Abstract
Lentiviral RNA genomes contain structural elements that play critical roles in viral replication. Although structural features of 5′-untranslated regions have been well characterized, attempts to identify important structures in other genomic regions by Selective 2′-Hydroxyl Acylation analyzed by Primer Extension (SHAPE) have led to conflicting structural and mechanistic conclusions. Previous approaches accounted neither for sequence heterogeneity that is ubiquitous in viral populations, nor for selective constraints operating at the protein level. We developed an approach that augments SHAPE with phylogenetic analyses and applied it to investigate structure in coding regions (cRNA) within the HIV and SIV envelope genes. Analysis of single-genome SHAPE data with phylogenetic information from diverse lentiviral sequences argues against the conservation of a putative global gp120 RNA structure but points to the existence of core RNA sub-structures. Our findings establish a framework for considering sequence heterogeneity and protein function in de novo RNA structure inference approaches.
Keywords: HIV, SIV, RNA structure, evolution, SHAPE, phylogenetics
1. Introduction
Human immunodeficiency virus type 1 (HIV-1) is among the fastest evolving viruses, incorporating approximately one nucleotide substitution every two to three replication cycles, and displaying remarkable heterogeneity at both the inter- and intra-host levels (Salemi 2013). The relatively short (approximately ten kilobases) genome of HIV-1 is packed with information: nine protein-coding genes (with up to three overlapping reading frames), a large number of transcription regulatory sequences, and a plethora of functionally important protein-binding sites. These properties make HIV-1 an ideal system for large-scale evolutionary inferences of the interplay of protein and RNA structure (Sanjuan and Borderia 2011). However, despite promising work in nuclear magnetic resonance spectroscopy (NMR), crystallography, and chemical probing (Felden 2007; Sukosd 2015; Mathews, Turner, and Watson 2016), we have only recently begun to scratch the surface of knowledge of the intricate biochemical interactions conserved across large regions of viral genomes (Felden 2007; Sukosd 2015; Mathews, Turner, and Watson 2016).
Efforts to characterize the lentiviral RNA structural landscape focused initially on the non-coding 5′-untranslated region (UTR) of the viral genome. Combinations of chemical reactivity-based nucleotide accessibility mapping, mutagenesis, and biochemical approaches applied to both recombinant RNAs (Harrison and Lever 1992; Baudin 1993; Sassetti, and Parslow 1995; McBride and Panganiban 1996; Clever, Miranda, and Parslow 2002; Abbink and Berkhout 2003; Damgaard et al. 2004; Paillart et al. 2004; Wilkinson et al. 2008; Watts et al. 2009) and genomic RNAs isolated from viruses and transfected cells (Paillart et al. 2004; Wilkinson et al. 2008; Watts et al. 2009; Lu et al. 2011a) led to more than twenty proposed secondary structure models (Lu et al. 2011a). Although early studies typically reported probability scales or multiple models that satisfied or partially satisfied the chemical probing data (Harrison and Lever 1992; Baudin 1993; Clever, Sassetti, and Parslow 1995; McBride and Panganiban 1996; Clever, Miranda, and Parslow 2002; Abbink and Berkhout 2003; Damgaard et al. 2004; Paillart et al. 2004), more recent studies that employ selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) have led to proposals for unique RNA secondary structures. Indeed, SHAPE has been proposed to afford structures with >95% accuracy for some RNAs (Deigan et al. 2009). However, it is now clear from NMR and in-gel chemical probing studies that RNAs, including the HIV-1 5′-UTR, can exist as an equilibrium mixture of conformers with different secondary structures, and this may explain discrepancies among models derived from bulk chemical reactivity analyses (Lu, Heng, and Summers 2011b; Kenyon et al. 2013; Keane et al. 2015; Keane and Summers 2016).
In order to more broadly characterize the lentiviral RNA structural landscape, several research groups (Watts et al. 2009; Pollom et al. 2013) have collaborated to measure RNA structure across full-length HIV-1 and SIV genomes using SHAPE. With this single-nucleotide resolution method, Pollom et al. (2013) correctly identified local structures within extensively studied non-coding regions and went on to propose a functional role for global RNA structures across large protein-coding regions. While the importance of HIV-1 non-coding RNA (ncRNA) structures in the regulation of viral replication is well-appreciated (Berkhout 2000; Damgaard et al. 2004; D’Souza and Summers 2005; Lu et al. 2011a; Kuzembayeva et al. 2014), a large proportion of the predicted RNA structures within coding regions (cRNA) remain structurally and functionally uncharacterized (Watts et al. 2009; Pollom et al. 2013; Lavender, Gorelick, and Weeks 2015), and the validity of the proposed structures is debated (Knoepfel and Berkhout 2013). The ambiguity in the inferred cRNA structures may be due to heterogeneity in RNA sequence and structure (Zhu et al. 2013; Sukosd 2015). Alternatively, RNA structural segments could exist simultaneously as an equilibrium mixture of structural conformers, which would confound SHAPE analysis, as appears to have been the case for the HIV-1 5′-UTR structure (Lu, Heng, and Summers 2011b; Deforges, Chamond, and Sargueil 2012; Kenyon et al. 2013; Keane et al. 2015). The uncharacterized cRNA structures may also represent a new class of regulatory elements constrained evolutionarily by selective pressures at both the protein and RNA levels. Although the recently proposed coupling of SHAPE with next-generation sequencing (SHAPE-seq) has expanded on the original method to study multiple structures within diverse virus populations (Mortimer et al. 2012), it is unable to incorporate information on protein evolutionary constraints in the identification of conserved cRNA structures.
Phylogenetic methods have been a mainstay in RNA structure inference or corroboration, for example, see Seetin and Mathews (2012) for a review, and references (Hofacker et al. 1998; Poon et al. 2010; Knoepfel and Berkhout 2011; Zanini and Neher 2013; Rollins et al. 2014; Mueller, Das, and Berkhout 2016) for studies focused specifically on various regions of the HIV-1 genome. The evolutionary signal derives from the dependence of evolutionary rates at individual sites on their degree of constraint in the secondary RNA structure (Shapiro et al. 2007); for example, bases that are paired in the RNA structure are expected to evolve more slowly relative to unpaired bases, although substitutions that maintain or restore canonical or weakly canonical pairings occur at higher rates than other mutations in stem regions (Kosakovsky Pond et al. 2007) (see Supplementary Fig. S1). Numerous algorithms take these patterns and codon position (a proxy for protein-level constraints) into consideration, thereby combining RNA interactions and protein-level conservation. For example, RNA-Decoder (Pedersen et al. 2004a,b), uses a generative model of RNA sequences, wherein the secondary structure is modeled by a stochastic context-free grammar, and the evolution of sites is governed by a di-nucleotide or single-nucleotide model, depending on the predicted structural category. However, current phylogenetic methods do not fully model evolutionary constraints jointly and do not permit a full joint inference of the RNA structure and evolutionary model parameters (e.g., it is typically specified a priori and fixed during the analysis), even in the face of significant sequence variation (Shapiro et al. 2007).
Considering the limitations specific to the SHAPE and phylogenetic RNA structure prediction methods, it is not surprising that many SHAPE-derived structures within the genome of the commonly used laboratory strain HIV-1NL4-3 did not agree with measures of evolutionary conservation across HIV-1 group M reference subtypes (Watts et al. 2009). Contradictory findings point to the need to understand the source of disagreement, specifically as it pertains to identifying structures that warrant further exploration using biophysical investigative methods. To that end, we compared the performance of different ways of incorporating RNA structure prediction data into evolutionary analyses of a rapidly evolving gene (env) in HIV and SIV populations within and among infected hosts. The gp120 gene fragment of env offers an attractive target for the evolutionary analysis in the context of RNA structure: it evolves rapidly due to strong selective pressures (this is desirable for phylogenetic rate inference), large and informative data sets are available for within- and between-host analyses, and many previous analyses have focused on this gene.
We found that by combining prediction data from SHAPE and phylogenetic methods, core conserved cRNA structures could be identified that may be important to gp120 function and evolutionary conservation and that would otherwise be lost when searching for a single global structure within a heterogeneous viral population.
2. Results
2.1 Phylogenetic inferences indicate significant contribution of RNA structure to HIV and SIV gp120 evolution
RNA structure imposes a set of a priori constraints on individual sites in multiple sequence alignments (MSA) of viral sequences. For instance, sites that lie within paired RNA regions are expected to evolve slower (due to the need to maintain Watson–Crick pairing) than sites that are unpaired. We used different definitions of RNA structure and conservation information (see Table 1) to partition sites within MSA of the env gene of HIV and SIV into two to six non-overlapping sets and endowed each set with its own evolutionary rate parameters during phylogenetic analysis. We also considered two control models, which do not use any structural information. The first (random-partition) model divided sites into a fixed number of groups at random, and the second (gamma-distributed) model is the standard rate-variation model in phylogenetic inference, where the evolutionary rate at each site is drawn from a gamma distribution with four discrete rates.
Table 1.
Description of data partition strategies. Details for each partition model and categorization strategy can be found in Section 4.
| Model | Classes | Site partitioning | Source |
|---|---|---|---|
| SHAPE reactivity | 4 | Scaled ([0–1]) reactivity, log2 transformed, and binned into quartiles | Site-specific values determined by Pollom et al. (2013) and Watts et al. (2009) using single HIV-1NL4-3 and SIVMAC239 reference genomes |
| Pairing status within predicted structure | 2 |
|
Final structure refers to the final lowest free energy Pollom et al. (2013) HIV and SIV predicted structures, originally obtained by incorporating SHAPE reactivities as pseudo-free energy restraints in conjunction with nearest neighbor parameters in RNAstructure (Mathews 2014) |
| Relative position within helices of predicted structure | 4 |
|
Categories previously described by Mimouni et al. (2009) were assigned to corresponding sites within the final Pollom et al. (2013) HIV and SIV structures. Helices consisting of fewer than four interacting residues were discarded. |
| Codon position within predicted structure | 6 | Codon positions (1, 2, and 3) within paired (P) and unpaired (U) regions | Category was assigned according to the final Pollom et al. (2013) HIV and SIV predicted structures |
| Pairing probability (PPROB) | 4 | Scaled ([0–1]) values, log2 transformed, and binned into quartiles | Site-specific values were determined by Pollom et al. (2013) and Watts et al. (2009) using RNA-Decoder (Pedersen et al. 2004a,b) for HIV and SIV reference sequence alignments. |
| ‘SHAPE + PPROB’ (Combined Model) | 4 | Structural stability
|
Sites were re-assigned as part of this study according to categorized concordance between original SHAPE and PPROB values (e.g., ‘most structurally stable’ refers to sites with both low [near 0] SHAPE reactivity and high [near one] PPROB). Sites with discordant assignments were partitioned separately (5) and discarded during statistical analyses. |
We next fitted these partitioned models to the data in the Bayesian coalescent phylogenetic reconstruction framework (Drummond et al. 2005) and used (log) Bayes Factors (Supplementary Table S1) to compare model fits. The combined partition approach (assigning sites based on both SHAPE and pairing probability data) provided the best model fit for all data sets, whereas the randomly generated partition model ranked last, as expected. The ranking of other structural partition models varied depending on the data set. Importantly, most of these structure-only models (e.g., SHAPE data only) performed worse than the standard (structure-agnostic) gamma variation model, indicating that rate variation patterns learned from the MSA were more informative than those supplied from structural sources only. The only exception for the majority of data sets was the partition based on relative nucleotide position within helices of the minimal free-energy structure predicted by Pollom et al. (2013). However, the significantly better fit for the combined approach indicates that either the definition of helices alone is insufficient to capture relevant rate variation, or that a significant proportion of these helices are not accurately annotated. Additionally, for the HIV and SIV reference (or among-host) data sets and all but one SIV-infected macaque data set, the codon partition model (incorporating nucleotide position within the codon in the previously predicted structure) outperformed the SHAPE model, whereas the opposite was true for HIV-infected patients, for which the SHAPE model was significantly better. This finding suggests that protein-level selective constraints, ignored by the SHAPE method, should also be considered when evaluating the evolution and/or conservation of RNA structure, particularly among highly heterogeneous sequences.
2.2 Core RNA structures within HIV and SIV gp120 are conserved, but SHAPE-indicated global structure is not
Evolutionary selective pressure to preserve functional RNA secondary structure can be estimated using a variety of molecular evolutionary analyses. As previously mentioned, nucleotide sites involved in forming intra-molecular base pairs tend to exhibit less genetic variability relative to unpaired positions (Stephan 1996; Innan and Stephan 2001)—a pattern that has been reported for ncRNA structures in several different RNA viruses (Braun, Clements, and Gonda 1987; Garcia et al. 1996; Rodriguez-Alvarado and Roossinck 1997; Simmonds and Smith 1999). Transition/transversion ratios (κ) have also been reported to differ between paired and unpaired positions in ncRNAs—because κ > 1 for most nucleotide sequences, compensatory transitions occurring on the opposite paired strand in order to maintain Watson–Crick (WC) pairings are expected to further increase κ (Knies et al. 2008). Because of the increased thermostability of the WC guanosine (G)–cytosine (C) interaction, increased G + C content in structured regions of RNA has also been reported (Schultes, Hraber, and LaBean 1997; Piskol and Stephan 2008; Smit, Knight, and Heringa 2009).
We evaluated to what extent inferred evolutionary parameter values (using the structure-agnostic approach) from different partitioning models conformed to prior expectations. We expected (see Supplementary Fig. S1) each measure of RNA structural stability (with larger values indicating more RNA structural constraints) to be:
Negatively correlation with evolutionary rates.
Positively correlated with κ.
Positively correlated with GC content.
Additionally, a common approximation for the effects of protein level selection, made to reduce the computational complexity inherent in fitting more realistic codon-substitution models, is to assume that third codon positions are selectively neutral (because a large proportion of them are synonymous), and that first and second codon positions is where natural selection promoting (or suppressing) non-synonymous substitutions makes its mark (Kimura 1983). While this model is clearly insufficiently realistic, it does provide an expectation that evolutionary rates will be suppressed in first and second codon positions relative to the third codon position (as on an average selection is expected to be purifying), with similar expectations described above distinguishing paired and unpaired codon positions (Supplementary Fig. S1).
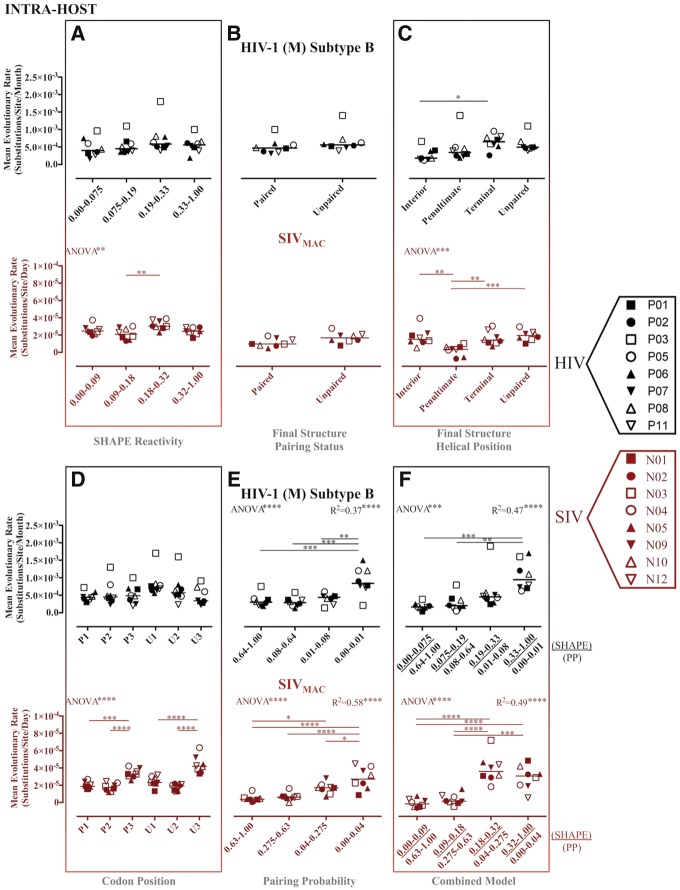
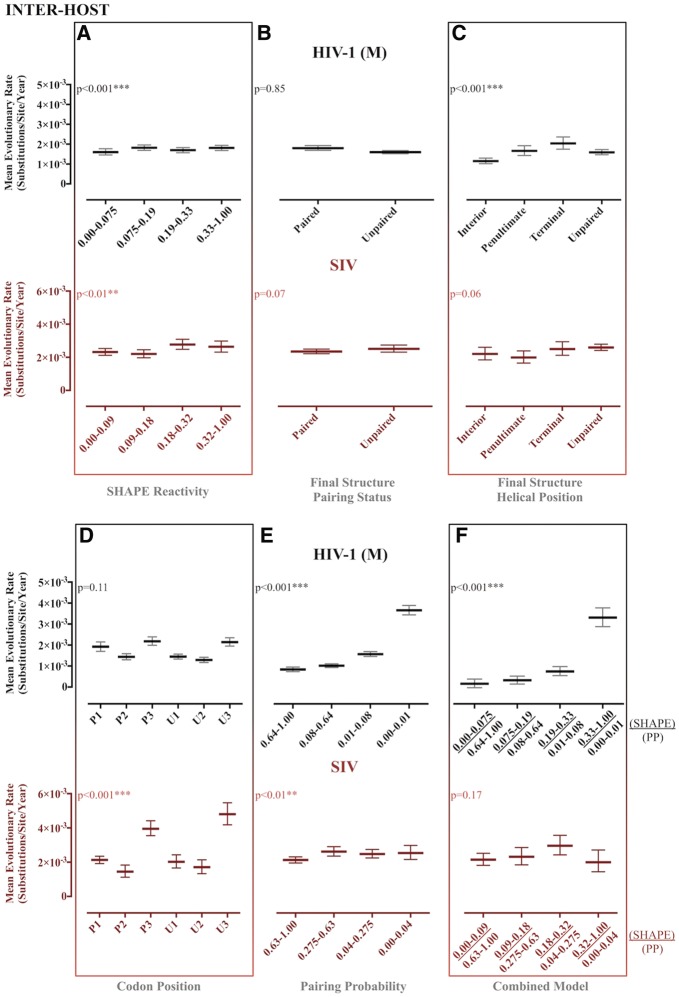
Although we observed some significant differences between partitioned site categories using the original structure-based partitions (i.e., SHAPE and PPROB alone), expected trends were not reliably recapitulated (Figs 1–6 and Table 2). Evolutionary patterns were often inconsistent across both HIV and SIV inter- and intra-host data sets and were not suggestive of the presence of RNA structure when compared with results of the combined partitioning approach (Figs 1 and 2). Despite significant differences in evolutionary rate between partitioned categories, expected patterns were only observed for the final structure helical position partition scheme (Mimouni et al. 2009) in the SIV intra-host data set (Fig. 2C), the PPROB partition scheme in all data sets except for SIV reference (Figs 1E and 2E), and the combined approach scheme for all data sets except for SIV reference (Figs 1F and 2F). However, significant differences in variance across the PPROB and combined data partition categories imply that more sophisticated methods of partitioning sites into categories, or including structural constraint as a continuous covariate, may improve the relationship between observed and expected patterns in evolutionary parameter estimates (data not shown).
Figure 1.
Maximum likelihood estimates of evolutionary rates for partitioned intra-host HIV-1 (subtype B) and SIVMAC gp120 RNA sequences. Maximum likelihood estimation of evolutionary rates in substitutions/site/time (months for HIV, days for SIV) from each of eight HIV-1-infected patients or SIV-infected macaques was performed in HYPHY (Pond et al. 2005) for internal branches of a fixed phylogeny. Individual subject-specific maximum clade credibility trees used as the fixed tree (topology and branch lengths scaled in time) were obtained from the posterior distribution of a Bayesian analysis of the same data sets. Rates were estimated for individual site partitions according to SHAPE reactivity (A), final structure pairing status (B), helical position within the final structure (C) codon positions (1, 2, and 3) within paired (P) and unpaired (U) regions according to the final structure (D), pairing probabilities (Pprob) (E), and the nucleotides identified as concordant between SHAPE reactivity and Pprob (F). Error bars represent ±1 SD. *P value < 0.05, **P value < 0.01, ***P value < 0.001 using one-way ANOVA with post-test multiple comparisons (all) and linear trend (A, E, and F). SHAPE reactivity values were obtained from Pollom et al. (2013), whereas PPROB were obtained from Watts et al. (HIV) (2009) and Pollom et al. (SIV) (2013). The final structure referred to in the graph was derived by Pollom et al. (2013) using SHAPE reactivity constraints in RNAstructure (Mathews 2014) thermodynamic folding.
Table 2.
Recovery of expected evolutionary trends under various RNA-structure informed partitioned site models.
| Model | Evolutionary rate | Ts/Tv ratio | G + C content |
|---|---|---|---|
| SHAPE reactivity | SIVw, SIVb, HIVb | SIVw, HIVb, HIVw | HIVw, HIVb, SIVw, SIVb |
| Final structure pairing status | HIVb, | HIVw, HIVb, SIVw. SIVb | |
| Final structure helical position | SIVw, HIVw SIVb, HIVb | SIVw, HIVb | HIVw, HIVb, SIVw. SIVb |
| Codon position within predicted structure | SIVw, SIVb | SIVw, SIVb, HIVb | HIVw, HIVb, SIVw. SIVb |
| Pairing probability | HIVw, HIVb, SIVw, SIVb | SIVb, HIVb | HIVwa, HIVb, SIVwa. SIVb |
| Combined model | HIVw, HIVb, SIVw, SIVb | HIVw, HIVb, SIVw | HIVw, HIVb, SIVw. SIVb |
SIVw, within-host SIV data; HIVw, within-host HIV data; SIVb, between-host SIV data; HIVb, between-host HIV data.
Entries in plain text indicate that evolutionary measures were influenced by partitioning (significant statistical test, P < 0.05, see text for the description of tests). For entries in bold, the expected trend (Supplementary Fig. S1) was also recovered.
Inverse trend compared with expectation.
Figure 2.
Maximum likelihood estimates of evolutionary rates for partitioned reference HIV-1 group M and SIV gp120 RNA reference. Maximum likelihood estimation of evolutionary rates in substitutions/site/year for HIV-1 group M and SIV reference sequences (LANL HIV Sequence Database) was performed in HYPHY (Pond et al. 2005) for internal branches of a fixed phylogeny. Reference-specific maximum clade credibility trees used as the fixed tree (topology and branch lengths scaled in time) were obtained from the posterior distribution of a Bayesian analysis of the same data sets. Rates were estimated for individual site partitions according to SHAPE reactivity (A), final structure pairing status (B), helical position within the final structure (C) codon positions (1, 2, and 3) within paired (P) and unpaired (U) regions according to the final structure (D), pairing probabilities (Pprob) (E), and the nucleotides identified as concordant between SHAPE reactivity and Pprob (F). Error bars represent Wald 95% confidence intervals. *P value < 0.05, **P value < 0.01, ***P value < 0.001 using profile likelihood analysis. SHAPE reactivity values were obtained from Pollom et al. (2013), whereas Pprob were obtained from Watts et al. (HIV) (2009) and Pollom et al. (SIV) (2013). The final structure referred to in the graph was derived by Pollom et al. (2013) using SHAPE reactivity constraints in RNAstructure (Mathews 2014) thermodynamic folding.
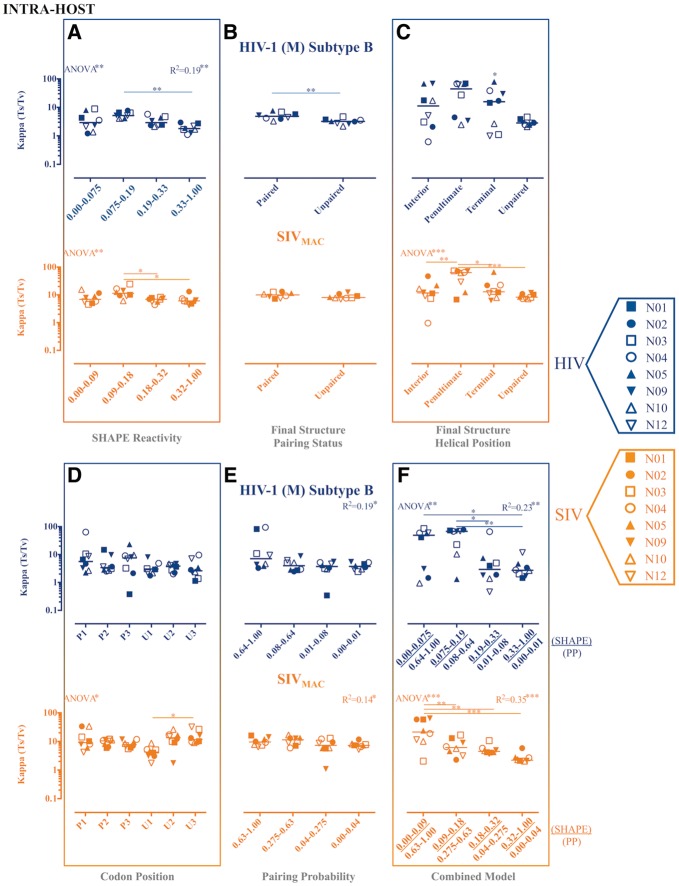
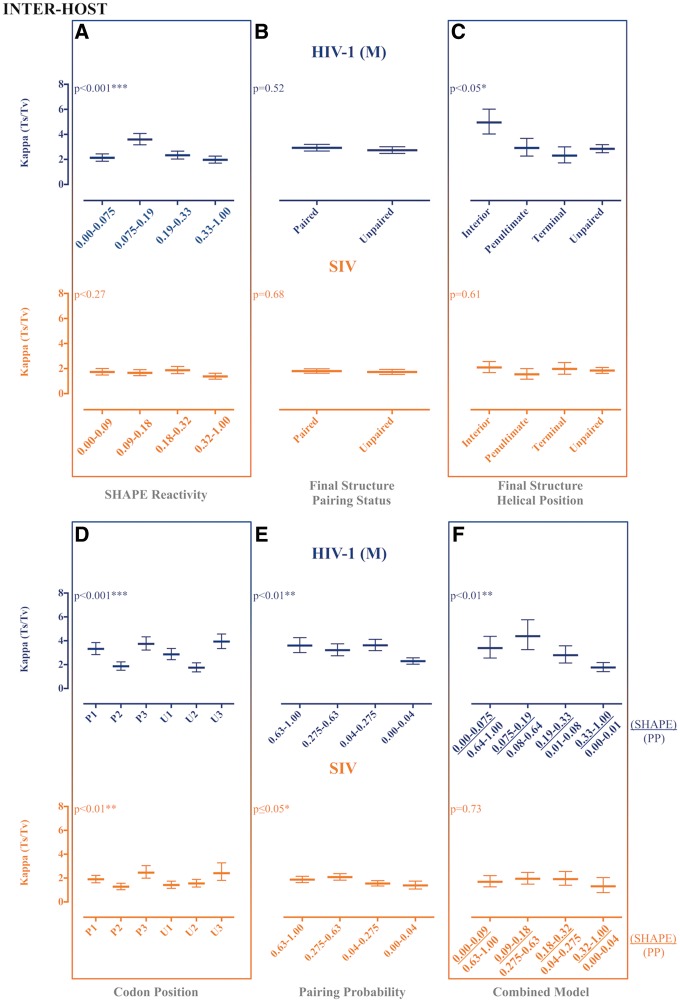
As with evolutionary rates, significant differences in Ts/Tv were inferred across data categories using the various partitioning schemes (Figs 3 and 4), although expected patterns were only observed for the final structure helical position partition scheme in the SIV intra-host data set (Fig. 4C), the PPROB partition scheme in all intra-host data (Figs 3E and 4E), and the combined approach scheme for all intra-host data (Fig. 3F and 4F).
Figure 3.
Maximum likelihood estimates of transition transversion rate ratios (Ts/Tv) for partitioned intra-host HIV-1 (subtype B) and SIVMAC gp120 RNA sequences. Maximum likelihood estimation of the kappa parameter, or Ts/Tv, from each of eight HIV-1-infected patients or SIV-infected macaques was performed in HYPHY (Pond et al. 2005) for internal branches of a fixed phylogeny. Individual subject-specific maximum clade credibility trees used as the fixed tree (topology and branch lengths scaled in time) were obtained from the posterior distribution of a Bayesian analysis of the same data sets. Ts/Tv were estimated for individual site partitions according to SHAPE reactivity (A), final structure pairing status (B), helical position within the final structure (C) codon positions (1, 2, and 3) within paired (P) and unpaired (U) regions according to the final structure (D), pairing probabilities (Pprob) (E), and the nucleotides identified as concordant between SHAPE reactivity and Pprob (F). Error bars represent ±1 SD. *P value < 0.05, **P value < 0.01, ***P value < 0.001 using one-way ANOVA with post-test multiple comparisons (all) and linear trend (A, E, and F). SHAPE reactivity values were obtained from Pollom et al. (2013), whereas Pprob were obtained from Watts et al. (HIV) (2009) and Pollom et al. (SIV) (2013). The final structure referred to in the graph was derived by Pollom et al. (2013) using SHAPE reactivity constraints in RNAstructure (Mathews 2014) thermodynamic folding.
Figure 4.
Maximum likelihood estimates of transition transversion rate ratios (Ts/Tv) for partitioned reference HIV-1 group M and SIV gp120 RNA reference sequences. Maximum likelihood estimation of the kappa parameter, or Ts/Tv, for HIV-1 group M and SIV reference sequences (LANL HIV Sequence Database) was performed in HYPHY (Pond et al. 2005) for internal branches of a fixed phylogeny. Reference-specific maximum clade credibility trees used as the fixed tree (topology and branch lengths scaled in time) were obtained from the posterior distribution of a Bayesian analysis of the same data sets. Ts/Tv were estimated for individual site partitions according to SHAPE reactivity (A), final structure pairing status (B), helical position within the final structure (C) codon positions (1, 2, and 3) within paired (P) and unpaired (U) regions according to the final structure (D), pairing probabilities (Pprob) (E), and the nucleotides identified as concordant between SHAPE reactivity and Pprob (F). Error bars represent Wald 95% confidence intervals. *P value < 0.05, **P value < 0.01, ***P value < 0.001 using profile likelihood analysis. SHAPE reactivity values were obtained from Pollom et al. (2013), whereas PPROB were obtained from Watts et al. (HIV) (2009) and Pollom et al. (SIV) (2013). The final structure referred to in the graph was derived by Pollom et al. (2013) using SHAPE reactivity constraints in RNAstructure (Mathews 2014) thermodynamic folding.
Significant differences in G + C content among partition categories, and between G + C and A + U content within the same category, were prevalent due to small variation within and among sequence alignments, so only the absence of significance (denoted as ‘ns’) has been highlighted and deviation from expected patterns described herein (Supplementary Figs S2 and S3). In terms of statistical significance and expected patterns, G + C content results differed drastically from those of the evolutionary rate and Ts/Tv. Intra-host data exhibited expected patterns, which were significant, for the SHAPE, codon, and combined approach partitioning schemes for both HIV and SIV (Supplementary Fig. S2A, D, and F). Furthermore, significant differences were observed between final structure pairing status partition categories, and, although G + C and A + U contents were not significantly different among nucleotides categorized as paired, this result may be expected, given the elevated A content in lentiviruses. For both HIV and SIV, deviation from the expected pattern was characterized by significantly greater A + U than G + C content in the more stable nucleotide categories. HIV inter-host reference data similarly exhibited the expected trend for the SHAPE partition scheme (Supplementary Fig. S3A), though not for the codon or combined partition schemes, as a similar elevated A + U content, relative to G + C, was observed for the more stable nucleotide categories in both schemes (Supplementary Fig. S3D and F). For SIV reference sequences, however, none of the partition schemes appeared to exhibit the expected pattern (Supplementary Fig. S3). A similar GC and AU composition in more stable regions may be explained by a unique pressure within gp120 to maintain more flexible helices, which would not be a phenomenon unique to HIV (Xia and Holcik 2009), although codon bias cannot be ruled out (Van Hemert 1995; Jenkins and Holmes 2003). This structural relaxation may, for example, allow for more rapid transitions between multiple functional structures.
2.3 Regression analysis of combined SHAPE and pairing probability information allows for identification of core conserved RNA structures
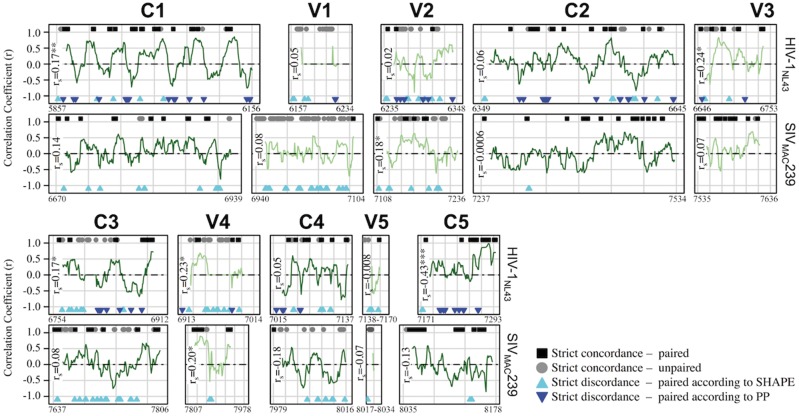
The fraction of nucleotides identified in this study as concordant between SHAPE and pairing probability (PPROB) varied widely along the length of the HIV-1NL4-3 and SIVMAC239 genomes used in Watts et al. (2009) and Pollom et al. (2013). Despite similar observations by Watts et al. and Pollom et al., evidence has been presented using SHAPE, as well as PPROB, in favor of highly unstructured RNA within the variable loop (Vx) regions of gp120 (Watts et al. 2009), particularly V1 and V2 (Sukosd 2015). We, therefore, chose the region of gp120 to further assess this hypothesis and compare the two approaches within the individual Vx (V1-V5) and Cx (C1-V5) domains using correlation analysis (Fig. 5). Using the Spearman correlation analysis, we found that the methods achieved considerable agreement (rs>0.39) only in the C5 region and only for HIV-1NL4-3. However, consistent with the findings of Suskosd et al. (Sukosd 2015), a relatively large fraction (26% for HIV and 35% for SIV) of sites within V1 were in strict concordance, in the direction of structural instability (high SHAPE reactivity and low PPROB). Furthermore, a substantial fraction (5% for HIV and 17% for SIV) of sites within this region were also characterized by low SHAPE reactivity but also low PPROB, suggesting that previous estimates using SHAPE reactivity alone may have underestimated the extent of flexibility within V1 when considering the virus as a population shaped by sequence heterogeneity. However, it is also important to keep in mind the possibility of the presence of specific structures identified by SHAPE due to other factors, such as physiological conditions, as discussed elsewhere.
Figure 5.
Correlation analysis of SHAPE reactivity and pairing probability for HIV-1NL4-3 and SIVMAC239 gp120 reference sequences. Normalized (0–1) SHAPE reactivity values and RNA-Decoder-derived pairing probabilities (PPROB) for individual nucleotides throughout gp120 (A) were obtained from Pollom et al. (2013) and Watts et al. (2009). A running correlation analysis (fifteen nucleotide window) was used to assess the level of agreement between the two methods and to identify conserved RNA structures within constant (‘C’, light green lines) and variable (‘V’, dark green lines) regions. In addition to plotted correlation coefficients (r), individual sites identified as exhibiting strict concordance or discordance between the two methods were indicated along the x-axis at y + 1 and y−1, respectively. Strict paired concordance (black square) was defined as ≤25th percentile of the total gp120 normalized SHAPE reactivity distribution (HIV = 0.013, SIV = 0.026) and ≥25th percentile of the total gp120 normalized PPROB distribution (HIV = 0.73, SIV = 0.65). Strict unpaired concordance (grey circle) was defined as ≥75th percentile of the total gp120 normalized SHAPE reactivity distribution (HIV = 0.059, SIV = 0.084) and ≤25th percentile of the total gp120 Pprob normalized distribution (HIV = 0.02, SIV = 0.05). Strict discordance refers to the combination of these thresholds, with each of the two colored triangles representing base pairing according to one method and flexibility according to the other. Spearman correlation coefficient (rs) was estimated for all values within each gp120 region. *P value ≤ 0.01, **P value ≤ 0.001, ***P value ≤ 0.0001.
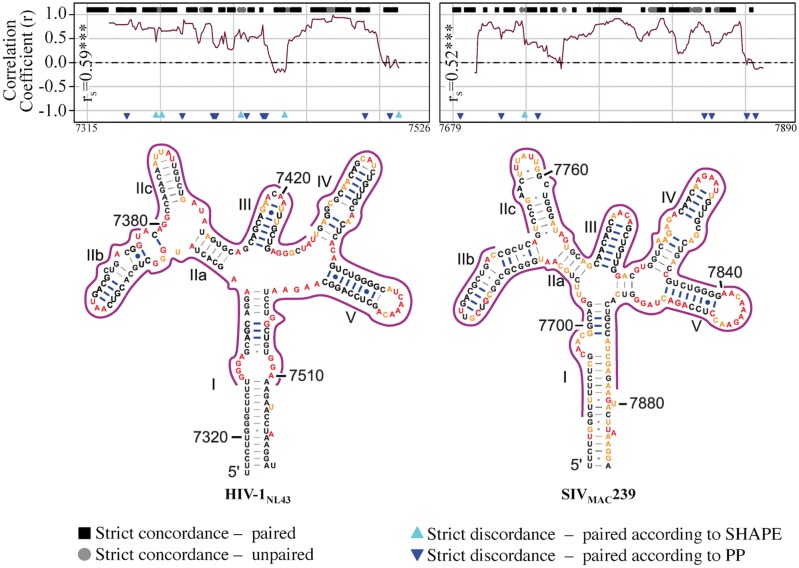
The consistent appearance of discordant sites, primarily characterized by both low SHAPE reactivity and low PPROB, would also seem to suggest a high rate of false positives, in the context of viral genetic heterogeneity, attributed by the SHAPE method. Evidence in support of this supposition is presented herein following assessment of the level of agreement between the two methods for the well-characterized 5SL structure of RRE (Chen, Le, and Maizel 2000). Significant agreement and a high degree of coverage of concordance between both methods were observed for HIV-1NL4-3 and SIVMAC239 sequences, despite slightly different overall RRE 5SL structure compositions between the two viral strains (Fig. 6). Exceptions were small stretches (two to five nucleotides) within stem-loops IV and IIB of HIV-1NL4-3 SIVmac239, respectively. Although the results of this combined approach cannot be used to differentiate the relative stabilities of conformationally flexible structures, such as the functionally relevant 5SL and 4SL RREs (Sherpa et al. 2015), the parallel assessment of both methods using correlation analysis was able to accurately identify a multistructural, multifunctional RNA element as exhibiting characteristics of a stable and conserved local RNA structure. Expanding upon the predictive power of this approach, despite little domain-wide agreement within the large nucleotide stretch of C1, each 15-bp window corresponding to a peak in correlation coefficient (up to 0.5<rs<0.8) was attributed to nucleotide pairing according to both methods. Similar local patterns could be found for the 3′ end of C3 leading into the 5′ region of V4, as well as the 3′ end of C5, suggesting the presence of conserved substructures, masked by the global analysis of their corresponding Vx or Cx region.
Figure 6.
Correlation analysis of SHAPE reactivity and pairing probability for HIV-1NL4-3 and SIVMAC239 rev-response element (RRE) reference sequences. Normalized (0–1) SHAPE reactivity values and RNA-Decoder-derived pairing probabilities (PPROB) for individual nucleotides throughout the rev-response element (RRE) were obtained from Pollom et al. (2013) and Watts et al. (2009). A running correlation analysis (fifteen nucleotide window) was used to assess the level of agreement between the two methods and to validate the use of this combined approach. In addition to plotted correlation coefficients (r), individual sites identified as exhibiting strict concordance or discordance between the two methods were indicated along the x-axis at y + 1 and y−1, respectively. Strict paired concordance (black squares) was defined as ≤25th percentile of the total RRE normalized SHAPE reactivity distribution (HIV = 0.01, SIV = 0.02) and ≥25th percentile of the total RRE normalized PPROB distribution (HIV = 0.75, SIV = 0.66). Strict unpaired concordance (grey circles) was defined as ≥75th percentile of the total RRE normalized SHAPE distribution (HIV = 0.05, SIV = 0.096) and ≤25th percentile of the total RRE normalized PPROB distribution (HIV = 0.07, SIV = 0.04). Strict discordance refers to the combination of these thresholds, with each of the two colored triangles representing base pairing according to one method and flexibility according to the other. Sites corresponding to r > 0.30 were outlined in green in the 2D RRE structure obtained from Pollom et al. (2013). Spearman correlation coefficient (rs) was estimated for all HIV and SIV values. ***P value ≤ 0.0001.
3. Discussion
Although recent studies have predicted the existence of highly structured RNA within large coding regions of the HIV-1NL4-3 and SIVMAC239 RNA genomes (Watts et al. 2009; Pollom et al. 2013; Lavender, Gorelick, and Weeks 2015), the influence of RNA sequence and structure heterogeneity, as well as protein selective constraints, on preservation of global, or even local, structures within these regions not been thoroughly investigated using evolutionary metrics. In this study, we utilized previously published chemical probing data in the form of SHAPE reactivity and an extensive collection of HIV-1 and SIV gp120 sequences in order to investigate appositeness of a global gp120 RNA structure and the utility of a combined approach of SHAPE and phylogenetic methods in identifying core conserved protein-coding RNA (cRNA) structures.
Studies seeking to understand the interplay between selective forces operating upon RNA structure have postulated that one optimal or ‘average’ RNA structure is an accurate reflection of the underlying molecular mechanics. This approach may be entirely reasonable for organisms that have slow rates of evolution and low intra-host genetic heterogeneity. However, fast evolving viruses such as HIV-1 exist as a genetically heterogeneous population, each potentially with its own distinct structural variant, and that population itself changes over time within the host population as well as the host (Drummond, Pybus, and Rambaut 2003). Therefore, is it hardly surprising that the chemical method, SHAPE (Merino et al. 2005), which examines single molecular substrates, and a phylogenetic tool (RNA-Decoder) (Pedersen et al. 2004b), which relies on extracting conservation patterns from multiple sequence alignments, often disagree in their predictions. Because each method takes into account differing aspects of RNA structure formation and preservation, we viewed them as complementary sources of classification: if the methods agree at a particular site, there is stronger evidence that the site is paired or unpaired. Indeed, when we classified the sites into RNA structure groups using information from SHAPE and RNA-Decoder jointly, the induced partitioning was in good agreement with the published structure of the 5SL RRE—a functionally essential component of viral nuclear export (Sukosd 2015). Our studies do not attribute functions to either of these discrete structures, but that fact that both substructures are evolutionarily conserved across divergent strains of HIV-1 implies that the conformational heterogeneity is biologically important. Assuming that the combined methods approach is generalizable to other regions of gp120, we identified several promising candidates for future investigation of physiological relevance (i.e., mutagenesis studies), such as the C5 region of HIV-1, where the predictions of SHAPE and RNA-Decoder are in strong agreement. Although our results are concordant with the hypothesis of Pollom et al. (2013) that global HIV-1 and SIV RNA secondary structure features are unlikely to be as conserved as the RRE, core structures can provide information as to the general function of more complex structures within the gene and can be overlooked when using analyses that target global structures.
If core RNA structure(s) modulate sequence evolution, then one can expect to see detectable differences in evolutionary model parameters between sites binned according to different levels of RNA conservation. We found that evolutionary model parameter variation across categories of sites designated according to concordance between these two methods ranked highest in terms of the goodness of fit for both the within-host and highly divergent inter-host HIV and SIV data when compared with a variety of other structurally informed and uninformed models. This finding implies that RNA conservation imposes evolutionary constraints on HIV and SIV gp120. The degree of RNA conservation within this genetic region has been heavily debated; however, given the temporally, spatially, and contextually varied selective forces operating on HIV-1 gp120, it would be naïve to expect that any global RNA structure would provide a simple and consistent explanation for inferred evolutionary rates. Therefore, we were encouraged by the finding that the combined approach-based partitioning of sequence data, rather than SHAPE partitioning alone, more closely resembled the expected patterns in evolutionary parameters indicative of RNA function, although less evident in the more divergent data sets. One explanation for this finding is that the structure proposed by Pollom et al. (2013) might contain inaccuracies induced by non-physiological conditions employed during chemical probing. In this regard, the dependence of RNA secondary structures on small changes in preparatory conditions is well known (Casiano-Negroni, Sun, and Al-Hashimi 2007; Lee et al. 2013). Another possible explanation is that the SHAPE-predicted structure was inaccurate due to low, undirected (no SHAPE data) minimum free energy accuracy, which has been shown to be strongly correlated for ribosomal RNA (rRNA) (Sukosd et al. 2013). It is also possible that some disagreements between biochemical and phylogenetic approaches reflect real functional heterogeneity in RNA structure (Zhu et al. 2013; Sukosd 2015). RNA segments could exist simultaneously as an equilibrium mixture of structural conformers, one or several of which may include, for example, a larger proportion of single-stranded regions, resulting in lower confidence levels for SHAPE in pairing, depending on the predominant structure for preparation conditions. In any of the above scenarios, reliable detection of smaller conserved core structural elements, as was shown for RRE in this study can aid in more efficient scanning of genomic material for putative structural regions that are shared by multiple structures. These core structures may be easier to conserve during adaptation to immune or other selective pressure, and more functionally constrained. In this scenario, no global or ‘average’ structure need be evolutionarily conserved (Lu, Heng, and Summers 2011b; Deforges, Chamond, and Sargueil 2012; Kenyon et al. 2013; Zhu et al. 2013; Keane et al. 2015; Sherpa et al. 2015).
As new viral species are continually being discovered and sequenced, considering viral sequence and structure heterogeneity is imperative in the study of de novo RNA structure determination and analysis of evolutionary constraints imposed by these structures. We have demonstrated that frequently used chemical probing and phylogenetic methods alone cannot capture this heterogeneity that is characteristic of rapidly evolving RNA viruses and are misleading during preliminary investigations of conserved RNA structure. Although more sophisticated models are needed to understand the evolution of complex cRNA structures among more divergent lineages, chemical evaluation of a single reference sequence combined with evolutionary inference from a heterogeneous sequence population together provide a reliable and feasible approach to identifying core RNA structural segments within large genomic regions.
4. Materials and methods
4.1 Sequence data
One hundred and fifty-three HIV-1 group M partial gp120 sequences (HXB2 coordinates 7,023–7,592), representing fifty-six subtypes and common circulating recombinant forms (CRFs), were obtained from the Los Alamos National Laboratory (LANL) HIV Sequence Database (https://www.hiv.lanl.gov/). Forty SIV partial gp120 sequences (SIVMAC239 coordinates 6,706–8,049), representing seven strains from chimpanzee (SIVcpz) and rhesus macaque (SIVMAC) hosts, were similarly obtained from the LANL HIV Sequence Database.
HIV-1 intra-host gp120 sequences were reported previously (Shankarappa et al. 1999) as isolated from longitudinal peripheral blood mononuclear cell (PBMC) samples of eight HIV-1 (subtype B)-infected patients from the MACS cohort collected over the course of infection prior to or during anti-retroviral treatment. SIV intra-host gp120 sequences used in this study were obtained from several different tissues within eight of twelve treatment-naïve rhesus macaques intravenously inoculated with the SIVMAC251 viral swarm, as described previously (Rife et al. 2016). A detailed description of the single genome sequencing approach and alignment methods used to obtain intra-host SIV gp120 sequence alignments can be found in Rife et al. (2016). GenBank accession numbers for all HIV and SIV sequence data used herein are reported in the Supplementary Methods. Final sequences were obtained following quality control screening described in Supplementary Methods, and final alignments are available at https://github.com/rifebd88/HIV-SIV-RNA-structure-manuscript.git.
4.2 Data partitioning
Sequence data for each subject were partitioned according to site-specific values corresponding to SHAPE reactivity (Merino et al. 2005) and RNA-Decoder (Pedersen et al. 2004a,b)-derived pairing probability categories, as well as pairing status within the final structure that was obtained and determined by Pollom et al. (2013) and Watts et al. (2009). SHAPE reactivities and pairing probabilities were normalized (0–1) based on minimum and maximum values, log2 transformed, and divided into quartiles. Final pairing status (paired vs. unpaired) was derived from the final lowest free energy-predicted Pollom et al. (2013) HIV and SIV structures, originally obtained by incorporating SHAPE reactivities as pseudo-free energy restraints in conjunction with nearest neighbor parameters in RNAstructure (Mathews 2014) (available from http://rna.urmc.rochester.edu/RNAstructure.html). Sites within the Pollom et al. (2013) structure corresponding to different intra-helical stacking positions, as previously determined by Mimouni et al. (2009) to experience varying degrees of selective pressure, were also analyzed with respect to the previously described evolutionary parameters and included terminal, penultimate, and internal helical base pairs.
4.3 Redefinition of structural partitions
In addition to categorization of nucleotide sites based on SHAPE reactivities and pairing probabilities individually, sites were assigned to the four following concordant categories corresponding to (top to bottom) progressively increasing SHAPE flexibility and corresponding decreasing probability of intra-molecular base pairing based on agreement between the two approaches and a similar normalization and transformation process as described earlier:
Most structurally stable: low (near 0) SHAPE reactivity and high (near one) pairing probability
Medium–high structural stability: medium–low SHAPE reactivity and medium–high pairing probability
Low–medium structural stability: medium–high SHAPE reactivity and medium–low pairing probability, and
Least structurally stable: high (near 1) SHAPE reactivity and low (near 0) pairing probability
Remaining nucleotide sites were considered discordant and discarded for corresponding analyses. Evolutionary parameters were estimated for these categories in order to investigate the shared impact of both chemical and evolutionary data on the resulting criteria indicative of RNA secondary structure.
4.4 Phylogenetic tree reconstruction
Phylogenetic resolution and informativeness of individual patient-specific sequence alignments, as well as individual alignment partitions, was assessed using likelihood mapping (Strimmer and von Haeseler 1997) in IQ-TREE (Nguyen et al. 2015) (available from http://www.iqtree.org/). Greater than 30% allocation of quartets (groups of four randomly sampled sequences) to fully resolved sub-trees using likelihood mapping indicated sufficient resolution for reliable interpretation of phylogenetic tree topology (Supplementary Table S2).
Maximum likelihood tree reconstruction was performed for all HIV-1 patient sequences in RaxML v8.0.25 (Stamatakis 2014) using the general time-reversible (GTR) model of nucleotide substitution (Tavare 1986) and gamma distribution (Γ) of rate variation across sites in order to evaluate potential inter-patient cross-contamination prior to further phylogenetic analysis (Supplementary Fig. S4A). Rapid bootstrap (Stamatakis, Hoover, and Rougemont 2008) values >0.95 (obtained with 1,000 replicates) at each patient-specific node indicated the absence of cross-contamination. A ML tree was also reconstructed for all SIV intra-host sequences, but animal-specific clustering of sequences was not expected, as all animals were infected with the same viral swarm (Supplementary Fig. S4B).
Because the sequence data were heterochronous, ML tree reconstruction was also performed for individual subject and reference sequence alignments as above for temporal resolution evaluation based on information provided by tip dates (Supplementary Fig. S5). Linear regression analysis of the relationship of root-to-tip genetic distance within the ML tree and sampling time for each sequence indicated a significantly positive slope (data available upon request) and was interpreted as sufficient temporal resolution for molecular clock rooting of the phylogeny within the Bayesian framework (described below). Following maximum likelihood analysis, a Bayesian coalescent analysis was performed in BEAST v1.8.2 (Drummond et al. 2012) (available from http://beast.bio.ed.ac.uk) in order to account for varying sampling times in branch length and evolutionary rate estimation. The posterior distribution of sampled trees and related parameters was summarized using maximum clade credibility (MCC) trees. Additional information regarding ML and Bayesian tree reconstruction and linear regression analysis can be found in Supplementary Methods.
As expected, patient-specific phylogenetic resolution was sufficient for ML and Bayesian tree reconstruction, whereas relatively low resolution was observed for data partition categories owing to the small number of nucleotides assigned to each partition (Supplementary Table S3). Therefore, evolutionary parameters for individual partitions were inferred using a fixed tree topology (MCC tree) and branch lengths obtained from the Bayesian evolutionary analysis incorporating all sites. Although the presence of CRFs in the HIV subtype reference alignment violates the assumption of a single phylogeny (MCC tree), 90% of sequences in the alignment consisted of >95% of the partial gp120 assignable to a single major subtype, based on information obtained from the LANL HIV Sequence Database (https://www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html) (Carr et al. 2001; Koulinska et al. 2001; Delgado et al. 2002; Montavon et al. 2002; Wilbe et al. 2002; Tovanabutra et al. 2003; Casado et al. 2005; Thomson et al. 2005; Powell et al. 2007a,b; Guimarães et al. 2008; Yamaguchi et al. 2008; Niama et al. 2009; Fernández-García et al. 2010), suggesting a negligible impact on parameter estimation.
4.5 Bayesian inference of best-fitting partition model and evolutionary parameters
A subset (200) of the sampled trees from each subject-specific Bayesian tree reconstruction incorporating all sites was used as an empirical tree distribution for additional Bayesian analysis of fit of the previously described partition models. Two additional models, incorporating a Bayesian estimate of a gamma rate distribution partition (four categories), representing a structurally uninformed model, and a randomly generated gamma distribution partition model, were also used for comparison as controls. We refer to the model without structure-based partitions as the ‘uninformed’ model; however, it is important to note that several assumptions were still made (e.g., similar models of nucleotide substitution across sites). For the randomly generated partition model, nucleotide positions were assigned random values according to a gamma distribution (rgamma function in R stats package; https://stat.ethz.ch/R-manual/R-devel/library/stats/html/00Index.html). Values were binned according to quartiles, similar to the other partition models. Each partition category was allowed individual nucleotide substitution models and parameters (e.g., kappa) and evolutionary rates, according to a relaxed molecular clock. Evolutionary rates among sites within a partition category were not allowed to vary.
4.6 Maximum likelihood parameter estimation along internal branches of the MCC trees
Maximum likelihood estimates of evolutionary rates and transition–transversion rate ratios (kappa, or Ts/Tv) for data partitions were estimated using the HKY85 model along internal branches of the MCC tree in order to exclude potential transient substitutions closer to sampling time. Nucleotide frequencies were assumed to behave similarly along all branches, and, therefore, empirical frequencies from the original alignment were used to assess G + C content among individual partitions. Inferences of evolutionary rate and Ts/Tv were derived using a purpose-written HYPHY (Pond, Frost, and Muse 2005) (http://www.hyphy.org/) script (available from https://github.com/spond/pubs/tree/master/HIV-RNA). For each partition, a nucleotide substitution model was fitted using the following assumptions: 1) Strict molecular clock with separate rates on terminal and internal branches (chronological time was read from the input MCC tree), 2) Terminal and internal branches were assigned separate transition/transversion ratio parameters, 3) Nucleotide frequencies were estimated by counts from each partition, with no difference between internal and terminal branches, 4) Confidence intervals (CIs) were estimated by profile likelihood, as intervals do not assume normality and perform better for small sample sizes than Wald CIs (Cole, Chu, and Greenland 2014).
These evolutionary parameters were also analyzed for final pairing status assignments to individual codon positions in order to evaluate evolutionary patterns between primarily synonymous (third codon position) and non-synonymous (first and second codon positions) sites, thereby evaluating the relationship of protein and RNA structure evolution.
4.7 Relationship of SHAPE reactivity and pairing probability
SHAPE reactivity values and pairing probabilities (Pedersen et al. 2004a,b) for individual nucleotides throughout HIV-1NL4-3 and SIVMAC239 gp120 were obtained from Pollom et al. (2013) and Watts et al. (2009), respectively. Values were normalized (0–1), and SHAPE values were subtracted from 1 for visualization of the representation of agreement between both methods as positive correlation. A running correlation analysis (15-nucleotide window) was used to assess quantitatively the level of agreement between the two methods and to identify potentially conserved RNA structures within the constant (Cx; C1–C5) and variable (Vx; V1–V5) regions. A similar procedure was utilized for the well-characterized Rev-Response Element (RRE) 5 stem loop (5SL) structural conformations (Chen, Le, and Maizel 2000; Sherpa et al. 2015) in order to validate the use of this combined approach. Individual sites corresponding to high levels of concordance or discordance, using normalized data quartile cutoffs, between the two methods were also identified in order to evaluate contributions to regions of low-level (rs<0.39) correlation.
4.8 Statistical analysis
A detailed description of the statistical analyses used in this study can be found in Supplementary Methods.
Supplementary data
Supplementary data are available at Virus Evolution online.
Supplementary Material
Acknowledgements
We are thankful to Sara Palmer and Melissa M. Norstrom for their suggestions regarding single genome sequencing experiments and to David J. Nolan for carrying out these experiments. We would also like to thank Carla Mavian for help with the creation of figures and Gary Fogel for insightful discussions. Lastly, we would like to acknowledge the organizers and attendees of the 20th International Bioinformatics Workshop on Virus Evolution and Molecular Epidemiology, with whom we had the opportunity to discuss the findings herein.
Funding
This work was supported in part by the National Institutes of Health (P50 GM103297 and R01 NS063897 to M.S.; R01 GM093939 to S.K.P.; F31 MH109398 to B.R.M. and M.S.; and R01 GM42561 to M.F.S.).
Conflict of interest: None declared.
References
- Abbink T. E., Berkhout B. (2003) ‘A Novel Long Distance Base-Pairing Interaction in Human Immunodeficiency Virus Type 1 RNA Occludes the Gag Start Codon’, The Journal of Biological Chemistry, 278: 11601–11. [DOI] [PubMed] [Google Scholar]
- Baudin F. (1993) ‘Functional Sites in the 5′ Region of Human Immunodeficiency Virus Type 1 RNA Form Defined Structural Domains’, Journal of Molecular Biology, 229: 382–97. [DOI] [PubMed] [Google Scholar]
- Berkhout B. (2000) ‘Multiple Biological Roles Associated with the Repeat (R) Region of the HIV-1 RNA Genome’, Advances in Pharmacology (San Diego, Calif.), 48: 29–73. [DOI] [PubMed] [Google Scholar]
- Braun M. J., Clements J. E., Gonda M. A. (1987) ‘The Visna Virus Genome: Evidence for a Hypervariable Site in the Env Gene and Sequence Homology among Lentivirus Envelope Proteins’, Journal of Virology, 61: 4046–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr J. K. et al. (2001) ‘The AG Recombinant IbNG and Novel Strains of Group M HIV-1 Are Common in Cameroon’, Virology, 286: 168–81. [DOI] [PubMed] [Google Scholar]
- Casado G. et al. (2005) ‘Identification of a Novel HIV-1 Circulating ADG Intersubtype Recombinant Form (CRF19_Cpx) in Cuba’, Journal of Acquired Immune Deficiency Syndromes (1999), 40: 532–7. [DOI] [PubMed] [Google Scholar]
- Casiano-Negroni A., Sun X., Al-Hashimi H. M. (2007) ‘Probing Na(+)-Induced Changes in the HIV-1 TAR Conformational Dynamics Using NMR Residual Dipolar Couplings: New Insights into the Role of Counterions and Electrostatic Interactions in Adaptive Recognition’, Biochemistry, 46: 6525–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. H., Le S. Y., Maizel J. V. (2000) ‘Prediction of Common Secondary Structures of RNAs: A Genetic Algorithm Approach’, Nucleic Acids Research, 28: 991–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clever J., Sassetti C., Parslow T. G. (1995) ‘RNA Secondary Structure and Binding Sites for Gag Gene Products in the 5′ Packaging Signal of Human Immunodeficiency Virus Type 1’, Journal of Virology, 69: 2101–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clever J. L., Miranda D., Parslow T. G. (2002) ‘RNA Structure and Packaging Signals in the 5′ Leader Region of the Human Immunodeficiency Virus Type 1 Genome’, Journal of Virology, 76: 12381–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. R., Chu H., Greenland S. (2014) ‘Maximum Likelihood, Profile Likelihood, and Penalized Likelihood: A Primer’, American Journal of Epidemiology, 179: 252–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza V., Summers M. F. (2005) ‘How Retroviruses Select Their Genomes’, Nature Reviews. Microbiology, 3: 643–55. [DOI] [PubMed] [Google Scholar]
- Damgaard C. K. et al. (2004) ‘RNA Interactions in the 5′ Region of the HIV-1 Genome’, Journal of Molecular Biology, 336: 369–79. [DOI] [PubMed] [Google Scholar]
- Deforges J., Chamond N., Sargueil B. (2012) ‘Structural Investigation of HIV-1 Genomic RNA Dimerization Process Reveals a Role for the Major Splice-Site Donor Stem Loop’, Biochimie, 94: 1481–9. [DOI] [PubMed] [Google Scholar]
- Deigan K. E. et al. (2009) ‘Accurate SHAPE-Directed RNA Structure Determination’, Proceedings of the National Academy of Sciences of the United States of America, 106: 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado E. et al. (2002) ‘Identification of a Newly Characterized HIV-1 BG Intersubtype Circulating Recombinant Form in Galicia, Spain, Which Exhibits a Pseudotype-like Virion Structure’, Journal of Acquired Immune Deficiency Syndromes, 29: 536–43. [DOI] [PubMed] [Google Scholar]
- Drummond A., Pybus O. G., Rambaut A. (2003) ‘Inference of Viral Evolutionary Rates from Molecular Sequences’, Advances in Parasitology, 54: 331–58. [DOI] [PubMed] [Google Scholar]
- Drummond A. J. et al. (2005) ‘Bayesian Coalescent Inference of past Population Dynamics from Molecular Sequences’, Molecular Biology and Evolution, 22: 1185–92. [DOI] [PubMed] [Google Scholar]
- Drummond A. J. (2012) ‘Bayesian Phylogenetics with BEAUti and the BEAST 1.7’, Molecular Biology and Evolution, 29: 1969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felden B. (2007) ‘RNA Structure: Experimental Analysis’, Current Opinion in Microbiology, 10: 286–91. [DOI] [PubMed] [Google Scholar]
- Fernández-García A. et al. (2010) ‘Identification of a New HIV Type 1 Circulating BF Intersubtype Recombinant Form (CRF47_BF) in Spain’, AIDS Research and Human Retroviruses, 26: 827–32. [DOI] [PubMed] [Google Scholar]
- Garcia M. et al. (1996) ‘Heterogeneity in the Haemagglutinin Gene and Emergence of the Highly Pathogenic Phenotype among Recent H5N2 Avian Influenza Viruses from Mexico’, Journal of General Virology, 77: 1493–504. [DOI] [PubMed] [Google Scholar]
- Guimarães M. L. et al. (2008) ‘Identification of Two New CRF_BF in Rio De Janeiro State, Brazil’, Aids (London, England), 22: 433–5. [DOI] [PubMed] [Google Scholar]
- Harrison G. P., Lever A. M. (1992) ‘The Human Immunodeficiency Virus Type 1 Packaging Signal and Major Splice Donor Region Have a Conserved Stable Secondary Structure’, Journal of Virology, 66: 4144–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I. L. et al. (1998) ‘Automatic Detection of Conserved RNA Structure Elements in Complete RNA Virus Genomes’, Nucleic Acids Research, 26: 3825–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H., Stephan W. (2001) ‘Selection Intensity against Deleterious Mutations in RNA Secondary Structures and Rate of Compensatory Nucleotide Substitutions’, Genetics, 159: 389–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins G. M., Holmes E. C. (2003) ‘The Extent of Codon Usage Bias in Human RNA Viruses and Its Evolutionary Origin’, Virus Research, 92: 1–7. [DOI] [PubMed] [Google Scholar]
- Keane S. C., Summers M. F. (2016) ‘NMR Studies of the Structure and Function of the HIV-1 5′-Leader’, Viruses, 8: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane S. C. et al. (2015) ‘RNA Structure. Structure of the HIV-1 RNA Packaging Signal’, Science (New York, N.Y.), 348: 917–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon J. C. et al. (2013) ‘In-Gel Probing of Individual RNA Conformers within a Mixed Population Reveals a Dimerization Structural Switch in the HIV-1 Leader’, Nucleic Acids Research, 41: e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. (ed.). (1983) The Neutral Theory of Molecular Evolution New York: Cambridge University Press. [Google Scholar]
- Knies J. L. et al. (2008) ‘Compensatory Evolution in RNA Secondary Structures Increases Substitution Rate Variation among Sites’, Molecular Biology and Evolution, 25: 1778–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfel S. A., Berkhout B. (2011) ‘Phylogenetic Screen for Important RNA Structure Motifs in the HIV-1 Genome’, Frontiers of Retrovirology, 8: P40. [Google Scholar]
- Knoepfel S. A., Berkhout B. (2013) ‘On the Role of Four Small Hairpins in the HIV-1 RNA Genome’, RNA Biology, 10: 540–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond S. L. et al. (2007) ‘Evolutionary Model Selection with a Genetic Algorithm: A Case Study Using Stem RNA’, Molecular Biology and Evolution, 24: 159–70. [DOI] [PubMed] [Google Scholar]
- Koulinska I. N. et al. (2001) ‘A New Human Immunodeficiency Virus Type 1 Circulating Recombinant Form from Tanzania’, AIDS Research and Human Retroviruses, 17: 423–31. [DOI] [PubMed] [Google Scholar]
- Kuzembayeva M. et al. (2014) ‘Life of Psi: How Full-Length HIV-1 RNAs Become Packaged Genomes in the Viral Particles’, Virology, 454-455: 362–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavender C. A., Gorelick R. J., Weeks K. M. (2015) ‘Structure-Based Alignment and Consensus Secondary Structures for Three HIV-Related RNA Genomes’, PLoS Computational Biology, 11: e1004230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. et al. (2013) ‘Influence of Dimethylsulfoxide on RNA Structure and Ligand Binding’, Analytical Chemistry, 85: 9692–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K. et al. (2011a) ‘NMR Detection of Structures in the HIV-1 5′-Leader RNA That Regulate Genome Packaging’, Science (New York, N.Y.), 334: 242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K., Heng X., Summers M. F. (2011b) ‘Structural Determinants and Mechanism of HIV-1 Genome Packaging’, Journal of Molecular Biology, 410: 609–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews D. H. (2014) ‘RNA Secondary Structure Analysis Using RNAstructure’, Current Protocols in Bioinformatics, 46: 12 6 1–25. [DOI] [PubMed] [Google Scholar]
- Mathews D. H., Turner D. H., Watson R. M. (2016) ‘RNA Secondary Structure Prediction’, Current Protocols in Nucleic Acid Chemistry, 67: 11 2 1–2 19. [DOI] [PubMed] [Google Scholar]
- McBride M. S., Panganiban A. T. (1996) ‘The Human Immunodeficiency Virus Type 1 Encapsidation Site Is a Multipartite RNA Element Composed of Functional Hairpin Structures’, Journal of Virology, 70: 2963–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino E. J. et al. (2005) ‘RNA Structure Analysis at Single Nucleotide Resolution by Selective 2′-Hydroxyl Acylation and Primer Extension (SHAPE)’, Journal of the American Chemical Society, 127: 4223–31. [DOI] [PubMed] [Google Scholar]
- Mimouni N. K. et al. (2009) ‘An Analysis of Structural Influences on Selection in RNA Genes’, Molecular Biology and Evolution, 26: 209–16. [DOI] [PubMed] [Google Scholar]
- Montavon C. et al. (2002) ‘Identification of a New Circulating Recombinant Form of HIV Type 1, CRF11-Cpx, Involving Subtypes a, G, J, and CRF01-AE, in Central Africa’, AIDS Research and Human Retroviruses, 18: 231–6. [DOI] [PubMed] [Google Scholar]
- Mortimer S. A. et al. (2012) ‘SHAPE-Seq: High-Throughput RNA Structure Analysis’, Current Protocols in Chemical Biology, 4: 275–97. [DOI] [PubMed] [Google Scholar]
- Mueller N., Das A. T., Berkhout B. (2016) ‘A Phylogenetic Survey on the Structure of the HIV-1 Leader RNA Domain That Encodes the Splice Donor Signal’, Viruses, 8: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. T. et al. (2015) ‘IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies’, Molecular Biology and Evolution, 32: 268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niama F. R. et al. (2009) ‘CRF45_AKU, a Circulating Recombinant from Central Africa, Is Probably the Common Ancestor of HIV Type 1 MAL and HIV Type 1 NOGIL’, AIDS Research and Human Retroviruses, 25: 1345–53. [DOI] [PubMed] [Google Scholar]
- Paillart J. C. et al. (2004) ‘First Snapshots of the HIV-1 RNA Structure in Infected Cells and in Virions’, The Journal of Biological Chemistry, 279: 48397–403. [DOI] [PubMed] [Google Scholar]
- Pedersen J. S. et al. (2004a) ‘An Evolutionary Model for Protein-Coding Regions with Conserved RNA Structure’, Molecular Biology and Evolution, 21: 1913–22. [DOI] [PubMed] [Google Scholar]
- Pedersen J. S. (2004b) ‘A Comparative Method for Finding and Folding RNA Secondary Structures within Protein-Coding Regions’, Nucleic Acids Research, 32: 4925–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskol R., Stephan W. (2008) ‘Analyzing the Evolution of RNA Secondary Structures in Vertebrate Introns Using Kimura’s Model of Compensatory Fitness Interactions’, Molecular Biology and Evolution , 25: 2483–92. [DOI] [PubMed] [Google Scholar]
- Pollom E. et al. (2013) ‘Comparison of SIV and HIV-1 Genomic RNA Structures Reveals Impact of Sequence Evolution on Conserved and Non-Conserved Structural Motifs’, PLoS Pathogens, 9: e1003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond S. L., Frost S. D., Muse S. V. (2005) ‘HyPhy: Hypothesis Testing Using Phylogenies’, Bioinformatics (Oxford, England), 21: 676–9. [DOI] [PubMed] [Google Scholar]
- Poon A. F. et al. (2010) ‘Phylogenetic Analysis of Population-Based and Deep Sequencing Data to Identify Coevolving Sites in the Nef Gene of HIV-1’, Molecular Biology and Evolution, 27: 819.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell R. L. et al. (2007a) ‘Circulating Recombinant Form (CRF) 37_Cpx: An Old Strain in Cameroon Composed of Diverse, Genetically Distant Lineages of Subtypes a and G’, AIDS Research and Human retroviruses, 23: 923–33. [DOI] [PubMed] [Google Scholar]
- Powell R. L. (2007b) ‘Identification of a Novel Circulating Recombinant Form (CRF) 36_Cpx in Cameroon That Combines Two CRFs (01_AE and 02_AG) with Ancestral Lineages of Subtypes a and G’, AIDS Research and Human retroviruses, 23: 1008–19. [DOI] [PubMed] [Google Scholar]
- Rife B. D. et al. (2016) ‘Evolution of Neuroadaptation in the Periphery and Purifying Selection in the Brain Contribute to Compartmentalization of Simian Immunodeficiency Virus (SIV) in the Brains of Rhesus Macaques with SIV-Associated Encephalitis’, Journal of Virology, 90: 6112–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Alvarado G., Roossinck M. J. (1997) ‘Structural Analysis of a Necrogenic Strain of Cucumber Mosaic Cucumovirus Satellite RNA in Planta’, Virology, 236: 155–66. [DOI] [PubMed] [Google Scholar]
- Rollins C. et al. (2014) ‘Thermodynamic and Phylogenetic Insights into hnRNP A1 Recognition of the HIV-1 Exon Splicing Silencer 3 Element’, Biochemistry, 53: 2172–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salemi M. (2013) ‘The Intra-Host Evolutionary and Population Dynamics of Human Immunodeficiency Virus Type 1: A Phylogenetic Perspective’, Infectious Disease Reports, 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan R., Borderia A. V. (2011) ‘Interplay between RNA Structure and Protein Evolution in HIV-1’, Molecular Biology and Evolution, 28: 1333–8. [DOI] [PubMed] [Google Scholar]
- Schultes E., Hraber P. T., LaBean T. H. (1997) ‘Global Similarities in Nucleotide Base Composition among Disparate Functional Classes of Single-Stranded RNA Imply Adaptive Evolutionary Convergence’, RNA, 3: 792–806. [PMC free article] [PubMed] [Google Scholar]
- Seetin M. G., Mathews D. H. (2012) ‘RNA Structure Prediction: An Overview of Methods’, Methods in Molecular Biology (Clifton, N.J.), 905: 99–122. [DOI] [PubMed] [Google Scholar]
- Shankarappa R. et al. (1999) ‘Consistent Viral Evolutionary Changes Associated with the Progression of Human Immunodeficiency Virus Type 1 Infection’, Journal of Virology, 73: 10489–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro B. A. et al. (2007) ‘Bridging the Gap in RNA Structure Prediction’, Current Opinion in Structural Biology, 17: 157–65. [DOI] [PubMed] [Google Scholar]
- Sherpa C. et al. (2015) ‘The HIV-1 Rev Response Element (RRE) Adopts Alternative Conformations That Promote Different Rates of Virus Replication’, Nucleic Acids Research, 43: 4676–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P., Smith D. B. (1999) ‘Structural Constraints on RNA Virus Evolution’, Journal of Virology, 73: 5787–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit S., Knight R., Heringa J. (2009) ‘RNA Structure Prediction from Evolutionary Patterns of Nucleotide Composition’, Nucleic Acids Research, 37: 1378–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014) ‘RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies’, Bioinformatics (Oxford, England), 30: 1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J. (2008) ‘A Rapid Bootstrap Algorithm for the RAxML Web Servers’, Systematic Biology, 57: 758–71. [DOI] [PubMed] [Google Scholar]
- Stephan W. (1996) ‘The Rate of Compensatory Evolution’, Genetics, 144: 419–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer K., von Haeseler A. (1997) ‘Likelihood-Mapping: A Simple Method to Visualize Phylogenetic Content of a Sequence Alignment’, Proceedings of the National Academy of Sciences of the United States of America, 94: 6815–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukosd Z. (2015) ‘Full-Length RNA Structure Prediction of the HIV-1 Genome Reveals a Conserved Core Domain’, Nucleic Acids Research, 43: 10168–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukosd Z. et al. (2013) ‘Evaluating the Accuracy of SHAPE-Directed RNA Secondary Structure Predictions’, Nucleic Acids Research, 41: 2807–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavare S. (1986) ‘Some probabilistic and statistical problems in the analysis of DNA sequences’, Lectures on Mathematics in the Life Sciences (AMS), 17: 57–86. [Google Scholar]
- Thomson M. M. et al. (2005) ‘Identification of a Novel HIV-1 Complex Circulating Recombinant Form (CRF18_Cpx) of Central African Origin in Cuba’, AIDS, 19: 1155–63. [DOI] [PubMed] [Google Scholar]
- Tovanabutra S. et al. (2003) ‘A New Circulating Recombinant Form, CRF15_01B, Reinforces the Linkage between IDU and Heterosexual Epidemics in Thailand’, AIDS Research and Human Retroviruses, 19: 561–7. [DOI] [PubMed] [Google Scholar]
- Van Hemert A. M. (1995) ‘Dilution of Effect: A Systematic Bias in the Randomized Controlled Trial’, Journal of Psychosomatic Research, 39: 933–5. [DOI] [PubMed] [Google Scholar]
- Watts J. M. et al. (2009) ‘Architecture and Secondary Structure of an Entire HIV-1 RNA Genome’, Nature, 460: 711–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbe K. et al. (2002) ‘Identification of Two CRF11-Cpx Genomes and Two Preliminary Representatives of a New Circulating Recombinant Form (CRF13-Cpx) of HIV Type 1 in Cameroon’, AIDS Research and Human Retroviruses, 18: 849–56. [DOI] [PubMed] [Google Scholar]
- Wilkinson K. A. et al. (2008) ‘High-Throughput SHAPE Analysis Reveals Structures in HIV-1 Genomic RNA Strongly Conserved across Distinct Biological States’, PLoS Biology, 6: e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X., Holcik M. (2009) ‘Strong Eukaryotic IRESs Have Weak Secondary Structure’, PLoS One, 4: e4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi J. et al. (2008) ‘Identification of New CRF43_02G and CRF25_Cpx in Saudi Arabia Based on Full Genome Sequence Analysis of Six HIV Type 1 Isolates’, AIDS Research and Human Retroviruses, 24: 1327–35. [DOI] [PubMed] [Google Scholar]
- Zanini F., Neher R. A. (2013) ‘Quantifying Selection against Synonymous Mutations in HIV-1 Env Evolution’, Journal of Virology, 87: 11843–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. Y. et al. (2013) ‘Transient RNA Structure Features Are Evolutionarily Conserved and Can Be Computationally Predicted’, Nucleic Acids Research, 41: 6273–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.