Abstract
Lineage-specific duplicated genes likely contribute to the phenotypic divergence in closely related species. However, neither the frequency of duplication events nor the degree of selection pressures immediately after gene duplication is clear in the speciation process. Here, using Illumina DNA-sequencing reads from Arabidopsis halleri, which has multiple closely related species with high-quality genome assemblies (A. thaliana and A. lyrata), we succeeded in generating orthologous gene groups in Brassicaceae. The duplication frequency of retained genes in the Arabidopsis lineage was ∼10 times higher than the duplication frequency inferred by comparative genomics of Arabidopsis, poplar, rice and moss (Physcomitrella patens). The difference of duplication frequencies can be explained by a rapid decay of anciently duplicated genes. To examine the degree of selection pressure on genes duplicated in either the A. halleri-lyrata or the A. halleri lineage, we examined positive and purifying selection in the A. halleri-lyrata and A. halleri lineages throughout the ratios of nonsynonymous to synonymous substitution rates (KA/KS). Duplicate genes tended to have a higher proportion of positive selection compared with non-duplicated genes. Interestingly, we found that functional divergence of duplicated genes was accelerated several million years after gene duplication compared with immediately after gene duplication.
Keywords: plant evolution, gene duplication, Arabidopsis, selection pressure, functional divergence
1. Introduction
Plant genomes have experienced more gene duplication events than most other eukaryotes.1 These duplications are largely classified into whole genome duplications (WGDs) and small-scale duplications (SSDs). WGD events are concentrated in the Cretaceous-Paleogene extinction period.2 SSDs such as retroduplication and tandem duplication have occurred continuously at a high rate during plant evolution, indicating that SSDs may be a key factor for plant speciation or phenotypic differences in close relatives.3–8 SSDs tend to be lineage-specific in land plants.4 There is a clear functional bias between WGD and SSD in plants.4,9 SSDs tend to be associated with stress responses,4,8–10 and are likely important for adaptive evolution to rapidly changing environments in close relatives.
SSD genes functionally diverge through either neo-functionalization or sub-functionalization, which may contribute to speciation.11,12 SSD genes can also retain functional redundancy through the long-term fate of becoming pseudogenes,13,14 dosage selection15,16 or avoiding developmental errors.17–19 Functional divergence of duplicated genes can be inferred by selection pressures based on the non-synonymous substitution rate (KA) versus the synonymous substitution rate (KS). Genes undergoing positive selection (KA/KS > 1) and purifying selection (KA/KS < 1) are associated with functional divergence and constraint, respectively.20 Most earlier studies compared selection pressures in WGDs with those in SSDs, or selection pressures in anciently duplicated genes (several hundred million years [MY] ago) in plants.21–24 There are little data with which to examine selection pressures in recent SSDs in plants. Because selection pressure likely varies during the speciation process, it is important to examine the selection pressures immediately after gene duplications among close relatives.
Recently, there are many plant genomes assembled by Illumina DNA-sequencing reads.25 However, SSDs tend to be underestimated in contigs generated by only Illumina DNA-sequencing.26,27 It is likely that genomes generated by BAC are useful to examine SSDs. However, construction of such a library takes much budget and time to generate highly qualified genomes. In the present study, we tried to generate reliable orthologous genes by only paired-end Illumina DNA-sequencing reads using available genomes of close relatives. We then inferred SSDs in a species after the divergence of closed species throughout the reading depth of Illumina DNA sequencing.
The Arabidopsis lineage seems to be the best lineage to examine recent SSDs in plants because Arabidopsis has two BAC library-based genomes in Arabidopsis thaliana and Arabidopsis lyrata.28,29 In particular, A. thaliana has a large amount of functional data for its annotated genes. To comprehensively examine SSDs in Arabidopsis, we investigated an additional Arabidopsis species—A. halleri. The divergence time between A. lyrata and A. halleri is supposed to be ∼2 MYA,30 and the divergence time between either A. halleri or A. lyrata and A. thaliana is ∼10–11 MYA.31 Among recently diverged three Arabidopsis species, there are phenotypic variations such as self-compatibility and heavy-metal tolerance. A. thaliana has self-compatibility, but the others are self-incompatible.32–34 Therefore, A. halleri and A. lyrata have large petals to attract pollinator insects, and the anthers are separated from the stigma to avoid autopollination.35A. halleri is known as a Zn/Cd hyper-accumulator33,34 and some wild populations of A. lyrata are tolerant of serpentine soils, which are characterized by a high heavy-metal content,36 while A. thaliana is not.
A. halleri has highly variable morphologies with respect to leaf, flower colour and development of stolons among the subspecies. To examine SSDs in the Arabidopsis lineage, we focused on A. halleri subsp. gemmifera which grows in the Russian Far East, northeastern China, Korea, Taiwan and Japan across lowland and highland areas. We collected the plants in Mt. Ibuki, Shiga, Japan. We then extracted the plant DNAs, and performed the paired-end Illumina DNA sequencing. After generating contigs from the Illumina DNA-sequencing reads, we inferred A. halleri orthologous genes against the A. thaliana and A. lyrata genes. The number of SSDs was inferred both from phylogenetic analysis of orthologous genes and the reading depth of Illumina DNA sequencing. As outgroup species for the three Arabidopsis species, we used five non-Arabidopsis species in Brassicaceae for which genome sequences are already determined. There is trans-specific polymorphism in Arabidopsis; in particular, some genes have undergone gene flow in Arabidopsis.37 Here, excluding genes that have undergone gene flow in Arabidopsis, we generated three sets of orthologous gene groups (OGGs) among five non-Arabidopsis species, A. thaliana, A. lyrata and A. halleri in Brassicaceae (Fig. 1). Each OGG represent genes derived from one ancestral gene in each speciation event. By identifying the genes in each OGG, we examined the evolutionary process in the Arabidopsis lineage. Although a BAC library-based genome for A. halleri is unavailable, there is an available A. halleri genome based on long-insert mate paired reads and short-insert paired-end reads in Illumina DNA sequencing.38,39 The available A. halleri genome was originated from A. halleri subsp. gemmifera which was the same subspecies in our plant material. The collection site was in Tada mine, Hyogo, Japan. Microsatellite analyses suggested that our plant material collected from Mt. Ibuki, Shiga, Japan was genetically closed to the plant with the available genome.40 Thus, it is likely that A. halleri genes inferred from the available genome are similar to our inferred A. halleri genes. To validate our overall results, we also examined SSDs based on the available A. halleri genome,
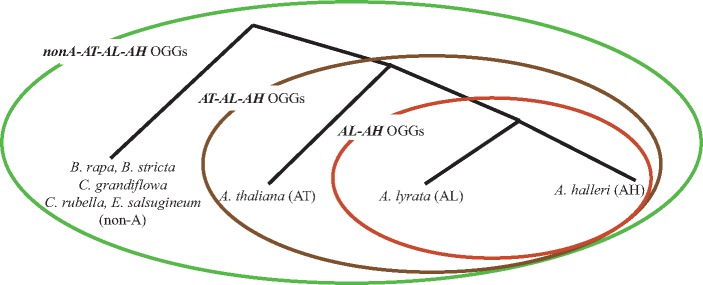
Figure 1.
Three sets of OGGs among non-Arabidopsis species, A. thaliana, A. lyrata and A. halleri. OGGs between A. lyrata and A. halleri were defined as AL–AH OGGs. There were 25,833, 26,428 and 26,007 AL–AH OGGs based on Illumina paired-end DNA-sequencing reads without any pseudogene-like genes, Illumina paired-end DNA-sequencing reads including pseudogene-like genes and the available A. halleri genome, respectively. OGGs among A. thaliana, A. lyrata and A. halleri were defined as AT–AL–AH OGGs. There were 22,105, 22,684 and 21,537 AT–AL–AH OGGs based on Illumina paired-end DNA-sequencing reads without any pseudogene-like genes, Illumina paired-end DNA-sequencing reads including pseudogene-like genes and the available A. halleri genome, respectively. OGGs among non-Arabidopsis species (B. rapa, B. stricta, C. grandiflora, C. rubella, E. salsugineum), A. thaliana, A. lyrata and A. halleri were defined as nonA-AT–AL–AH OGGs. There were 17,669 and 17,925 non-A-AT–AL–AH OGGs based on Illumina paired-end DNA-sequencing reads without any pseudogene-like genes and the available A. halleri genome, respectively.
In a previous report, selection pressures of genes duplicated after the divergence of A. thaliana and A. lyrata were examined in the A. thaliana lineage.23 These lineage-specific duplicated genes tend to have undergone positive selection. Here, we examined the selection pressures in duplicated genes in the other lineage after the split between A. thaliana and A. lyrata. This lineage was further split into the A. halleri–lyrata lineage and A. halleri lineage (Fig. 2). We then examined whether gene duplications in either the A. halleri–lyrata or A. halleri lineage contributed to functional divergence in the retained duplicate genes.
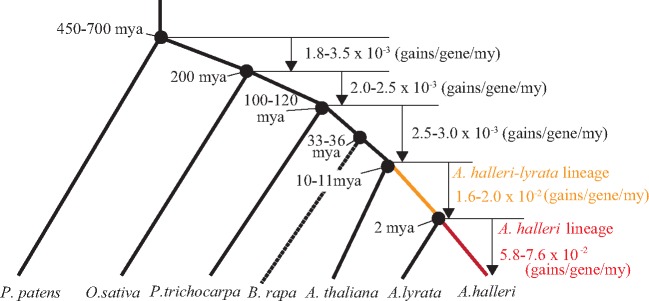
Figure 2.
Gain rates through gene duplication in land plants. The divergence times among moss (P. patens), rice (O. sativa), poplar (P. trichocarpa), B. rapa, A. thaliana, A. lyrata and A. halleri were taken from previous reports.30,31,72–75 Gene gains through gene duplication were estimated for each branch. The gene gains in two branches were estimated in this study. One was the A. halleri-lyrata lineage, which represents the evolutionary process of the A. halleri lineage between the divergence of A. thaliana and the divergence of A. lyrata. The other was the A. halleri lineage, which represents the evolutionary process of the A. halleri lineage after the divergence of A. lyrata. Gene gains were divided by ancestral gene numbers and branch lengths corresponding to times (MY). The rate of gene gain was defined as the number of genes gained through gene duplication per gene per MY.
Duplicated genes may have undergone purifying selection in the Arabidopsis lineage. The gene dosage hypothesis proposes that duplicate genes can be fixed for increased gene dosage, keeping redundant functions among duplicated genes. In A. halleri, the expression of HEAVY METAL ATPASE 4 (HMA4) has been enhanced by cis-regulatory mutations and increased gene copy number for metal hyperaccumulation.41 This is an example of increased gene dosage by gene duplication. Consequently, duplicated genes with purifying selection tend to be associated with increased gene dosage.
To understand the contribution of duplicated genes to the speciation process in Arabidopsis within the last 10 MY, we first examined the frequency of gene duplications in the A. halleri–lyrata and A. halleri lineages. We then examined the relationship between gene duplication and selection pressure based on the KA/KS ratio. Finally, we examined the over-represented functional categories of genes under positive and purifying selection.
2. Materials and methods
2.1. Sampling and illumina paired-end DNA-sequencing analysis
Arabidopsis halleri subsp. gemmifera is one of the three subspecies of A. halleri, and grows in the Russian Far East, northeastern China, Korea, Taiwan and Japan.42 In 2008, a leaf sample was collected from an individual of A. halleri subsp. gemmifera on Mt. Ibuki, Shiga, Japan. DNA was extracted from the leaf using a DNeasy Plant Mini Kit (QIAGEN, Venlo, The Netherlands). A 300-bp paired-end DNA library was constructed according to the paired-End Genomic DNA Sample Preparation Kit (Illumina, San Diego, CA, USA), and 93-bp paired-end reads were obtained from the Illumina GAIIx sequencer.
A total of 44.5 Gb reads were generated by Illumina DNA paired-end sequencing. Using the Trim Galore (www.bioinformatics.babraham.ac.uk) software, sequences with low-quality base calls (Phred score < 20) were trimmed off from the 3ʹ end of the reads. Adapter sequences started with ‘AGATCGGAAGAGC’ were completely removed from the reads. Approximately 28% of reads had either low-quality scores or adapters and were trimmed by Trim Galore. When a paired-end read was completely removed by this procedure, the other read was used as a single-end read. Given that mitochondrial and chloroplast genomes have much higher copy numbers than the nuclear genome, sequencing reads mapped to either the mitochondrial or chloroplast genome in A. thaliana (https://www.arabidopsis.org, TAIR10) or A. lyrata (http://genome.jgi.doe.gov, FilteredModels6) by BLASTN version 2.2.29 (e-value < 1.0) were excluded from the following procedures.43 Assuming that the genome size of A. halleri was 255 Mb,44 the sequencing coverage was estimated to be ∼135× (34.4/0.255 Gb).
2.2. Gene sequences of Brassica rapa, Boechera stricta, Capsella grandiflora, Capsella rubella, Eutrema salsugineum, A. thaliana and A. lyrata
Gene sequences in B. rapa (BrapaFPsc_277_v1.3), B. stricta (Bstricta_278_v1.2), C. grandiflora (Cgrandiflora_266_v1.1), C. rubella (Crubella_183_v1.0) and E. salsugineum (Esalsugineum_173_v1.0) were collected from Phytozome (https://phytozome.jgi.doe.gov, version 10). Gene sequences in A. thaliana and A. lyrata were collected from TAIR (https://www.arabidopsis.org/, version 10, TAIR10_cds_20110103) and JGI (http://genome.jgi.doe.gov, FilteredModels6), respectively.
2.3. Assembly of A. halleri genes orthologous to A. lyrata genes based on illumina paired-end DNA-sequencing reads
We first generated A. halleri contigs from a total of 44.5 Gb Illumina DNA-sequencing reads using several assembly software tools such as Platanus 1.2.4,45 ABySS 1.5.2,46 SOAP-denovo247 and CLC Genomics Workbench 7.0.3 (https://www.qiagenbioinformatics.com/) software. A higher proportion of genes in closely related species tend to be mapped to reliable assembled sequences. As the closest species, 32,670 annotated A. lyrata genes were mapped to each type of assembled DNA segment by the gmap (version 2014-08-20) software with default parameters, which takes into account exon–intron junctions.48 The A. halleri contigs generated by the ABySS software with 63 K-mer size (N50 size = 5,331 bp) had the highest mapping rate to the 32,670 annotated A. lyrata genes with >80% coverage (Supplementary Table S1). Some of the matched regions were truncated by terminal codons. When coding sequences up to the terminal codon had >80% similarity to and 80% coverage for an A. lyrata gene, the coding sequence was defined as the orthologous A. halleri gene sequence to an A. lyrata gene. Following this procedure, we generated 22,727 orthologous gene pairs between A. halleri and A. lyrata. However, an A. lyrata gene sequence was frequently mapped to different A. halleri contigs depending on the region of the A. lyrata gene because the whole sequence of each A. halleri gene was split into several contigs. To further identify A. halleri genes orthologous to A. lyrata genes, we re-assembled the A. halleri contigs based on the mapping information of A. lyrata genes. To collect additional A. halleri genes orthologous to A. lyrata, unmapped A. lyrata genes were re-mapped to the assembled DNA segments by TBLASTN version 2.2.29 (e-value < 1 × 10−5).43 However, the contigs mapped separately by A. lyrata genes may not be orthologous syntenic regions. To focus on only syntenic contigs between A. halleri and A. lyrata, we defined syntenic contigs to which more than two A. lyrata genes within less than five spacer genes in A. lyrata genome were mapped. When an A. lyrata gene was separately mapped to two syntenic contigs between A. lyrata and A. halleri, the contigs were concatenated following the direction of the A. lyrata gene. The A. lyrata gene was then mapped to the concatenated contigs. We thus obtained an additional 3,106 A. halleri gene sequences with >80% similarity and 80% coverage against A. lyrata genes. Finally, we succeeded in identifying 25,833 (79.1%) A. halleri genes orthologous to 32,670 A. lyrata genes. Given that all identified A. halleri genes were paired with A. lyrata genes, a total of 25,833 pairs were defined as A. lyrata–A. halleri (AL–AH) OGGs.
In the above procedures, we did not identify pseudogene-like A. halleri genes which are orthologous to A. lyrata genes. Consequently, duplication frequency may be underestimated in either the A. lyrata–A. halleri or the A. halleri lineage. Therefore, we also identified 26,428 orthologous A. halleri regions truncated by terminal codons with >80% similarity and 80% coverage against A. lyrate genes.
These analysis procedures and findings are summarized in Supplementary Figure S1.
2.4. Validation of A. halleri lineage-specific duplicated genes by droplet digital PCR
To calculate the read coverage of each A. halleri gene, the mapped count was divided by the number of genes to which a read was mapped by BOWTIE2 version 2.2.3.49 Reads per kilobase of exon model per million (RPKM) values were calculated for each A. halleri gene. For 12 A. halleri genes whose copy numbers were known (Supplementary Table S2), we designed primer pairs using the following parameters in the Primer3Plus software:44 primer length of 18–24 bases, amplicon length of 70–150 bp, Tm value of 57–63°C and GC composition of 40–60%.50 To obtain enough genomic DNA for droplet digital PCR (ddPCR), we mixed the genomic DNAs from four individuals of A. halleri from Mt. Ibuki. The genomic DNA was sonicated with the Covaris M220 system (25 s in frequency sweeping mode). The concentration of the sonicated genomic DNA sample was 6 ng/μl. The peak size of sonicated DNA fragments was 2,382 bp according to the Fragment Analyzer system (Advanced Analytical, Ankeny, IA, USA) with the High Sensitivity Genomic DNA Analysis Kit (Advanced Analytical). Each ddPCR reaction was performed with ddPCR EvaGreen Supermix (Bio-Rad, Hercules, CA, USA). Each reagent was divided into ∼20,000 droplets with a droplet generator (Bio-Rad QX-200). Cycled droplets were measured with a QX200 droplet reader (Bio-Rad).
To find A. halleri genes whose DNA concentrations were robustly inferred by ddPCR, we first identified uniquely mapped regions (>80 bp) in A. halleri genes from the Illumina DNA-sequencing reads. Among the A. halleri genes with uniquely mapped regions, we manually chose 50 A. halleri genes with a variety of RPKMs. We designed a pair of primers for each gene in the Primer3Plus software using the parameters described earlier. To examine the specificity of the targeted DNAs, we performed real-time PCR analysis using the protocol for the Mx3000P qPCR System (Agilent Technologies). The PCR analyses were performed using SsoFast EvaGreen Supermix (Bio-Rad) and the products were analysed using the Mx3000P multiplex quantitative PCR system (Agilent Technologies). Specific and multiple reactions should result in a single and multiple melting peaks corresponding to the PCR products. Of the 50 primer pairs, 25 showed a clear raised curve for all three replicates. Thus, for the copy number assay by ddPCR, we used 13 primer pairs for unknown copy number genes, 10 pairs for single-copy genes in a broad range of plant species and 2 pairs for a three-copy gene (Supplementary Table S2).
2.5. Assembly of A. halleri genes orthologous to A. lyrata genes based on the available draft A. halleri genome
Coding genes of A. halleri were generated on the draft A. halleri genome using long-insert mate paired reads and short-insert paired-end reads in Illumina DNA sequencing.38 On the genome, we used annotated coding genes. We performed BLAST search between the annotated coding A. halleri and A. lyrata genes by BLASTP. The best matching pairs were defined to be AL–AH OGGs with more than >80% similarity to and 80% coverage at amino acid level. We succeeded in identifying 26,007 (79.6%) A. halleri genes which were orthologous to 32,670 A. lyrata genes. Out of 26,007 orthologous pairs of A. halleri and A. lyrata genes, 22,105 (22,105/26,007 = 85%) A. lyrata genes were inferred in the AL–AH OGGs based on only Illumina paired-end DNA-sequencing reads. The number of specifically identified A. halleri genes based on Illumina paired-end DNA sequencing and the available A. helleri genome was 3,728 and 3,902, respectively (Supplementary Fig. S2). Thus, either method had a particular advantage to infer the orthologous genes.
A. halleri genes duplicated in the A. lyrata lineage were identified with the Inparanoid algorithm51 Given the best-match pair, A1 and B1, an A. halleri gene that had a smaller sequence distance to A1 (or B1) than the distance between A1 and B1, was added to the AL–AH OGG containing A1 and B1 (seeds). This process was continued until all qualified A. halleri genes were assigned to seed pairs of the genes.
2.6. Selection pressures in the A. halleri-lyrata and A. halleri lineages
To infer selection pressure in the A. halleri–lyrata lineage, we focussed on orthologous groups that followed the speciation process among the genes of the five non-Arabidopsis species (B. rapa, B. stricta, C. grandiflora, C. rubella, E. salsugineum), A. thaliana, A. lyrata and A. halleri. In each orthologous group, a multiple alignment was made to match the coding regions with the computer programme MAFFT v7.215 with default parameters.52 Using the phylogenetic tree and multiple alignment, we constructed the common ancestral sequences among A. thaliana, A. halleri and A. lyrata, and the common ancestral sequence between A. halleri and A. lyrata with codeml (model = 1, NSsites = 0) in PAML. For each pair of ancestral sequences, the synonymous (KS) and non-synonymous substitution rates (KA) were calculated using yn00 (code = 0, weighting = 0, commonf3x4 = 0) in PAML. To determine whether the KA/KS ratio was significantly <1, two maximum likelihood values were calculated with the KA/KS ratio fixed at 1 and with the KA/KS ratio as a free parameter. The ratio of the maximum likelihood values was then compared with the χ2 distribution. A P-value representing the deviation of the two models was then calculated for the A. halleri-lyrata lineage.
To infer selection pressure in the A. halleri lineage, we generated a tree file for the A. thaliana, A. lyrata and A. halleri genes. In each of the orthologous groups, a multiple alignment was made to match the coding regions by the computer programme MAFFT.52 Using the tree file and multiple alignment file, we constructed the common ancestral sequences among A. thaliana, A. halleri, and A. lyrata with codeml (model = 1, NSsites = 0) in PAML. Although we used a representative A. halleri gene sequence to infer the ancestral sequence, proper A. halleri genes had sequence variation when they were lineage-specific duplicated after the split from A. lyrata. From the Illumina DNA-sequencing reads mapped to A. halleri genes by BOWTIE2 version 2.2.3 with default parameters,49 the sequence variation was examined in each A. halleri gene by the Picard software with default parameters (http://broadinstitute.github.io/picard/). When a variable sequence was observed in the A. halleri genes, the codon sequence including the variable nucleotide was concatenated into the representative A. halleri genes. Alignments between A. halleri genes including codons with variable nucleotides and the ancestral sequence were modified by adding codons of the ancestral sequence against concatenated codons including variable nucleotides. KA and KS were calculated in each pair of ancestral and A. halleri gene sequences including variable sequences using yn00 in PAML. To determine whether the KA/KS ratio was significantly <1, two maximum likelihood values were calculated with the KA/KS ratio fixed at 1 and with the KA/KS ratio as a free parameter using codeml in PAML. The ratio of the maximum likelihood values was then compared with the χ2 distribution. A P-value representing the deviation of the two models was then calculated for each A. halleri gene. These analysis procedures are summarized in Supplementary Figure S3.
2.7. Inference of over-represented gene ontologies
Assuming that A. halleri and A. lyrata genes have similar GO assignments in A. thaliana in the same OGG, we obtained gene ontology (GO) assignments for the A. thaliana genes from The Arabidopsis Information Resource (www.arabidopsis.org). Among the top three GO categories (cellular components, molecular functions, and biological processes), we analysed only biological processes. For each GO category in the biological processes category, the expected ratio was inferred to be the ratio between the number of genes assigned to the GO category and the number of genes not assigned to the GO category among all annotated A. thaliana genes. In each GO category, the observed ratio in each category of OGGs was compared with the expected ratio by a χ2 test for different categories of OGGs. The ratios were processed in the R software environment (www.r-project.org) and normalized among different arrays using Z-scores calculated using genescale in the R library. A heatmap was generated using heatmap.2 in the R library. To correct for multiple testing, the false discovery rate (FDR) was estimated by the R-library Q-VALUE software. The null hypothesis was rejected if FDR values were < 0.01.
3. Results and discussion
3.1. Duplication frequency in the A. halleri-lyrata lineage
To examine the frequency of duplication events in the A. halleri-lyrata lineage (Fig. 2), we first used 25,833 AL–AH OGGs based on Illumina paired-end DNA-sequencing reads. We searched for an orthologous A. thaliana gene for each AL–AH OGG by BLASTP searches between AL and AH OGG protein sequences and annotated A. thaliana protein sequences.43 When both the A. lyrata and A. halleri genes in an AL–AH OGG had the best hit to the same A. thaliana gene, the A. thaliana gene was considered an orthologous candidate gene. Of 25,833 AL–AH OGGs, 93.3% (24,019/25,833) had orthologous candidate genes in A. thaliana. Among these, we searched for orthologous groups that were consistent with the species tree. To do this, the synonymous substitution rates (KS) were calculated among the A. thaliana, A. lyrata, and A. halleri genes in each orthologous group using yn00 in PAML.53 When the KS between A. lyrata and A. halleri was lower than both the KS between A. thaliana and A. lyrata and the KS between A. thaliana and A. halleri, we assumed that the topology of the orthologous group was consistent with the species tree. Among the 24,019 AL–AH OGGs, 22,602 (22,602/24,019 = 94.1%) had the same topology as the species tree. In the 22,602 AL–AH OGGs, 16,704 and 2,370 A. thaliana genes were uniquely and multiply assigned to AL–AH OGGs, respectively. To examine the duplication frequency in A. halleri-lyrata lineage, we considered two evolutionary scenarios for the 2,370 A. thaliana genes multiply assigned to 5,898 OGGs. One is that gene duplication events occurred in the A. halleri-lyrata lineage. That is, the assigned A. thaliana gene is orthologous to the AL–AH OGGs. The other is that gene loss events occurred in the A. thaliana lineage after gene duplication in the common ancestor among A. thaliana, A. lyrata and A. halleri. In this case, the assigned A. thaliana gene is not orthologous to the AL–AH OGGs. To examine these two possible scenarios, we collected AL–AH OGGs to which an A. thaliana gene was assigned as an orthologous gene. We then calculated the KS between the A. thaliana gene and AL–AH OGG, and searched for the closest pair of the AL–AH OGG against the A. thaliana gene. When the KS between the chosen AL-AH OGG and the other AL-AH OGG was lower than the KS in the closest pair, the other AL–AH OGG was defined as sharing the A. thaliana gene as an orthologous gene. Of the 5,898 AL–AH OGGs, 497 had no orthologous A. thaliana genes and 5,401 had 2,370 orthologous A. thaliana genes. In the following analyses, 22,105 (16,704 + 5,401) AL–AH OGG with 19,074 (16,704 + 2,370) orthologous A. thaliana genes were defined as A. thaliana–A. lyrata–A. halleri (AT–AL–AH) OGGs (Supplementary Table S3). These analysis procedures are summarized in Supplementary Figure S4.
In A. lyrata-halleri lineage, 22,105 AT–AL–AH OGGs were derived from the 19,074 genes, which represent the number of genes in the common ancestor of A. thaliana, A. lyrata and A. halleri. Thus, 3,031 (22,105−19,074) gains through gene duplication were supposed to have occurred in the A. halleri-lyrata lineage. The gain rate (total gene duplication gains during a given time period divided by the estimated duration per gene) was inferred to be 1.8–2.0 × 10−2 (3,031 gains/19,074 genes/8–9 MY) in the A. halleri-lyrata lineage. Additionally, we examined 26,428 AL–AH OGGs including pseudogene-like A. halleri genes (Supplementary Table S4), and identified 22,684 AT–AL–AH OGGs derived from the 19,318 orthologous A. thaliana genes. The gain rate was inferred to be 1.9–2.2 × 10−2 (3,366 gains/19,318 genes/8–9 MY) in the A. halleri-lyrata lineage. The gain rate in OGGs including pseudogene-like genes (Fig. 4C) is similar to OGGs without pseudogene-like genes, indicating that addition of pseudogene-like A. halleri genes did not affect duplication frequency in the A. halleri-lyrata lineage.
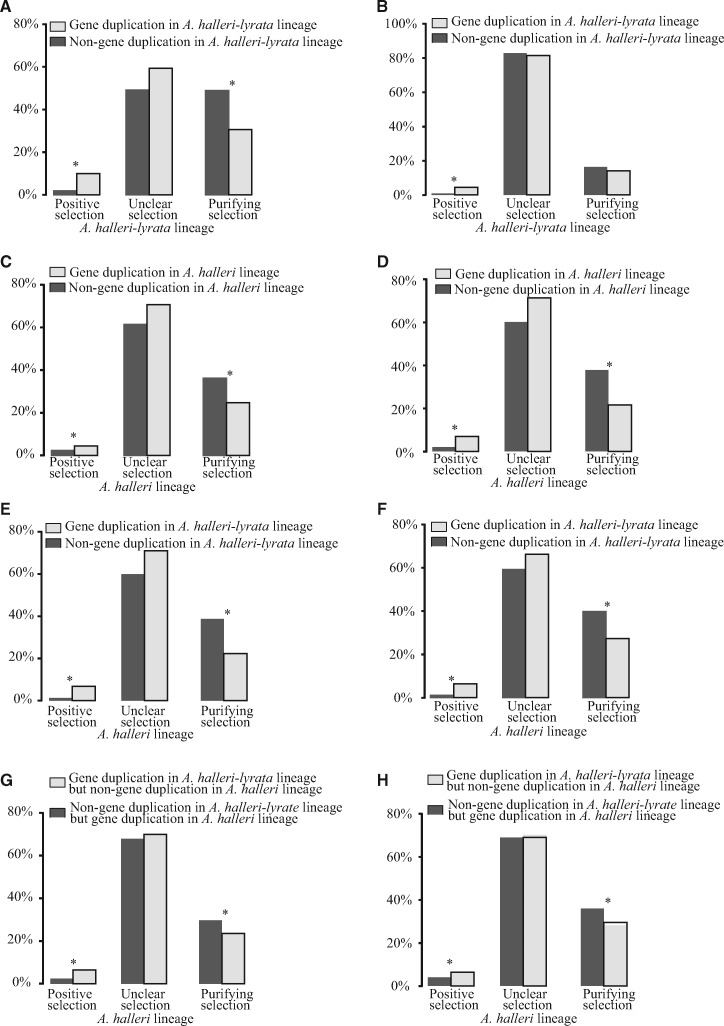
Figure 4.
Gene duplication and selection pressure in the A. halleri-lyrata and A. halleri lineages. Genes were classified as being under positive selection (KA/KS > 1), unclear selection or purifying selection (KA/KS < 1) in the A. halleri-lyrata and A. halleri lineages. Asterisks (*) represent significant differences by the chi-square test (P < 0.05). (A) The relationship between gene duplication and selection pressure in the A. lyrata-halleri lineage in 17,669 OGGs base on Illumina paired-end DNA-sequencing reads. (B) The relationship between gene duplication and selection pressure in the A. lyrata-halleri lineage in 17,925 OOGs based on the available A. halleri genome. (C) The relationship between gene duplication and selection pressure in the A. halleri lineage in 22,105 AT–AL–AH OGGs based on Illumina paired-end DNA-sequencing reads. (D) The relationship between gene duplication and selection pressure in the A. halleri lineage in 21,537 AT–AL–AH OOGs based on the available A. halleri genome. (E) The relationship between gene duplication in the A. lyrata-halleri lineage and selection pressure in the A. halleri lineage in 22,105 AT–AL–AH OGGs based on Illumina paired-end DNA-sequencing reads. (F) The relationship between gene duplication in the A. lyrata-halleri lineage and selection pressure in the A. halleri lineage in 21,537 AT–AL–AH OOGs based on the available A. halleri genome. (G) Comparison of selection pressure in the A. halleri lineage between gene duplication in only the A. lyrata-halleri lineage and gene duplication in only the A. halleri lineage in OGGs base on Illumina paired-end DNA-sequencing reads. (H) Comparison of selection pressure in the A. halleri lineage between gene duplication in only the A. lyrata-halleri lineage and gene duplication in only the A. halleri lineage in OGGs based on the available A. halleri genome.
Second, we used 26,007 AL–AH OGGs based on the available A. halleri genome. Of 26,007 AL–AH OGGs, 91.0% (23,682/26,007) had orthologous candidate genes in A. thaliana (Supplementary Table S5). Among the 23,682 AL–AH OGGs, 21,984 (21,984/23,682 = 92.8%) had the same topology as the species tree. In the 21,984 AL–AH OGGs, 16,800 and 2,058 A. thaliana genes were uniquely and multiply assigned to AL–AH OGGs, respectively. Since 2,058 A. thaliana genes multiply assigned to 5,183 OGGs, 5,183 OGGs were classified into 446 OGGs which had no orthologous A. thaliana genes and 4,737 OGGs which had 2,058 orthologous A. thaliana genes. Following the above procedure, 21,537 (16,800 + 4,737) AL–AH OGG with 18,858 (16,800 + 2,058) orthologous A. thaliana genes were defined as AT–AL–AH OGGs. The gain rate was inferred to be 1.6–1.8 × 10−2 (2,679 gains/18,858 genes/8–9 MY) in the A. halleri-lyrata lineage. Taken together, the gain rate without pseudogene-like A. halleri genes was 1.6–2.0 × 10−2 in the A. halleri-lyrata lineage (Fig. 2).
In our previous study, we inferred the gain rate of gene duplication in the lineage leading to Arabidopsis after the divergence of moss.4 Using the same method, we re-estimated the gain rates in three times periods—after the divergence of moss (Physcomitrella patens), rice (Oryza sativa) and poplar (Populus trichocarpa). The gain rates were 1.8–3.5 × 10−3 in the three branches, ∼10 times lower than in the A. halleri-lyrata lineage (Fig. 2). One explanation for this gain rate is that even though many genes were fixed and retained, a large number of them did not survive in the long run. This explanation is consistent with the gradual decay of paralog synonymous substitution rates observed in several eukaryotes over time.54,55
Note that the effect of tandemly duplicated genes on these gain rates needs to be considered because tandemly duplicated genes have undergone gene conversion, which leads to identical sequences among tandemly duplicated genes in the same species.56 Consequently, our procedure may have failed to identify some orthologous A. halleri genes among the A. lyrata tandemly duplicated genes. In such cases, our method would have missed the duplication event in the A. halleri-lyrata lineage, meaning our gain rate is underestimated in the A. lyrata-halleri lineage. Thus, tandemly duplicated genes did not disturb the trend of a higher gain rate in A. halleri-lyrata in comparison with other land plants.
3.2. Duplication frequency in the A. halleri lineage
The copy numbers of the A. halleri genes were unclear from AT–AL–AH OGGs. However, the absolute copy number of an A. halleri gene could be experimentally inferred by the absolute DNA concentration of the gene by ddPCR.57 First, to examine the relationship between copy number and DNA concentration, we focussed on known three-copy genes, HMA441 and MTP1,58 and 10 singleton genes that share a single copy in a broad range of plants.59 The concentration of HMA4 was 627.5 ± 40.39 and 626.75 ± 39.38 copies/μl (four replicates for each of the two primer pairs, average of each primer pair ± 95% CI). The concentration of MTP1 was 605.0 ± 22.39. Conversely, the concentration of the 10 singleton genes was 193.30 ± 9.06 copies/μl (four replicates for each of the 10 primer pairs, average of all 10 primer pairs ± 95% CI). The average DNA concentration was ∼3.2 times higher for the three-copy genes compared with the single-copy genes (Fig. 3A), indicating that the copy number corresponded to the DNA concentration inferred by ddPCR.
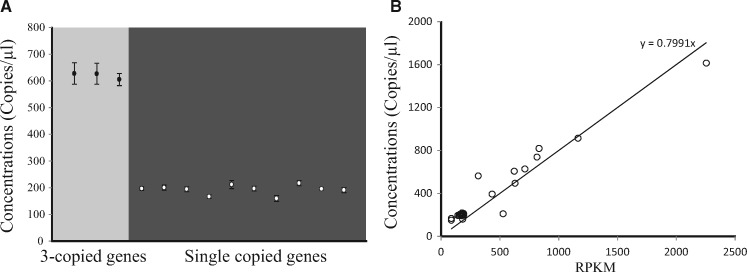
Figure 3.
Relationship between the Illumina DNA-sequencing read depth and the copy number inferred by ddPCR. (A) The Y-axis represents copy numbers per μl inferred by ddPCR. Black circles (gray background) and open circles (black background) indicate three-copy genes and single-copy genes, respectively. All points and error bars represent averages of four replicates and 95% CIs. (B) Each dot represents an A. halleri gene. The X-axis represents the Illumina DNA-sequencing read depth, which is the number of reads per 1 Kbp per 1 million reads. The Y-axis represents copy numbers per μl, which were inferred by ddPCR. The regression line was calculated with the simple formula Y = αX; α was inferred by the least squares method.
The copy numbers of the A. halleri genes could also be inferred by the reading depth of the Illumina DNA-sequencing reads. The reading depth was defined as the RPKM. To examine whether RPKM values corresponded with copy numbers, we focussed on 25 A. halleri genes that had a wide variety of RPKMs (see section 2). For these genes, the copy number estimated by digital droplet PCR (ddPCR) was compared with the RPKM. Consequently, we found that RPKM was significantly correlated with concentration (Fig. 3B, R2 = 0.94). This result indicated that RPKM values could be used an index of copy numbers for A. halleri genes.
To infer duplication frequencies by RPKM values, we focussed on 285 AT–AL–AH OGGs with single-copy genes in a broad range of plant species such as Arabidopsis, Carica, Populus, Vitis, Oryza, Selaginella and Physcomitrella.59 The RPKM values of the A. halleri genes showed a normal distribution (Supplementary Fig. S5A). To call duplicated genes, the top 5% (309) of RPKMs was defined as the threshold for duplicated genes. That is, A. halleri genes with an RPKM < 309 were defined as non-duplicated genes, A. halleri genes with RPKM values from 309 to 618 (309 × 2) were defined as two-copy genes, and A. halleri genes with RPKM values of 618–927 (309 × 3) were defined as three-copy genes. Following this rule, in 22,105 AT–AL–AH OGGs based on Illumina paired-end DNA-sequencing reads, we identified 2,488 multiply duplicated A. halleri genes and 3,378 gain events through gene duplications in the A. halleri lineage (Supplementary Table S3). The gain rate (total gains from gene duplication during a given time period divided by the estimated duration per gene) was 7.6 × 10−2 (3,378 gains/22,105 genes/2 MY).
However, the gain events may have been over-estimated because duplication events occurring in the A. halleri-lyrata lineage caused an increase of RPKMs. Therefore, excluding A. halleri genes that were duplicated in the A. halleri-lyrata lineage, we counted the number of gain events in the A. halleri lineage. Among the currently retained A. halleri genes, we focussed on 16,946 A. halleri genes that had not undergone gene duplication in the A.halleri-lyrata lineage. These 16,946 genes had undergone 1,958 gain events through gene duplication in the A. halleri lineage. The re-estimated gain rate was 5.8 × 10−2 (1,958 gains/16,946 genes/2 MY). The gain rate (5.8–7.6 × 10−2) of duplicate genes in the A. halleri lineage was approximately four times higher than the gain rate (1.6–2.0 × 10−2) in the A. halleri-lyrata lineage (Fig. 2). As mentioned earlier, the gain rate difference can be explained by a rapid decay of duplicated genes. However, this gain rate did not include gene duplications derived from pseudogenes which were on the earlier process of decayed genes. To determine the decayed effect in the A. halleri lineage, we focussed on 22,684 AT–AL–AH OGGs including pseudogene-like genes (Supplementary Table S4). Out of 22,684 OGGs, the gain rate in the A. halleri lineage was 1.2 × 10−1 (5,665 gains/22,684 genes/2 MY). Thus, the gain rate including pseudogenes-like A. halleri genes was approximately two times higher than the gain rate (7.6 × 10−2) without any pseudogene-like A. halleri genes. This result indicates that pseudogene-like A. halleri genes accelerated the gain rates in the A. halleri lineage. To determine whether pseudogenes tended to have a higher duplication rate in comparison with non-pseudogene-like genes, we fosused on 1,159 AT–AL–AH OGGs composing only pseudogene-like A. halleri genes among 22,684 AT–AL–AH OGGs including pseudogene-like genes. We then identified 713 pseudogene-like A. halleri genes experiencing gene duplications in either the A. halleri-lyrata or the A. halleri lineage. In the case of 22,105 AT–AL–AH OGGs without any pseudogene-like A. halleri genes, we identified 6,752 A. halleri genes experiencing gene duplications in either the A. halleri-lyrata or the A. halleri lineage. The proportion (713/1,159 = 62%) of duplicated genes in pseudogene-like A. halleri genes was significantly higher than that (6,752/22,105 = 31%) of duplicated genes in the other A. halleri genes (P-value = 1.9 × 10−107 by χ2 test, Table 1), indicating that most of pseudogenes tended to appear via gene duplication in Arabidopsis. Taken together, it is likely that most of recently duplicated genes in Arabidopsis may be on the process of decayed genes but some of duplicated genes significantly contributed to functional divergence among Arabidopsis species.
Table 1.
Comparison of gene duplication to non-gene duplication ratio in either A. halleri-lyrata or A. halleri lineage between OGGs including pseudogene-like A. halleri genes and OGGs without any pseudogene-like A. halleri genes
| Gene duplication in either A. halleri- lyrata or A. halleri lineage (D) | No gene duplication in either A. halleri- lyrata or A. halleri lineage (N) | D/N ratio | P value (χ2 test) | |
|---|---|---|---|---|
| OGGs derived from only pseudogenes | 713 | 446 | 1.60 | 1.9 × 10−107 |
| OGGs without any pseudogenes | 6,752 | 15,353 | 0.44 |
Using 21,537 AT–AL–AH OGGs based on the available A. halleri genome, we identified 1,248 gain events in 838 genes (Supplementary Table S5). The gain rate was 2.9 × 10−2 (1,248 gains/21,537/2MY). The gain rate was approximately half in comparison with the gain rate (5.8–7.6 × 10−2) inferred by RPKMs. Therefore, copy number information inferred by ddPCR was compared with the number of A. halleri lineage-specific duplicated genes. Most of OGGs had single-copy genes in A. halleri although various duplication frequencies were inferred by ddPCR (Supplemental Fig. S5B). These results indicate that some of gene duplication tended to be missed in a genome assembly based only on Illumina short reads. This shows that the reading depth of raw Illumina reads may be informative for inferring the number of recently duplicated genes.
3.3. Selection pressures in the A. halleri-lyrata lineage
We found that 23–30% of OGGs had undergone gene duplications in the Arabidopsis lineage at high gain rates (1.6–7.6 × 10−2 gains/gene/MY), which were 10 times higher than the rates inferred by comparative genomics among Arabidopsis, poplar, rice and moss (Fig. 2). Therefore, we were interested in investigating the functional divergence of duplicated genes in Arabidopsis. To infer the functional divergence of duplicated genes in the A. halleri-lyrata lineage, we tried to infer the ancestral sequences of the most recent common ancestor of A. thaliana, A. lyrata and A. halleri. To define the node of the most recent common ancestor among A. thaliana, A. lyrata, and A. halleri, we used an orthologous non-Arabidopsis gene as an outgroup sequence for each of AT–AL–AH OGGs and performed BLASTP searches of AT–AL–AH protein sequences against all five non-Arabidopsis (B. rapa, B. stricta, C. grandiflora, C. rubella, E. salsugineum) protein sequences.43 When the best-hit non-Arabidopsis gene was the same for the A. thaliana, A. lyrata and A. halleri genes, the non-Arabidopsis gene was considered a candidate orthologous gene to the AT–AL–AH OGG. For each of candidate genes, we searched for candidate orthologous groups that were consistent with the species tree. To do this, we generated a phylogenetic tree by the neighbour-joining method using the PAUP software (set outroot = mono, dset distance = hky).60,61 When the topology of the gene tree was the same as that of the species tree, we assumed that the topology of the orthologous group was consistent with the species tree. There were 22,105 and 21,537 AT–AL–AH OGGs based on Illumina paired-end DNA-sequencing reads and the available A. halleri genomes, respectively. Out of 22,105 and 21,537 AL–AH OGGs, we identified 17,669 and 17,925 orthologous groups that followed the speciation process among non-Arabidopsis, A. thaliana, A. lyrata, and A. halleri genes, respectively (Supplementary Table S3). These OGGs were defined as non-A-AT–AL–AH OGGs. These analysis procedures are summarized in Supplementary Figure S6.
Based on the phylogenetic tree in each non-A-AT–AL–AH OGG, we inferred the ancestor sequences of all nodes using codeml in the PAML package,53 and calculated KA and KS in the A. halleri-lyrata lineage. First, among the 17,669 OGGs base on Illumina paired-end DNA-sequencing reads, we found 481, 8,313 and 8,875 genes with positive selection (KA/KS > 1), purifying selection (KA/KS < 1) and unclear selection respectively, by the maximum likelihood approach (Supplementary Table S3). To address whether the duplicated genes contributed to functional divergence in the A. halleri-lyrata lineage, the proportions of positive selection and purifying selection in 1,782 duplicated genes were compared with those in 15,887 non-duplicated genes. In this test, the null model was the ratio of duplicated genes to non-duplicated genes without any particular selection pressure in the A. halleri-lyrata lineage. The proportions of positive selection in the duplicated genes was significantly higher than those in the non-duplicated genes (P-value = 2.8 × 10−57 by χ2 test, Fig. 4A). The proportion of purifying selection in the non-duplicated genes was significantly higher than in the duplicated genes in the A. halleri-lyrata lineage (P-value = 1.2 × 10−33 by χ2 test, Fig. 4A). Furthermore, we did the same analysis in 17,925 OOGs based on the available A. halleri genome (Supplementary Table S5). We found the same trend for only positive selection in the OOGs (P-value = 3.0 × 10−36 for positive selection, P-value = 0.05 for purifying selection by χ2 test, Fig. 4B). These results indicate that gene duplication induced functional divergence in the A. halleri-lyrata lineage.
3.4. Selection pressure in the A. halleri lineage
To infer selection pressure in the A. halleri lineage, we generated ancestral sequences of the A. lyrata and A. halleri genes using A. thaliana genes as outgroups. When an A. halleri gene did not have any sequence variation in the Illumina DNA-sequencing reads, KS and KA were simply calculated by comparing the ancestral sequence to the representative A. halleri gene sequence. When sequence variation for a representative A. halleri gene sequence was identified from the Illumina DNA-sequencing reads, KS and KA were separately calculated for the varied sequences (see Materials and Methods, Supplementary Fig. S4). Among the 22,105 AT–AL–AH OGGs based on Illumina paired-end DNA-sequencing reads, we found 568, 7,717, and 13,820 genes with positive selection (KA/KS > 1), purifying selection (KA/KS < 1) and unclear selection, respectively, in the A. halleri lineage by the maximum likelihood approach (Supplementary Table S3). To examine the relationship between the effect of gene duplication and selection pressures in the A. halleri lineage, the proportions of positive selection and purifying selection in 2,488 duplicated genes were compared with those in 19,617 non-duplicated genes. In this test, the null model was the ratio of duplicated genes to non-duplicated genes in the A. halleri lineage. As observed in the A. halleri-lyrata lineage, the proportions of positive selection in the duplicated genes were significantly higher than those in the non-duplicated genes (P-value = 2.3 × 10−6 for positive selection by χ2 test, Fig. 4C), while the proportion of purifying selection in the non-duplicated genes was significantly higher than in the duplicated genes (P-value = 1.9 × 10−26 by χ2 test, Fig. 4C). We did the same analysis in 21,537 AT–AL–AH OOGs based on the available A. halleri genome (Supplementary Table S5). We found the same trend in the OOGs (P-value = 9.9 × 10−15 for positive selection, P-value = 2.2 × 10−18 for purifying selection by χ2 test, Fig. 4D). These results indicate that gene duplication induced functional divergence in the A. halleri lineage as well.
Functional divergence may not occur immediately after gene duplication. To determine whether gene duplication in the A. halleri-lyrata lineage affected selection pressures in the A. halleri lineage, we classified the 22,105 AT–AL–AH OGGs based on Illumina paired-end DNA-sequencing reads into 5,159 OGGs with gene duplications and 16,946 OGGs without any gene duplications, focusing on the A. halleri-lyrata lineage. The proportions of positive selection and purifying selection in the duplicated genes in the A. halleri lineage were compared with those in the non-duplicated genes. In this test, the null model was the ratio of duplicated genes to non-duplicated genes in the A. halleri-lyrata lineage. The proportion of positive selection in the duplicated genes was significantly higher than those in the non-duplicated genes (P-value = 1.3 × 10−72 for positive selection by χ2 test, Fig. 4E). The proportion of purifying selection in the non-duplicated genes was significantly higher than in the duplicated genes (P-value = 1.9 × 10−85 by χ2 test, Fig. 4E). We did the same analysis in 21,537 based on the available A. halleri genome. We again found the same trend (P-value = 4.8 × 10−81 for positive selection, P-value = 1.1 × 10−39 for purifying selection by χ2 test, Fig. 4F). These results indicate that gene duplication in the A. halleri-lyrata lineage contributed to functional divergence in the A. halleri lineage.
To determine whether gene duplications in the A. halleri-lyrata or A. halleri lineage contributed to functional divergence in the A. halleri lineage, we focussed on two categories of OGGs—genes not duplicated in the A. halleri-lyrata lineage but duplicated in the A. halleri lineage (1,593 OGGs), and genes duplicated in the A. halleri-lyrata lineage but not duplicated in the A. halleri lineage (4,264 OGGs). The proportions of positive selection and purifying selection in the A. halleri lineage were compared in the two categories. In this test, the null model was the ratio of the two categories of OGGs (1,593 and 4,264 OGGs). The proportions of positive selection in genes duplicated only in the A. halleri-lyrata lineage were significantly higher than in genes duplicated only in the A. halleri lineage (P-value = 7.3 × 10−9 for positive selection by χ2 test, Fig. 4G). Conversely, the proportion of purifying selection in genes duplicated only in the A. halleri-lyrata lineage was significantly lower than in genes duplicated only in the A. halleri lineage (P-value = 2.9 × 10−8 by χ2 test, Fig. 4G). We did the same analysis in 21,537 based on based on the available A. halleri genome. We found the same trend (P-value = 0.04 for positive selection, P-value = 0.02 for purifying selection in the available genome by χ2 test, Fig. 4H). These results indicate that gene duplication in the A. halleri-lyrata lineage was the main determinant of the elevated proportion of positive selection in the A. halleri lineage.
3.5. Functional bias of genes under positive or purifying selection in the A. halleri-lyrata and A. halleri lineages
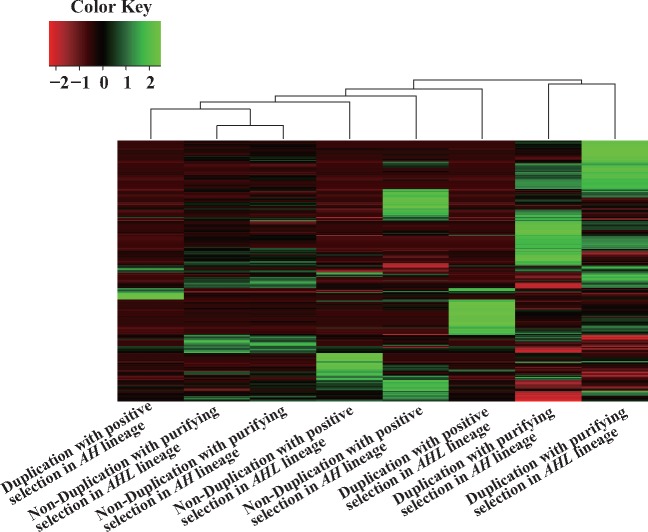
We found that gene duplication contributed to functional divergence in comparison with non-duplicated genes in Arabidopsis but many duplicated genes had been retained with purifying selection, which induces an increase of gene dosage. Thus, we were interested in investigating the functional bias in duplicated and non-duplicated genes with positive/purifying selection in the A. halleri-lyrata and A. halleri lineages. OGGs based on Illumina paired-end DNA-sequencing reads were classified into duplicated and non-duplicated genes in the A. halleri-lyrata and A. halleri lineages. The OGGs were then further classified by positive and purifying selection. This gave a total of eight classes. In each class, we examined the degree of over-representation in 1,504 GO categories compared with the number of GO categories assigned to A. thaliana genes (see Section 2) (Fig. 5). Although two classes with no duplication and purifying selection were clustered into one group, the other six classes were not essentially clustered with each other. These results indicate that the evolutionary direction of A. halleri was quite different in the A. halleri-lyrata and A. halleri lineages with respect to either gene dosage by gene duplication or functional divergence by positive selection.
Figure 5.
Over-represented functional categories. The X-axis represents different kinds of OGGs. OGGs were classified into duplicated genes and non-duplicated genes in the A. halleri-lyrata (AHL) and A. halleri (AH) lineages. The OGGs were then further classified by positive or purifying selection. The Y-axis represents 1,504 GO categories (biological processes) assigned to A. thaliana genes belonging to the OGGs. The key shows the relationship between colour and the z-score of over-representation in the GO categories. Red and green indicate low and high over-representation, respectively. The ratio of observed gene numbers in selected OGGs to expected gene numbers inferred from the data for all annotated genes was calculated in each GO category.
To examine the kinds of genes associated with phenotypic differences among A. thaliana, A. lyrata and A. halleri, we identified significantly over-represented GO categories in OGGs with gene duplication and purifying selection, OGGs with gene duplication and positive selection and OGGs with non-duplication and positive selection in the A. halleri–A. lyrata and A. halleri lineages. OGGs with non-duplication and purifying selection were disregarded in the analysis because such genes tended to have the same functions in A. halleri, A. lyrata and A. halleri. From the GO categories assigned to the A. thaliana genes belonging to the OGGs, over-represented GO categories were identified (Supplementary Table S6; see Section 2, FDR < 0.01).
When we focussed on metal and zinc tolerance or accumulation among A. thaliana, A. lyrata and A. halleri, both A. lyrata and A. halleri had a higher tolerance to metal and zinc than A. thaliana.36 In particular, both metal and zinc tend to be accumulated in A. halleri compared with A. lyrata.62,63 This observation suggests that metal tolerance is enhanced in the A. halleri–A. lyrata lineage, and that metal accumulation is enhanced in the A. halleri lineage. Genes associated with metal and zinc transporters/responses were highly duplicated with purifying selection in the A. halleri-lyrata lineage, indicating that the dosages of genes associated with metal responses and transporters had been enhanced in the A. halleri-lyrata lineage. Furthermore, genes associated with metal and zinc responses were highly duplicated with purifying selection in the A. halleri-lyrata lineage, indicating that the dosages of genes associated with metal and zinc responses had been enhanced in the A. halleri-lyrata lineage. Thus, tolerance or accumulation of metal and zinc was enhanced in the A. halleri-lyrata and A. halleri lineages through gene duplication.
Genes associated with the reproductive system, cell cycle, various developments, various metabolites, epigenetics, metabolites, abiotic and biotic responses were subject to positive selection in either the A. halleri-lyrata or A. halleri lineage (Supplementary Table S6). However, we do not have any clear idea why such genes were subject to positive selection. Additionally, genes associated with various ubiquitous processes tended to be duplicated in either the A. halleri-lyrata or the A. halleri lineage with purifying selection (Supplementary Table S6). These over-represented functional categories may contribute to phenotypic differences between A. thaliana and either A. lyrata or A. halleri through high dosages. However, we do not know the phenotypic differences associated with these functional categories. These duplicated genes might have been retained because increased gene dosages associated with these functional categories were not too disadvantageous for A. halleri. To avoid gaining novel functions, these genes may be under purifying selection. In the future, these duplicated genes may be lost if disadvantageous functions appear. Indeed, the A. halleri-lyrata and A. halleri lineages have extraordinarily high retention rates of duplicated genes in comparison with earlier plant lineages. These observations indicate that most of the duplicated genes in the A. halleri-lyrata and A. halleri lineages may be lost in future evolution.
3.6. Concluding remarks
In this analysis, we generated 25,833 A. halleri genes that were orthologous to 79.1% of the annotated A. lyrata genes from contigs generated from only paired-end reads of Illumina DNA-sequencing. On the other hands, we inferred 26,007 AH-AL OOGs based on the available A. halleri genome. Out of 32,670 A. lyrata genes, 79.6% were identified as orthologous genes to A. halleri genes. Thus, our method inferring orthologous genes is compatible to a method to infer orthologous genes based on the available genome. However, it has significant limitations for examining gene loss, exon shuffling and heterozygosity because it does not infer any new genes in the genomes. Nevertheless, the number of duplicated genes inferred by reading depth of Illumina DNA-sequencing reads is likely to be more reliable in comparison with duplicated genes identified on scaffolds generated by Illumina DNA-sequencing reads (Supplementary Fig. S5B). Therefore, this procedure is useful for inferring lineage-specific duplicated genes resulting from such events as SSDs.
The gain rates based on 25, 833 AL–AH OGGs based on both Illumina paired-end DNA-sequencing reads and the available A. halleri genome were 1.6–2.0 × 10−2 per gene per MY in the A. halleri-lyrata lineage (Fig. 2). Furthermore, using the only mapping coverage of the Illumina DNA-sequencing reads, the gain rate was inferred to be 5.7–7.6 × 10−2 in the A. halleri lineage because gene duplication tended to be missed in a genome assembly based on Illumina short reads. That is, the gain rates were inferred to be 1.6–2.0 and 5.7–7.6 × 10−2 per gene per MY in the A. halleri-lyrata and A. halleri lineages, respectively. The gain rate in the A. halleri lineage was approximately four times higher than in the A. halleri-lyrata lineage. Using our previous data, we re-estimated the gain rates in the three time periods after the divergence of mosses, rice and poplar (Fig. 2). The inferred gain rates (1.8–3.0 × 10−3) were ∼10 times lower than in the Arabidopsis lineage (Fig. 2). Thus, gain rates tend to increase as the evolutionary period gets younger. One explanation for this gain rate difference is that duplicated genes tend to rapidly decay over time. This explanation is supported by a higher rate of pseudogenization in recently duplicated genes in comparison with non-duplicated genes (Table 1). Also, several previous reports showed that younger duplicated genes tended to be relaxed compared with older duplicated genes.54,55,64–67 That is, most anciently duplicated genes tend not to be retained in current species. Consequently, the gain rates inferred in earlier evolutional periods tend to decrease.
To investigate the functional divergence of duplicated genes in the A. halleri-lyrata and A. halleri lineages, we identified OGGs under either positive or purifying selection in these lineages based on the ratio of nonsynonymous and synonymous substitution rates (KA/KS). Interestingly, the proportions of positive and purifying selection tended to increase and decrease, respectively, when gene duplication occurred in either the A. halleri-lyrata or A. halleri lineage. This result indicates that gene duplication tends to enhance functional divergence in comparison with non-duplicated genes in the Arabidopsis lineage. In contrast, the general observation is that duplicated genes tend to have less functional divergence in yeasts, plants and mammals.24,68,69 This is because functionally important genes are more likely to be retained as duplicates.68 This contradictory relationship might derive from the duplication ages. Most previous analyses have examined recently observed selection pressures in anciently duplicated genes. When functional divergence was examined in recently duplicated genes, the duplicated genes tended to have higher functional divergence than singletons.70 Together, these results suggest duplicated genes tend to have higher functional divergence immediately after duplication than singletons.
How long gene duplication accelerates functional divergence remains an open question. To address this, we examined whether gene duplication in the A. halleri-lyrata lineage (2–10 MYA) accelerated functional divergence in the A. halleri lineage (<2 MYA). Interestingly, we found that gene duplication in the A. halleri-lyrata lineage enhanced the proportion of positive selection in the A. halleri lineage (Fig. 4E–H). This result indicated that the functional divergence of duplicated genes was accelerated several MY after gene duplication. If gene duplication is too deleterious for a gene, the gene tends to be lost immediately after duplication. If not, duplicated genes may be retained for a long period without functional divergence because functional divergence may be evolutionarily disadvantageous. Therefore, immediately after duplication, most duplicated genes might be under functional constraints in comparison with genes duplicated several MY earlier. Indeed, many recently duplicated genes have functional redundancy in A. thaliana17,18 and in mammals.71 These young duplicated genes tend to be less functionally constrained than singletons, and may have the potential to obtain an essential function to survive in new environments.
Finally, we examined the kinds of genes that were duplicated and/or under positive selection in the A. halleri-lyrata and A. halleri lineages. Different functional categories tended to have experienced gene duplication and/or selection pressure in the A. halleri-lyrata and A. halleri lineages. For example, A. halleri is known as a heavy metal hyper-accumulator with high metal tolerance. A. lyrata is tolerant of heavy-metal ions in the soil to some degree but A. thaliana is not. Genes related to heavy-metal tolerance and accumulation tended to be highly duplicated with purifying selection in the A. halleri-lyrata and A. halleri lineages. Earlier studies reported that metal tolerance was enhanced by increasing gene dosage through gene duplication.41 Our results supported this trend at a genomic scale. Taken together, the results of our study reveal that lineage-specific duplicated genes have contributed to species-specific evolution in Arabidopsis.
4. Availability
Illumina DNA-sequencing data (DRA004564) have been deposited in the DDBJ Sequence Read Archive (https://trace.ddbj.nig.ac.jp/dra/). Contig sequences (BFAE01000001-BFAE01344622) assembled from the Illumina DNA-sequencing data have been deposited in the DDBJ Mass Submission System. A. halleri gene sequences determined in this study are included in Supplementary Tables S3 and S4.
Supplementary Material
Acknowledgements
We thank Kiyomi Imamura, Makiko Tosaka, Taiji Kikuchi, Terumi Horiuchi, Kanako Onizuka and Miu Kubota for Illumina DNA-sequencing analyses and ddPCR analyses. We also thank the National Institute of Genetics of the Research Organization of Information and Systems for providing excellent supercomputer services. This work was supported by the Research for Evolutional Science and Technology (CREST) programme ‘Creation of essential technologies to utilize carbon dioxide as a resource through the enhancement of plant productivity and the exploitation of plant products’ of the Japan Science and Technology Agency (JST) (JPMJCR11B3 to K.H. and S.I.M.); Grants-in-Aid for Scientific Research (15K14421, 15H02433, K.H. and S.I.M.) and research grants from the Takeda Science Foundation, the Sumitomo Foundation, the Kurume Research Park and the Asahi Glass Foundation (to K.H.).
Conflict of interest
None declared.
Supplementary data
Supplementary data are available at DNARES online.
References
- 1. Lockton S., Gaut B. S.. 2005, Plant conserved non-coding sequences and paralogue evolution, Trends Genet., 21, 60–5. [DOI] [PubMed] [Google Scholar]
- 2. Vanneste K., Baele G., Maere S., Van de Peer Y.. 2014, Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary, Genome Res., 24, 1334–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hanada K., Vallejo V., Nobuta K., et al. 2009, The functional role of pack-MULEs in rice inferred from purifying selection and expression profile, Plant Cell, 21, 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hanada K., Zou C., Lehti-Shiu M. D., Shinozaki K., Shiu S. H.. 2008, Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli, Plant Physiol., 148, 993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fortna A., Kim Y., MacLaren E., et al. 2004, Lineage-specific gene duplication and loss in human and great ape evolution, PLoS Biol., 2, E207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rostoks N., Borevitz J. O., Hedley P. E., et al. 2005, Single-feature polymorphism discovery in the barley transcriptome, Genome Biol., 6, R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clark R. M., Schweikert G., Toomajian C., et al. 2007, Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana, Science, 317, 338–42. [DOI] [PubMed] [Google Scholar]
- 8. Rizzon C., Ponger L., Gaut B. S.. 2006, Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice, PLoS Comput Biol., 2, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rodgers-Melnick E., Mane S. P., Dharmawardhana P., et al. 2012, Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus, Genome Res., 22, 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leister D. 2004, Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene, Trends Genet., 20, 116–22. [DOI] [PubMed] [Google Scholar]
- 11. Ohno S. 1970, Evolution by Gene Duplication. Springer-Verlag: New York. [Google Scholar]
- 12. Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L., Postlethwait J.. 1999, Preservation of duplicate genes by complementary, degenerative mutations, Genetics, 151, 1531–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zou C., Lehti-Shiu M. D., Thibaud-Nissen F., Prakash T., Buell C. R., Shiu S. H.. 2009, Evolutionary and expression signatures of pseudogenes in Arabidopsis and rice, Plant Physiol., 151, 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zheng D., Gerstein M. B.. 2007, The ambiguous boundary between genes and pseudogenes: the dead rise up, or do they? Trends Genet., 23, 219–24. [DOI] [PubMed] [Google Scholar]
- 15. Conant G. C., Wolfe K. H.. 2008, Turning a hobby into a job: how duplicated genes find new functions, Nat. Rev. Genet., 9, 938–50. [DOI] [PubMed] [Google Scholar]
- 16. Kondrashov F. A. 2012, Gene duplication as a mechanism of genomic adaptation to a changing environment, Proc. Biol. Sci., 279, 5048–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hanada K., Kuromori T., Myouga F., Toyoda T., Li W. H., Shinozaki K.. 2009, Evolutionary persistence of functional compensation by duplicate genes in Arabidopsis, Genome Biol. Evol., 1, 409–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hanada K., Sawada Y., Kuromori T., et al. 2011, Functional compensation of primary and secondary metabolites by duplicate genes in Arabidopsis thaliana, Mol. Biol. Evol., 28, 377–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nowak M. A., Boerlijst M. C., Cooke J., Smith J. M.. 1997, Evolution of genetic redundancy, Nature, 388, 167–71. [DOI] [PubMed] [Google Scholar]
- 20. Hanada K., Kuromori T., Myouga F., Toyoda T., Shinozaki K.. 2009, Increased expression and protein divergence in duplicate genes is associated with morphological diversification, PLoS Genet., 5, e1000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alvarez-Ponce D., Fares M. A.. 2012, Evolutionary rate and duplicability in the Arabidopsis thaliana protein-protein interaction network, Genome Biol. Evol., 4, 1263–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Warren A. S., Anandakrishnan R., Zhang L.. 2010, Functional bias in molecular evolution rate of Arabidopsis thaliana, BMC Evol. Biol., 10, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang J., Marowsky N. C., Fan C.. 2013, Divergent evolutionary and expression patterns between lineage specific new duplicate genes and their parental paralogs in Arabidopsis thaliana, PLoS One, 8, e72362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang L., Gaut B. S.. 2011, Factors that contribute to variation in evolutionary rate among Arabidopsis genes, Mol. Biol. Evol., 28, 2359–69. [DOI] [PubMed] [Google Scholar]
- 25. Wendel J. F., Jackson S. A., Meyers B. C., Wing R. A.. 2016, Evolution of plant genome architecture, Genome Biol. , 17, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. DeBarry J. D., Kissinger J. C.. 2014, A survey of innovation through duplication in the reduced genomes of twelve parasites, PLoS One, 9, e99213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sin K., Street N., Lundeberg J., Arvestad L.. 2012, Improved gap size estimation for scaffolding algorithms, Bioinformatics, 28, 2215–22. [DOI] [PubMed] [Google Scholar]
- 28. Arabidopsis_Genome_Initiative 2000, Analysis of the genome sequence of the flowering plant Arabidopsis thaliana, Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 29. Hu T. T., Pattyn P., Bakker E. G., et al. 2011, The Arabidopsis lyrata genome sequence and the basis of rapid genome size change, Nature Genetics, 43, 476–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beilstein M. A., Nagalingum N. S., Clements M. D., Manchester S. R., Mathews S.. 2010, Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana, Proc. Natl. Acad. Sci. U. S. A., 107, 18724–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moghe G. D., Hufnagel D. E., Tang H., et al. 2014, Consequences of whole-genome triplication as revealed by comparative genomic analyses of the wild radish raphanus raphanistrum and three other Brassicaceae species, Plant Cell, 26, 1925–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koenig D., Weigel D.. 2015, Beyond the thale: comparative genomics and genetics of Arabidopsis relatives, Nat. Rev. Genet., 16, 285–98. [DOI] [PubMed] [Google Scholar]
- 33. Kramer U. 2010, Metal hyperaccumulation in plants, Ann. Rev. Plant Biol., 61, 517–34. [DOI] [PubMed] [Google Scholar]
- 34. Verbruggen N., Hermans C., Schat H.. 2009, Molecular mechanisms of metal hyperaccumulation in plants, New Phytol., 181, 759–76. [DOI] [PubMed] [Google Scholar]
- 35. Shimizu K. K., Purugganan M. D.. 2005, Evolutionary and ecological genomics of Arabidopsis, Plant Physiol., 138, 578–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Turner T. L., Bourne E. C., Von Wettberg E. J., Hu T. T., Nuzhdin S. V.. 2010, Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils, Nat. Genet., 42, 260–3. [DOI] [PubMed] [Google Scholar]
- 37. Novikova P. Y., Hohmann N., Nizhynska V., et al. 2016, Sequencing of the genus Arabidopsis identifies a complex history of nonbifurcating speciation and abundant trans-specific polymorphism, Nat. Genet, 48, 1077–82. [DOI] [PubMed] [Google Scholar]
- 38. Briskine R. V., Paape T., Shimizu-Inatsugi R., et al. 2017, Genome assembly and annotation of Arabidopsis halleri, a model for heavy metal hyperaccumulation and evolutionary ecology, Mol. Ecol. Resour., 17, 1025–36. [DOI] [PubMed] [Google Scholar]
- 39. Akama S., Shimizu-Inatsugi R., Shimizu K. K., Sese J.. 2014, Genome-wide quantification of homeolog expression ratio revealed nonstochastic gene regulation in synthetic allopolyploid Arabidopsis, Nucleic Acids Res., 42, e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sato Y., Kudoh H.. 2014, Fine-scale genetic differentiation of a temperate herb: relevance of local environments and demographic change, AoB Plants, 6, plu070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hanikenne M., Talke I. N., Haydon M. J., et al. 2008, Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4, Nature, 453, 391–5. [DOI] [PubMed] [Google Scholar]
- 42. Al-Shehbaz I. A., O'Kane S. L. Jr. 2002, Taxonomy and phylogeny of Arabidopsis (Brassicaceae). In The Arabidopsis Book/American Society of Plant Biologists, Vol. 1. [DOI] [PMC free article] [PubMed]
- 43. Boratyn G. M., Camacho C., Cooper P. S., et al. 2013, BLAST: a more efficient report with usability improvements, Nucleic Acids Res., 41, W29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Johnston J. S., Pepper A. E., Hall A. E., et al. 2005, Evolution of genome size in Brassicaceae, Ann. Bot., 95, 229–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kajitani R., Toshimoto K., Noguchi H., et al. 2014, Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads, Genome Res., 24, 1384–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Simpson J. T., Wong K., Jackman S. D., Schein J. E., Jones S. J., Birol I.. 2009, ABySS: a parallel assembler for short read sequence data, Genome Res., 19, 1117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Luo R., Liu B., Xie Y., et al. 2012, SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler, Gigasci., 1, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu T. D., Watanabe C. K.. 2005, GMAP: a genomic mapping and alignment program for mRNA and EST sequences, Bioinformatics, 21, 1859–75. [DOI] [PubMed] [Google Scholar]
- 49. Langdon W. B. 2015, Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks, BioData Min., 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J. A.. 2007, Primer3Plus, an enhanced web interface to Primer3, Nucleic Acids Res., 35, W71–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Remm M., Storm C. E., Sonnhammer E. L.. 2001, Automatic clustering of orthologs and in-paralogs from pairwise species comparisons, J. Mol. Biol., 314, 1041–52. [DOI] [PubMed] [Google Scholar]
- 52. Katoh K., Misawa K., Kuma K., Miyata T.. 2002, MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform, Nucleic Acids Res., 30, 3059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang Z. 2007, PAML 4: phylogenetic analysis by maximum likelihood, Mol. Biol. Evol., 24, 1586–91. [DOI] [PubMed] [Google Scholar]
- 54. Lynch M., Conery J. S.. 2000, The evolutionary fate and consequences of duplicate genes, Science, 290, 1151–5. [DOI] [PubMed] [Google Scholar]
- 55. Bu L., Katju V.. 2015, Early evolutionary history and genomic features of gene duplicates in the human genome, BMC Genomics, 16, 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gao L. Z., Innan H.. 2004, Very low gene duplication rate in the yeast genome, Science, 306, 1367–70. [DOI] [PubMed] [Google Scholar]
- 57. Heredia N. J., Belgrader P., Wang S., et al. 2013, Droplet Digital PCR quantitation of HER2 expression in FFPE breast cancer samples, Methods, 59, S20–3. [DOI] [PubMed] [Google Scholar]
- 58. Drager D. B., Desbrosses-Fonrouge A. G., Krach C., et al. 2004, Two genes encoding Arabidopsis halleri MTP1 metal transport proteins co-segregate with zinc tolerance and account for high MTP1 transcript levels, Plant J., 39, 425–39. [DOI] [PubMed] [Google Scholar]
- 59. Duarte J. M., Wall P. K., Edger P. P., et al. 2010, Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels, BMC Evol. Biol., 10, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wilgenbusch J. C., Swofford D.. 2003, Inferring evolutionary trees with PAUP*, Curr. Protoc. Bioinformatics, Chapter 6, Unit 6 4. [DOI] [PubMed] [Google Scholar]
- 61. Saitou N., Nei M.. 1987, The neighbor-joining method: a new method for reconstructing phylogenetic trees, Mol. Biol. Evol., 4, 406–25. [DOI] [PubMed] [Google Scholar]
- 62. Willems G., Drager D. B., Courbot M., Gode C., Verbruggen N., Saumitou-Laprade P.. 2007, The genetic basis of zinc tolerance in the metallophyte Arabidopsis halleri ssp. halleri (Brassicaceae): an analysis of quantitative trait loci, Genetics, 176, 659–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Frerot H., Faucon M. P., Willems G., et al. 2010, Genetic architecture of zinc hyperaccumulation in Arabidopsis halleri: the essential role of QTL x environment interactions, New Phytol., 187, 355–67. [DOI] [PubMed] [Google Scholar]
- 64. Vishnoi A., Kryazhimskiy S., Bazykin G. A., Hannenhalli S., Plotkin J. B.. 2010, Young proteins experience more variable selection pressures than old proteins, Genome Res., 20, 1574–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jordan I. K., Wolf Y. I., Koonin E. V.. 2004, Duplicated genes evolve slower than singletons despite the initial rate increase, BMC Evol. Biol., 4, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Alba M. M., Castresana J.. 2005, Inverse relationship between evolutionary rate and age of mammalian genes, Mol. Biol. Evol., 22, 598–606. [DOI] [PubMed] [Google Scholar]
- 67. Wolf Y. I., Novichkov P. S., Karev G. P., Koonin E. V., Lipman D. J.. 2009, The universal distribution of evolutionary rates of genes and distinct characteristics of eukaryotic genes of different apparent ages, Proc. Natl. Acad. Sci. U. S. A., 106, 7273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Davis J. C., Petrov D. A.. 2004, Preferential duplication of conserved proteins in eukaryotic genomes, PLoS Biol., 2, E55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yang J., Gu Z., Li W. H.. 2003, Rate of protein evolution versus fitness effect of gene deletion, Mol. Biol. Evol., 20, 772–4. [DOI] [PubMed] [Google Scholar]
- 70. Satake M., Kawata M., McLysaght A., Makino T.. 2012, Evolution of vertebrate tissues driven by differential modes of gene duplication, DNA Res., 19, 305–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lan X., Pritchard J. K.. 2016, Coregulation of tandem duplicate genes slows evolution of subfunctionalization in mammals, Science, 352, 1009–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rensing S. A., Lang D., Zimmer A. D., et al. 2008, The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants, Science, 319, 64–9. [DOI] [PubMed] [Google Scholar]
- 73. Heckman T. M., Kauffmann G.. 2011, The coevolution of galaxies and supermassive black holes: a local perspective, Science, 333, 182–5. [DOI] [PubMed] [Google Scholar]
- 74. Tuskan G. A., Difazio S., Jansson S., et al. 2006, The genome of black cottonwood, Populus trichocarpa (Torr. & Gray), Science, 313, 1596–604. [DOI] [PubMed] [Google Scholar]
- 75. Wolfe K. H., Gouy M., Yang Y. W., Sharp P. M., Li W. H.. 1989, Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data, Proc. Natl. Acad. Sci. U. S. A., 86, 6201–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.