Abstract
During reduced energy intake, skeletal muscle maintains homeostasis by rapidly suppressing insulin-stimulated glucose utilization. Loss of this adaptation is observed with deficiency of the fatty acid transporter CD36. A similar loss is also characteristic of the insulin-resistant state where CD36 is dysfunctional. To elucidate what links CD36 to muscle glucose utilization, we examined whether CD36 signaling might influence insulin action. First, we show that CD36 deletion specific to skeletal muscle reduces expression of insulin signaling and glucose metabolism genes. It decreases muscle ceramides but impairs glucose disposal during a meal. Second, depletion of CD36 suppresses insulin signaling in primary-derived human myotubes, and the mechanism is shown to involve functional CD36 interaction with the insulin receptor (IR). CD36 promotes tyrosine phosphorylation of IR by the Fyn kinase and enhances IR recruitment of P85 and downstream signaling. Third, pretreatment for 15 min with saturated fatty acids suppresses CD36-Fyn enhancement of IR phosphorylation, whereas unsaturated fatty acids are neutral or stimulatory. These findings define mechanisms important for muscle glucose metabolism and optimal insulin responsiveness. Potential human relevance is suggested by genome-wide analysis and RNA sequencing data that associate genetically determined low muscle CD36 expression to incidence of type 2 diabetes.
Introduction
CD36 (SR-B2) has high affinity for long-chain fatty acids (FA) and facilitates tissue FA uptake in rodents (1,2) and humans (3,4). The protein also transduces signaling initially documented to mediate its role in immunity and atherosclerosis (5–7). However, accumulating evidence supports the importance of CD36 signaling in regulating metabolic pathways such as FA oxidation (8), fatty taste perception (9,10), eicosanoid formation (11), and chylomicron production (12), among others.
CD36 is important for muscle metabolic adaptation (7). In wild-type mice, fasting causes muscle to reduce glucose utilization, whereas in CD36−/− mice, muscle glucose utilization persists despite high circulating FA causing hypoglycemia and increasing risk of sudden death (13). CD36−/− mice have accelerated depletion of glycogen stores during exercise and fail to increase muscle FA oxidation (14). Also, beneficial changes in substrate utilization and muscle performance induced by exercise training are not observed in CD36−/− mice (15).
The mechanisms associating CD36 to regulation of muscle glucose metabolism are unknown. CD36 signaling was shown to regulate FA oxidation by directly modulating AMPK activation (8,14). We examined whether it might influence insulin action on muscle glucose utilization. Using a mouse with conditional deletion of CD36 in muscle and primary-derived human myotubes, we show that CD36 regulates insulin stimulation of glucose metabolism. CD36 interacts with the insulin receptor (IR) and augments its insulin-induced phosphorylation by the kinase Fyn. Saturated FA rapidly dissociate Fyn, transducing pathway inhibition. The findings provide insight into mechanisms important for optimal muscle insulin responsiveness.
Research Design and Methods
Study Approval
All protocols for animal experiments in this study were approved by the Washington University in St. Louis Animal Studies Committee.
Reagents
Chemicals were from Sigma-Aldrich (St. Louis, MO), pRK5 c-Fyn plasmid, a gift from F. Giancotti (16), was from Addgene (plasmid #16032). Sources for antibodies are listed in Supplementary Table 1.
Generation of Skeletal Muscle-Specific CD36−/− Mice
C57BL/6 Cd36-floxed (Cd36fl/fl) mice (17) were crossed with mice expressing human skeletal actin–reverse tetracycline-controlled transactivator (HSA-rtTA) Cre (18) for more than five generations. Skeletal muscle Cd36 deletion (smCd36−/−) was induced by giving 8-week-old Cre-positive and Cre-negative (Cd36fl/fl) mice one intraperitoneal doxycycline injection (100 mg/kg) then doxycycline (2 g/L) in drinking water for 7 days, followed by a washout period of >7 days. The mice were used for studies between 10 and 18 weeks, a range that includes mice fed a high-fat diet (HFD) for 5 weeks (Surwit, D12331; Research Diets). Male mice were used for most studies unless indicated, but key findings on muscle insulin signaling and glucose disposal were reproduced in females. Combined male/female data are presented as indicated.
Intragastric Palm Oil and Glucose Tolerance Test
Palm kernel oil, blended at 57°C 1:1 with fat-free milk, or milk (vehicle), was administered to 10- to 15-week-old mice intragastrically (200 μL/mouse) after a 16-h fast. Glucose (2 g/kg) was given intraperitoneally 2 h later, and its clearance monitored in tail blood.
Cells and Treatments
Human skeletal muscle myotubes (HSMMs; Lonza), cultured and differentiated per Lonza’s instructions, were treated with lipofectamine RNAiMAX and 20 nmol/L CD36 (small interfering [si]RNA s2647 [siCD36_1], siRNA s2646 [siCD36_2]; Ambion) or nonspecific siRNAs and used 72 h after transfection. CHO cells with stable expression of human IR and human CD36 (CHO/IR/+CD36) and CHO/IR vector controls (11) were transiently transfected where indicated (lipofectamine LTX; Life Technologies) and used 48 h later. C2C12 myoblasts were cultured and differentiated as previously described (8). Unless indicated, all cells were serum-starved (16 h) before studies in low-glucose DMEM with 2 mmol/L l-glutamine, 100 μmol/L minimum essential medium nonessential amino acids, 100 units/mL penicillin, and 100 μg/mL streptomycin. All FA were added with BSA (2:1) for 15 min before insulin.
Quantitative Real-time PCR
RNA (TRIzol; Invitrogen) was subjected to cDNA reverse transcription and quantitative real-time PCR (ABI Prism 7000; Applied Biosystems) using Power SYBR Green PCR Master Mix and a 7500 Fast Real-Time PCR System (Applied Biosystems). Values (ΔΔCt) were normalized to 36B4 expression. Primers are listed in Supplementary Table 2.
Glucose Metabolism
Glycolysis in HSMMs and CHO cells was measured using Seahorse XF96 (Agilent) following the manufacturer’s protocols. Cells were seeded (3.4 × 104 cells/well) in XF96 microplates and switched 24 h later to serum-free medium (16 h) before experiments. For muscle explants, mice were euthanized by CO2, and dissected diaphragms were placed in warm (37°C) Seahorse XF medium with 2 mmol/L l-glutamine, 1 mmol/L sodium pyruvate, and 10 mmol/L glucose. Sections (∼2 × 2 × 1 mm) were transferred into XF24 islet capture microplates, seeded using islet capture screens, and processed (19).
For glucose uptake (20), cells were incubated with 1 μCi/mL 2-deoxy-d-[3H]glucose (2DG) in PBS and 0.1 mmol/L 2DG, washed with cold PBS containing 20 mmol/L 2DG, solubilized in 1% SDS, and counted (TriCarb 1600 TR; Packard). For glycogen (21), cells were incubated with d-[3H]glucose (1 μCi/mL) and 100 nmol/L insulin in PBS plus 0.2% BSA and 10 mmol/L glucose. Carrier glycogen in NaOH was added to lysates (35% weight [w]/volume [v]) before boiling (30 min, 95°C) and ethanol (95%) precipitation (−20°C). After centrifugation (12,000g for 10 min), ethanol-washed water-resuspended pellets were counted. Wheat germ agglutinin (WGA) staining adjusted for cell number (22).
Lipids
Ceramides and diacylglycerols were measured by the Biomolecular Analysis Core at Washington University in St. Louis by liquid chromatography-tandem mass spectroscopy, as previously described (23). Muscles were homogenized in PBS (w/v: 1:4), and 50 μL homogenates plus internal standards (ceramides 17:0 and diacylglycerols 15:0–15:0) lipids were extracted. Peak area ratios of analytes to internal standards were used for analysis.
Plasma and muscle triglycerides (TG) were measured using the L-Type Triglyceride M kit and HR Series NEFA-HR (2) kit from Wako Diagnostics.
Positron Emission Tomography
Positron emission tomography imaging of fed mice used [18F]-2-fluoro-2-deoxy-d-glucose (FDG) to assess glucose metabolism (24). Uptake/transport of tracer was visualized and quantified by summing uptake kinetics up to 60 min after the tracer injection.
Histology
Tissue sections were processed as previously described (25). Slides were scanned (NanoZoomer 2.0 HT; Hamamatsu, Bridgewater, NJ), and images were analyzed to yield muscle cross-sectional area (CSA) using Visiomorph (Broomfield, CO).
Proximity Ligation Assay
For proximity ligation assay (PLA; Duolink In Situ Hybridization kit; Sigma-Aldrich), cells on coverslips were incubated with or without insulin, washed with PBS, fixed with ice-cold methanol (−20°C) for 20 min, and blocked for 1 h in PBS with 0.05% Tween 20, 1% BSA, and 5% goat serum. After incubation for 16 h with goat anti-CD36 (R&D Systems) and rabbit anti-IRβ (Cell Signaling) antibodies, processing followed the manufacturers’ instructions. For tissues, formalin-fixed paraffin-sections were deparaffinized and processed as above. Imaging used Nikon Eclipse TE2000-U, a Photometrics CoolSNAP cf camera and MetaMorph 6.2r6 (Molecular Devices).
Coimmunoprecipitation and Immunofluorescence
Tissues and cells were lysed (30–60 min) in ice-cold buffer (20 mmol/L Tris-HCL [pH 7.5], 150 mmol/L NaCL, 1% Triton X-100, 60 mmol/L octyl β-d-glucopyranoside, 200 μmol/L sodium orthovanadate, 50 mmol/L NaF, 1 mmol/L phenylmethylsulfonyl fluoride, and 1.0 µg/mL protease inhibitor mix) and cleared lysates (10,000g for 10 min) assayed for protein content (BCA 23225; Pierce Biotech). For immunoprecipitation (IP), cells or tissues were lysed in IP buffer containing 1% Anapoe-C12E8 (Anatrace), 0.1% Triton X-100, 150 mmol/L NaCl, 5 mmol/L MgCl2, and 25 mmol/L HEPES (pH 7.5) with phosphatase and protease inhibitors. Primary antibodies or isotype-matched IgG, prebound (30 min) to Dynabeads protein G magnetic beads (Thermo Fisher Scientific) were incubated overnight (4°C) with equal lysate proteins, washed with cold IP buffer, and boiled (5 min) in SDS sample buffer. Immunofluorescent microscopy was performed as previously described (8).
In-Cell Western Assay
Cells in black-walled 96-well microplates (Corning) at confluence were serum-starved, treated as indicated, fixed with 3% paraformaldehyde, and blocked (1 h, PBS containing 0.05% Tween 20, 1% BSA, and 5% goat serum). Incubation (16 h) with monoclonal anti–phosphorylated (p)AKT(S) antibody (Cell Signaling) was followed by washing (PBS, Tween 0.05%), incubation (60 min) with horseradish peroxidase–anti-rabbit antibody, washing, then incubation (30 min, room temperature) with 3,3′,5,5′-tetramethylbenzidine (TMB) liquid substrate (Sigma-Aldrich) before adding 1 N NaOH and reading absorbance at 450 nmol/L. To normalize signals, cells were incubated (60 min) with WGA Alexa Fluor 680 (Invitrogen), rinsed (PBS), and fluorescence was measured at 700 nmol/L (LI-COR). Backgrounds for TMB (primary antibody omitted) and WGA Alexa Fluor 680 staining (WGA omitted) were subtracted from the data.
Statistics
Analyses were performed with GraphPad Prism 7.2 software (GraphPad Software). Unpaired two-tailed Student t tests and two-way ANOVA, with the Bonferroni correction for multiple comparisons, were used as appropriate. Data are presented as the means ± SE unless otherwise indicated. A P value <0.05 was considered significant.
Results
CD36 Deletion in Skeletal Muscle
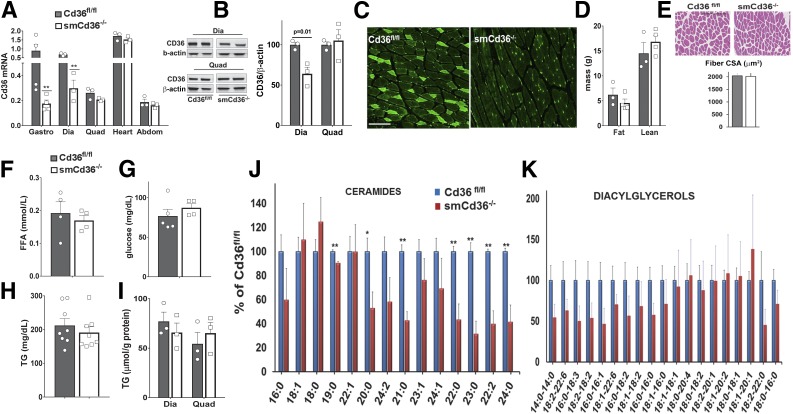
We generated the smCd36−/− mouse using has-rtTA Cre (18). Adult (8-week-old) Cre-positive and floxed controls were administered doxycycline (detailed under research design and methods) and used after a minimum 7-day washout period. Muscle CD36 expression was assayed for all cohorts. Typically, CD36 mRNA (Fig. 1A) and protein (Fig. 1B) were reduced by 50–60% in the oxidative (slow-twitch) diaphragm and the metabolically mixed (slow and fast twitch fibers) gastrocnemius, and no reduction was observed in the heart, as expected (Fig. 1A). No reduction was observed in mixed quadriceps and abdominal muscles (rectus abdominus), where CD36 expression is normally low (Fig. 1A and B). This is consistent with minor mosaicism of the expression cassette (18). Residual CD36 expression in the diaphragm and gastrocnemius is likely accounted for by blood vessel CD36, because immunostained myotubes appeared CD36 depleted (Fig. 1C). All studies were conducted with diaphragm or gastrocnemius, with quadriceps or rectus abdominus used as negative controls.
Figure 1.
Characterization of the mouse with conditional muscle CD36 deletion. A: Induction of smCd36−/− using the HSA-rtTA Cre reduced Cd36 gene expression in the predominantly slow-twitch oxidative diaphragm (Dia) and in the mixed (slow-fast twitch, oxidative-glycolytic) gastrocnemius (Gastro) but not in the mixed quadriceps (Quad) or abdominal muscle (rectus abdominus, Abdom). CD36 expression in the heart, as expected, was not reduced. Tissues were obtained from smCd36−/− and floxed littermate controls (Cd36fl/fl). All mice were given doxycycline, followed by a 7-day washout period (see research design and methods for details). Cd36 gene expression by quantitative PCR was normalized to 36B4 and to Cd36 in gastrocnemius of Cd36fl/fl mice set as 1 (n = 3–4 males/genotype). **P < 0.01. B: Representative Western blots show reduced CD36 protein in diaphragm (Dia) but not in quadriceps (Quad) muscle. Graph: means ± SE of CD36/β-actin expression (n = 3/genotype, males). C: Immunostaining of CD36 in gastrocnemius sections of Cd36fl/fl and smCd36−/− mice show CD36 depletion in muscle fibers of smCd36−/−. Representative images of five to six fields (n = 3/genotype, males). Scale bar: 100 μm. D: Body composition of Cd36fl/fl and smCd36−/− mice. Shown are means ± SE of fat and lean mass (n = 4/genotype, males). E: Muscle CSA for Cd36fl/fl and smCd36−/− mice. Hematoxylin and eosin–stained gastrocnemius sections were scanned and analyzed. Representative areas are shown (scale bar: 100 μm). Graph shows means ± SE of 228 individual muscle fibers for Cd36fl/fl and 359 for smCd36−/− (n = 3/genotype, males). Fasting plasma levels of free FA (FFA) (F), glucose (G), and TG (H) in Cd36fl/fl and smCd36−/− mice. Shown are means ± SE (n = 4–8/genotype, n = 3–4 females, 4–5 males). I: Diaphragm (Dia) and quadriceps (Quad) TG content (means ± SE (n = 3/genotype, males). J: Reduced muscle content of ceramide species in diaphragm of smCd36−/− mice. The decrease correlates with CD36 knockdown and is not observed in quadriceps (Supplementary Fig. 1A) (n = 4/genotype, males). *P < 0.05 **P < 0.01. K: Diacylglycerol species in diaphragm (see also Supplementary Fig. 1B) (n = 4/genotype, males).
CD36 Deletion Reduces Muscle Ceramides
Chow-fed smCd36−/− mice and Cd36fl/fl littermates weighed the same (data not shown) and had comparable fat and lean body mass (Fig. 1D). Muscle morphology and fiber CSA was unaltered (Fig. 1E). Fasting plasma levels of glucose, TG, and unesterified FA (Fig. 1F–H) were similar. Diaphragm and quadriceps TG content was equivalent for both groups (Fig. 1I). However, most ceramide species measured were reduced (Fig. 1J), and several diacylglycerol species (Fig. 1K) trended lower in smCd36−/− diaphragms; however, no changes were measured in quadriceps, where CD36 knockdown is not observed (Supplementary Fig. 1A and B).
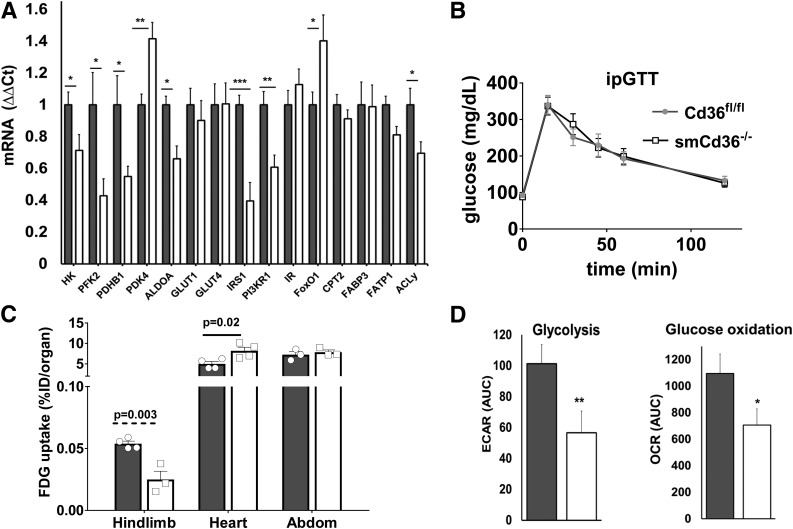
Reduced Glucose Uptake by Muscle
CD36 deletion reduced muscle expression of glucose metabolism and insulin signaling genes (Fig. 2A). Expression of FA metabolism genes (Fig. 2A) and mitochondrial genes and proteins (Supplementary Fig. 1C and D) was unchanged.
Figure 2.
Muscle CD36 deletion alters glucose metabolism in vivo. A: Cd36 deficiency in gastrocnemius reduced expression of genes for insulin signaling (insulin receptor substrate-1 [IRS1] and phosphatidylinositol 3-kinase regulatory-α [PI3KR1]) and glucose metabolism (hexokinase [HK], phosphofructokinase-2 [PFK2], pyruvate dehydrogenase B1 [PDHB1], aldolase A [ALDOA], and ACLy) while pyruvate dehydrogenase kinase 4 (PDK4) and FOXO1 (FoxO1) increased. FA metabolism genes, FA transport protein 1 (FATP1), carnitine palmitoyl transferase 2 (CPT2), and FA binding protein 3 (FABP3) were unchanged. Mitochondrial genes (Cox2, Porin1, and CPT1B) (Supplementary Fig. 1C) and protein levels of electron transfer chain complexes (C1–C5) (Supplementary Fig. 1D) were also unaltered. Gene expression by quantitative PCR normalized to 36B4. Shown are means ± SE (n = 7–14 mice/genotype, males). *P < 0.05, **P < 0.01, and ***P < 0.001 compared with Cd36fl/fl mice. B: Glucose disposal is not affected by muscle CD36 depletion. Intraperitoneal glucose tolerance test (ipGTT) for Cd36fl/fl and smCd36−/− mice (n = 4 males and 3 females/genotype). Shown are means ± SE of glucose as percentage of basal level (see also Supplementary Fig. 2A–C). C: FDG uptake is suppressed in hind-limb muscle but not in heart or abdominal (Abdom) muscle of smCd36−/− mice compared with Cd36fl/fl. Data are percentages of injected dose (%ID) per organ shown as means ± SE of cumulative tracer uptake at 60 min (n = 4/genotype, females). D: Insulin-stimulated glucose oxidation and glycolysis is suppressed in diaphragm explants from smCd36−/− mice. Glucose oxidation and glycolysis were assayed by Seahorse XF24. Shown are means ± SE AUC (n = 3/genotype, males, five technical replicates). OCR, oxygen consumption rate. *P < 0.05 and **P < 0.01 compared with Cd36fl/fl.
Glucose tolerance was similar between genotypes (Fig. 2B). Although females from both genotypes had slightly better glucose disposal than males, there were no sex-driven genotype-related differences (Supplementary Fig. 2A–C). In vivo uptake of FDG measured in hind-limb muscles was reduced in smCd36−/− mice, whereas heart glucose uptake increased (Fig. 2C), suggesting that less glucose uptake by skeletal muscle makes more available for the heart. Glucose uptake by abdominal muscle, where CD36 expression was not decreased (Fig. 1A), was unchanged (Fig. 2C).
Glucose metabolism assayed using muscle explants ex vivo showed insulin stimulation of glycolysis (extracellular acidification rate [ECAR]) and glucose oxidation (oxygen consumption rate) was diminished in explants from smCd36−/− mice compared with explants from Cd36fl/fl controls (Fig. 2D).
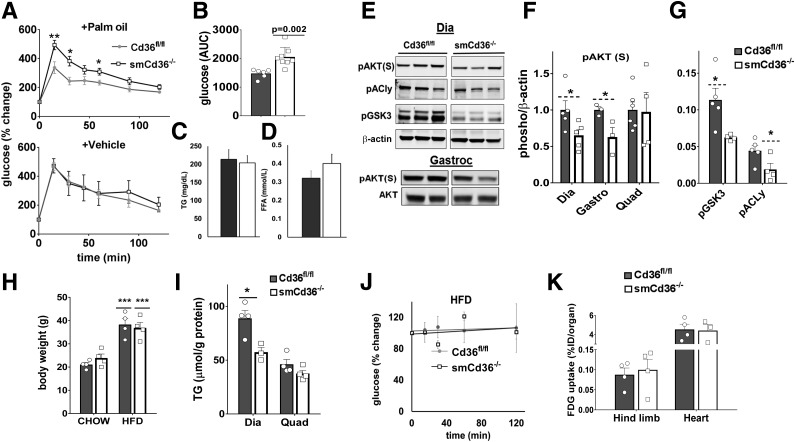
Impaired Disposal of Postprandial Glucose
Postprandial glucose disposal was tested during absorption of a fat-rich meal to engage mixed and oxidative muscles where CD36 expression is deleted. The mice were given an intragastric bolus of palm oil with skim milk (1:1), and 2 h later, during peak absorption (26) (Supplementary Fig. 1E), intraperitoneal (i.p.) glucose and its blood clearance were monitored. Glucose disposal was impaired by 40% (Fig. 3A, top panel), and the area under the curve (AUC) (Fig. 3B) was reduced (P = 0.002) in smCd36−/− compared with Cd36fl/fl controls. Clearance was similar when intragastric skim milk was given as a vehicle control (Fig. 3A, bottom panel). Plasma TG (Fig. 3C) and free FA (Fig. 3D) levels determined after glucose injection did not differ between groups.
Figure 3.
Skeletal muscle CD36 is important for postprandial glucose disposal. A: Glucose disposal after a fatty meal is impaired in smCd36−/− mice. Cd36fl/fl and smCd36−/− mice were given intragastric palm oil (1:1 with skim milk) or skim milk (vehicle). Glucose was administered i.p. 2 h later, and glucose disposal was measured in tail blood. Shown are means ± SE of % change in glucose related to level before i.p. glucose injection (n = 6–8/genotype). *P < 0.05 and **P < 0.01 compared with Cd36fl/fl controls (n = 6–8/genotype, 1–2 females, 6 males). B: AUC (means ± SE) for glucose disposal in mice given palm oil. Plasma TG (C) and free FA (FFA) (D) in mice after palm oil feeding. Cd36fl/fl (n = 3 males and 3 females) and smCd36−/− (n = 8, 5 males and 3 females) mice were given intragastric palm oil or skim milk (vehicle), followed 2 h later by glucose i.p. and blood collection. Shown are means ± SE of plasma TG and free FA. E: Muscle insulin signaling after a fatty meal is impaired in smCd36−/− mice. Representative immunoblots of diaphragm (Dia) and gastrocnemius (Gastroc) muscle from Cd36fl/fl and smCd36−/− mice. Mice were given palm oil as in A, followed 2 h later by i.p. insulin, and tissues were harvested after 15 min. Quantification of pAKT in diaphragm (Dia), gastrocnemius (Gastro), and quadriceps (Quad) (F) and of pGSK3 and pACLy in diaphragm (G) (n = 4–5/genotype, 2–3 males, 2 females). *P < 0.05 compared with Cd36fl/fl controls. H–K: Muscle CD36 knockdown does not protect against HFD-induced insulin resistance. H: Weight gain at the end of 5 weeks of chow or HFD feeding. Groups were initially weight matched (n = 4/diet/genotype, males). ***P < 0.001. Weight (means ± SE) HFD compared with chow controls. I: TG content (means ± SE) of diaphragm (Dia) and quadriceps (Quad) (n = 4/genotype, males). *P < 0.05 compared with Cd36fl/fl controls. J: Intraperitoneal insulin tolerance test (ipITT) for Cd36fl/fl and smCd36−/− mice. Mice fasted for 4 h were given 0.75 IU/kg insulin, and blood glucose was monitored. Shown are means ± SE of glucose as percentage of basal before insulin (n = 4/genotype, males, representative of two experiments). (See also Supplementary Fig. 2D and E.) K: FDG uptake is similar in hind limb and heart of Cd36fl/fl and smCd36−/− mice fed the HFD. Shown are means ± SE of organ tracer uptake at 60 min, expressed as percentage of injected dose (%ID) per organ (n = 4/genotype, males, representative of two experiments).
To examine whether the impaired glucose disposal in smCd36−/− mice reflects diminished muscle insulin signaling, mice were given the palm oil, then 2 h later insulin i.p., and diaphragms were harvested after 15 min. Diaphragm and gastrocnemius, but not quadriceps, of smCd36−/− mice had attenuated insulin-stimulated AKT phosphorylation (Fig. 3E and F). Phosphorylation of AKT targets glycogen synthase kinase 3 (GSK3α/βS21/9) and acyl citrate lyase (ACLyS455) (Fig. 3E and G) was also diminished. These data suggest CD36 deletion reduced insulin signaling in oxidative muscle during the high-fat meal.
SmCd36−/− Mice Are Not Protected From High-Fat Feeding
The mice were challenged with an HFD for 5 weeks, and effects on glucose metabolism were examined. Weight gain by smCd36−/− and Cd36fl/fl mice was similar (Fig. 3H). TG content was reduced in diaphragm but not quadriceps of smCd36−/− mice compared with controls (Fig. 3I), but both groups had similarly impaired insulin-induced glucose clearance (Fig. 3J), irrespective of sex (Supplementary Fig. 2D and E). Muscle glucose uptake in vivo was similar (Fig. 3K). Overall, CD36 deletion did not protect against HFD-induced muscle insulin resistance.
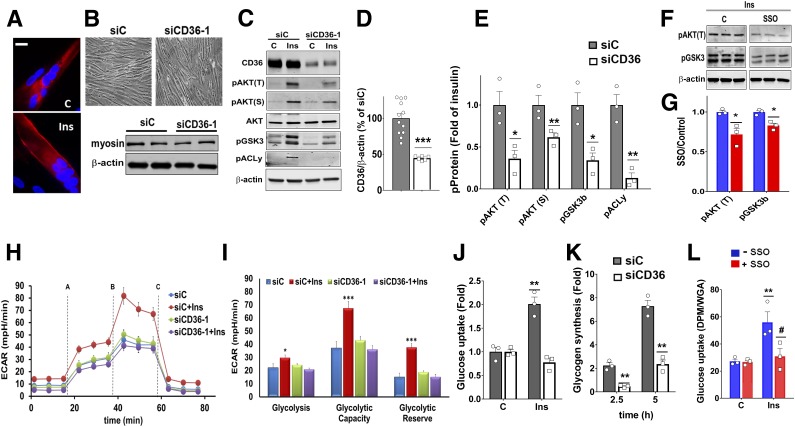
Cell-Autonomous Regulation by CD36 of Insulin Signaling and Glucose Metabolism
The findings with smCd36−/− mice suggested that Cd36 deletion suppresses insulin signaling. To determine whether this effect is cell autonomous, we examined whether it can be reproduced in primary-derived HSMMs. CD36 is highly expressed in these cells and relocates to the plasma membrane after insulin (Fig. 4A), as reported for muscle and cardiomyocytes (22,27). Treatment with siRNA did not alter myotube morphology or myosin heavy-chain content (Fig. 4B) but reduced CD36 level by 60% (Fig. 4C and D). Insulin addition to HSMMs induced robust phosphorylation of AKTT308, S473 and its targets GSK3S21/9 and ACLyS45. However, the insulin response was suppressed in CD36-depleted myotubes (Fig. 4C and E). A second siRNA yielded similar data (Supplementary Fig. 3A and B). In contrast, expression of CD36 in HEK293 cells enhanced insulin signaling compared with controls (Supplementary Fig. 3C and D), suggesting CD36 regulation is not limited to myotubes.
Figure 4.
CD36 modulates insulin signaling in HSMMs. A: Insulin (Ins) induces membrane CD36 translocation in differentiated HSMMs. Myotubes serum-starved for 16 h were incubated with or without insulin (100 nmol/L, 15 min) and processed for immunostaining: red, mouse monoclonal anti-CD36; blue, DAPI (nuclei). Images are representative of multiple fields from three experiments. C, control. Scale bar: 10 μm. B: Representative bright-field images of differentiated myotubes after treatment with CD36-targeted siRNA (siCD36-1) or with a nonspecific control siRNA (siC). Western blot shows expression of myosin heavy-chain and β-actin in siC and siCD36 myotubes, representative of two experiments. C: CD36 depletion decreases insulin (Ins) signaling in myotubes. Immunoblot shows insulin signaling in siC and siCD36 HSMM. Insulin (100 nmol/L) was added for 5 min. Blots were probed for CD36, pAKTT308, S473, total AKT, pGSK3 α/βS21/9 (pGSK3), pACLyS455, and β-actin. Blots are representative of four experiments. D: Quantification of CD36 knockdown normalized to β-actin. Shown are means ± SE of five experiments, as percentage of siC. ***P < 0.001. E: Quantification of insulin-induced pAKTT308, S473, pGSK3βS21/9 (bottom band), and pACLyS455. Shown are means ± SE of fold change compared with insulin-treated siC myotubes (n = 4 experiments). *P < 0.05, **P < 0.01. F: Acute CD36 inhibition by SSO suppresses insulin signaling: HSMMs were serum-starved and incubated with or without SSO (20 μmol/L, 15 min), followed by insulin as in C. Immunoblots of cell lysates (triplicates from different wells), representative of two experiments. G: Quantification of insulin-induced pAKTT308 and pGSK3βS21/9 with or without SSO. Shown are means ± SE from two experiments with triplicate samples. *P < 0.05 vs. controls (no SSO). H: Glycolysis was measured (Seahorse XF96 analyzer) from the ECAR in siCD36 or siC HSMMs. The following were added when indicated: (A) glucose ± insulin, (B) oligomycin, and (C) 2DG. Shown are means ± SE of 32 wells from one experiment representative of two more. I: Glycolytic measurements (means ± SE) from three experiments. *P < 0.05, ***P < 0.001 compared with respective unstimulated siC. J: CD36 depletion suppresses insulin-induced glucose uptake. siCD36 or siC HSMMs were incubated (60 min) with 0.1 mmol/L 2DG (1 μCi/mL) in PBS in the presence or absence of insulin. Disintegrations per minute (DPM) were normalized by WGA staining. Shown are means ± SE of four experiments. **P < 0.01 vs. untreated siC. K: CD36 depletion suppresses insulin-induced glycogen synthesis. siCD36 and siC HSMMs were incubated (2.5 or 5 h) with [3H]glucose (1 μCi/mL) in presence or absence of insulin. Insulin-induced fold change in radiolabel incorporation, normalized by WGA staining, is shown. Means ± SE of three experiments, three to five replicates per experiment. **P < 0.01 compared with siC. L: SSO (as in F) inhibits glucose uptake by HSMMs. Shown are means ± SE of three experiments. **P < 0.01 compared with untreated controls, #P < 0.05 compared with insulin-treated controls.
We next examined whether acute CD36 inhibition was sufficient to blunt insulin signaling. The FA analog sulfosuccinimidyl-oleate (SSO) is a widely used irreversible and specific inhibitor of CD36 (28,29). Pretreating myotubes for 15 min with SSO (20 μmol/L) effectively reduced insulin-induced pAKT and pGSK3 (Fig. 4F and G).
CD36 depletion in HSMMs did not alter basal glycolysis but diminished insulin-stimulated glycolytic activity, capacity, and reserve (Fig. 4H and I). Insulin-stimulated glucose uptake (Fig. 4J) and incorporation into glycogen (Fig. 4K) were suppressed. Glucose uptake was also reduced by 15-min SSO pretreatment (Fig. 4L) in line with the reduced insulin signaling (Fig. 4F and G). CD36 expression also enhanced insulin-stimulated glycolysis in CHO cells expressing the IR (CHO/IR/+CD36) (11), as shown in Supplementary Fig. 3E.
Saturated FA Suppress Insulin Signaling via CD36
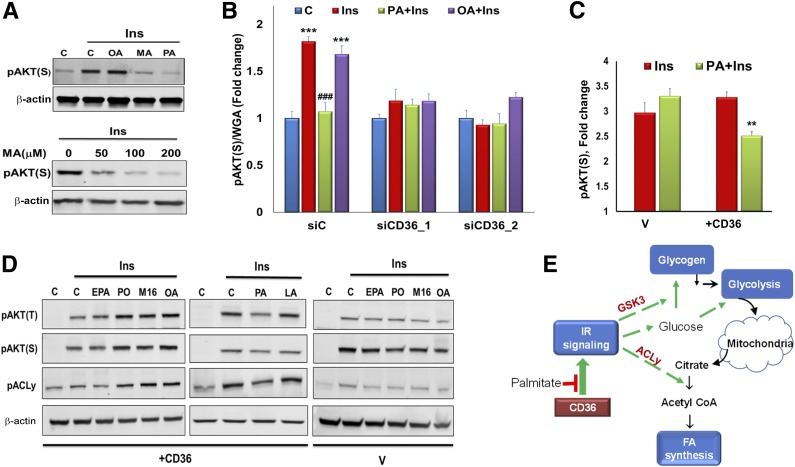
CD36 is a high-affinity receptor for saturated and unsaturated FA (30), so we examined whether it transduces FA effects on insulin signaling. HSMMs were pretreated (15 min) with the saturated FA palmitic acid (PA, 16:0) and myristic acid (MA, 14:0) or with monounsaturated oleic acid (OA, 18:1), complexed to BSA (2:1 molar ratio). Cells were then tested for insulin-stimulated (5 min) AKT phosphorylation. Interestingly, PA or MA suppressed insulin-induced pAKT but no suppression occurred with OA (Fig. 5A).
Figure 5.
CD36 mediates FA effects on insulin (Ins) signaling in myotubes. A: Saturated MA and PA but not monounsaturated OA suppress insulin signaling. Top: HSMMs were incubated 15 min with OA, PA, or MA (200 μmol/L at 2:1 BSA), followed by addition of insulin (100 nmol/L, 5 min). Cells were lysed, and pAKT(S) and β-actin were detected by immunoblotting. Data are representative of two experiments. Bottom: HSMMs were incubated with different MA concentrations, followed by insulin, lysed, and immunoblotted. Data are representative of two experiments. C, control. B: CD36 mediates suppression of insulin signaling by PA. ICW assay showing effect of CD36 depletion on insulin signaling with and without FA. HSMMs were incubated with OA or PA (2:1 BSA, 200 μmol/L, 15 min), followed by incubation with insulin (100 nmol/L, 5 min), fixation, and processing for ICW. pAKTS473 quantification normalized by WGA staining. Shown are means ± SE (n = 24 samples per condition, representative of three experiments). ***P < 0.001 vs. no insulin, ###P < 0.001 vs. insulin-treated siC. C: CD36/IR/+CD36 (+CD36) or vehicle (V) control CHO/IR cells were incubated with PA, followed by insulin, fixed, and processed for ICW. Graph shows pAKTS473 quantification normalized by WGA staining. Shown are means ± SE for n = 24 samples per condition, representative of two experiments. **P < 0.01 vs. insulin-stimulated +CD36 cells. D: Unsaturated FA do not suppress insulin signaling. Vector (V) or +CD36 CHO cells were treated with EPA, PO, M16, OA, PA, or LA, followed by insulin (100 nmol/L, 5 min). Cells were lysed and immunoblotted as indicated. Data are representative of two experiments. E: Schematic summary of key steps in regulation of glucose metabolism by insulin. Activation of IR leads to Akt phosphorylation and enhanced glucose uptake. Insulin induces phosphorylation of GSK3, which promotes glycogen synthesis. Glucose metabolized via glycolysis can be further oxidized in the citric acid cycle. Insulin signaling also promotes FA synthesis by enhancing phosphorylation of the key lipogenic enzyme ACLy, which converts citrate to cytosolic acetyl-CoA.
FA effects on insulin signaling were further validated using an in-cell Western (ICW) assay (Fig. 5B) sensitive for phosphorylated proteins (31). In control myotubes, insulin increased pAKT, and this was suppressed by pretreatment with PA but not OA. However, insulin stimulation and PA suppression were absent in CD36-depleted HSMMs, consistent with CD36 dependence of these effects (Fig. 5B). PA also reduced insulin-induced pAKT in CHO/IR/+CD36 cells but not in vector controls (Fig. 5C).
Additional FA types tested (Fig. 5D) included polyunsaturated FA (PUFA), eicosapentaenoic acid (EPA, 20:5), and linoleic acid (LA, 18:2), monounsaturated palmitoleic acid (PO, 16:1), a product of lipogenesis with positive effects on insulin responsiveness (32), and the nonmetabolized C16 FA analog β-β′-tetramethyl-hexadecanedioic acid (M16), an insulin sensitizer in rodents (33). In CHO/IR/+CD36 cells, 15-min pretreatment with EPA and LA did not alter insulin-stimulated phosphorylation of AKT or ACLy, whereas PO, OA, and M16 enhanced phosphorylation, and PA, as before, was inhibitory (Fig. 5D). These data show CD36 mediates differential effects of FA on insulin signaling, negative for PA and MA, and positive for PO, OA, and M16.
CD36 Enhances Tyrosine Phosphorylation of IR
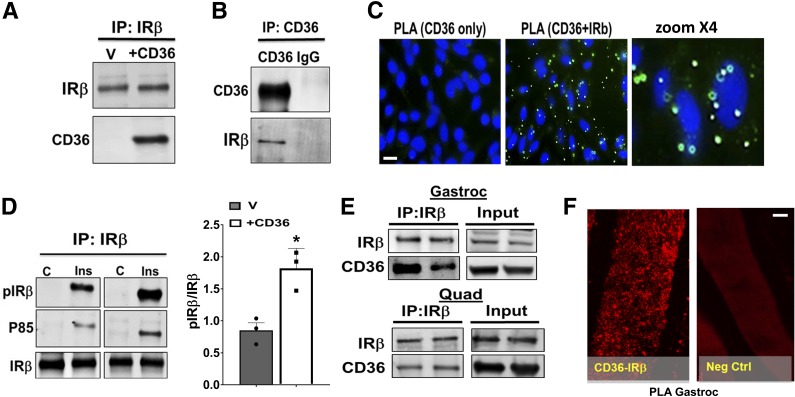
Insulin signaling is initiated by tyrosine phosphorylation of IR. CD36 signal transduction involves interaction with specific signaling protein clusters (5,8–10), so we explored whether CD36 might functionally interact with IR. Immunoprecipitates (IPs) of IRβ, the regulatory IR subunit, from CHO/IR/+CD36 cells pulled down CD36 (Fig. 6A), and conversely, CD36 IPs pulled down IRβ (Fig. 6B). Proximity ligation assays for in situ visualization of protein interaction (34) showed the typical amplification patterns suggestive of CD36-IRβ proximity (Fig. 6C).
Figure 6.
CD36 interacts with IRβ and enhances its tyrosine phosphorylation. A and B: CD36 and IRβ co-IP. A: CHO/IR/CD36 cells (+CD36) or empty vector controls (V) were lysed, and equal concentrations of lysate protein were used to IP IRβ. B: Reciprocal IP using anti-CD36 antibody. Data are representative of three experiments. C: CHO/IR/+CD36 cells were stained and processed for PLA. CD36-only staining (no anti-IRβ antibody) was used as the negative control. Nuclei were visualized by DAPI (blue). Data are representative of three experiments. Scale bar: 10 μm. D: CD36 expression facilitates IRβ tyrosine phosphorylation and P85 recruitment. Vector or +CD36 cells were incubated with or without insulin (100 nmol/L, 5 min) and lysed. Equal lysate protein was used to IP IRβ, and IPs were probed for pY100 (pIRβ), P85, and IRβ. Quantification of insulin-induced pIRβ/IRβ data are means ± SE from three experiments. *P < 0.05. E: Equal amounts of lysate protein (3.5 mg/sample) from gastrocnemius (Gastroc) and quadriceps (Quad) muscles were subjected to IRβ IP and resolved by SDS-PAGE. The IPs were immunoblotted for CD36 and IRβ. Data are representative of two experiments. F: Proximity ligation assay of gastrocnemius (Gastroc) paraffin-embedded sections stained with anti-CD36 and anti-IRβ antibodies. Combined CD36 and IRβ staining resulted in appearance of typical punctate patterns of amplified DNA. Nonspecific IgGs, isotype-matched to CD36 and IRβ antibodies, were used as negative controls. Images are representative of three mice. Scale bar: 35 μm.
CD36 expression enhanced insulin-induced tyrosine phosphorylation of IRβ and recruitment of the phosphatidylinositide 3-kinase (PI3K) catalytic subunit P85 (Fig. 6D), supporting functional relevance of the CD36-IR interaction. We also examined whether CD36 interacts with IRβ in vivo. IPs of IRβ from mice gastrocnemius and quadriceps muscles pulled down CD36, consistent with interaction (Fig. 6E), and further validation was obtained using PLA, which showed the amplification pattern expected for interacting proteins (Fig. 6F).
Fyn Recruited by CD36 Phosphorylates IR
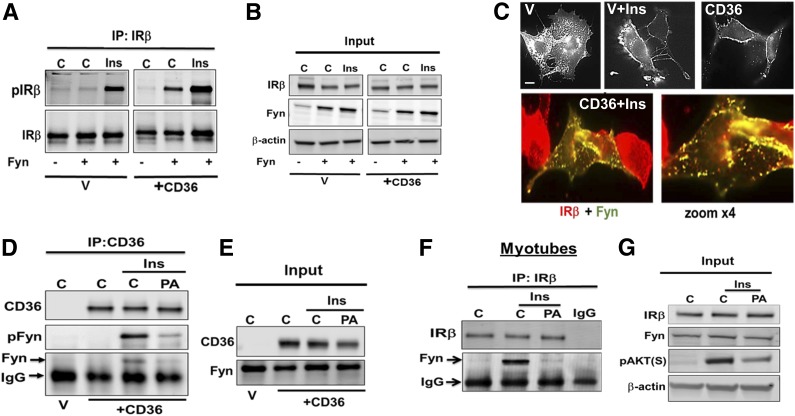
The mechanism for CD36 effect on IR phosphorylation was investigated next. Src tyrosine kinases mediate most CD36 signaling, and Fyn, in particular, was implicated in its metabolic effects (8,35). Insulin treatment of CHO/IR cells transiently transfected with Fyn resulted in recovery of more pIRβ (Fig. 7A). In contrast, IRβ phosphorylation was enhanced even without insulin when CHO/IR stably expressed CD36 (Fig. 7A), suggesting that some of the overexpressed Fyn was IRβ associated. In addition, more pIRβ was measured in these cells when insulin was added (Fig. 7A and B). These data suggested that Fyn phosphorylates IRβ and that CD36 recruits Fyn to the IR. Consistent with this, insulin treatment or CD36 expression each recruited Fyn to the plasma membrane (Fig. 7C), and cells expressing CD36 and Fyn showed extensive insulin-stimulated colocalization of Fyn and IRβ (Fig. 7C, bottom).
Figure 7.
CD36 facilitates Fyn recruitment to plasma membrane and interaction with IRβ. A and B: Combined Fyn and CD36 expression enhances insulin-stimulated IRβ phosphorylation. CHO/IR+CD36 or vector (V) control (C) cells were transiently transfected with or without Fyn. Cells were incubated with or without insulin (Ins; 100 nmol/L, 5 min), lysed, and equal cell lysate protein was used to IP IRβ. Data are representative of three experiments. C: Top panels: V or +CD36 CHO/IR cells transiently transfected with Fyn were incubated with or without insulin, fixed, and stained with anti-Fyn antibody. Bottom panel: +CD36 cells transiently transfected with Fyn were incubated with insulin, fixed, and imaged for Fyn and IRβ. Image shows overlay between IRβ (red) and Fyn (green). Image magnification shows distinct punctate yellow structures, suggesting IRβ-Fyn colocalization. Data are representative of two experiments. Scale bar: 10 μm. D and E: PA suppresses Fyn association with CD36 and IRβ. +CD36 or V CHO/IR cells were preincubated with PA (200 μmol/L, 2:1 BSA, 15 min), then with insulin (100 nmol/L, 5 min), and lysates were used to IP CD36. Resolved IPs and inputs were immunoblotted for CD36, Fyn, and pFynY416. F and G: Differentiated C2C12 myotubes, treated as in D, were processed for IP of IRβ, and IPs were probed for IRβ and Fyn. Data are representative of two experiments.
Saturated FA Dissociate Fyn From IR
We showed in Fig. 5 that saturated FA inhibit CD36 action to enhance insulin signaling, so we examined how PA affects Fyn phosphorylation of IRβ with or without insulin. CD36 IPs from CHO/IR/+CD36 cells treated with insulin contained pFynY416 (Fig. 7D), but pFynY416 was absent in IPs of cells pretreated (15 min) with PA. These data suggested insulin promotes interaction of activated Fyn with CD36, and this is disrupted by PA (Fig. 7D and E). Similarly, PA pretreatment abolished Fyn recovery in IPs of IRβ from C2C12 myotubes, which express high endogenous IRβ (Fig. 7F and G).
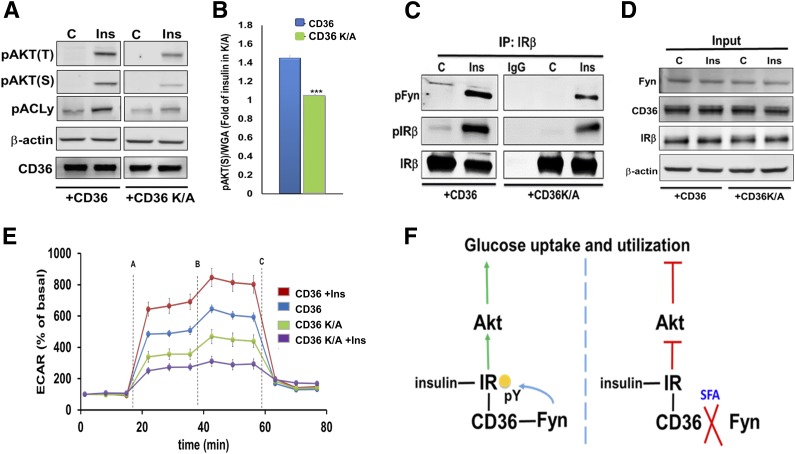
To further test Fyn’s mediation of CD36’s effect on insulin signaling, we used CHO/IR cells stably expressing the CD36 mutant CD36K/A (11,25). The COOH-terminal cytosolic segment of CD36 is required for Fyn-mediated effects, and this mutant has two COOH-terminal lysines substitution with alanine that impair CD36-induced Fyn signaling (11,25). Cells expressing CD36K/A had diminished insulin signaling compared with native CD36 (Fig. 8A and B) and reduced insulin’s ability to activate Fyn and phosphorylate IRβ (Fig. 8C and D). CD36K/A cells also did not show insulin stimulation of glycolysis (Fig. 8E). Together, the data in Figs. 7 and 8 suggest that insulin induces CD36-Fyn phosphorylation of IRβ, further stimulating glucose utilization. In contrast, PA dissociates the Fyn-CD36-IRβ complex, reducing IRβ phosphorylation and insulin-stimulated glucose metabolism (Fig. 8F).
Figure 8.
COOH-terminal CD36 mutation suppresses IRβ-Fyn interaction and blunts insulin signaling. A: CHO/IR/+CD36 and CHO/IR/+CD36K/A cells were incubated with insulin and cell lysates and immunoblotted for the indicated proteins. Data are representative of two experiments. B: CHO/IR/+CD36 and CHO/IR/+CD36K/A cells were incubated with or without insulin, fixed, and processed for ICW assay. Quantification of pAKTS473 normalized by WGA. Shown are means ± SE of 24 samples per condition, representative of three experiments. ***P < 0.001 vs. +CD36. C and D: +CD36 and +CD36K/A cells were incubated with or without insulin (100 nmol/L, 5 min), lysed, and lysates were used for IP of IRβ. IP (C) and cell lysates (D) were immunoblotted as indicated. Data are representative of two experiments. E: Insulin-induced glycolysis is reduced in the CD36K/A mutant. ECAR was measured in +CD36 and +CD36K/A cells. The following compounds were added when indicated: (A) glucose/glucose + insulin, (B) oligomycin, and (C) 2DG. Shown are means ± SE of 32 wells from one experiment representative of two more. F: Proposed mechanism for regulation of glucose metabolism by CD36 and FA. Left panel: Insulin stimulates IR signaling and Akt phosphorylation to drive glucose uptake and utilization. CD36 association with IR promotes Fyn recruitment to IR to increase its tyrosine phosphorylation (pY). This enhances IR signaling and muscle glucose utilization. Right panel: Saturated FA (SFA) trigger Fyn dissociation from CD36 and IR, reducing IR phosphorylation and signaling and diminishing glucose uptake. Thus, CD36 enhances glucose metabolism in skeletal muscle and its saturated FA sensing spares glucose, when FA and glucose are both available.
Low Muscle CD36 Expression Associates With Insulin Resistance and Type 2 Diabetes in Humans
To explore potential human relevance of current findings, we examined whether genetically determined CD36 expression associates with the incidence of insulin resistance or type 2 diabetes (T2D). We applied PrediXcan analysis to a “discovery” cohort of 4,702 patients from Vanderbilt University’s BioVU genomic resource. We found significant associations (P = 10−3 to 10−7) between the genetic component of muscle CD36 expression and disease status. Notably, decreased CD36 expression in muscle associated with increased risk of T2D (Supplementary Table 3).
A similar analysis used the genome-wide association study summary statistics data from the Meta-Analyses of Glucose and Insulin-related traits Consortium. Decreased CD36 expression was found to associate with higher HOMA-insulin resistance (HOMA-IR) or insulin resistance (P = 0.03) (Supplementary Fig. 4A). Furthermore, the single nucleotide polymorphism within the cis region of CD36 (±1 mega base pairs of transcription start site) showed a distribution of P values significantly different from that expected by chance (Supplementary Fig. 4A). This gene region, from which gene expression estimation was generated, contains the rs17236824 variant that was highly significant for association with HOMA-IR (Supplementary Fig. 4B). These data linked genetically reduced CD36 expression with incidence of insulin resistance or T2D.
Discussion
Major findings of this study are that the FA transporter CD36, which facilitates muscle FA uptake and oxidation, also enhances insulin action to stimulate muscle glucose utilization. CD36 functionally interacts with the IR to enhance its phosphorylation, P85 recruitment, and downstream signaling to promote glucose metabolism. These findings show that metabolic actions of CD36 are more complex than would be expected from its FA uptake function. We previously reported that CD36 signaling regulates oxidation of exogenous FA during fasting by modulating AMPK activation (8). Here we show that CD36 influences postprandial glucose metabolism by modulating insulin action. This dual role of CD36 signaling would influence energy adaptation and homeostasis.
The conditional deletion of CD36 in skeletal muscle showed that CD36 is required for optimal insulin stimulation of glucose metabolism. The systemic phenotype of the smCd36−/− was subtle, likely a limitation of the mosaic effect of the deletion. Still, this mouse provided opportunity to compare glucose metabolism in muscles with and without CD36 depletion from the same animal. We could document specific effect of Cd36 deletion to reduce ceramides and expression of genes of insulin and glucose metabolism. We also showed diminished glucose uptake by muscle in vivo and reduced insulin-stimulated glycolysis and glucose oxidation in muscle explants ex vivo. Together with the findings in myotubes and CHO/IR cells, these data provide strong support for physiological regulation of the IR pathway by CD36 signaling.
Sensitivity of the CD36-IR interaction to inhibition by saturated FA, especially PA, might be relevant to muscle glucose sparing, because PA, the main product of lipogenesis, is preferentially stored as TG by adipose tissue and released during fasting or exercise (36). These data suggest that PA might feedback in adipose tissue to limit lipogenesis by suppressing glucose uptake and that the data provide mechanistic insight into the glucose-FA cycle in muscle. This cycle initially proposed that FA metabolites inhibit muscle glucose utilization (37). Our data implicate membrane signaling as a major FA inhibition site, consistent with a primary role of membrane glucose transport (38,39). Saturated and unsaturated FA transduce markedly different CD36-mediated signaling (8,35), but the mechanisms for the distinctive effects are unclear. They might involve differential FA effects on CD36 localization in plasma membrane subdomains or restrictive properties of the lipid transport tunnel identified inside CD36 (40). CD36 regulation of IR (Fig. 8) and of AMPK (8) might explain CD36 influence on muscle fuel choice between FA and glucose. However, the mechanistic details of how this is accomplished, including downstream effects on transporter translocation, nutrient intracellular trafficking, and targeting to mitochondria (41–44), remain to be elucidated.
Our findings support a beneficial homeostatic role of muscle CD36. The associations identified between genetically determined low CD36 and insulin resistance (HOMA-IR) and T2D (Supplementary Table 3 and Supplementary Fig. 4) are consistent with this, although they are likely to reflect complex causality. Genetic CD36 variants are relatively common and often affect CD36 expression (45,46). These variants have been associated with risk of metabolic syndrome (47) and diabetes or insulin resistance (48,49). Dysfunctional interaction of CD36 with IR, Fyn, and P85 might contribute to its role in the etiology of metabolic disease.
Supplementary Material
Article Information
Acknowledgments. The authors thank Dr. Jacob Bar-Tana (Hebrew University, Jerusalem) for the M16 and Meghan Lam (Washington University in St. Louis) for technical support.
Funding. This study was supported by National Institute of Diabetes and Digestive and Kidney Diseases grants R01-DK-33301 and R01-DK-111175 and by pilot and feasibility awards to D.S. from P30-DK056341 (Nutrition Obesity Research Center), P30-DK020579 (Diabetes Research Center), and Longer Life Foundation (2017-007). This work was also supported by the Hope Center Alafi Neuroimaging Laboratory and National Institutes of Health Shared Instrumentation Grant (S10-RR-027552).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. D.S. designed experiments, obtained and analyzed data, and wrote the manuscript. P.D. conducted experiments and data analysis. T.P., M.J.-S., N.-H.S., C.R.F., and K.I.S. helped conduct the experiments and analyze data. E.P. performed animal breeding and genotyping. K.L.H. conducted histological analysis. I.J.G. reviewed the manuscript. E.R.G. analyzed human genetic data. N.A.A. designed experiments, analyzed data, and wrote the manuscript. D.S. and N.A.A. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db17-1226/-/DC1.
References
- 1.Coburn CT, Knapp FF Jr, Febbraio M, Beets AL, Silverstein RL, Abumrad NA. Defective uptake and utilization of long chain fatty acids in muscle and adipose tissues of CD36 knockout mice. J Biol Chem 2000;275:32523–32529 [DOI] [PubMed] [Google Scholar]
- 2.Glatz JF, Luiken JJ, Bonen A. Membrane fatty acid transporters as regulators of lipid metabolism: implications for metabolic disease. Physiol Rev 2010;90:367–417 [DOI] [PubMed] [Google Scholar]
- 3.Hames KC, Vella A, Kemp BJ, Jensen MD. Free fatty acid uptake in humans with CD36 deficiency. Diabetes 2014;63:3606–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka T, Nakata T, Oka T, et al. Defect in human myocardial long-chain fatty acid uptake is caused by FAT/CD36 mutations. J Lipid Res 2001;42:751–759 [PubMed] [Google Scholar]
- 5.Silverstein RL, Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci Signal 2009;2:re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kazerounian S, Duquette M, Reyes MA, et al. Priming of the vascular endothelial growth factor signaling pathway by thrombospondin-1, CD36, and spleen tyrosine kinase. Blood 2011;117:4658–4666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abumrad NA, Goldberg IJ. CD36 actions in the heart: lipids, calcium, inflammation, repair and more? Biochim Biophys Acta 2016;1861:1442–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samovski D, Sun J, Pietka T, et al. Regulation of AMPK activation by CD36 links fatty acid uptake to β-oxidation. Diabetes 2015;64:353–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laugerette F, Passilly-Degrace P, Patris B, et al. CD36 involvement in orosensory detection of dietary lipids, spontaneous fat preference, and digestive secretions. J Clin Invest 2005;115:3177–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Besnard P, Passilly-Degrace P, Khan NA. Taste of fat: a sixth taste modality? Physiol Rev 2016;96:151–176 [DOI] [PubMed] [Google Scholar]
- 11.Kuda O, Jenkins CM, Skinner JR, et al. CD36 protein is involved in store-operated calcium flux, phospholipase A2 activation, and production of prostaglandin E2. J Biol Chem 2011;286:17785–17795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abumrad NA, Davidson NO. Role of the gut in lipid homeostasis. Physiol Rev 2012;92:1061–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pietka TA, Sulkin MS, Kuda O, et al. CD36 protein influences myocardial Ca2+ homeostasis and phospholipid metabolism: conduction anomalies in CD36-deficient mice during fasting. J Biol Chem 2012;287:38901–38912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McFarlan JT, Yoshida Y, Jain SS, et al. In vivo, fatty acid translocase (CD36) critically regulates skeletal muscle fuel selection, exercise performance, and training-induced adaptation of fatty acid oxidation. J Biol Chem 2012;287:23502–23516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manio MC, Matsumura S, Masuda D, Inoue K. CD36 is essential for endurance improvement, changes in whole-body metabolism, and efficient PPAR-related transcriptional responses in the muscle with exercise training. Physiol Rep 2017;5:e13282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mariotti A, Kedeshian PA, Dans M, Curatola AM, Gagnoux-Palacios L, Giancotti FG. EGF-R signaling through Fyn kinase disrupts the function of integrin alpha6beta4 at hemidesmosomes: role in epithelial cell migration and carcinoma invasion. J Cell Biol 2001;155:447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cifarelli V, Ivanov S, Xie Y, et al. CD36 deficiency impairs the small intestinal barrier and induces subclinical inflammation in mice. Cell Mol Gastroenterol Hepatol 2017;3:82–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarthy JJ, Srikuea R, Kirby TJ, Peterson CA, Esser KA. Inducible Cre transgenic mouse strain for skeletal muscle-specific gene targeting. Skelet Muscle 2012;2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shintaku J, Guttridge DC. Analysis of aerobic respiration in intact skeletal muscle tissue by microplate-based respirometry. Methods Mol Biol 2016;1460:337–343 [DOI] [PubMed] [Google Scholar]
- 20.Kozma J, Bartók G, Szentirmai A. Fructose-2,6-bisphosphate level and beta-lactam formation in Penicillium chrysogenum. J Basic Microbiol 1993;33:27–34 [DOI] [PubMed] [Google Scholar]
- 21.Fleig WE, Nöther-Fleig G, Steudter S, Enderle D, Ditschuneit H. Regulation of insulin binding and glycogenesis by insulin and dexamethasone in cultured rat hepatocytes. Biochim Biophys Acta 1985;847:352–361 [DOI] [PubMed] [Google Scholar]
- 22.Samovski D, Su X, Xu Y, Abumrad NA, Stahl PD. Insulin and AMPK regulate FA translocase/CD36 plasma membrane recruitment in cardiomyocytes via Rab GAP AS160 and Rab8a Rab GTPase. J Lipid Res 2012;53:709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan M, Sidhu R, Fujiwara H, et al. Identification of Niemann-Pick C1 disease biomarkers through sphingolipid profiling. J Lipid Res 2013;54:2800–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burkart EM, Sambandam N, Han X, et al. Nuclear receptors PPARbeta/delta and PPARalpha direct distinct metabolic regulatory programs in the mouse heart. J Clin Invest 2007;117:3930–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sundaresan S, Shahid R, Riehl TE, et al. CD36-dependent signaling mediates fatty acid-induced gut release of secretin and cholecystokinin. FASEB J 2013;27:1191–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drover VA, Ajmal M, Nassir F, et al. CD36 deficiency impairs intestinal lipid secretion and clearance of chylomicrons from the blood. J Clin Invest 2005;115:1290–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luiken JJ, Dyck DJ, Han XX, et al. Insulin induces the translocation of the fatty acid transporter FAT/CD36 to the plasma membrane. Am J Physiol Endocrinol Metab 2002;282:E491–E495 [DOI] [PubMed] [Google Scholar]
- 28.Harmon CM, Abumrad NA. Binding of sulfosuccinimidyl fatty acids to adipocyte membrane proteins: isolation and amino-terminal sequence of an 88-kD protein implicated in transport of long-chain fatty acids. J Membr Biol 1993;133:43–49 [DOI] [PubMed] [Google Scholar]
- 29.Coort SL, Willems J, Coumans WA, et al. Sulfo-N-succinimidyl esters of long chain fatty acids specifically inhibit fatty acid translocase (FAT/CD36)-mediated cellular fatty acid uptake. Mol Cell Biochem 2002;239:213–219 [PubMed] [Google Scholar]
- 30.Pepino MY, Kuda O, Samovski D, Abumrad NA. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu Rev Nutr 2014;34:281–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aguilar HN, Zielnik B, Tracey CN, Mitchell BF. Quantification of rapid myosin regulatory light chain phosphorylation using high-throughput in-cell Western assays: comparison to Western immunoblots. PLoS One 2010;5:e9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mozaffarian D, Cao H, King IB, et al. Circulating palmitoleic acid and risk of metabolic abnormalities and new-onset diabetes. Am J Clin Nutr 2010;92:1350–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalderon B, Mayorek N, Ben-Yaacov L, Bar-Tana J. Adipose tissue sensitization to insulin induced by troglitazone and MEDICA 16 in obese Zucker rats in vivo. Am J Physiol Endocrinol Metab 2003;284:E795–E803 [DOI] [PubMed] [Google Scholar]
- 34.Söderberg O, Leuchowius KJ, Gullberg M, et al. Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods 2008;45:227–232 [DOI] [PubMed] [Google Scholar]
- 35.El-Yassimi A, Hichami A, Besnard P, Khan NA. Linoleic acid induces calcium signaling, Src kinase phosphorylation, and neurotransmitter release in mouse CD36-positive gustatory cells. J Biol Chem 2008;283:12949–12959 [DOI] [PubMed] [Google Scholar]
- 36.Mittendorfer B, Liem O, Patterson BW, Miles JM, Klein S. What does the measurement of whole-body fatty acid rate of appearance in plasma by using a fatty acid tracer really mean? Diabetes 2003;52:1641–1648 [DOI] [PubMed] [Google Scholar]
- 37.Randle PJ. Regulatory interactions between lipids and carbohydrates: the glucose fatty acid cycle after 35 years. Diabetes Metab Rev 1998;14:263–283 [DOI] [PubMed] [Google Scholar]
- 38.Hue L, Taegtmeyer H. The Randle cycle revisited: a new head for an old hat. Am J Physiol Endocrinol Metab 2009;297:E578–E591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roden M, Price TB, Perseghin G, et al. Mechanism of free fatty acid-induced insulin resistance in humans. J Clin Invest 1996;97:2859–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neculai D, Schwake M, Ravichandran M, et al. Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36. Nature 2013;504:172–176 [DOI] [PubMed] [Google Scholar]
- 41.Glatz JF, Luiken JJ. From fat to FAT (CD36/SR-B2): understanding the regulation of cellular fatty acid uptake. Biochimie 2017;136:21–26 [DOI] [PubMed] [Google Scholar]
- 42.Jaldin-Fincati JR, Pavarotti M, Frendo-Cumbo S, Bilan PJ, Klip A. Update on GLUT4 vesicle traffic: a cornerstone of insulin action. Trends Endocrinol Metab 2017;28:597–611 [DOI] [PubMed] [Google Scholar]
- 43.Campbell SE, Tandon NN, Woldegiorgis G, Luiken JJ, Glatz JF, Bonen A. A novel function for fatty acid translocase (FAT)/CD36: involvement in long chain fatty acid transfer into the mitochondria. J Biol Chem 2004;279:36235–36241 [DOI] [PubMed] [Google Scholar]
- 44.Yoshida Y, Jain SS, McFarlan JT, Snook LA, Chabowski A, Bonen A. Exercise- and training-induced upregulation of skeletal muscle fatty acid oxidation are not solely dependent on mitochondrial machinery and biogenesis. J Physiol 2013;591:4415–4426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghosh A, Murugesan G, Chen K, et al. Platelet CD36 surface expression levels affect functional responses to oxidized LDL and are associated with inheritance of specific genetic polymorphisms. Blood 2011;117:6355–6366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love-Gregory L, Kraja AT, Allum F, et al. Higher chylomicron remnants and LDL particle numbers associate with CD36 SNPs and DNA methylation sites that reduce CD36. J Lipid Res 2016;57:2176–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Avery CL, He Q, North KE, et al. A phenomics-based strategy identifies loci on APOC1, BRAP, and PLCG1 associated with metabolic syndrome phenotype domains. PLoS Genet 2011;7:e1002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corpeleijn E, van der Kallen CJ, Kruijshoop M, et al. Direct association of a promoter polymorphism in the CD36/FAT fatty acid transporter gene with type 2 diabetes mellitus and insulin resistance. Diabet Med 2006;23:907–911 [DOI] [PubMed] [Google Scholar]
- 49.Leprêtre F, Vasseur F, Vaxillaire M, et al. A CD36 nonsense mutation associated with insulin resistance and familial type 2 diabetes. Hum Mutat 2004;24:104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.