Dr. Pablo Lamata is a Wellcome Trust Senior Fellow in Basical Biomedical Science and a Reader in Computational Cardiology at King's College of London. His research interest focuses in the combination of imaging and computational modelling technologies to improve the management of cardiovascular diseases. His team (http://cmib.website/) develops solutions to stratify subjects according to the remodelling of cardiac anatomy, to characterise the performance of the heart during diastole, and to assess non-invasively the pressure driving blood flow in the central circulatory system. He is the coordinator of the EU consortium “Personalised In-Silico Cardiology” that develops modelling methodologies to optimize clinical protocols, from data acquisition to device parameters and intervention choices. Dr. Lamata has more than 15 years of experience in the development and clinical adoption of image analysis, physiological modelling, and surgical simulation and navigation solutions. He was a Marie Curie Fellow at Siemens, he obtained his PhD from the Universidad Politécnica de Madrid, and he received his MSc degree from the Universidad de Zaragoza. You can follow him on twitter: @pablolamata.

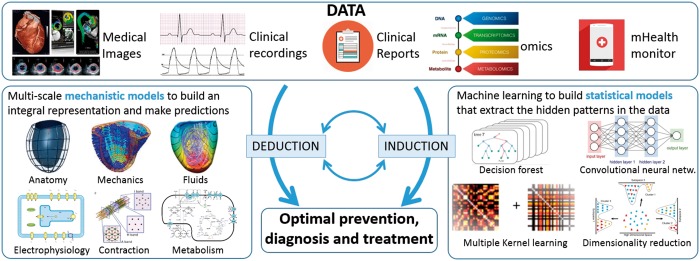
Engineers and researchers are boosting human induction and deduction skills assisted by computers, and this is one of the main drivers of progress in cardiovascular sciences and health care. This claim builds on two core ideas: machine learning1 as the vehicle for inductive reasoning, and computational cardiac models2 as the tool for deductive reasoning, both illustrated in Figure 1.
Figure 1.
Conceptual representation of the synergy between mechanistic and statistical computational models as the pathway towards precision medicine.
Machine learning refers to the ability of computers to gain knowledge without being explicitly programmed for it: the computer extracts patterns from the data and thus ‘learns’ a statistical model that will perform a given task (i.e. disease classification and prediction3 or segment the myocardium in an image4). The logic process followed here is inductive, since the machine makes broad generalizations from specific observations it has learned from. The availability of new sources of data (i.e. omics or continuous monitoring systems), the digitalization of the health record, and the recent advances in machine learning technology now offer the opportunity to reveal new patterns and new signatures of cardiovascular health and disease.1
On the other hand, computational cardiac models are the representations of our knowledge of the physiology of the heart pump and circulatory system governed by fundamental laws of physics and biochemistry. Computers are explicitly programmed to represent this knowledge into mechanistic models and use them to perform a given task (i.e. estimate myocardial stiffness,5 identify the fibrosis patterns that lead to persistent re-entrant drivers in atrial fibrillation,6 or compute the risk of drug toxicity7). The logic process followed here is deductive, since the machine reaches conclusions based on the concordance of multiple premises and assumptions. The availability of rich anatomical and functional data and the recent advances in computational cardiology technology now offer, through the process of model personalization, two opportunities: the ability to present an integral and cohesive diagnostic picture of the patient and the possibility to simulate and predict the evolution of a condition or the impact of a treatment.2,8
Both statistical and mechanistic models are thus rendering very encouraging perspectives and expectations, but these should be handled with caution.
Machine learning, as a tool for inductive reasoning, does not provide us with a conclusive proof of causal connections. Accordingly, the patterns observed in the data will not necessarily continue to exist in the future (or simply in other studies or individual cases). This translates into the specific problem of the generalizability of the model learned, which is in compromise with its complexity (i.e. in essence, the number of features or parameters to be learned from the data). As a consequence, if the problem to be solved requires a large number of features in order to make accurate predictions, then many more training examples are needed to ensure a valid generalization.
Computational cardiac models, as tools for deductive reasoning, rely on the validity of the premises and assumptions they are built on. Models are always a simplification of the reality, we simply cannot capture the entire complexity of the natural world. Consequently, the biomarkers and predictions extracted from models will always have a degree of uncertainty caused by the impact of assumptions and of the limitations of the data inputted into the model. And similarly with statistical models, mechanistic models present a compromise between complexity and uncertainty: the larger the number of model parameters the problem requires to make accurate predictions, the richer the dataset from an individual patient is required to ensure a valid personalized estimation.
An exciting perspective is the combination of inductive and deductive reasoning of statistical and mechanistic models. Machine learning can be informed by the features learned from mechanistic models.9 Our knowledge or the cardiovascular system is vast and should definitely guide the identification of valid data pieces to build the optimal statistical model. Revisiting the physical principles that underpin key clinical decisions can improve the accuracy and precision of well-recognized diagnostic biomarkers used in clinical guidelines, such as the pressure gradient caused by aortic stenosis.10
On the other hand, statistical models can be built from a population of mechanistic models,11 and mechanistic explanations can originate from, or can validate, the findings of statistic models—an example is the identification of the anatomical signature of heart failure to predict response to cardiac resynchronization therapy.12 Arguably the most attractive area of synergy between statistical and mechanistic models is the redefinition of patient classes, with early success stories in hypertrophic cardiomyopathy based on the electrocardiogram phenotype,13 in heart failure with preserved ejection fraction,14 or the proposal of the three main mechanistic substrates that explain the potential impact of a resynchronization therapy in heart failure.15
Guidelines and quality assurance in this emerging field is an important consideration. Medical devices that do not have any precedent are much more challenging to get regulatory approval, and there are early cases of success in this process such as the prediction of the fractional flow reserve using mechanistic simulations.16 Earlier in the research process, analysis standards and reporting guidelines help the field to mature as a discipline, where an excellent example is the Comprehensive In vitro Proarrhythmia Assay initiative.17
My personal vision is that the most significant impact will be the solution of the tension between clinical guidelines and the actual decisions with each individual patient: machine learning, equipped with the right population data, will identify the most relevant existing cases (similar to the individual) to trigger the inductive reasoning; computational cardiac models, equipped with the right data of the individual patient, will identify of the most sensible set of plausible parameters (those that explain the data of the subject) to trigger the deductive reasoning; and their combination will be the actual pathway towards the concept of the precision medicine,18 where computational tools will assist the cardiologists in the definition of the optimal diagnosis and therapy planning for the individual patient.
Acknowledgements
This work was supported by the Wellcome/EPSRC Centre for Medical Engineering at King's College London [g.a. 203148/Z/16/Z] and by the EU ITN Personalised In-Silico Cardiology [g.a. 764738]. P.L. holds a Wellcome Senior Fellowship [g.a. 209450/Z/17/Z].
Conflict of interest: none declared.
References
- 1. Deo RC. Machine learning in medicine. Circulation 2015;132:1920–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Trayanova NA. Whole-heart modeling: applications to cardiac electrophysiology and electromechanics. Circ Res 2011;108:113–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Austin PC, Tu JV, Ho JE, Levy D, Lee DS.. Using methods from the data-mining and machine-learning literature for disease classification and prediction: a case study examining classification of heart failure subtypes. J Clin Epidemiol 2013;66:398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Poudel RPK, Lamata P, Montana G. Recurrent fully convolutional neural networks for multi-slice MRI cardiac segmentation. In: Zuluaga M, Bhatia K, Kainz B, Moghari M, Pace D (eds), Reconstruction, Segmentation, and Analysis of Medical Images. RAMBO 2016, HVSMR 2016. Lecture Notes in Computer Science, Vol. 10129. Cham: Springer, 2017.
- 5. Xi J, Lamata P, Niederer S, Land S, Shi W, Zhuang X, Ourselin S, Duckett SG, Shetty AK, Rinaldi CA, Rueckert D, Razavi R, Smith NP.. The estimation of patient-specific cardiac diastolic functions from clinical measurements. Med Image Anal 2013;17:133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zahid S, Cochet H, Boyle PM, Schwarz EL, Whyte KN, Vigmond EJ, Dubois R, Hocini M, Haïssaguerre M, Jaïs P, Trayanova NA.. Patient-derived models link re-entrant driver localization in atrial fibrillation to fibrosis spatial pattern. Cardiovasc Res 2016;110:443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mirams GR, Cui Y, Sher A, Fink M, Cooper J, Heath BM, McMahon NC, Gavaghan DJ, Noble D.. Simulation of multiple ion channel block provides improved early prediction of compounds’ clinical torsadogenic risk. Cardiovasc Res 2011;91:53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lamata P, Cookson A, Smith N.. Clinical diagnostic biomarkers from the personalization of computational models of cardiac physiology. Ann Biomed Eng 2016;44:46–57. [DOI] [PubMed] [Google Scholar]
- 9. Cabrera LR, Berte B, Cochet H, Jais P, Ayache N, Sermesant M.. Model-based feature augmentation for cardiac ablation target learning from images. IEEE Trans Biomed Eng 2018. https://ieeexplore.ieee.org/abstract/document/8322060/. [DOI] [PubMed] [Google Scholar]
- 10. Donati F, Myerson S, Bissell MM, Smith NP, Neubauer S, Monaghan MJ, Nordsletten DA, Lamata P.. Beyond Bernoulli: improving the accuracy and precision of noninvasive estimation of peak pressure drops. Circ Cardiovasc Imaging 2017;10:e005207.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Britton OJ, Bueno-Orovio A, Ammel K, Van Lu HR, Towart R, Gallacher DJ, Rodriguez B.. Experimentally calibrated population of models predicts and explains intersubject variability in cardiac cellular electrophysiology. Proc Natl Acad Sci USA 2013;110:E2098–E2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Warriner DR, Jackson T, Zacur E, Sammut E, Sheridan P, Hose DR, Lawford P, Razavi R, Niederer SA, Rinaldi CA, Lamata P.. An asymmetric wall-thickening pattern predicts response to cardiac resynchronization therapy. JACC Cardiovasc Imaging 2018. http://imaging.onlinejacc.org/content/early/2018/03/09/j.jcmg.2018.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lyon A, Ariga R, Mincholé A, Mahmod M, Ormondroyd E, Laguna P, Freitas N, de Neubauer S, Watkins H, Rodriguez B.. Distinct ECG phenotypes identified in hypertrophic cardiomyopathy using machine learning associate with arrhythmic risk markers. Front Physiol 2018;9:213.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shah SJ, Katz DH, Selvaraj S, Burke MA, Yancy CW, Gheorghiade M, Bonow RO, Huang C-C, Deo RC.. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation 2015;131:269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lumens J, Tayal B, Walmsley J, Delgado-Montero A, Huntjens PR, Schwartzman D, Althouse AD, Delhaas T, Prinzen FW, Gorcsan J.. Differentiating electromechanical from non–electrical substrates of mechanical discoordination to identify responders to cardiac resynchronization therapy. Circ Cardiovasc Imaging 2015;8:e003744.. [DOI] [PubMed] [Google Scholar]
- 16. Nørgaard BL, Leipsic J, Gaur S, Seneviratne S, Ko BS, Ito H, Jensen JM, Mauri L, Bruyne B, De Bezerra H, Osawa K, Marwan M, Naber C, Erglis A, Park S-J, Christiansen EH, Kaltoft A, Lassen JF, Bøtker HE, Achenbach S; NXT Trial Study Group. Diagnostic performance of noninvasive fractional flow reserve derived from coronary computed tomography angiography in suspected coronary artery disease. J Am Coll Cardiol 2014;63:1145–1155. [DOI] [PubMed] [Google Scholar]
- 17. Cavero I, Holzgrefe H.. CiPA: ongoing testing, future qualification procedures, and pending issues. J Pharmacol Toxicol Methods 2015;76:27–37. [DOI] [PubMed] [Google Scholar]
- 18. Antman EM, Loscalzo J.. Precision medicine in cardiology. Nat Rev Cardiol 2016;13:591–602. [DOI] [PubMed] [Google Scholar]