Abstract
We describe phenotypic characterization of dli-1, the Caenorhabditis elegans homolog of cytoplasmic dynein light intermediate chain (LIC), a subunit of the cytoplasmic dynein motor complex. Animals homozygous for loss-of-function mutations in dli-1 exhibit stochastic failed divisions in late larval cell lineages, resulting in zygotic sterility. dli-1 is required for dynein function during mitosis. Depletion of the dli-1 gene product through RNA-mediated gene interference (RNAi) reveals an early embryonic requirement. One-cell dli-1(RNAi) embryos exhibit failed cell division attempts, resulting from a variety of mitotic defects. Specifically, pronuclear migration, centrosome separation, and centrosome association with the male pronuclear envelope are defective in dli-1(RNAi) embryos. Meiotic spindle formation, however, is not affected in these embryos. DLI-1, like its vertebrate homologs, contains a putative nucleotide-binding domain similar to those found in the ATP-binding cassette transporter family of ATPases as well as other nucleotide-binding and -hydrolyzing proteins. Amino acid substitutions in a conserved lysine residue, known to be required for nucleotide binding, confers complete rescue in a dli-1 mutant background, indicating this is not an essential domain for DLI-1 function.
INTRODUCTION
Cytoplasmic dynein is a large multisubunit complex composed of two motor proteins, the heavy chains, and several associated subunits that have been named light, light intermediate, and intermediate chains. These subunits have no sequence homology; instead, the names reflect their relative molecular weights. Cytoplasmic dynein is the major microtubule minus-end–directed motor protein and has been implicated in a variety of cellular processes, during both interphase and mitosis. Membranous vesicle transport, endoplasmic reticulum-to-Golgi transport, and axonal retrograde transport are all dynein-dependent processes (Lacey and Haimo, 1992; Dillman et al., 1996; Presley et al., 1997).
Dynein's mitotic roles are numerous. Function blocking antibody experiments against heavy chain in vertebrate cells have shown the motor protein is required for proper spindle formation and centrosome separation (Vaisberg et al., 1993). Similar results were observed in Drosophila heavy chain mutants with additional observations suggesting dynein is required to maintain centrosome association with the nuclear envelope (Robinson et al., 1999). In both Aspergillus nidulans and Neurospora crassa, dynein heavy chain mutations reveal a role for the complex in nuclear migration (Plamann et al., 1994; Xiang et al., 1995). Other recent work suggests dynein may mediate microtubule binding at the kinetochore (Wordeman and Mitchison, 1995).
The one-cell Caenorhabditis elegans embryo is also an excellent model system for investigating the roles of dynein, its subunits, and the proteins with which it interacts. When cytoplasmic dynein heavy chain (dhc-1) was eliminated through RNA-mediated gene interference (RNAi), all of the above-mentioned phenotypes were observed (Gonczy et al., 1999). Pronuclear migration and centrosome separation did not occur and multiple female pronuclei were formed, suggesting defects in meiosis. Additionally, centrosomes were dissociated from the male pronuclei in 15% of these embryos.
The variety of roles dynein plays during mitosis suggests complex regulation must be at work to ensure proper timing and coordination with mitotic events. Numerous lines of evidence point to the dynein subunits as key players in this regulation. For example, the intermediate chain (IC) subunit has been shown to directly interact with the p150glued subunit of dynactin in vertebrate cell extracts (Karki and Holzbaur, 1995; Vaughan and Vallee, 1995). Dynactin is an adapter molecule that links dynein to vesicular cargo and has been shown to be required for dynein-dependent vesicle motility (Gill et al., 1991). RNAi against the C. elegans homologs of p150glued and another dynactin subunit, p50/dynamintin, also revealed a role in nuclear migration and centrosome separation (Skop and White, 1998; Gonczy et al., 1999). IC diversity, as a result of alternative splicing, gives rise to tissue-specific distribution as well as differential subcellular localization (Nurminsky et al., 1998). Function blocking antibody experiments directed against IC suggest it is only required for a subset of dynein functions; spindle assembly is disrupted as in the dynein heavy chain experiment, but centrosome separation is not affected (Gaglio et al., 1997). Additional function blocking experiments against IC suggest it is also required for proper meiotic spindle formation (Palazzo et al., 1999).
Multiple cytoplasmic dynein light chains have been identified and are also responsible for linking dynein to specific cargo or regulatory molecules. LC8, for example, has been shown to be required for proper nuclear migration in A. nidulans (Beckwith et al., 1998). LC8 has also been shown to directly interact with neuronal nitric-oxide synthase and the Bcl-2 family protein Bim and therefore is implicated in neurotransmitter function and the apoptotic pathway (Jaffrey and Snyder, 1996; Puthalakath et al., 1999). Mutations in another light chain family member, roadblock/LC7, reveal roles in axonal transport and mitosis in Drosophila (Bowman et al., 1999).
Interestingly, the heavy, intermediate, and light chains all have homologs in axonemal dynein. Light IC (LIC), however, is specific to cytoplasmic dynein and is the least well characterized subunit of the complex. No alleles of LIC have previously been described. Recent work has shown there are two rat light intermediate chains (Tynan et al., 2000). Each LIC forms a homodimer and binds the dynein complex mutually exclusive of one another. Additionally, LIC1, but not LIC2, has been shown to bind pericentrin, a conserved component of the centrosome (Purohit et al., 1999; Tynan et al., 2000). Pericentrin has been observed to translocate along microtubules toward the centrosomes in a dynein-dependent manner (Young et al., 2000). These data suggest LIC, like the intermediate and light chains, is responsible for linking dynein to specific cargo. Recently, antibodies against human LIC were generated and found to localize to the nuclear periphery and to the microtubule-organizing center in HeLa cells (Bielli et al., 2001). The latter of these observations is consistent with the observed pericentrin interaction.
Pepetide sequence analysis of rat LIC revealed no overall homology to known proteins; however, a putative P-loop sequence was identified near its N terminus with extended sequence similarity to the ATP-binding cassette (ABC) transporter family of ATPases (Gill et al., 1994; Hughes et al., 1995). The P-loop, also known as the Walker A box, is one of three conserved motifs making up the nucleotide-binding domain found in numerous nucleotide-binding proteins, including ATPases and kinases, as well as proteins whose functions are not fully understood (Walker et al., 1982). LIC, however, lacks the other two motifs and its ATPase activity has not been assayed.
Here, we report the first isolated alleles of dynein LIC and show it is required for dynein's mitotic roles. Maternal rescue allows loss-of-function homozygotes to develop to mid-larval stages before cell division failure begins. However, an early embryonic mitotic requirement is revealed through RNAi analysis.
MATERIALS AND METHODS
Strains and Alleles
C. elegans strains were cultured and maintained at 20°C according to standard procedures (Brenner, 1974). Mutagenesis of wild-type, N2 Bristol animals was performed with 50 mM ethyl methanesulfornate (Brenner, 1974). Mutagenized adults were individually transferred to a Petri plate and allowed to lay eggs. When the F1 progeny had grown to adults (∼4 d later), 10 F1 animals were randomly selected and transferred to individual Petri plates. The F2 generation was then scored under a dissecting microscope for a protruding vulva/sterile phenotype. Four to five wild-type siblings of these F2 sterile animals were cloned to obtain heterozygous strains. Approximately 10,000 haploid genome (5000 F1 plates) were screened, and 60 independent strains were isolated. The three dli-1 alleles ku266, ku275, and ku287 were isolated in this manner. Each allele was outcrossed at least five times. Mapping strains used were unc-30(e191), unc-31(e169) dpy-4(e1166), the deficiency sDf22, and the translocation balancer DnT1 (Riddle et al., 1997).
Genetic Analysis and Molecular Cloning
dli-1 was mapped relative to the cloned markers unc-31 and unc-30 on linkage group IV. The strain unc-31 dpy-4/ku266 was constructed and a standard three-factor recombination analysis was performed. Twenty-four recombinants were obtained: 10/12 Dpy-nonUnc and 2/12 Unc-nonDpy recombinants segregated ku266. The deficiency sDf22 was shown to uncover ku266, placing the locus left of the lev-1 gene. This genetic distance between unc-31 and lev-1 is represented by 12 cosmids interrupted by one sequencing gap near the unc-31 locus. Cosmids covering the region between unc-31 and lev-1 were mixed with plasmid DNA carrying the dominant marker SUR-5::GFP (100 ng/μl) in pools of three (15 ng/μl each) and injected into the strain unc-31 ku266/unc-30 (Mello et al., 1991). SUR-5::GFP served as a visible transgenic marker for scoring progeny of injected animals. The F1 generation was scored for transgenic fertile animals with the unc-31 phenotype to identify rescued homozygous ku266 animals. The minimal rescuing subclone pJHY10 of cosmid C39E9 contains 3.5 kb of upstream sequence and the entire 1.5 kb of coding sequence followed by ∼450 bases of 3′ untranslated region. This subclone was generated by inserting the 5.5-kb XhoI/MscI restriction fragment from C39E9 into the BlueScript SK+ vector (Stratagene, La Jolla, CA).
Molecular Analysis and RNA Interference
To identify molecular lesions in the 3 dli-1 alleles, polymerase chain reaction of whole-worm lysates was performed (Barstead et al., 1991), and sequences generated and compared against wild-type sequence similarly obtained. The dli-1 cDNAs yk102f4, yk303f8, and yk448c3 were sequenced with the use of T3- and T7-specific primers. The three cDNAs were shown to be full length by identification of both the predicted start and stop codons. Point mutations in the putative P-loop domain in pJHY10 were generated with the use of the QuickChange site-directed mutagenesis kit (Stratagene).
The cDNAs yk102f4 and yk22b3 were used as templates for generation of double-stranded RNA for dli-1 and dhc-1 RNAi analysis, respectively. yk102f4 is ∼2 kb in length and encodes a full-length cDNA for dli-1. yk22b3 is ∼3 kb and encodes the 3′-most end of dhc-1, including part of exon 10 and the remaining five exons. The methods of Fire et al. (1998) were used for generation and injection of double-stranded RNA. Double-stranded RNA for dli-1 and dhc-1 were injected at equimolar ratios into either wild-type animals or the heterozygous strain strain unc-31 ku266/unc-30 [only for dli-1(RNAi)]. All cDNAs were provided by Yuji Kohara (National Institute of Genetics, Mishima, Japan).
Video Microscopy and Immunocytochemistry
Embryos were dissected from adults in a minimal volume of M9 buffer with the use of a sterile 20-gauge syringe to cut the adults open. Embryos were then transferred with a mouth pipette to a 2% agar pad on a microscope slide in 5–10 μl of M9 buffer and covered with a coverslip. Time-lapsed movies of one-cell embryos were made with the use of OpenLab 2.0 software (Improvision, Coventry, United Kingdom), collecting one Nomarski image per second on a Zeiss Axioskop. These files were then saved as QuickTime movie files for further analysis. All embryos were harvested 26–30 h after RNAi injections, and all embryos were observed and movies recorded at 23°C.
Immunocytochemistry was performed on single cell embryos harvested as described above. Embryos were placed on subbed poly-l-lysine slides in a minimal volume of M9 buffer and flash frozen in liquid nitrogen. Embryos were permeabilized with the use of the freeze-crack method and rehydrated through series of MeOH/phosphate-buffered saline washes before blocking with goat serum for 0.5 h (Epstein and Shakes, 1995). Immunocytochemistry performed on gonads to visualize sheath cells with the use of antibodies to CEH-18 followed the protocol described by Greenstein et al. (1994).
Primary antibodies were used at the following concentrations: 1:100 rabbit anti-DHC-1 (Gonczy et al., 1999), 1:50 rabbit anti-ZYG-9 (Matthews et al., 1998), 1:100 mouse monoclonal anti-Drosophila α-tubulin (4A-1; M. Fuller, Stanford University, Stanford, CA), and 1:200 anti-CEH-18 (Greenstein et al., 1994). Secondary antibodies were used at the following concentrations: 1:100 tetramethylrhodamine B isothiocyanate-conjugated goat anti-rabbit and 1:100 Cy-2-conjugated donkey anti-mouse (Jackson Immunoresearch, West Grove, PA).
For DNA staining, embryos were incubated for 10 min with 0.1 μg/ml 4′,6-diamidino-3-phenylindole dihydrochloride (DAPI). Embryos were then mounted in ∼10 μl of n-propylgalate (Sigma, St. Louis, MO) in 80% glycerol. Fluorescent images were taken with a Zeiss Axioskop equipped with a Hamamatsu digital camera and deconvolution was performed with the use of OpenLab 2.0 software (Improvision).
RESULTS
Isolation of dli-1 Alleles and Molecular Cloning of dli-1
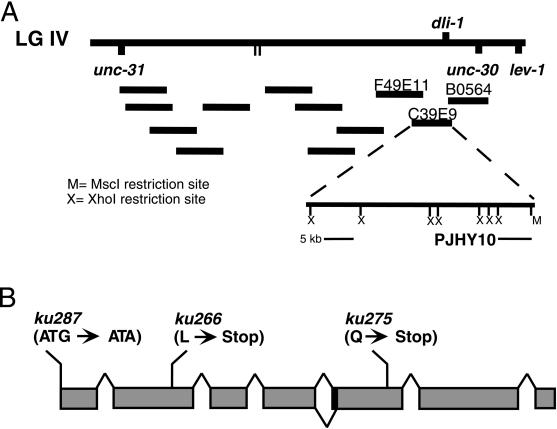
An ethyl methanesulfornate-induced F1 clonal screen of ∼10,000 haploid genomes was performed to isolate mutants with a protruding vulva and sterile phenotype (Fay and Han, 2000; see MATERIALS AND METHODS). Three mutants with similar vulva phenotypes mapped to the same genetic interval on linkage group IV between unc-31 and dpy-4. Noncomplementation tests between these three mutants (ku266, ku275, and ku287) indicated they are allelic. When trans to one another, the various allelic combinations result in the same protruding vulva and sterile phenotype seen in the three homozygous mutants. Further mapping with the ku266 allele placed it between the cloned genes unc-31 and lev-1 in a region spanned by 12 cosmids (Figure 1; see MATERIALS AND METHODS).
Figure 1.
Map position and genetic structure of dli-1. (A) Top line shows a portion of the genetic map from linkage group IV with relative positions of relevant genetic markers. Below are the three cosmids injected as a pool that confer rescue in the dli-1 alleles. The cosmid C39E9 confers rescue alone as does the large XhoI deletion and the subclone pJHY10. (B) Structure of the dli-1 gene showing locations of lesions within the three alleles and the two splice variants as determined by cDNA sequence comparison.
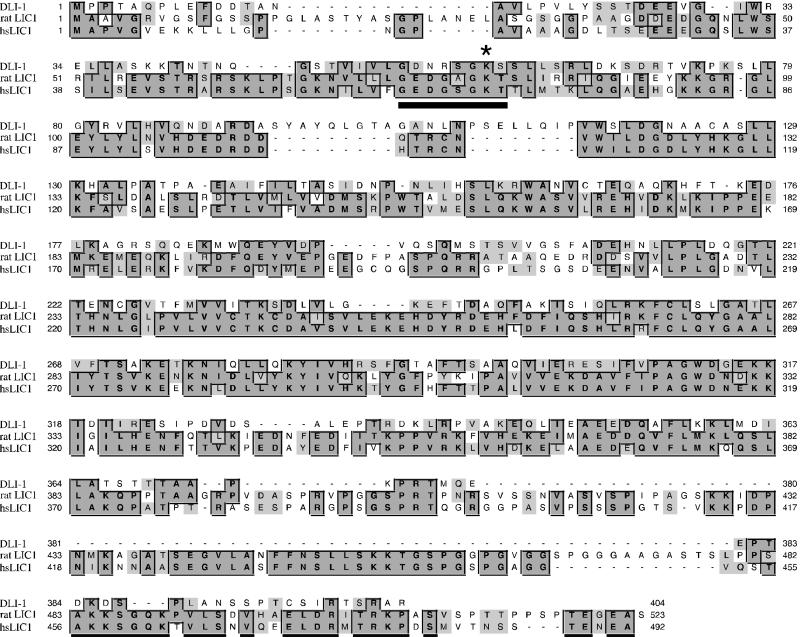
To identify the affected locus, germline transformation rescue was performed by injecting pools of overlapping cosmids from this region into the gonad arms of ku266 heterozygotes as described in MATERIALS AND METHODS (Figure 1A). We were able to rescue both the protruding vulva and sterile phenotypes with the single cosmid C39E9. A large XhoI deletion of this cosmid (CJHY2), containing only one predicted open reading frame (C39E9.14), was also able to rescue the phenotypes in all four transgenic lines obtained, and a minimal rescuing subclone (pJHY10) from the CJHY2 deletion construct was generated, which contains a 5.5-kb XhoI/MscI restriction fragment (Figure 1). An NCBI Blast search with the translation of this predicted open reading frame found homology with human, rat, and chick LIC with 33% sequence identity and 51% sequence similarity throughout the entire lengths of the proteins (Figure 2). Two vertebrate LICs have been identified (Tynan et al., 2000). LIC1 and LIC2 are ∼75% identical but show mutually exclusive binding to the dynein motor complex. Additionally, LIC1 specifically binds pericentrin, a conserved component of the centrosome. C39E9.14 shares the same degree of similarity with LIC1 and LIC2, and it is the only predicted gene in the C. elegans genome with homology to the LICs; therefore, after C. elegans nomenclature convention, we have given it the name dli-1 for cytoplasmic dynein light intermediate chain-1.
Figure 2.
Alignment between DLI-1, rat LIC1, and human LIC1. Dark shaded boxes show identical residues in two or more of the three proteins shown. Light gray boxes highlight similar residues. There is 33% sequence identity throughout the full length of the proteins and 51% similarity. The underlined residues constitute the putative P-loop sequence and the asterisk highlights the conserved lysine residue that is mutated in our rescuing constructs to either alanine or asparginine. Sequences were aligned with the use of Clustal W.
To confirm the mutant phenotypes are the result of mutations in the dli-1 gene, the entire dli-1 open reading frame was sequenced from the three alleles and compared with wild-type sequence. Two of the three alleles, ku266 and ku275, result from premature stop codons in the second and fifth exons, respectively (Figure 1B). ku287 results from a point mutation in the start codon (ATG to ATA). The next in-frame methionine is found in the fourth exon. Because the three alleles phenocopy one another, are not exacerbated when placed in trans to a deficiency, and based on the likely instability of messages produced by the introduction of premature stop codons (Pulak and Anderson, 1993), it is likely all three alleles are strong loss-of-function or null mutations.
Sequences of three full-length cDNAs were generated and compared against one another, revealing two distinct splice variants that differ only by 6 bp at the beginning of the fifth exon (Figure 1B). The two variants are found in both mixed stage and embryonic-derived cDNA libraries and will not be further addressed. With the exception of this predicted splice variant, the three cDNAs confirm the intron/exon boundaries predicted by Genefinder for C39E9.14 (GI:3874851). Each of the three cDNAs contains 20–70 bases of 5′ sequence before the predicted start codon, and none of the cDNAs contain an SL1 trans splice leader sequence, suggesting dli-1 is not processed by trans splice machinery (Blumenthal, 1997).
dli-1 Mutants Exhibit Failed Cell Divisions in a Number of Postembryonic Lineages
Numerous postembryonic cell lineages in dli-1 mutants exhibit stochastic failed cell division attempts. Lineages affected include the vulva precursor cells (VPCs) and their descendents, male tail-specific cells, hypodermal cells, and cells of the gonad, both germline and somatic. All of the affected lineages in wild-type animals begin or continue divisions through the third and fourth (final) larval stage. Early larval lineages are not affected in dli-1 mutants, suggesting successful cell divisions in the homozygous null mutants occurs until maternal protein is depleted (Fay and Han, 2000).
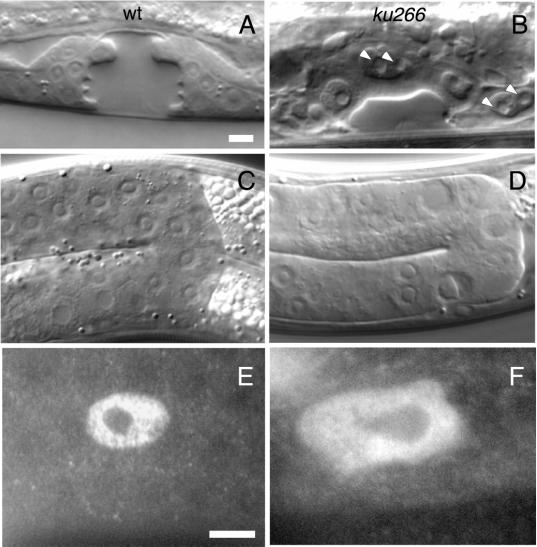
The vulva in C. elegans is derived from three of six equipotent cells, the VPCs, each of which undergoes three rounds of divisions (beginning early in the third larval stage) to generate a total of 22 vulval-specific cells (Sulston and Horvitz, 1977). To address the nature of the aberrant vulva morphology observed in the three alleles, we performed lineage analysis for the first two rounds of VPC divisions in 15 ku266 animals. Numerous stochastic failed cell division attempts were observed. One, two, or all three VPCs failed the first division in some animals, whereas in other animals, cells completed the first round of division, and failed division attempts were only observed during the second round. In each case, cells were observed to undergo nuclear envelope breakdown (NEB), chromatin then appeared to condense, but ∼20 min later, when two daughter nuclei should appear, only a single nucleus reformed. These nuclei were large and misshapen and often appeared to contain multiple nucleoli, suggesting single polyploid nuclei (Figure 3B, arrowheads). Also, several cells were seen to not attempt division at all. Similar observations were made for male tail-specific cell division attempts. These failed divisions were not cell lethal. Some cells that had failed division attempts were seen to attempt division in the next round. Additionally, cells that resulted from failed divisions were capable of expressing lineage-specific markers.
Figure 3.
dli-1 alleles undergo stochastic failed cell divisions in many postembryonic lineages. (A and C) Wild-type C. elegans at late L4, the last larval stage before the adult molt. (B and D) Similarly staged ku266 animals. (A) Wild-type late L4 vulva. Only 12 of 22 cells are visible in this plane of focus. (B) ku266 late L4 vulva. The enlarged nuclei are the result of failed divisions. Several of these cells contain multiple nucleoli (arrowheads) indicative of failed chromosome segregation. (C) Gonad of a wild-type animal composed of many syncitial germline nuclei. (D) Germline nuclei in ku266 animals also have abnormal morphology and are probably polyploid as a result of failed mitotic division in the distal end of the gonad arm. (E) Wild-type somatic sheath cell nucleus visualized by staining with anti-CEH-18 antibodies. (F) Sheath cell nucleus of a ku266 animal. Like all other late postembryonic lineages observed, the somatic sheath cells also appear to undergo failed division attempts, resulting in too few cells (average of 6 per gonad arm compared with 10 in wild-type) with abnormal nuclear morphology. Bar, ∼10 μm.
The sterility of dli-1 mutants is also likely a result of failed mitotic divisions. The gonad in C. elegans hermaphrodites consists of two U-shaped arms that terminate proximally in a structure called the spermatheca and are joined by the uterus, which opens, via the vulva, to the outside of the animal (Schedl, 1997). L1 animals possess a four-cell gonad primordium consisting of two somatic and two germline precursor cells (Sulston et al., 1983). In hermaphrodites, by L2, 10 somatic blast cells form a central somatic gonad primordium. During L3 and L4, these blast cells divide to produce the sheath, spermatheca, and uterus. Additionally, during late L3 and through adulthood, a distal pool of nuclei within the syncitial gonad divide mitotically. The resulting nuclei then enter meiosis, cellularize, and undergo gametogenesis. During L4 these cells differentiate as spermatocytes, and after the L4-to-adult molt, these cells begin differentiation as oocytes (Schedl, 1997). The sheath of each gonad arm is composed of five pairs of sheath cells, which are involved in regulating meiotic progression, ooctye maturation, and ovulation of mature oocytes. Laser ablation of sheath cells results in endomitotic germ cells and reduction of distal mitosis (McCarter et al., 1997). Failed division attempts are obvious in the distal end of gonads in dli-1 mutants, resulting in enlarged germline nuclei (Figure 3D). Additionally, ku266 gonads stained with antibodies against the POU domain containing homeobox protein CEH-18 (a gift from D. Greenstein, Vanderbilt University, Nashville, TN), which is expressed only in the somatic gonad cells, shows the nuclei of the sheath cells are large and have irregular morphology compared with wild-type gonads similarly stained (Figure 3, E and F). Additionally, ku266 gonad arms contain on average six sheath cells (n = 15) compared with 10 in the wild type. Loss of sheath cells could also contribute to irregular morphology and the large size (as a result of endomitosis) of germ cells in dli-1 mutants.
Point Mutations in a Putative P-Loop Domain Can Rescue dli-1 Phenotype
Previously reported sequence analysis of chick and rat LIC suggests a conserved P-loop domain near the N terminus of the protein (Gill et al., 1994; Hughes et al., 1995). The highest degree of similarity was found with the ABC transporter family of ATPases. C. elegans DLI-1 sequence also shares similarity in this region (Figure 2, underlined). On the basis of this observed similarity with ABC transporters, it was suggested LIC may be an ATPase (Gill et al., 1994; Hughes et al., 1995); however, no experimental evidence has been published in support of this suggestion.
Numerous lines of evidence suggest that a conserved lysine residue within the P-loop of ATPases and other proteins such as kinases, which also contain P-loops, is essential for the role of this domain (Azzaria et al., 1989; Stephens et al., 1995; Deyrup et al., 1998; Wu and Horvitz, 1998). An example is the C. elegans proapoptotic ABC transporter-like gene ced-7. Arginine substitutions for the conserved lysine residue in one of two P-loop domains in CED-7 fail to rescue a ced-7 cell corpse engulfment defect (Wu and Horvitz, 1998). To test the necessity of this putative P-loop domain in DLI-1, point mutations were constructed in the minimal rescuing subclone pJHY10, generating both the conserved K59R (pJHY11) and nonconserved K59A (pJHY14) substitutions. Both constructs completely rescued the vulva cell division and sterility phenotypes in the two alleles tested, ku266 and ku287. In each case, rescued animals homozygous for the two alleles were cultured for several generations and the efficiency of this rescue was not observed to diminish. These data, therefore, suggest this region is not essential for DLI-1 function.
DLI-1 Is Required for Pronuclear Migration
Double-stranded RNA generated against a gene of interest has been shown to specifically and potently disrupt that gene's function in C. elegans (Fire et al., 1998). Assuming that maternal protein from the heterozygous parent of dli-1 mutants allows homozygous progeny to develop normally until that message is depleted, we performed RNAi against dli-1 in both wild-type and dli-1 heterozygous animals to observe the earliest phenotype.
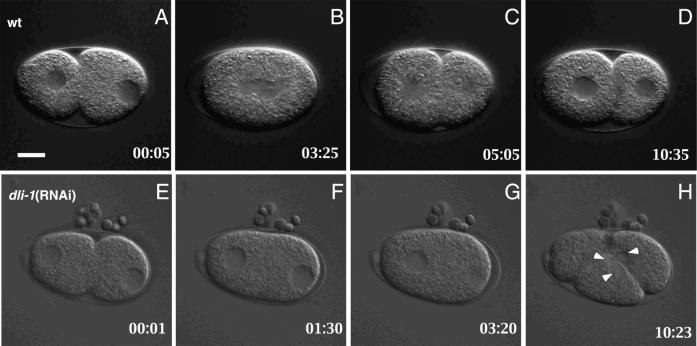
In uninjected wild-type embryos, shortly after fertilization the two meiotic divisions are completed, usually resulting in the extrusion of two polar bodies at the anterior end of the embryo (Albertson, 1997). The resulting female pronucleus is positioned slightly off the anterior cortex, whereas the male pronucleus, which contributes the single centrosome, is tightly associated with the posterior cortex (Figure 4A). A pseudocleavage furrow partially bisects the one-cell embryo. After duplication, the two daughter centrosomes associated with the male pronuclear envelope separate and move to opposite sides of the pronucleus while remaining associated with envelope (reviewed in Strome, 1993). The female and male pronuclei then begin migration toward one another. The female pronucleus moves slightly faster than the male and accelerates once it reaches the neck of the pseudocleavege furrow. Consequently, the two pronuclei meet in the posterior end of the embryo. After regression of the pseudocleavage furrow, the fused nuclei along with the two associated centrosomes move anteriorly to the center of the embryo. As the pronuclei undergo nuclear envelope breakdown, the centrosomes rotate to align themselves along the anterior-posterior axis of the embryo and the bipolar spindle is formed (Figure 4, B and C). Chromosome segregation follows, the embryo divides into two daughter cells, and nuclear envelopes are reformed (Figure 4D).
Figure 4.
Comparison of pronuclear migration in wild-type and dli-1(RNAi) embryos. All images are oriented with anterior to the left. (A–D) Wild-type embryo; female (anterior) and male (posterior) pronuclei begin migration before regression of the pseudocleavage furrow. The two pronuclei meet and fuse before nuclear envelope breakdown. (E–H) In the majority (16/19) of dli-1(RNAi) embryos harvested from injected wild-type mothers (shown) and 100% of embryos harvested from injected dli-1 heterozygous mothers (n = 19), pronuclear migration does not occur. (F) At the completion of pseudocleavage furrow regression, neither pronucleus has initiated migration. (G) At a time point equivalent to that in B in which the wild-type embryo is in metaphase, no pronuclear migration is apparent, and the male pronucleus has undergone NEB before female pronuclear NEB. (H) Multiple nuclei were formed in all dli-1(RNAi) embryos observed (arrowheads). Timestamps are relative to completion of anterior cytoplasmic contractions. In a wild-type embryo almost immediately after this, the female pronucleus initiates posteriorward migration. Bar, ∼10 μm.
RNAi against C. elegans dhc-1 results in early embryonic arrest, resulting from failed cell divisions beginning in the one-cell stage C. elegans embryo (Gonczy et al., 1999). Specifically, under Nomarski optics, male and female pronuclear migrations are not observed, suggesting this motor protein is required for both migration events. Also, a majority of dhc-1(RNAi) embryos contained multiple female pronuclei and aberrant polar body formation, suggesting defects in female meiotic divisions.
RNAi against dli-1 reveals it is also required for pronuclear migration. Wild-type animals injected with double-stranded RNA directed against dli-1 are 100% sterile 30 h after injection (n > 30). Embryos were first scored for number of female pronuclei and pronuclear migration defects. Unlike dhc-1(RNAi), 19/19 dli-1(RNAi) embryos generated a single female pronucleus (Figure 4E and Table 1). In one of these embryos, polar body extrusion appeared defective, resulting in one enlarged polar body. Although completion of meiosis (as scored by the formation of a single female pronucleus) appeared mostly unaffected, the majority of these embryos underwent failed pronuclear migration. Male pronuclei migration was not observed in any of the 19 embryos. Complete failure of female pronuclear migration was observed in 12/19 embryos (Figure 4, E–H). For these 12 embryos, both male and female pronuclei remained stationary through pseudocleavage furrow regression. Shortly after completion of this event, the male pronucleus underwent NEB, followed within 1–2 min by female NEB. Such asynchronous NEB events are common in mutants that exhibit failed pronuclear migration (Gonczy et al., 1999). The remaining seven embryos showed varying degrees of female pronuclear migration. The female pronucleus in four of these seven embryos migrated only to the midline of the embryo, to the point of pseudocleavage furrow constriction, and then halted. Thus, we scored these as failed migration events (Table 1). In the remaining three embryos, the pronucleus did successfully migrate to the posterior end; however, the rate of migration was considerably decreased and the male and female pronuclei never fused before NEB.
Table 1.
Summary of time-lapse differential interference contrast and immunocytochemistry analysis
| N2 | dhc-1(RNAi) | dli-1(RNAi) | dli-1(RNAi) in dli-1/+ | |
|---|---|---|---|---|
| A. Multiple female pronuclei | 0 /20 | 11 /14 | 0 /19 | 0 /19 |
| B. Failed pronuclear migration | 0 /20 | 14 /14 | 16 /19 | 19 /19 |
| C. Failed centrosome separation | 0 /20 | 5 /5 | 10 /17 | 6 /8 |
| D. Centrosomes dissociated from male pronucleus | 0 /20 | n /d | 5 /17 | 3 /8 |
| E. Failure to form meiotic spindle | 0 /15 | 7 /12 | 1 /11 | 1 /10 |
We attempted to increase the penetrance of the observed phenotypes by performing RNAi in a dli-1 heterozygous strain. As seen in the wild type, 100% of animals were sterile within 30 h after injection (n = 20). In 19/19 embryos a single female pronucleus formed as in the wild-type, but 100% of these embryos failed to undergo either female or male pronuclear migration (Figure 4 and Table 1).
In all 38 embryos, proper cell division did not occur. Cleavage furrows were initiated in each embryo after NEB, but these furrows were never able to completely bisect the embryos. Additionally, an ectopic cleavage furrow was often initiated at the posterior cortex these embryos near the nucleating microtubules. As in dhc-1(RNAi) embryos, multiple nuclei reformed as the dli-1(RNAi) embryos progressed to interphase, suggesting defects in chromosome segregation, or alternatively as a result of failed metaphase chromosome congression (Figure 4H, arrowheads).
Meiotic Spindle Formation Is not Affected by dli-1(RNAi)
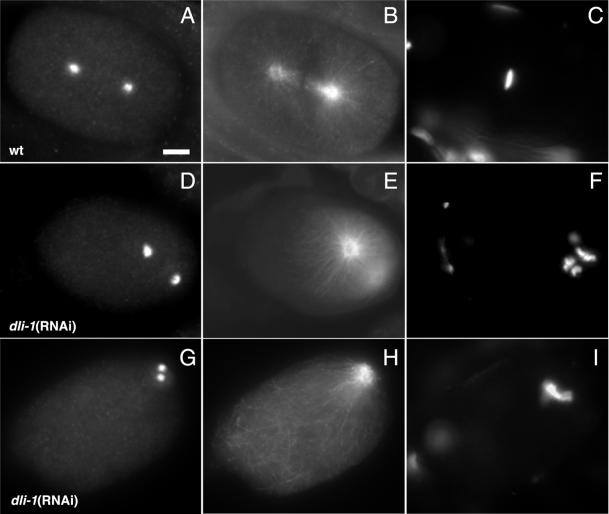
Dynein heavy chain and the IC subunit have been shown to be necessary for meiotic spindle formation (Palazzo et al., 1999). Because 100% of dli-1(RNAi) embryos formed a single female pronucleus, we wanted to investigate meiosis in greater detail. Therefore, dli-1 and dhc-1(RNAi) embryos were stained with anti-α-tubulin and DAPI to observe meiotic spindles and chromosomes. In wild-type embryos, after fertilization, the two meiotic divisions proceed with the formation of an acentrosomal meiotic spindle at each round surrounding the condensed metaphase meiotic chromosomes (Figure 5, A–C). During each meiotic division, one set of oocyte chromosomes is extruded into a polar body. When scored for meiotic spindle assembly, 19/21 observed dli-1(RNAi) embryos formed a meiotic spindle structure indistinguishable from wild-type embryos (10/11 embryos from wild-type injection and 9/10 embryos from dli-1 heterozygous injections; Figure 5, D–F, and Table 1). In the dli-1(RNAi) embryo from a wild-type injected mother in which a meiotic spindle was not observed, oocyte metaphase chromosomes were scattered along the anterior cortex of the embryo and no polar body was visible. In the other embryo (from an injected heterozygous mother), one polar body had been extruded anteriorly, and the monovalent meiosis II chromosomes were tightly condensed at the apex of the anterior cortex. In contrast, of 12 dhc-1(RNAi) embryos observed, only three had formed wild-type meiotic spindles with metaphase chromosomes condensed at their centers. Two additional embryos formed meiotic spindles, but the oocyte chromosomes were misplaced and scattered outside of the structure. The remaining seven dhc-1(RNAi) embryos failed to form meiotic spindles, and their oocyte chromosomes were randomly scattered throughout the anterior end of the embryos (Figure 5, G–I, and Table 1).
Figure 5.
Meiotic spindles are formed in dli-1(RNAi) embryos, but not in all dhc-1(RNAi) embryos observed. All embryos are oriented with anterior to the left. (A, D, and G) Anti-tubulin staining to reveal microtubules. (B, E, and H) DAPI staining to reveal DNA. (A–C) Wild-type embryo at meiosis II. A barrel-shaped spindle surrounds the female meiotic chromosomes. One polar body has already been extruded (B and C, arrow). The male meiotic chromosomes are tightly condensed at the posterior end of the embryo. (D–F) dli-1(RNAi) embryo slightly later in meiosis II. A barrel-shaped meiotic spindle has also formed around the condensed meiotic chromosomes. The spindle has rotated to the anterior/posterior axis to complete the meiosis II division. A single polar body has also already been extruded (E and F, arrow). (G–I) dhc-1(RNAi) embryo. No meiotic spindle is observable and the female meiotic chromosomes are scattered throughout the embryo. Bar, ∼10 μm.
DLI-1 Is Required for Centrosome Separation and Attachment to Pronuclear Envelope
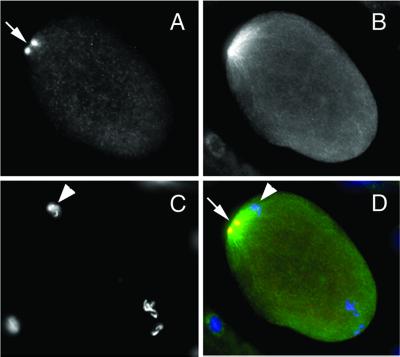
To more accurately observe the effects that depletion of the dli-1 gene product has on centrosome separation and spindle orientation, we stained dli-1(RNAi) embryos with antibodies to ZYG-9, a conserved centrosome protein (a gift from L. Matthews, Cornell University, Ithaca, NY) and α-tubulin (a gift from M. Fuller, Stanford University, Stanford, CA). Gönczy et al. (1999) reported 100% of dhc-1(RNAi) one-cell embryos exhibited failed centrosome separation. We were able to faithfully duplicate these results (n = 5). Unlike dhc-1(RNAi) embryos, 7 of 17 dli-1(RNAi) embryos from injected wild-type mothers, just before NEB or at metaphase had undergone centrosome separation, but these centrosomes were never observed to undergo full rotation, centration, or alignment along the anterior/posterior axis (Figure 6, D–F). The remaining 10 dli-1(RNAi) embryos were phenotypically indistinguishable from dhc-1(RNAi) embryos. Although centrosomes had duplicated, they had not separated after NEB and progression into metaphase (Figure 6, G–I). Similar phenotypes were observed in embryos from injected heterozygous mothers. Six of eight observed embryos had undergone failed centrosome separation. In similar experiments, Gönczy et al. (1999) observed 15% of centrosomes in dhc-1(RNAi) embryos were disassociated from the male pronuclei. This observation was also made in Drosophila heavy chain mutants (Robinson et al., 1999). Similarly, centrosomes were dissociated from the male pronuclei in 5/17 (30%) dli-1(RNAi) embryos from injected wild-type mothers and 3/8 embryos from heterozygous injections had dissociated centrosomes (Figure 7).
Figure 6.
DLI-1 is required for centrosome separation in the one-cell C. elegans embryo. Cytological analysis of metaphase embryos stained for the centrosomal component ZYG-9, tubulin, or DNA as indicated. All images are oriented with anterior to the left. (A–C) Wild-type embryo at metaphase. (A) Anti-ZYG-9 stains the two centrosomes that have rotated and aligned along an anterior-posterior axis. (B) Centrosomes nucleate microtubules forming the bipolar spindle visualized by anti-tubulin staining. (C) DAPI staining shows chromosomes condensed to the metaphase plate. (D–F) dli-1(RNAi) embryo in which centrosomes did separate. (D) Centrosomes have separated but have failed to centrate. This panel represents the most successful degree of rotation and spindle alignment observed. (E and F) Centrosomes do nucleate microtubules and chromosomes are condensed but have not yet congressed to a metaphase plate. (G–I) dli-1(RNAi) embryo exhibiting the most severe centrosome separation phenotype. (G) Some embryos (10/17) from wild-type injections failed to separate centrosomes and therefore formed no bipolar spindle. (H) Microtubules are nucleated and chromosomes have condensed and congressed to a metaphase plate (I). Embryos from heterozygous injections (7/8) were phenotypically identical to the embryo in G–I. Bar, ∼10 μm.
Figure 7.
Centrosomes are dissociated from the male pronucleus in 32% of dli-1(RNAi). In 5/17 embryos from wild-type–injected mothers and 3/8 embryos from dli-1 heterozygous injected mothers, centrosomes (arrows) were dissociated from the male pronucleus (arrowheads). (A–D) Embryo from a dli-1 heterozygous-injected mother showing centrosomes as visualized by anti-ZYG-9 staining (A), microtubules visualized by anti-tubulin staining (B), DAPI staining to reveal DNA (C), and merged (D). Bar, ∼10 μm.
DHC-1 Is Correctly Localized in dli-1 RNAi Embryos
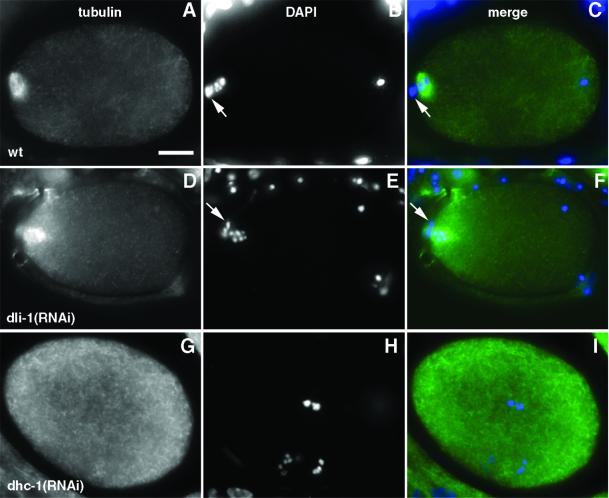
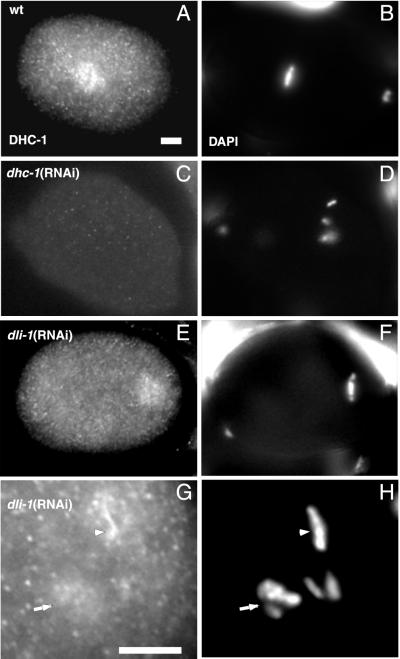
The phenotypic similarity between dli-1(RNAi) and dhc-1(RNAi) embryos raises the question of whether the dli-1(RNAi) phenotype is directly related to the function of the gene product or the result of ablating DHC-1 function through disruption of the cytoplasmic dynein complex itself. To address this question, dli-1(RNAi) embryos were stained with antibodies to C. elegans DHC-1 (a gift from P. Gönczy and A. Hyman, EMBL, Heidelberg, Germany). In wild-type embryos, DHC-1 is distributed throughout the cytoplasm in a punctate manner, localizes to the cell cortex, and is enriched at the periphery of both the male and female pronuclei before NEB (Gonczy et al., 1999). During prometaphase, DHC-1 is enriched on both sides of congressing chromosomes, and at metaphase and anaphase it is additionally enriched on the spindle on both sides of the metaphase plate (Figure 8A) and between segregating chromosomes, respectively. Compared with wild-type and dhc-1(RNAi) embryos, DHC-1 appears to localize correctly in dli-1(RNAi) embryos. We were unable to faithfully reproduce the pronuclear envelope staining pattern in wild-type embryos; however, metaphase and anaphase DHC-1 staining clearly shows DHC-1 localized to metaphase chromosomes and spindles. Therefore, we focused on metaphase and prometaphase staining in dli-1(RNAi) embryos to score for correct DHC-1 localization. In several dli-1(RNAi) embryos, due to failed centrosome separation, nothing similar to a bipolar spindle is formed and therefore spindle staining could not be observed. However, in these embryos DHC-1 still clearly localizes to prometaphase and metaphase chromosomes (Figure 8G). Additionally, when centrosome separation does occur, DHC-1 can be seen to localize in a pattern similar to the metaphase spindle staining seen in wild-type embryos (compare Figure 8, A and E). These data suggest the phenotypes observed in dli-1(RNAi) embryos are specific to loss of DLI-1 and not strictly a result of disrupting the cytoplasmic dynein complex.
Figure 8.
DHC-1 localizes correctly in dli-1(RNAi) embryos. (A) DHC-1 localization to the mitotic spindle in a wild-type metaphase embryo. (B) The same wild-type embryo stained with DAPI to reveal metaphase chromosomes. (C and D) dli-1(RNAi) embryo also at metaphase. DHC-1 localizes to either side of the condensed metaphase chromosomes and the mitotic spindle. Centrosomes have apparently separated enough in this embryo for a mitotic spindle to form. (E and F) dhc-1(RNAi) embryo also at metaphase. No DHC-1 staining is detectable. (G and H) Posterior end of a dli-1(RNAi) embryo as chromosomes are condensing at metaphase. When centrosomes fail to separate and no spindle is formed, DHC-1 staining is still observed on metaphase (arrowhead) and prometaphase (arrow) chromosomes. Bars, ∼10 μm.
DISCUSSION
We describe phenotypic characterization of the first mutant alleles of cytoplasmic dynein LIC, which we have named dli-1. Unlike the dynein motor protein and other subunits of the dynein complex, LIC is restricted to the cytoplasmic dynein complex and has no homolog in axonemal dynein (reviewed in King, 2000). This suggests LIC may be required for discrete aspects of dynein's function. Support for a role in mitosis is found in the recent observation that LIC directly interacts with pericentrin, a conserved centrosome component (Purohit et al., 1999).
The three loss-of-function alleles we have isolated in C. elegans also suggest a requirement for the dli-1 gene product during mitosis. Homozygous animals are sterile and many postembryonic cell lineages undergo stochastic failed division attempts. Affected cell lineages are those that continue division attempts through late larval stages in wild-type animals. Early larval lineages appear unaffected in dli-1 mutants. These observations suggest enough maternal product or message is available during embryogenesis and early larval development to allow proper cell division to occur.
The precise role that LIC plays during cell division is still not understood. Previous analysis of LIC peptide sequences led to the speculation that LIC may be an ATPase and serve a regulatory role for dynein's function through this activity (Gill et al., 1994; Hughes et al., 1995). This suggestion is based on identification of a putative P-loop domain within the protein sequence. The P-loop (or Walker A box) is one of three conserved motifs found in numerous nucleotide-hydrolyzing proteins, including the ABC transporter family of ATPases, and is defined by the consensus sequence GXXXXGK(S/T) (Walker et al., 1982). Although this consensus is present in all LIC sequences found to date, the other two motifs (the Walker B box and the signature motif) are not present. Additionally, previous work has shown a conserved lysine residue (corresponding to K59 in dli-1) within the P-loop sequence is required for proper nucleotide binding and therefore ATPase activity (Wu and Horvitz, 1998). Amino acid substitutions at K59 in our rescuing construct to either alanine or the conservative arginine residue are still able to confer complete rescue in the dli-1 alleles for multiple generations of homozygous animals. This suggests that nucleotide binding is not required for DLI-1 function and that DLI-1 is not an ATPase.
To circumvent maternal rescue and observe the earliest phenotype for depletion of DLI-1, we have used RNAi to specifically inhibit function of the dli-1 gene product. As expected, a much earlier requirement for dli-1, than observed in our mutants was revealed. The resulting phenotypes suggest DLI-1 is required for many of dynein's mitotic functions, but potentially not for dynein's role in meiotic spindle formation. When RNAi is performed against the C. elegans dynein heavy chain dhc-1, numerous female pronuclei are formed in one-cell embryos (Gonczy et al., 1999). Additionally, when RNAi is performed against two subunits of the dynactin complex, p150glued or p50/dynamintin, multiple female pronuclei are also observed, although at a much smaller percentage compared with dhc-1(RNAi) (Gonczy et al., 1999). In all dli-1(RNAi) embryos observed, whether the injected mother was wild type or heterozygous, only a single female pronucleus is formed. Additionally, anti-tubulin antibody staining in early dli-1(RNAi) embryos shows meiotic spindles are formed and meiotic chromosomes are condensed and correctly localized within the spindle (Figure 5, D–F). However, meiotic spindles are only seen in 5/12 dhc-1(RNAi) embryos. Additionally, two of these five embryos show chromosomes scattered outside of the meiotic spindle. These data cannot be used as conclusive evidence that DLI-1 is not required for meiotic spindle formation whereas DHC-1 is, because these are static observations, and the dhc-1(RNAi) embryos could have been fixed before or in between meiotic spindle formation. An alternative explanation is that even in injected heterozygotes, dli-1(RNAi) is incompletely penetrant and only a small amount of protein is required for proper dynein function during meiosis. Defects in meiotic spindle formation, however, would account for missegregation of meiotic chromosomes and the formation of multiple female pronuclei in dhc-1(RNAi) embryos. Because LIC has been shown to interact with the centrosome component pericentrin, and meiotic spindles form without centrosomes, it is still an attractive possibility that meiotic spindle formation is not affected in dli-1(RNAi) embryos because DLI-1 is not required for meiotic spindle formation.
The pronuclear migration defects observed in dli-1(RNAi) embryos suggests LIC is required for this aspect of dynein motor function. The majority of dli-1(RNAi) embryos from wild-type injections underwent failed pronuclear migration (16/19), and 100% of embryos from heterozygous injections showed failed pronuclear migration Interestingly, alleles of most other subunits of the cytoplasmic dynein complex as well as components of the dynactin complex have been isolated in genetic screens for defective nuclear migration in A. nidulans and N. crassa (Plamann et al., 1994; Xiang et al., 1994). Although LIC mutants were not isolated in these screens, our RNAi data show dli-1 is required at least for pronuclear migration and potentially, therefore, in other nuclear migration events.
DLI-1 also appears to be required for centrosome separation. Although centrosome separation occurred in 9/25 dli-1(RNAi) embryos, the spindle that was formed failed to centrate and the centrosomes never completely migrated to opposite poles of the male pronucleus. Again, incomplete penetrance could explain the 9/25 embryos in which centrosomes did separate from one another. Additionally, in 32% (8/25) of dli-1(RNAi) embryos, centrosomes were dissociated from the male pronulcear envelope. This phenotype was also observed in 15% of dhc-1(RNAi) embryos. These data are not surprising because LIC has been shown to directly interact with the conserved centrosome component pericentrin. Loss of this interaction through dli-1(RNAi) may increase disassociation of centrosomes from the nuclear envelope.
Dynein LIC is required for the function of the dynein motor complex. However, like other subunits of this complex, a specific role has yet to be defined. Numerous light chain subunits are found associated with the dynein motor complex and have been shown to be required not only for certain dynein functions such as nuclear migration (Beckwith et al., 1998) but also for interacting with proteins that are not currently implicated in dynein's mitotic roles such as Bcl-2 (Puthalakath et al., 1999) and neuronal nitric-oxide synthase (Jaffrey and Snyder, 1996).
The observed interaction between LIC and pericentrin (Purohit et al., 1999) suggests a mechanism for at least two of the requirements we have shown for DLI-1: centrosome separation and centrosome association with the male pronucleus. The dynein motor complex could be localized to the microtubule-organizing centers through such an interaction, allowing it to provide the force necessary to separate centrosomes and align the bipolar spindle. This interaction, however, does not explain the pronuclear migration defect seen in dli-1(RNAi) embryos. Pronuclear migration defects are also seen after RNAi of dynein light chain (dlc-1) (Gonczy et al., 2000). Both observations, therefore, may result from mislocalization of the dynein motor complex.
It seems unlikely that LIC is playing only a structural role for the dynein motor complex. The protein has been shown to exist as numerous phosphorylated species, and this phosphorylation is regulated in a cell cycle-dependent manner (Gill et al., 1994; Hughes et al., 1995; Niclas et al., 1996). Mapping LIC's interaction domains and analyzing the regulation of its phosphorylation should begin to shed more light on the nature of its role within the dynein complex.
ACKNOWLEDGMENTS
We thank David Fay and Li Yan for isolating two of the dli-1 alleles. We thank Pierre Gönczy and Tony Hyman (EMBL, Heidelberg, Germany) for anti-DHC-1 antibodies, Lisa Matthews (Cornell University, Ithaca, NY) for anti-ZYG-9 antibodies, and David Greenstein (Vanderbilt University, Nashville, TN) for anti-CEH-18 antibodies. We also thank Yuji Kohara for providing cDNAs and Alan Coulson for cosmids. Some strains used in this work were obtained from the Caenorhabditis Genetics Center. We thank Dan Starr and David Fay for helpful discussions and comments on the manuscript. J.Y. was supported by a National Institutes of Health Predoctoral Training Grant. This work was supported by a grant from the Public Health Service (GM-47869) and Howard Hughes Medical Institute of which M.H. is an assistant investigator.
REFERENCES
- Albertson DG. C. elegans II. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 47–78. [Google Scholar]
- Azzaria M, Schurr E, Gros P. Discrete mutations introduced in the predicted nucleotide-binding sites of the mdr1 gene abolish its ability to confer multidrug resistance. Mol Cell Biol. 1989;9:5289–5297. doi: 10.1128/mcb.9.12.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barstead RJ, Kleiman L, Waterston RH. Cloning, sequencing, and mapping of an alpha-actinin gene from the nematode Caenorhabditis elegans. Cell Motil Cytoskeleton. 1991;20:69–78. doi: 10.1002/cm.970200108. [DOI] [PubMed] [Google Scholar]
- Beckwith SM, Roghi CH, Liu B, Ronald Morris N. The “8-kD” cytoplasmic dynein light chain is required for nuclear migration and for dynein heavy chain localization in Aspergillus nidulans. J Cell Biol. 1998;143:1239–1247. doi: 10.1083/jcb.143.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielli A, Thornqvist PO, Hendrick AG, Finn R, Fitzgerald K, McCaffrey MW. The small GTPase Rab4A interacts with the central region of cytoplasmic dynein light intermediate chain-1. Biochem Biophys Res Commun. 2001;281:1141–1153. doi: 10.1006/bbrc.2001.4468. [DOI] [PubMed] [Google Scholar]
- Blumenthal TaS, K. RNA Processing and Gene Structure. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 117–145. [PubMed] [Google Scholar]
- Bowman AB, Patel-King RS, Benashski SE, McCaffery JM, Goldstein LS, King SM. Drosophila roadblock and Chlamydomonas LC7: a conserved family of dynein-associated proteins involved in axonal transport, flagellar motility, and mitosis. J Cell Biol. 1999;146:165–180. [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyrup AT, Krishnan S, Cockburn BN, Schwartz NB. Deletion and site-directed mutagenesis of the ATP-binding motif (P-loop) in the bifunctional murine ATP-sulfurylase/adenosine 5′-phosphosulfate kinase enzyme. J Biol Chem. 1998;273:9450–9456. doi: 10.1074/jbc.273.16.9450. [DOI] [PubMed] [Google Scholar]
- Dillman III JF, Dabney LP, Pfister KK. Cytoplasmic dynein is associated with slow axonal transport. Proc Natl Acad Sci USA. 1996;93:141–144. doi: 10.1073/pnas.93.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein HF, Shakes DC. Caenorhabditis elegans: modern biological analysis of an organism. San Diego, CA: Academic Press; 1995. [Google Scholar]
- Fay DS, Han M. Mutations in cye-1, a Caenorhabditis elegans cyclin E homolog, reveal coordination between cell-cycle control and vulval development [In Process Citation] Development. 2000;127:4049–4060. doi: 10.1242/dev.127.18.4049. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans [see comments] Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gaglio T, Dionne MA, Compton DA. Mitotic spindle poles are organized by structural and motor proteins in addition to centrosomes. J Cell Biol. 1997;138:1055–1066. doi: 10.1083/jcb.138.5.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Cleveland DW, Schroer TA. Characterization of DLC-A and DLC-B, two families of cytoplasmic dynein light chain subunits. Mol Biol Cell. 1994;5:645–654. doi: 10.1091/mbc.5.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Schroer TA, Szilak I, Steuer ER, Sheetz MP, Cleveland DW. Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J Cell Biol. 1991;115:1639–1650. doi: 10.1083/jcb.115.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P, Echeverri G, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, Hannak E, Kirkham M, Pichler S, Flohrs K, Goessen A, Leidel S, Alleaume AM, Martin C, Ozlu N, Bork P, Hyman AA. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III [In Process Citation] Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Gonczy P, Pichler S, Kirkham M, Hyman AA. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenstein D, Hird S, Plasterk RH, Andachi Y, Kohara Y, Wang B, Finney M, Ruvkun G. Targeted mutations in the Caenorhabditis elegans POU homeo box gene ceh-18 cause defects in oocyte cell cycle arrest, gonad migration, and epidermal differentiation. Genes Dev. 1994;8:1935–1948. doi: 10.1101/gad.8.16.1935. [DOI] [PubMed] [Google Scholar]
- Hughes SM, Vaughan KT, Herskovits JS, Vallee RB. Molecular analysis of a cytoplasmic dynein light intermediate chain reveals homology to a family of ATPases. J Cell Sci. 1995;108:17–24. doi: 10.1242/jcs.108.1.17. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Snyder SH. PIN: an associated protein inhibitor of neuronal nitric oxide synthase. Science. 1996;274:774–777. doi: 10.1126/science.274.5288.774. [DOI] [PubMed] [Google Scholar]
- Karki S, Holzbaur EL. Affinity chromatography demonstrates a direct binding between cytoplasmic dynein and the dynactin complex. J Biol Chem. 1995;270:28806–28811. doi: 10.1074/jbc.270.48.28806. [DOI] [PubMed] [Google Scholar]
- King SM. The dynein microtubule motor. Biochim Biophys Acta. 2000;1496:60–75. doi: 10.1016/s0167-4889(00)00009-4. [DOI] [PubMed] [Google Scholar]
- Lacey ML, Haimo LT. Cytoplasmic dynein is a vesicle protein. J Biol Chem. 1992;267:4793–4798. [PubMed] [Google Scholar]
- Matthews LR, Carter P, Thierry-Mieg D, Kemphues K. ZYG-9, a Caenorhabditis elegans protein required for microtubule organization and function, is a component of meiotic and mitotic spindle poles. J Cell Biol. 1998;141:1159–1168. doi: 10.1083/jcb.141.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T. Soma-germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev Biol. 1997;181:121–143. doi: 10.1006/dbio.1996.8429. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niclas J, Allan VJ, Vale RD. Cell cycle regulation of dynein association with membranes modulates microtubule-based organelle transport. J Cell Biol. 1996;133:585–593. doi: 10.1083/jcb.133.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurminsky DI, Nurminskaya MV, Benevolenskaya EV, Shevelyov YY, Hartl DL, Gvozdev VA. Cytoplasmic dynein intermediate-chain isoforms with different targeting properties created by tissue-specific alternative splicing. Mol Cell Biol. 1998;18:6816–6825. doi: 10.1128/mcb.18.11.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo RE, Vaisberg EA, Weiss DG, Kuznetsov SA, Steffen W. Dynein is required for spindle assembly in cytoplasmic extracts of Spisula solidissima oocytes. J Cell Sci. 1999;112:1291–302. doi: 10.1242/jcs.112.9.1291. [DOI] [PubMed] [Google Scholar]
- Plamann M, Minke PF, Tinsley JH, Bruno KS. Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J Cell Biol. 1994;127:139–149. doi: 10.1083/jcb.127.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJ, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells [see comments] Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- Purohit A, Tynan SH, Vallee R, Doxsey SJ. Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J Cell Biol. 1999;147:481–492. doi: 10.1083/jcb.147.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthalakath H, Huang DC, O'Reilly LA, King SM, Strasser A. The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol Cell. 1999;3:287–296. doi: 10.1016/s1097-2765(00)80456-6. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Blumenthal T, Meyer BJ, Priess JR. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- Robinson JT, Wojcik EJ, Sanders MA, McGrail M, Hays TS. Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J Cell Biol. 1999;146:597–608. doi: 10.1083/jcb.146.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl T. Developmental genetics of the germ line. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 241–269. [PubMed] [Google Scholar]
- Skop AR, White JG. The dynactin complex is required for cleavage plane specification in early Caenorhabditis elegans embryos. Curr Biol. 1998;8:1110–1116. doi: 10.1016/s0960-9822(98)70465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens KM, Roush C, Nester E. Agrobacterium tumefaciens VirB11 protein requires a consensus nucleotide-binding site for function in virulence. J Bacteriol. 1995;177:27–36. doi: 10.1128/jb.177.1.27-36.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S. Determination of cleavage planes. Cell. 1993;72:3–6. doi: 10.1016/0092-8674(93)90041-n. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Tynan SH, Purohit A, Doxsey SJ, Vallee RB. Light intermediate chain 1 defines a functional subfraction of cytoplasmic dynein which binds to pericentrin [In Process Citation] J Biol Chem. 2000;275:32763–32768. doi: 10.1074/jbc.M001536200. [DOI] [PubMed] [Google Scholar]
- Vaisberg EA, Koonce MP, McIntosh JR. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan KT, Vallee RB. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J Cell Biol. 1995;131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L, Mitchison TJ. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis [see comments] J Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YC, Horvitz HR. The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell. 1998;93:951–960. doi: 10.1016/s0092-8674(00)81201-5. [DOI] [PubMed] [Google Scholar]
- Xiang X, Beckwith SM, Morris NR. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc Natl Acad Sci USA. 1994;91:2100–2104. doi: 10.1073/pnas.91.6.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Roghi C, Morris NR. Characterization and localization of the cytoplasmic dynein heavy chain in Aspergillus nidulans. Proc Natl Acad Sci USA. 1995;92:9890–9894. doi: 10.1073/pnas.92.21.9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A, Dictenberg JB, Purohit A, Tuft R, Doxsey SJ. Cytoplasmic dynein-mediated assembly of pericentrin and gamma tubulin onto centrosomes. Mol Biol Cell. 2000;11:2047–2056. doi: 10.1091/mbc.11.6.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]