civil wars in some regions of Africa are expected to play a major role in the emergence of different genetic clusters of the parasite. Mass human movements allow numerous and mixed parasite infections from different snails that come from the same or different parasite clusters [34].
Our study, in spite of its limitations, demonstrates an extent of genetic diversity of S. haematobium within humans that is comparable to that observed in the snail intermediate host. The study also suggests a relationship between clinical outcome and specific genetic clusters of the parasite as the two less clinically aggressive parasite clusters (in Zimbabwe and South Africa) have a relatively distant clustering from the parasite causing severe clinical disease in Egypt.
In conclusion, the current study presents original insights into the population genetics of S. haematobium. The highly variable random primers we have used were able to demonstrate the genetic diversity of S. haematobium within the human host from different geographical locations of Africa. They demonstrate that S. haematobium from three sampling locations are genetically diverse with the possible existence of two phylogenetic groups. A future study on a large geographic scale with more parasite isolates and more primers might characterize better the clonal lineages ofS. haematobium, which may have a major impact on management and control of urogenital schistosomiasis.
Conflict of interest
Authors declare that there is no conflict of interest.
References
- [1].Webster BL, Emery AM, Webster JP, Gouvras A, Garba A, Diaw O, et al. Genetic diversity within Schistosoma haematobium: DNA barcoding reveals two distinct groups. PLoS Negl Trop Dis. 2012;6:e1882. doi: 10.1371/journal.pntd.0001882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Van der Werf MJ, de Vlas SJ, Brooker S, Looman CW, Nagelkerke NJ, Habbema JD, Engels D. Quantification of clinical morbidity associated with schistosome infection in sub-Saharan Africa. Acta Trop. 2003;86:125–39. doi: 10.1016/s0001-706x(03)00029-9. [DOI] [PubMed] [Google Scholar]
- [3].Joubert PR, Pretorius SJ, Kruger FJ. Further studies on the susceptibility of Bullinus africanus to infection with Schistosoma haematobium. Ann Trop Med Parasitol. 1991;85:253–8. doi: 10.1080/00034983.1991.11812553. [DOI] [PubMed] [Google Scholar]
- [4].Fransden F. Studies of the relationship between Schistosoma and their intermediate hosts 1. The genus Bullinus and Schistosoma haematobium from Egypt. J Helminthol. 1979;35:15–29. doi: 10.1017/s0022149x0000568x. [DOI] [PubMed] [Google Scholar]
- [5].Meurs L, Mbow M, Vereecken K, Menten J, Mboup S. Polman. Epidemiology of mixed Schistosoma mansoni and Schistosoma haematobium infections in northern Senegal. Int J Parasitol. 2012;42:305–11. doi: 10.1016/j.ijpara.2012.02.002. [DOI] [PubMed] [Google Scholar]
- [6].Brouwer KC, Ndhlovu PD, Wagatsuma Y, Munatsi A, Shiff CJ. Epidemiological assessment of Schistosoma haematobium-induced kidney and bladder pathology in rural Zimbabwe. Acta Tropica. 2003;85:339–47. doi: 10.1016/s0001-706x(02)00262-0. [DOI] [PubMed] [Google Scholar]
- [7].Smith JB, Kamel IA, Elwi A, Lichtenberg FV. A quantitative postmortem analysis of Schistosoma haematobium in Egypt. l – Pathology and pathogenesis. Am J Trop Med Hyg. 1974;23:1054–71. doi: 10.4269/ajtmh.1974.23.1054. [DOI] [PubMed] [Google Scholar]
- [8].De Clerq D, Rollinson D, Diarra A, Sacko M, Coulibaly G, Landour A, etal Schistosomiasis in Dogon county Mali: identification and prevalence of the species responsible for infection in the local community. Trans Roy Soc Trop Med Hyg. 1994;88:653–6. doi: 10.1016/0035-9203(94)90212-7. [DOI] [PubMed] [Google Scholar]
- [9].Teesdale CB, Chitsulo L. Schistosomiasis in Malawi – a review. Trop Med Parasitol. 1985;36:1–6. [PubMed] [Google Scholar]
- [10].Edington GM. Schistosomiasis in Ghana with special reference to its pathology. Cent Afr J Med. 1994;6:45–57. [PubMed] [Google Scholar]
- [11].Ekanem EE, Asindi AA, Ejezie GC, Obong OE. Effect of Schistosoma haematobium infection on physical growth and school performance of Nigerean children. Cent Afr J Med. 1994;40:38–44. [PubMed] [Google Scholar]
- [12].Cooppan RM, Schutte CHJ, Mayet FGH, Dingle CE, Van Deventer JMG, Mosese PG. Morbidity from urinary schistosomiasis in relation to intensity of infection in the Natal province of South Africa. Am J Trop Med Hyg. 1986;35:765–76. doi: 10.4269/ajtmh.1986.35.765. [DOI] [PubMed] [Google Scholar]
- [13].Morand S, Manning SD, Woolhouse MEJ. Parasite-host coevolution and geographic patterns of parasite infectivity and host susceptibility. Proc R Soc Lond B Biol Sci. 1996;263:119–28. doi: 10.1098/rspb.1996.0019. [DOI] [PubMed] [Google Scholar]
- [14].Van den Broeck F, Geldof S, Polman K, Volckaert FA, Huyse T. Optimal sample storage and extraction procotols for reliable multilocus genotyping of the human parasite Schistosoma mansoni. Infect Genet Evol. 2011;11:1413–8. doi: 10.1016/j.meegid.2011.05.006. [DOI] [PubMed] [Google Scholar]
- [15].Golan R, Gower CM, Emery AM, Rollinson D, Webster JP. Isolation and characterization of the first polymorphic microsatellite markers for Schistosoma haematobium and their application in multiplex reactions of larval stages. Mol Ecol Resour. 2008;8:647–9. doi: 10.1111/j.1471-8286.2007.02031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Quan JH, Choi IW, Ismail HAHA, Mohamed AS, Jeong HG, Lee JS, et al. Genetic Diversity of Schistosoma haematobium eggs isolated from human urine in Sudan Korean. J Parasitol. 2015;53:271–7. doi: 10.3347/kjp.2015.53.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphism amplified by arbitrary primers, are useful as genetic markers. Nucleic Acid Res. 1991;18:6531–5. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–8. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Barral V, This P, Imbert-Establet D, Combes C, Delseny M. Genetic variability and evolution of the Schistosoma genome analysed by using random amplified polymorphic DNA markers. Mol Biochem Parasitol. 1993;59:211–21. doi: 10.1016/0166-6851(93)90219-n. [DOI] [PubMed] [Google Scholar]
- [20].Dias Neto EDSC, Rollinson D, Katz N, Pena SD, Simpson AJ. The random amplification of polymorphic DNA allows the identification of strains and species of schistosome. Mol Biochem Parasitol. 1993;57:83–8. doi: 10.1016/0166-6851(93)90246-t. [DOI] [PubMed] [Google Scholar]
- [21].Minchella DJ, Lewis FA, Sollenberger KM, Williams JA. Genetic diversity of Schistosoma mansoni: quantifying strain heterogeneity using a polymorphic DNA element. Mol Biochem Parasitol. 1994;68:307–13. doi: 10.1016/0166-6851(94)90175-9. [DOI] [PubMed] [Google Scholar]
- [22].Sire C, Durand P, Pointier JP, Theron A. Genetic diversity and recruitment pattern of Schistosoma mansoni in a Biomphalaria glabrata snail population: a field study using random-amplified polymorphic DNA markers. J Parasitol. 1999;85:436–41. [PubMed] [Google Scholar]
- [23].Shiff C, Brouwer KC, Clow L. Schistosoma haematobium: population genetics of S. haematobium by direct measurement of parasite diversity using RAPD-PCR. Exp Parasitol. 2000;96:47–51. doi: 10.1006/expr.2000.4548. [DOI] [PubMed] [Google Scholar]
- [24].LoVerde PT, DeWald J, Minchella DJ. Further studies of genetic variation in Schistosoma mansoni. J Parasitol. 1985;71:732–4. [PubMed] [Google Scholar]
- [25].Brouwer KC, Ndhlovu PD, Wagatsuma Y, Munatsi A, Shiff CJ. Urinary tract pathology attributed to Schistosoma haematobium: does parasite genetics play a role? Am J Trop Med Hyg. 2003;68:456–62. [PubMed] [Google Scholar]
- [26].Gasmelseed N, Karamino NE, Abdelwahed MO, Hamdoun AO, Elmadani AE. Genetic diversity of Schistosoma haematobium parasite IS NOT associated with severity of disease in an endemic area in Sudan. BMC Infect Dis. 2014;14:469–76. doi: 10.1186/1471-2334-14-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mott KER, Baltes J, Bambagha J, Baldassini B. Field studies of a reusable polyamide filter for detection of Schistosoma haematobium egg by urine filtration. Tropenmed Parasitol. 1982;33:227–8. [PubMed] [Google Scholar]
- [28].Nei M, Li WH. Mathematical models for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;74:5267–73. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Legendre P, Vaudor A. The R Package: multidimensional analysis, spatial analysis. Montreal: University of Montreal. 1991 [Google Scholar]
- [30].Van den Broeck F, Meurs L, Raeymaekers JAM, Boon N, Dieye TN, Volckaert FAM, et al. Inbreeding within human Schistosoma mansoni: do host-specific factors shape the genetic composition of parasite populations? Heredity. 2014;113:32–41. doi: 10.1038/hdy.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gower CM, Gabrielli AF, Sacko M, Dembele R, Golan R, Emery AM, et al. Population genetics of Schistosoma haematobium: development of novel microsatellite markers and their application to schistosomiasis control in Mali. Parasitol. 2011;138:978–94. doi: 10.1017/S0031182011000722. [DOI] [PubMed] [Google Scholar]
- [32].Glenn TC, Lance SL, McKee AM, Webster BL, Emery AM, Zerlotini A, et al. Significant variance in genetic diversity among populations of Schistosoma haematobium detected using microsatellite DNA loci from a genome-wide database. Parasit Vectors. 2013;6:300. doi: 10.1186/1756-3305-6-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gower CM, Gouvras AN, Lamberton PH, Deol A, Shrivastava J, Mutombo PN, et al. Population genetic structure of Schistosoma mansoni and Schistosoma haematobium from across six sub-Saharan African countries: implications for epidemiology, evolution and control. Acta Trop. 2013;128:261–74. doi: 10.1016/j.actatropica.2012.09.014. [DOI] [PubMed] [Google Scholar]
- [34].Dabo A, Durand P, Morand S, Diakite M, Langand J, Imbert-Establet D, et al. Distribution and genetic diversity of Schistosoma haematobium within its bulinid intermediate hosts in Mali. Acta Trop. 1997;24(66):15–26. doi: 10.1016/s0001-706x(97)00670-0. [DOI] [PubMed] [Google Scholar]
- [35].Minchella DJ, Sollenberger KM, Pereira de Sousa C. Distribution of schistosome genetic diversity within molluscan intermediate hosts. Parasitology. 1995;111:217–20. doi: 10.1017/s0031182000064970. [DOI] [PubMed] [Google Scholar]
- [36].Brouwer KC, Ndholvu P, Munatsi A, Shiff CJ. Genetic diversity of a population of Schistosoma haematobium derived from schoolchildren in East Central Zimbabwe. J Parasitol. 2001;87:762–9. doi: 10.1645/0022-3395(2001)087[0762:GDOAPO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]