Abstract
In spite of tolerance mechanisms, some individuals develop T-cell–mediated autoimmunity. Posttranslational modifications that increase the affinity of epitope presentation and/or recognition represent one means through which self-tolerance mechanisms can be circumvented. We investigated T-cell recognition of peptides that correspond to modified β-cell antigens in subjects with type 1 diabetes. Modified peptides elicited enhanced proliferation by autoreactive T-cell clones. Endoplasmic reticulum (ER) stress in insulinoma cells increased cytosolic calcium and the activity of tissue transglutaminase 2 (tTG2). Furthermore, stressed human islets and insulinomas elicited effector responses from T cells specific for modified peptides, suggesting that ER stress–derived tTG2 activity generated deamidated neoepitopes that autoreactive T cells recognized. Patients with type 1 diabetes had large numbers of T cells specific for these epitopes in their peripheral blood. T cells with these specificities were also isolated from the pancreatic draining lymph nodes of cadaveric donors with established diabetes. Together, these results suggest that self-antigens are enzymatically modified in β-cells during ER stress, giving rise to modified epitopes that could serve to initiate autoimmunity or to further broaden the antigenic repertoire, activating potentially pathogenic CD4+ T cells that may not be effectively eliminated by negative selection.
Introduction
Negative selection of T cells is essential to generate an appropriately self-tolerant repertoire (1,2). HLA-peptide/T-cell receptor (TCR) affinity is a primary determinant for negative selection, designating T cells with inappropriate self-recognition for deletion and/or diversion to a regulatory lineage (3,4). Nevertheless, individuals with appropriate generic risk and environmental exposure can develop T-cell–mediated autoimmunity. Mounting evidence suggests that recognition of posttranslationally modified epitopes circumvents tolerance mechanisms. In rheumatoid arthritis, conversion of arginine to citrulline by peptidyl arginine deiminase (PAD) generates neoepitopes that are presented by disease-associated HLA-DR proteins (5). Antibody responses against citrulline are remarkably specific and are used as a clinical diagnostic marker (6). Likewise, in celiac disease, conversion of glutamine to glutamate by tissue transglutaminase 2 (tTG2) increases gliadin peptide presentation by disease-susceptible HLA-DQ proteins (7). In both contexts, T cells with high affinity for PTM epitopes represent a potentially vast pool that can initiate or exacerbate autoimmunity.
Destruction of pancreatic β-cells causes type 1 diabetes. Significant overlap between risk factors associated with diabetes and other autoimmune diseases implies shared etiology (8). In particular, risk is associated with susceptible HLA class II haplotypes, which are thought to select a potentially autoreactive T-cell repertoire. However, the events that initiate an immune attack remain unclear. The appearance of autoantibodies predicts disease onset, implying underlying CD4+ T-cell reactivity against β-cell proteins (9,10). The hierarchical emergence of autoantibodies can be postulated to suggest multiple waves of autoimmune damage (11,12).
Epitope spreading, whereby the number of antigenic targets and the diversity of epitopes within these targets increase, has been described in human disease and mouse models of autoimmunity (13,14). In type 1 diabetes, these processes create a feed-forward loop that induces increasing inflammation and new T-cell specificities (15). Posttranslational modification (PTM) may represent one mechanism by which epitope spreading occurs. Indeed, published studies have demonstrated increased immunogenicity of β-cell peptides following PTM (16–19) and the formation of neo-epitopes through peptide fusion (20) or defective ribosomal initiation (21).
Recent reports have described PTM epitopes from GAD65 and insulin in patients with type 1 diabetes (17,18). Likewise, recent work demonstrates that antigens, including tyrosine phosphatase–related islet antigen 2 (IA-2), are processed naturally and presented as deamidated peptides on dendritic cells (17). Furthermore, peptides from the N-terminal domain of IA-2 are recognized in the context of HLA-DQB1*03:02 (DQ8) and can be studied through the use of HLA class II tetramers (22). Here we use these tools to investigate altered recognition of peptides derived from β-cell autoantigens restricted by DQ8 and the significance of such responses in established disease. In particular, we address whether HLA binding and TCR recognition are modulated through enzymatic peptide modification. We further investigate mechanisms through which PTM epitopes naturally arise in β-cells. Finally, we investigate whether T cells that recognize modified peptides are detectable within peripheral blood and among pancreatic draining lymph node (PLN) T cells from subjects with diabetes. Our results demonstrate that PTM occurs in β-cells undergoing endoplasmic reticulum (ER) stress, that HLA binding and TCR recognition are independently modulated, that T cells specific for modified epitopes are present in the blood and PLN of subjects with type 1 diabetes, and that these T cells exhibit a T helper 1 (Th1)–like phenotype.
Research Design and Methods
Human Subjects
Peripheral blood was collected from individuals with type 1 diabetes and healthy control subjects with DQ8 haplotypes after written consent was obtained. The study was approved by the Benaroya Research Institute Institutional Review Board. Subject attributes are summarized in Supplementary Tables 1 and 2. Islets were isolated from three de-identified cadaveric donors (Supplementary Table 3) in the University of Pittsburgh Islet Isolation Core and University of Louisville Clinical Islet Cell Laboratory, as described elsewhere (23).
Peptides
Peptides (Mimotopes) representing modified β-cell antigens were chosen through the use of a matrix based on predicted DQ8 binding (Supplementary Table 4). Prediction coefficients were derived by combining the results of Kwok et al. (24) and Chang and Unanue (25).
HLA Class II Protein and Tetramers
DQ8 proteins were purified from insect cell cultures as previously described (26,27). Monomers were loaded with 0.2 mg/mL peptide and incubated in the presence of 0.2 mg/mL n-dodecyl-β-maltoside and 1 mmol/L Pefabloc (Sigma-Aldrich) at 37°C for 72 h. Monomers were conjugated into tetramers using Invitrogen Streptavidin R PE (Thermo Fisher Scientific) at a molar ratio of 8:1.
Peptide Binding Competition
Peptide binding was measured by incubating increasing concentrations in competition with 0.02 μmol/L biotinylated GAD65253–265 (255F) in wells coated with DQ8 protein. After the wells were washed, residual biotin-GAD65253–265 was detected using europium-conjugated streptavidin (Perkin Elmer) and quantified using a Victor2D fluorometer (Perkin Elmer). Curves were simulated using Prism software (version 5.03; GraphPad Software Inc.) and the IC50 was calculated as the concentration required to displace 50% of the reference peptide.
In Vitro T-Cell Assays and Clone Isolation
Peripheral blood mononuclear cells (PBMCs) were isolated through the use of a Ficoll underlay, resuspended in medium (RPMI medium, 10% pooled human serum, 1% penicillin-streptomycin, 1% l-glutamine) at 4 × 106 cells/mL, and stimulated with peptides (20 mg/mL total) in 48-well plates for 14 days. Medium and interleukin (IL)-2 were added starting on day 7. Cells were stained with tetramers at 37°C for 75 min, followed by peridinin chlorophyll (PerCP) Mouse Anti-Human CD4 (BD Biosciences), CD3 allophycocyanin (APC) (eBioscience), and FITC Anti-Human CD25 Antibody (BioLegend) at 4°C for 15 min; run on a FACSCalibur cell analyzer (BD Biosciences); and analyzed using FlowJo software (Treestar Inc.). Clones were isolated by sorting single T cells that were tetramer-positive using a FACSAria cell sorter (BD Biosciences) and expanding in 96-well plates in the presence of 1 × 105 irradiated PBMCs and 2 µg/mL phytohemagglutinin (PHA; Remel Inc.). Media and IL-2 were added starting on day 10.
T-Cell Clone Maintenance and Characterization
Clones specific for modified peptides were maintained in supplemented RPMI medium and restimulated using either PHA (2 mg/mL; Remel Inc.) or concanavalin A (2.5 μmol/L; Sigma-Aldrich) every 2 weeks. To assess preferential recognition of modified peptides, 104 cells/well were plated with 1 × 105 irradiated DQ8+ PBMCs and stimulated in triplicate with 10 μg/mL peptide. After incubating at 37°C for 48 h and pulsing with 3[H]-thymidine (1 µCi/well), incorporation was measured 18 h later with a scintillation counter. To assess function, clones were activated with 50 ng/mL phorbol myristic acid and 1 µg/mL ionomycin for 30 min, followed by incubation with brefeldin A (eBioscience) at 37°C for 3 h. Cells were fixed in a fixation/permeabilization buffer (eBioscience); washed in permeabilization buffer; stained with IL-4 Alexa Fluor (AF) 488 (eBioscience), interferon (IFN)-γ AF700 (eBioscience), IL-10 BV421, IL-17A APC/cyanine (Cy) 7, and tumor necrosis factor-α PerCP-Cy5.5 (all BioLegend) at 4°C for 15 min; run on an LSR II flow cytometer (BD Biosciences); and analyzed using FlowJo software (Treestar Inc.). To assess cytokine secretion, clones were plated at 7.5 × 104 cells/well with 1.5 × 105 irradiated DQ8+ PBMCs and stimulated in triplicate wells with 10 μg/mL peptide. After incubation at 37°C for 48 h, supernatants were collected and IFN-γ, IL-4, and IL-10 concentrations were determined using Ready-SET-Go! ELISA kits (eBioscience).
Ex Vivo Tetramer Analysis
T-cell frequencies were assessed using our published approach (28). Briefly, 20–30 × 106 PBMCs were resuspended in 200 µL media, incubated with 50 nmol/L dasatinib at 37°C for 10 min, and stained with 20 μg/mL phycoerythrin-labeled tetramer at room temperature for 120 min. Cells were washed, incubated with anti-phycoerythrin magnetic beads (Miltenyi) at 4°C for 20 min, and enriched. A proportion (1%) of the cells was retained as a nonenriched sample. Enriched and nonenriched (precolumn) samples were stained with CD4 V500, CD14 PerCP-Cy5.5, and CD19 PerCP-Cy5.5 (eBioscience); CD45RA AF700 (BD Biosciences); and CXCR3 FITC, CCR6 BV421, and CCR4 BV605 (BioLegend) at 4°C for 15 min. After washing, cells were labeled with ViaProbe (BD Biosciences) and analyzed on a FACSCanto cell analyzer (BD Biosciences), gating on CD4+CD14−CD19−ViaProbe− cells. Frequencies were calculated as previously described (28).
Human Islet and βlox5 Cell Culture and Stress Induction
Primary human islets were dissociated into single-cell suspensions and then incubated in supplemented RPMI medium containing 5 μmol/L thapsigargin (Thaps; Sigma-Aldrich) or control at 37°C for 1 h. Cells were washed with the equivalent of 50,000 times the original buffer volume to remove residual Thaps. The human insulinoma βLox5 cell line (provided by Clayton Mathews, University of Florida) was maintained in an incubator, at 37°C in 5% CO2 humidified air, in DMEM (Invitrogen) supplemented with 10% heat-inactivated FBS (Mediatech, Inc.), 4 mmol/L l-glutamine (Gibco), 1 mmol/L nonessential amino acids (Gibco), 15 mmol/L HEPES (Gibco), 1 mmol/L sodium pyruvate (Gibco), and 100 units/mL penicillin/streptomycin (Gibco). βLox5 cells (passages 15–40) were cultured in 25-cm2 flasks (Greiner Bio-One) in supplemented DMEM containing 5 μmol/L Thaps or control at 37°C for 1 h. Cells were washed with the equivalent of 50,000 times the original buffer volume to remove residual Thaps and liberated with 0.05% trypsin-EDTA (Gibco).
Cell Lysate Preparation
Cells were lysed by sonication in 50 mmol/L Tris (pH 8.0), 137 mmol/L NaCl, 10% glycerol, 1% NP-40, 1 mmol/L NaF, 10 µg/mL leupeptin, 10 µg/mL aprotinin, 2 mmol/L Na3VO4, and 1 mmol/L phenylmethylsulfonyl fluoride. Protein concentration was determined through the use of a bicinchoninic acid protein assay (Thermo Fisher Scientific).
Western Blotting
Lysates were separated by SDS-PAGE with 10% polyacrylamide gels and transferred to polyvinylidene fluoride membranes. For ER stress markers, membranes were blocked in 5% BSA in Tris-buffered saline with Tween (TBST) for 1 h, then probed with anti–phosphorylated eIF2α (1:1,000; Cell Signaling Technology), anti–phosphorylated inositol-requiring enzyme 1 (IRE-1) (1:1,000; Abcam), anti-eIF2α (1:1,000; Cell Signaling Technology), or anti-IRE-1 (1:1,000; Cell Signaling Technology) overnight at 4°C. The membrane was then incubated with horseradish peroxidase (HRP)-conjugated goat antirabbit antibody (1:10,000; Jackson ImmunoResearch Laboratories) for 1 h. To detect peptidylarginine deiminase 2 (PAD2), membranes were blocked in 5% BSA in TBST for 1 h, then probed with anti-PAD2 (1:1,000; ProteinTech) at 4°C overnight. This was followed by HRP-conjugated goat antirabbit antibody (1:10,000; Jackson ImmunoResearch Laboratories) for 1 h. To detect citrulline, membranes were blocked in 5% milk in TBST for 1 h and probed with anticitrulline (1:1,000; Millipore-Sigma) at 4°C overnight, then probed by biotin-conjugated goat antirabbit antibody (1:5,000; Millipore) for 1 h and streptavidin-HRP (1:2,000; Invitrogen) for 1 h. Actin was detected with antiactin (1:10,000; Sigma-Aldrich) and goat antimouse antibody (1:10,000; Jackson ImmunoResearch Laboratories) for 1 h. Chemiluminescence was detected with Luminata Crescendo Western HRP Substrate (Millipore) and analyzed with a Fujifilm LAS-4000 imager and Multi Gauge software (Fujifilm Life Science).
Quantitative RT-PCR
mRNA was isolated with an RNeasy Kit (Qiagen), and cDNA was synthesized with an RT2 First Strand Kit (Qiagen). cDNA was quantified by quantitative RT-PCR (iCycler; Bio-Rad) with iQ SYBR Green Supermix (Bio-Rad). Cycling parameters were 95°C for 10 min then 40 cycles at 95°C for 10 s and 55°C for 30 s, using previously published primers for spliced and total X-box binding protein 1 (XBP-1) (29) and RPLO (rplo) (30). Data were normalized using the ΔΔCt method, where the amount of target, normalized to an endogenous reference and relative to a calibrator, is given by 2−ΔΔCt (where Ct is the cycle number of the detection threshold).
Ca2+ Tracing
βLox5 cells were seeded in glass-bottom plates and left overnight. Adherent cells were labeled with 1 μmol/L Fluo-4 (Invitrogen) at 37°C for 1 h then washed, and the intensity of Fluo-4 at 488 nm was monitored with live imaging at room temperature for 600 s using a 40× objective lens on an Olympus FluoView FV1000 microscope. At 100 s, cells were exposed to 5 μmol/L Thaps or control in supplemented DMEM. Data were analyzed with FV10-ASW imaging software.
tTG2 Activity Assay
Activity of tTG2 was measured by incorporating biotinylated pentylamine into endogenous proteins (31). Biotin levels in all cells pre-incubated with pentylamine-biotin were normalized by subtracting the absorbance of cells not incubated with pentylamine-biotin (representing endogenous biotin levels). Data are reported as the fold change in tTG2 activity compared with cells incubated with pentylamine-biotin alone.
T-Cell Assays With Islets and βlox5 Cells
WT51 cells (provided by Massimo Trucco, Allegheny-Singer Research Institute) were maintained in an incubator (37°C in 5% CO2 humidified air) in RMPI medium (Invitrogen) supplemented with 10% FBS (Mediatech, Inc.), 2 mmol/L l-glutamine (Gibco), and 100 units/mL penicillin/streptomycin (Gibco). T cells (2 × 104), WT51 cells (irradiated at 1,000 rad) as APCs (2 × 105), and β-cells (human islets, 1 × 104; βLox5 cells, 5 × 103) were combined in 200 µL supplemented RMPI medium in triplicate in 96-well round-bottom culture plates (Greiner Bio-One) and incubated at 37°C for 72 h. As experimental controls, T cells or WT51 cells were omitted. IFN-γ secretion was measured with a Human IFN-gamma DuoSet ELISA Kit (R&D Systems).
PLN-Derived T-Cell Assays
PLN were recovered from two donors (Supplementary Table 3). Tissues were collected through the National Disease Research Interchange and Vanderbilt University. Reactivity was tested in two ways. First, to generate lines, whole PLN cells were expanded in culture with peptides for two rounds of stimulation. The medium was HL-1 medium supplemented with 2 mmol/L l-glutamine, 5 mmol/L HEPES, 100 units/mL penicillin, and 100 μg/mL streptomycin, 0.1 mmol/L of each nonessential amino acid, 1 mmol/L sodium pyruvate (all from Lonza); 5% heat-inactivated human male AB serum (Omega Scientific); a blocking anti-Fas antibody (1 µg/mL; eBiosciences); anti-PD-1 antibody (1 µg/mL; BD Biosciences);and mifepristone (100 nmol/L; Invitrogen). Responsiveness was assessed as previously described (20). Briefly, expanded T cells were incubated (2.5 × 104 cells/well) with peptide (50 µg/mL) and autologous Epstein-Barr virus (EBV)–transformed B cells (10 × 104 cells/well) for 48 h. After 48 h of coculture, responses were detected by IFN-γ ELISA. Alternatively, lines were stained with antibodies to CD4, CD25, HLA-DR, and CD49d (all from BD Biosciences) as activation markers. Cells were detected using an LSR II flow cytometer (BD Biosciences) and analyzed with FlowJo software. Second, 30 amplified T-cell libraries (32) were generated with 4,000 PLN cells/well with 4.5 µg/mL PHA-P (Thermo Fisher Scientific), irradiated allogeneic feeders (1.5 × 105 cells/well), 40 units/mL IL-2 (Proleukin), IL-4, and IL-7 (all at 10 ng/mL) (PeproTech). After expansion, amplified lines were stimulated with peptides (50 µg/mL) and an irradiated, peptide-pulsed, autologous, splenic EBV-transformed B-cell line and cytokines, as described above. After 48 h of coculture, cells were stained and analyzed as described above.
Statistics
The Fisher exact test was used to compare T-cell responses between patients and control subjects. The Student t test was used to compare XBP-1 splicing, Ca2+ flux, tTG2 activity, the immunogenicity of control β-cells compared with Thaps-treated β-cells, and T-cell frequencies. Various levels of statistical significance are indicated as such: *P < 0.05; **P < 0.01; ***P < 0.001.
Results
Identification of Modified Peptides Presented by HLA-DQ8
Previous studies described preferential binding of negatively charged peptides by DQ8 (24,33) and identified deamidated peptides that CD4+ T cells recognize in vitro (22). Recent studies also documented the capacity for HLA-DQ proteins to present citrullinated peptides (34) and showed greater immune responses to these peptides (19). We used substituted peptides and a competition binding assay to confirm the presentation of deamidated and citrullinated peptides by DQ8 (Supplementary Fig. 1). Consistent with previous studies, specific glutamate substitutions and citrulline substitutions favored binding at certain positions. On the basis of these observations and previously characterized preferences (24,25), we predicted 129 citrullinated or deamidated motifs within β-cell antigens (Supplementary Table 5). Adding two to seven flanking residues to each side of each motif, we synthesized 15–20mer peptides and verified that 35 peptides bind DQ8 with appreciable affinity (Supplementary Table 6).
Preferential Recognition of Modified β-Cell Peptides
We then investigated the ability of these peptides to elicit CD4+ T-cell responses. PBMCs from subjects with type 1 diabetes were stimulated with peptides for 2 weeks and stained with tetramers. Five peptides exhibited high-affinity binding to DQ8 (Supplementary Fig. 1) and elicited positive responses in subjects with type 1 diabetes that were larger than those in HLA-matched control subjects (Supplementary Fig. 2). These epitopes are summarized in Table 1. Two peptides coincided with candidates reported by van Lummel et al. (17). However, their study did not determine whether these peptides are immunogenic. T-cell clones for each epitope were isolated from at least two different subjects. In total, we isolated 17 IA-2198–216–specific, 8 IA-2467–482–specific, 15 IA-2523–536–specific, 9 IA-2545–562–specific, and 10 islet amyloid polypeptide (IAPP)65–84–specific clones.
Table 1.
Sequences and binding affinities for antigenic modified IA-2 and IAPP peptides
| Peptide | Modification | Amino acid sequencea | Modified IC50 (µmol/L) | Wild-type IC50 (µmol/L) |
|---|---|---|---|---|
| IA-2198–216 | 207E, 213E | SLSYEPALLEPYLFHEFGS | 3.0 | n.b. |
| IA-2467–482 | 478E | AAEEYGYIVTDEKPLSb | 6.8 | n.b. |
| IA-2523–536 | 532E, 533E | QNLSLADVTEEAGLb | 2.8 | n.b. |
| IA-2545–562 | 548E, 551E, 556E | TGLEILETGVGEREEAAA | 16.8 | 21.4 |
| IAPP65–84 | 73Cit, 81Cit | VGSNTYGKXNAVEVLKXEPL | 23.1 | n.b. |
IC50, peptide concentration that displaces half of the reference peptide; n.b., nonbinding (>50 μmol/L in our assay).
aIn each sequence, the residue that modulates recognition when modified is boldface, and the top predicted minimal epitope is underlined. The Xs indicate citrulline.
bReported as a candidate HLA-DQ8 epitope by van Lummel et al. (17).
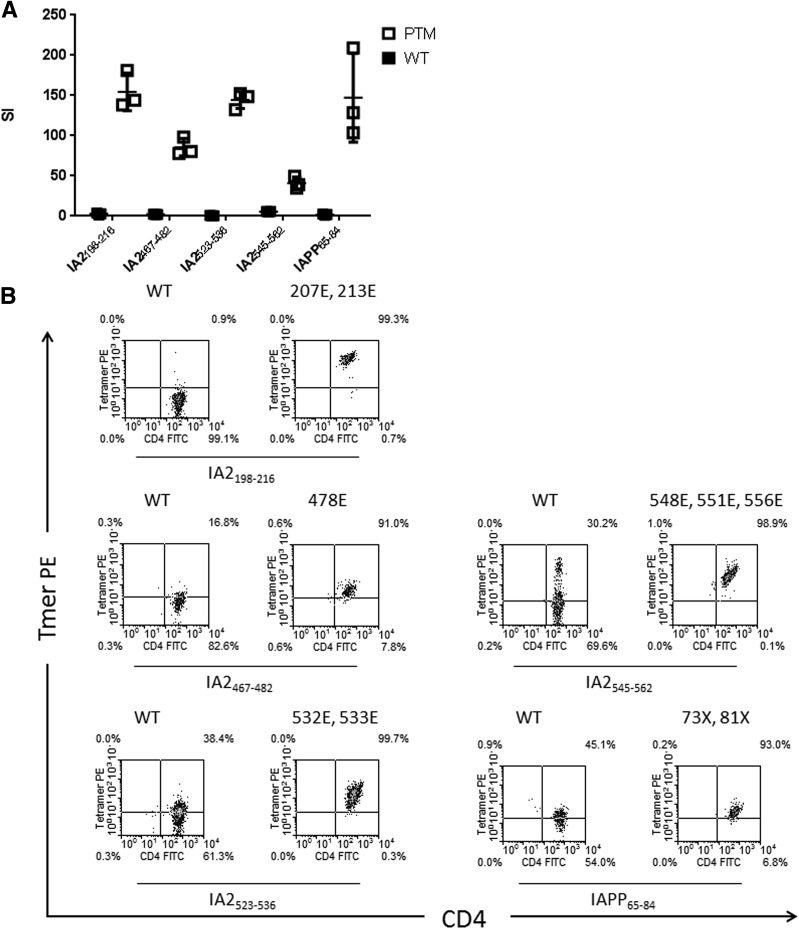
We next examined whether T cells respond to both unmodified and modified peptide, as observed by van Lummel et al. (17) for the insulin B30-13C epitope. Each T-cell clone exhibited proliferation above background only in response to the modified peptide (Fig. 1A). We then used HLA-DQ8 tetramers loaded with either unmodified or modified peptide to stain T-cell clones (Fig. 1B). Four unmodified peptides (IA-2198–216, IA-2467–482, IA-2523–536, and IAPP65–84) had no detectable binding to DQ8 (Table 1) and were not expected to generate usable tetramers. Consistent with this expectation, tetramers loaded with unmodified IA-2198–216 and IA-2467–482 could not stain T-cell clones. They were, however, effectively stained by tetramers loaded with deamidated IA-2198–216 and IA-2467–482. It is surprising that clones specific for IA-2523–536 and IAPP65–84 were weakly stained by tetramer loaded with unmodified peptide, implying that exogenous loading was successful despite a lack of high-affinity binding. However, IA-2523–536– and IAPP65–84-specific T-cell clones were stained more intensely by tetramers loaded with modified peptides. Wild-type and deamidated IA-2545–562 both had detectable binding to DQ8. Clones specific for this peptide were weakly stained by tetramer loaded with the unmodified peptide, implying that this TCR has some capacity to interact with wild-type peptide.
Figure 1.
Preferential recognition of posttranslationally modified peptides by T-cell clones from subjects with diabetes. A: Preferential recognition was confirmed by measuring the proliferative response of T-cell clones following stimulation with either modified peptide (white squares) or the corresponding unmodified peptide (black squares), as determined by [3H] thymidine incorporation. Data are represented as stimulation index (SI) values, calculated in triplicate by normalizing the proliferation of each clone based on [3H] thymidine incorporation in unstimulated wells. Horizontal lines represent means and error bars indicate SD. Each clone exhibited negligible proliferation (SI <3) in response to unmodified peptide and robust proliferation (SI >30) in response to modified peptide. B: T-cell clones specific for each modified epitope were also stained using tetramers loaded with modified peptide (each “E” indicates a glutamic acid modification at the indicated amino acid position and each “X” indicates a citrulline modification at the indicated amino acid position) or the corresponding unmodified version (wild-type peptide [WT]), and the mean fluorescence intensities of the staining were compared. Each clone was preferentially stained by modified peptide tetramers. Results shown are representative of T-cell clones isolated from multiple subjects with type 1 diabetes (n ≥ 2). FITC, fluorescein isothiocyanate; PE, phycoerythrin; Tmer, tetramer.
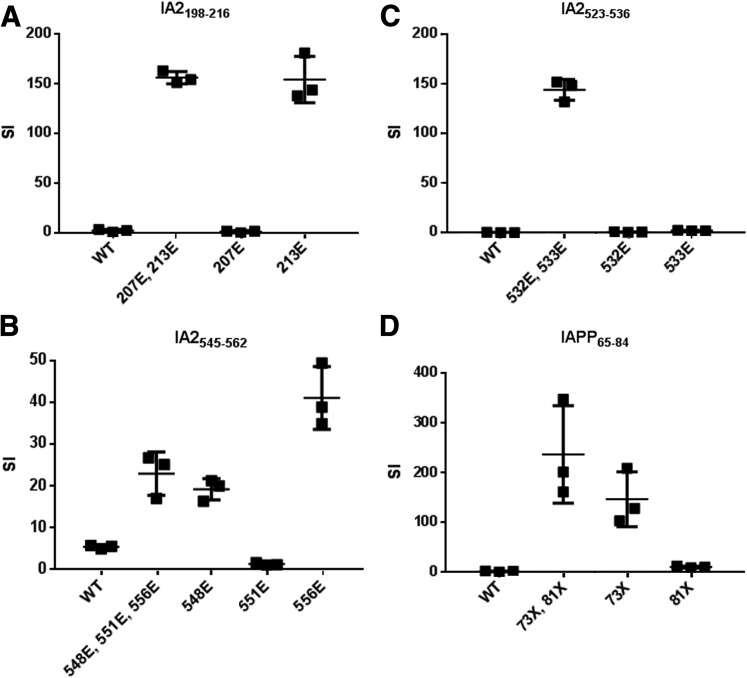
Three IA-2 peptides and IAPP contained multiple substitutions. To assess which modifications were necessary for recognition, peptides containing multiple, single, or no substitutions were used in binding and proliferation assays (Fig. 2 and Supplementary Table 6). For IA-2198–216, conversion of residue 213 to glutamate was required for binding but modification of residue 207 was dispensable. Accordingly, the IA-2213E peptide yielded maximal proliferation (Fig. 2A). For IA-2523–536, modification of residue 533 was required for binding, but a glutamate at residue 532 was also required for recognition (Fig. 2B). For IA-2545–562, the single substituted IA-2548E and IA-2556E peptides bound with affinity similar to that of IA-2548E, IA-2551E, and IA-2556E, but IA-2556E elicited the most proliferation (Fig. 2C), suggesting that modification of residues 548 and 551 is dispensable. For IAPP65–84, the single substituted IAPP73× peptide was able to bind and stimulated cells to proliferate (Fig. 2D), but to a lesser extent than the double-substituted peptide. In summary, these peptides include distinct residues required for DQ8 binding (e.g., IA-2213E and IA-2533), required for TCR recognition (e.g., IA-2532), and for which modification is dispensable.
Figure 2.
Single residues modulate recognition of posttranslationally modified peptides by T cells. A: For IA-2198–216, peptides that lacked a glutamate at residue 213 (wild type [WT] and 207E) could not elicit proliferation of the T-cell clone, indicating that this modification is required for recognition. B: For IA-2545–562, a peptide with only a glutamate at residue 556 elicited optimal proliferation of the T-cell clone, indicating that modification of residues 548 and 551 are not required for recognition. C: For IA-2523–536, peptides that lacked a glutamate at either residue 532 or residue 533 (WT, 532E, and 533E) could not elicit proliferation of the T-cell clone, indicating that both modifications are required for T-cell recognition. D: For IAPP65–84, peptides that lacked a citrulline at residue 73 (WT and 81X) could not elicit proliferation of the T-cell clone, indicating that this modification is required for recognition. The results shown are stimulation index (SI) values, calculated in triplicate by normalizing the proliferation of each representative clone based on [3H] thymidine incorporation in unstimulated wells. Horizontal lines show means and error bars indicate SDs. Results are representative of T-cell clones isolated from multiple subjects with type 1 diabetes (n ≥ 3). In panels A–C, each “E” indicates a glutamic acid modification at the indicated amino acid position. In panel D, each “X” indicates a citrulline modification at the indicated amino acid position.
CD4+ T Cells Specific for Modified Peptides Are Th1-Like
We investigated the functional phenotype of T-cell clones using ELISA assays to assess IL-4, IL-10, and IFN-γ secretion following stimulation with peptide. Only T cells stimulated with modified peptide produced appreciable amounts of cytokine (Supplementary Fig. 3). Some IL-4 and IL-10 were observed, but IFN-γ dominated the response. These data suggest a Th1-like phenotype, in agreement with previous observations (17,35).
β-Cell Stress Causes Recognition By PTM-Reactive T Cells
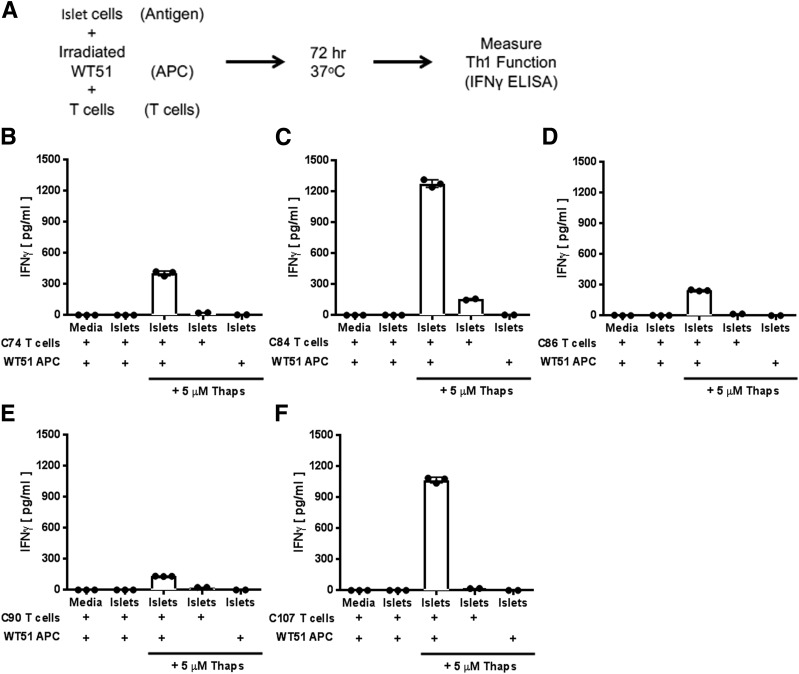
We next sought to determine whether PTM epitopes naturally arise in β-cells. We previously demonstrated that ER stress, to which β-cells are susceptible (36–41) and which is associated with β-cell loss in NOD mice and patients (42,43), increases tTG2 activity and recognition of murine islets and insulinomas by diabetogenic T cells (44). To determine whether ER stress leads to PTM in human cells, we used Thaps to induce ER stress in human islets. Islet cells from three individual donors (Supplementary Table 3) were incubated with Thaps for 1 h, and their immunogenicity was determined through T-cell assays. Briefly, islet cells (treated with Thaps or the control), irradiated WT51 cells as APCs, and T-cell clones were combined and incubated for 72 h (Fig. 3A). Each T-cell clone secreted large amounts of IFN-γ in response to Thaps-treated islet cells (Fig. 3B–F and Supplementary Table 7). Control-treated islet cells did not elicit IFN-γ, indicating that, as shown in previous murine studies (44), ER stress is required for modification and the release of antigens. It is important to note that Thaps-treated islet cells could not elicit IFN-γ secretion without antigen presentation by WT51 cells.
Figure 3.
Clones specific for modified antigens respond to primary human islet cells stressed by Thaps. A: Schematic representation of our experimental strategy for assessing T-cell responses to islet cells stressed by Thaps. Irradiated WT1 cells (DQ8+ APCs) were mixed with islet cells (as an antigen source) and cocultured with T-cell clones that recognize modified IA-2198–216 (B), IA-2467–482 (C), IA-2523–536 (D), IA-2545–562 (E), or IAPP65–84 (F), or omitted either Thaps or one cell type (as indicated) as experimental controls. Thaps-stressed islet cells elicited robust secretion of IFN-γ by T cells that was significantly higher than levels elicited by unstressed islet cells or medium, indicating that antigens generated by stressed islet cells can efficiently activate T cells that preferentially respond to modified peptides. Islets from three patients were tested, and data are the mean ± SD of triplicate measurements from one representative patient sample.
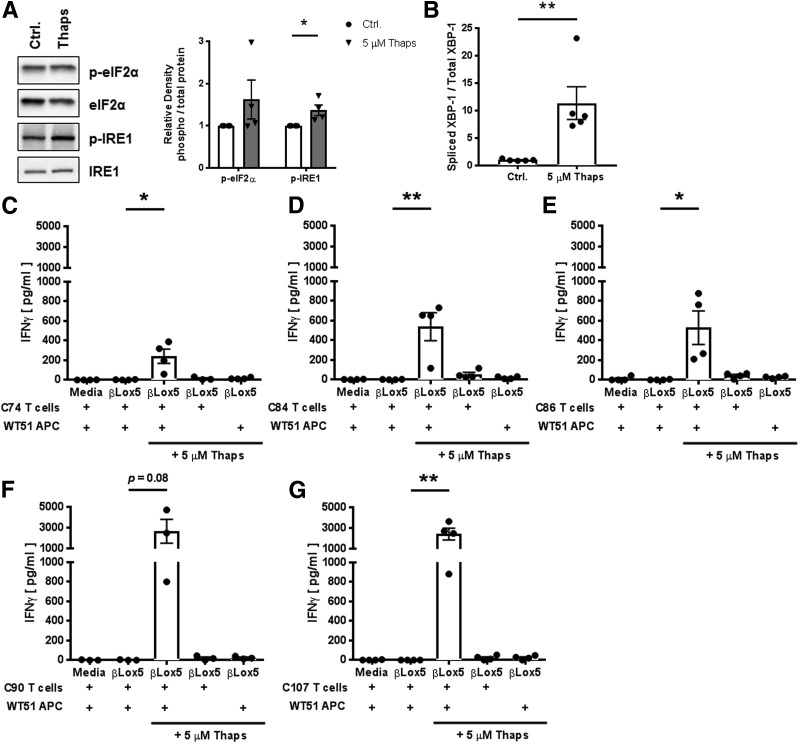
Primary human islet cells are an ideal cell type for use in examining the mechanisms of β-cell immunogenicity. However, it is difficult to obtain sufficient cell numbers to conduct biochemical assays. Therefore, to elucidate the mechanism by which ER stress increases immunogenicity, we used βLox5 insulinoma cells, a cell line derived from adult pancreatic islets (45). To confirm that βLox5 cells could be used to model primary human islet immunogenicity, βLox5 cells were incubated with Thaps for 1 h, and ER stress was measured by assessing the phosphorylation of unfolded protein response proteins eIF2α and IRE-1 by Western blotting, and the splicing of XBP-1 mRNA by quantitative PCR. Incubation with Thaps increased phosphorylation of eIF2α (1.6-fold) and IRE-1 (1.4-fold) (Fig. 4A), and increased splicing of XBP-1 (11.3-fold) (Fig. 4B), compared with control-treated cells. It is important to note that this dose did not lead to apoptosis, as determined by trypan blue exclusion. Second, to determine whether increased ER stress increased βLox5 immunogenicity, βLox5 cells were incubated with Thaps and their immunogenicity determined through T-cell assays. Each T-cell clone secreted IFN-γ in response to Thaps-treated β-cells (Fig. 4C–G). Control-treated βLox5 cells did not elicit IFN-γ, indicating that, as observed with primary islets (Fig. 3), ER stress is required for modification and release of antigens.
Figure 4.
ER stress in insulinoma cells leads to activation of T cells specific for modified antigens. A: ER stress induction in Thaps-treated βLox5 insulinoma cells was measured by assessing the phosphorylation of the unfolded protein response proteins eIF2α and IRE-1 by Western blotting. Bands for one representative blot are shown (left). Densitometry data (right) are the mean ± SEM of four independent experiments. White bars represent densitometry of bands from control-treated cells. Gray bars represent densitometry of bands from THAPS-treated cells. B: ER stress was assessed further by evaluating splicing of XBP-1 mRNA through the use of quantitative PCR. Spliced XBP-1 was significantly higher in Thaps-treated cells. Data are the mean ± SEM of five independent experiments. Irradiated WT1 cells (DQ8+ APCs) were mixed with βlox5 (as an antigen source) and cocultured with T-cell clones that recognize modified IA-2198–216 (C), IA-2467–482 (D), IA-2523–536 (E), IA-2545–562 (F), or IAPP65–84 (G), or omitted either Thaps or one cell type (as indicated) as experimental controls. Thaps-stressed βlox5 cells elicited robust secretion of IFN-γ by T cells that in most cases was significantly higher than levels elicited by unstressed βlox5 cells or the medium, indicating that antigens generated by stressed βlox5 cells can efficiently activate T cells that preferentially respond to modified peptides. Data are the mean ± SEM of four independent experiments. *P < 0.05; **P < 0.01. Ctrl., control; p-eIF2α, phosphorylated eIF2α; p-IRE-1, phosphorylated inositol-requiring enzyme 1.
β-Cell Stress Activates Ca2+-Dependent PTM Enzymes
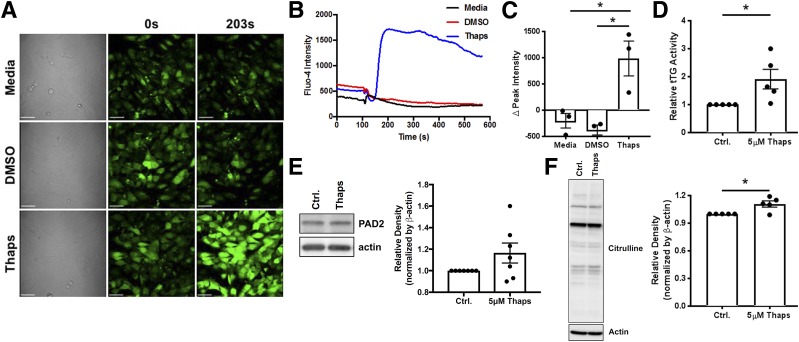
tTG2, the enzyme that deamidates proteins, and PAD2, the enzyme that citrullinates proteins, are Ca2+-dependent. To visualize whether ER stress increases cytosolic Ca2+, βLox5 cells were labeled with Fluo-4. Thaps significantly increased Fluo-4 fluorescence (P < 0.05), indicating increased cytosolic Ca2+ (Fig. 5A–C). We then evaluated tTG2 and PAD2 activity in βLox5 cells. Thaps (5 μmol/L) elicited 1.9-fold higher tTG2 activity (P < 0.05) than did control-treated cells (Fig. 5D), demonstrating that ER stress increased tTG2 activity. In addition, we confirmed that βLox5 cells express PAD2 (Fig. 5E). PAD2 expression did not significantly increase during 1 h of ER stress. However, Thaps treatment significantly increased the Ca2+-dependent activity of PAD2 (P < 0.05), as measured by the incorporation of citrulline residues in whole-cell lysates (Fig. 5F). Together, these data confirm our previous findings in murine β-cells (44) and demonstrate that ER stress in human β-cells activates Ca2+-dependent enzymes, which can generate modified epitopes for recognition by T cells.
Figure 5.
ER stress induces Ca2+ flux and increases tTG2 and PAD2 activity. A: To determine whether Thaps-induced ER stress leads to an increase in cytosolic Ca2+, βLox5 cells were labeled with the Ca2+ indicator Fluo-4 before incubation in medium alone (top row) or with DMSO (middle row) or Thaps (bottom row). B: A plot of the full time course of the Fluo-4 Ca2+ indicator clearly shows differences in Ca2+ flux elicited by Thaps treatment that are absent in the medium and the DMSO control (Ctrl.). C: The change in peak Fluo-4 intensity (from 0 to 203 s) was significantly higher for Thaps-treated cells vs. medium- or DMSO-treated cells. Data are the mean ± SEM of three independent experiments. D: Activity of tTG2 in Thaps- or DMSO-treated βLox5 cells was measured by the incorporation of biotinylated pentylamine into endogenous proteins. Cells incubated with 5 μmol/L Thaps exhibited 1.9-fold higher tTG2 activity than control-treated β-cells. Data are the mean ± SEM of five independent experiments. E: PAD2 expression in Thaps-treated βLox5 insulinoma cells was measured by Western blotting. Bands for one representative blot are shown (left). Densitometry data (right) are the mean ± SEM of seven independent experiments. F: Activity of PAD2 in Thaps- or control-treated βLox5 cells was measured by assessing the incorporation of citrulline residues in whole-cell lysates by Western blotting. Bands for one representative blot are shown (left). Densitometry data (right) are the mean ± SEM of five independent experiments. *P < 0.05.
T Cells Specific for Modified Autoantigens Are Elevated in Subjects With Diabetes
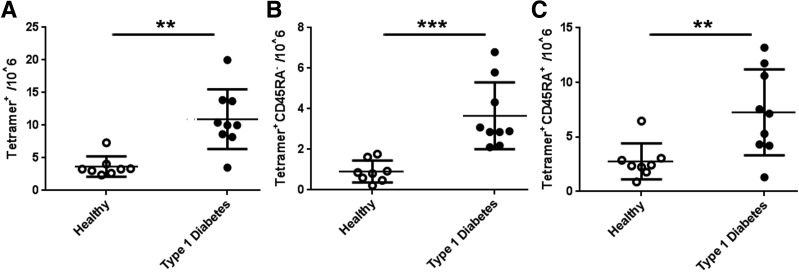
We next applied a direct enrichment approach (28) to measure the frequency of PTM epitope–specific T cells in subjects with type 1 diabetes and in control subjects. Given the scarcity of autoreactive T cells in peripheral blood (18), we combined five specificities to maximize our ability to characterize these rare cells. CD4+ T cells specific for modified antigens were found at significantly higher frequencies in subjects with diabetes than in control subjects (P < 0.005) (Fig. 6A). Use of CD45RA to distinguish antigen-experienced (CD45RA−) and naive T cells (CD45RA+) showed that patients had significantly more memory cells specific for modified antigen than did control subjects (P < 0.05) (Fig. 6B). Similar differences in naive cells were also observed (P < 0.005) (Fig. 6C). Thus, subjects with diabetes have significantly greater numbers of CD4+ T cells specific for modified IA-2 and IAPP than control subjects.
Figure 6.
CD4+ T cells specific for modified autoantigens are more frequent in subjects with diabetes. The frequency of CD4+ T cells in the peripheral blood of eight healthy control subjects and nine subjects with type 1 diabetes was determined by staining PBMCs directly ex vivo with tetramer and antibodies against various surface markers. A: The total frequency of PTM-specific CD4+ T cells was significantly higher in subjects with type 1 diabetes (mean ± SD 11 ± 4.4 cells/million [black circles]) than in autoantibody-negative control subjects (mean ± SD 3.7 ± 1.55 cells/million [white circles]). B: The memory frequency of PTM-specific CD4+ T cells was significantly higher in subjects with type 1 diabetes (mean ± SD 3.7 ± 1.5 cells/million [black circles]) than in autoantibody-negative control subjects (mean ± SD, 0.91 ± 0.49 cells/million [white circles]). C: The naive frequency of PTM-specific CD4+ T cells was significantly higher in subjects with type 1 diabetes (mean ± SD 7.3 ± 3.9 cells/million [black circles]) than in autoantibody-negative control subjects (mean ± SD 2.8 ± 1.5 cells/million [white circles]). **P < 0.01; ***P < 0.001.
We also examined the surface phenotype of antigen-specific memory cells. Six of nine patients exhibited tetramer-positive cells that were CD45RA−CXCR3+CCR4− (Supplementary Fig. 4). Two patients also had a small proportion of CD45RA−CXCR3−CCR4+ tetramer-positive cells. Considering these observations in combination with the observed cytokine profiles (Supplementary Fig. 3), we conclude that CD4+ T cells specific for modified β-cell antigens exhibit a Th1-like phenotype.
PTM-Specific T Cells Are Present in the PLN
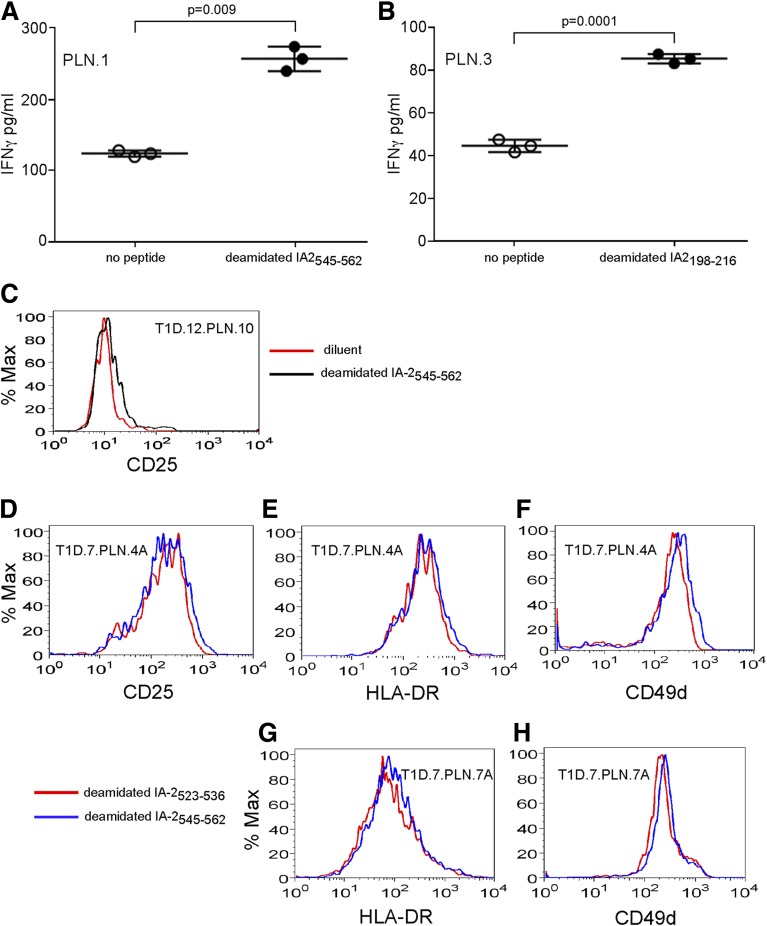
CD4+ T cells that recognize modified peptides are present in blood. However, the cells that are most relevant to disease should be found close to the affected tissue. Our recent work identified a citrullinated IAPP65–84-specific clone among T cells from the islets of a cadaveric donor with diabetes (23). To substantiate the relevance of T cells specific for modified β-cell antigens, we examined T cells from the PLN of two subjects with type 1 diabetes (Supplementary Table 3). For donor T1D.7, PLN cells were expanded during two rounds of stimulation with peptide. For donor T1D.12, amplified T-cell libraries were expanded with peptides. We tested reactivity by assaying for IFN-γ secretion or upregulation of activation markers in response to modified peptides. T-cell lines from donor T1D.7 secreted significant levels of IFN-γ in response to deamidated IA-2545–562 and IA-2198–216 (Fig. 7A and B). T-cell line PLN.10 from donor T1D.12 upregulated CD25 in response to IA-2545–562. After peptide stimulation with IA-2545–562, the PLN.4A and PLN.7A libraries upregulated multiple activation markers (Fig. 7C–H). These observations establish that CD4+ T cells that recognize deamidated IA-2 epitopes were present in the PLN of subjects with established disease.
Figure 7.
CD4+ T cells specific for modified autoantigens are present in the PLN of subjects with diabetes. T-cell lines expanded from the PLN of donor T1D.7 (23) or donor T1D.12 were tested for responsiveness to modified epitopes. A: The PLN.1 cell line from donor T1D.7 secreted IFN-γ in response to stimulation by an autologous EBV-transformed splenic B-cell line pulsed with deamidated IA-2545–562 peptide (black circles) that was significantly higher than the response to the control with no peptide (white circles). B: The PLN.3 cell line from donor T1D.7 secreted IFNγ in response to stimulation with deamidated IA-2198–216 peptide (black circles) was significantly higher than the response to the control with no peptide (white circles). C: The PLN.10 cell line from donor T1D.12 upregulated CD25 after 48 h of coculture with autologous splenic EBV-transformed B cells pulsed with deamidated IA-2545–562 (black line) but not with a diluent control (red line). Thirty amplified T-cell libraries were made from the PLN from donor T1D.7. After expansion, 2 of the 30 libraries showed CD4+ T-cell activation with deamidated IA-2545–562 peptide (blue lines) but not with deamidated IA-2523–536 peptide (red lines) as a comparison. After peptide stimulation, the PLN.4A library also showed upregulation of CD25 (D), HLA-DR (E), and CD49d (F). After peptide stimulation, the PLN.7A library showed upregulation of HLA-DR (G) and CD49d (H). Cells in the lymphocyte and CD4+ gates from flow cytometry are shown. Data from one of three similar experiments are shown. % Max, percentage of the maximum.
Discussion
In instances of stress and inflammation, atypical PTM arise and have been linked with autoimmunity. PTM have been hypothesized to have a role in type 1 diabetes, and published evidence increasingly supports this notion (16–19). Here we demonstrated the relevance of PTM epitopes derived from IA-2 and IAPP. T-cell clones mounted functional responses against the modified forms of these epitopes, and T cells specific for these epitopes were present at significantly higher frequencies in subjects with type 1 diabetes. It is important to note that T cells that recognize these epitopes were present in the PLN of subjects with established disease, further supporting their relevance. Functional characterization of PTM-specific CD4+ T cells indicated a Th1-like phenotype. Likewise, PBMCs from subjects with type 1 diabetes produced IFN-γ following stimulation with a pool of IA-2 and proinsulin epitopes (35). Thus, our results agree with previous observations of a Th1 phenotype and support the disease relevance of CD4+ T cells specific for posttranslationally modified antigens.
We acknowledge that only verifying the presence of citrullinated and deamidated peptides/proteins through the use of mass spectrometry would definitively prove that posttranslationally modified epitopes are produced by stressed β-cells. Although we did not comprehensively assess natural processing and presentation or verify peptide modification by mass spectrometry, T cells specific for these epitopes responded to stressed islets, suggesting that epitopes were generated corresponding to these modified peptides. These data, and our mechanistic findings with βLox5 cells, extend our previous observations from a murine model of diabetes (44), confirming that ER stress, which β-cells undergo as a result of normal secretory function (37–41) and which accompanies β-cell loss (43), generates sufficient cytosolic Ca2+ to activate tTG2 and PAD2. These data, then, support the hypothesis that the physiology of β-cells facilitates the generation of modified autoantigens (46) in mice and humans, suggesting that the pathways through which modified self-peptides are generated may be conserved among species.
It is notable that all four deamidated IA-2 peptides fall within the extracellular domain. The most widely published IA-2 epitopes fall within its transmembrane and intracellular domains (9), but recently published work demonstrated recognition of the extracellular domain (22). In light of the prognostic significance of autoantibodies directed against the extracellular domain of IA-2, T-cell responses against this domain may also have distinct significance (47). Modification of specific residues enhanced either peptide binding to HLA-DQ8 or interactions with TCR, leading to preferential recognition. These findings highlight the subtlety of the trimolecular interaction between peptide, MHC, and TCR and help to explain how seemingly modest PTMs can profoundly alter CD4+ T-cell responses.
Our study does have limitations. Beyond the epitopes that we studied, additional modified epitopes are likely to be recognized. Examining additional epitopes would provide a more comprehensive view of the role that such responses play in disease. Furthermore, responses to unmodified epitopes clearly play a role, as evidenced by a recent report that correlated insulin B10–23 responses with insulin autoantibodies (48). As in that study, we demonstrated elevated frequencies of CD4+ T cells that recognize our epitopes of interest in subjects with diabetes. Although we show that ER stress generates PTM-dependent immunogenicity in primary islets, our mechanistic experiments were conducted with insulinomas. Primary islets would be ideal for mechanistic studies, but the number of cells was inadequate for our purposes. An important unanswered question is in what stage of the disease process PTM-specific T cells appear. Future studies to address this question using samples from at-risk individuals are under consideration. In all, our findings establish a role for responses to modified epitopes in type 1 diabetes and suggest that such responses contribute to autoimmunity by activating pathogenic CD4+ T cells that are not effectively eliminated by negative selection.
Supplementary Material
Article Information
Acknowledgments. The authors thank Aru Arumaganthan (Benaroya Research Institute, Seattle, WA) for flow cytometry support, McKenzie Lettau and Jani Klein (Benaroya Research Institute, Seattle, WA) for assisting with subject recruitment, and members of W.W.K.’s laboratory (Benaroya Research Institute, Seattle, WA) for helpful discussions. The authors also thank the University of Massachusetts Medical School Flow Cytometry Core Facility.
Funding. This work was supported by JDRF (grant nos. 3-PDF-2014-213-A-N to M.L.M., 2-SRA-2016-149 to A.C.P., 2-SRA-2014-296-Q-R to J.D.P., and 17-2012-121 and 2-SRA-2014-297-Q-R to E.A.J.), the Leona M. and Harry B. Helmsley Charitable Trust (grant no. 2015PG-T1D057 to S.C.K.), the National Institutes of Health (grant nos. R21 AI126189 to S.C.K. and DK104211 and DK106755 to A.C.P.), the Human Islet Research Network (HIRN) Opportunity Pool Fund (National Institutes of Health grant no. U01 DK104162 to S.C.K.), U.S. Department of Veterans Affairs (grant no. BX000666 to A.C.P.), and the Vanderbilt Diabetes Research and Training Center (National Institutes of Health grant no. DK020593 to A.C.P.).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. M.L.M. performed βlox5 cell assays, prepared cells, performed cytokine release assays with T-cell clones, analyzed data, and cowrote the manuscript. J.W.M. identified peptides, isolated T-cell clones, performed tetramer assays, analyzed data, and cowrote the manuscript. I.-T.C. produced the HLA-DQ8 protein and provided technical support during ex vivo tetramer staining. M.E.D. and S.C.K. performed experiments with PLN-derived T cells. N.W.B. provided technical support during Western blotting. A.C.P., R.B., and D.M.H. provided crucial samples. C.J.G. selected and characterized subjects and contributed important ideas. W.W.K. developed experimental methods and contributed important ideas. J.D.P. worked with M.L.M. to develop methods for βlox5 cell assays, interpreted data, and contributed to the manuscript. J.D.P. edited the manuscript. E.A.J. designed the research, summarized data, provided technical training to J.W.M., and edited the manuscript. E.A.J. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db17-1166/-/DC1.
J.W.M. is currently affiliated with the Department of Immunology, University of Washington, Seattle, WA.
References
- 1.Griesemer AD, Sorenson EC, Hardy MA. The role of the thymus in tolerance. Transplantation 2010;90:465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramsdell F, Fowlkes BJ. Clonal deletion versus clonal anergy: the role of the thymus in inducing self tolerance. Science 1990;248:1342–1348 [DOI] [PubMed] [Google Scholar]
- 3.Williams CB, Engle DL, Kersh GJ, Michael White J, Allen PM. A kinetic threshold between negative and positive selection based on the longevity of the T cell receptor-ligand complex. J Exp Med 1999;189:1531–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong J, Obst R, Correia-Neves M, Losyev G, Mathis D, Benoist C. Adaptation of TCR repertoires to self-peptides in regulatory and nonregulatory CD4+ T cells. J Immunol 2007;178:7032–7041 [DOI] [PubMed] [Google Scholar]
- 5.Scally SW, Petersen J, Law SC, et al. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J Exp Med 2013;210:2569–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Venrooij WJ, Hazes JM, Visser H. Anticitrullinated protein/peptide antibody and its role in the diagnosis and prognosis of early rheumatoid arthritis. Neth J Med 2002;60:383–388 [PubMed] [Google Scholar]
- 7.van de Wal Y, Kooy Y, van Veelen P, et al. Selective deamidation by tissue transglutaminase strongly enhances gliadin-specific T cell reactivity. J Immunol 1998;161:1585–1588 [PubMed] [Google Scholar]
- 8.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Lorenzo TP, Peakman M, Roep BO. Translational mini-review series on type 1 diabetes: systematic analysis of T cell epitopes in autoimmune diabetes. Clin Exp Immunol 2007;148:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roep BO, Peakman M. Antigen targets of type 1 diabetes autoimmunity. Cold Spring Harb Perspect Med 2012;2:a007781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu L, Rewers M, Gianani R, et al. Antiislet autoantibodies usually develop sequentially rather than simultaneously. J Clin Endocrinol Metab 1996;81:4264–4267 [DOI] [PubMed] [Google Scholar]
- 12.Orban T, Sosenko JM, Cuthbertson D, et al.; Diabetes Prevention Trial-Type 1 Study Group . Pancreatic islet autoantibodies as predictors of type 1 diabetes in the Diabetes Prevention Trial-Type 1. Diabetes Care 2009;32:2269–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks-Worrell B, Gersuk VH, Greenbaum C, Palmer JP. Intermolecular antigen spreading occurs during the preclinical period of human type 1 diabetes. J Immunol 2001;166:5265–5270 [DOI] [PubMed] [Google Scholar]
- 14.Prasad S, Kohm AP, McMahon JS, Luo X, Miller SD. Pathogenesis of NOD diabetes is initiated by reactivity to the insulin B chain 9-23 epitope and involves functional epitope spreading. J Autoimmun 2012;39:347–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Herrath M, Sanda S, Herold K. Type 1 diabetes as a relapsing-remitting disease? Nat Rev Immunol 2007;7:988–994 [DOI] [PubMed] [Google Scholar]
- 16.Mannering SI, Harrison LC, Williamson NA, et al. The insulin A-chain epitope recognized by human T cells is posttranslationally modified. J Exp Med 2005;202:1191–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Lummel M, Duinkerken G, van Veelen PA, et al. Posttranslational modification of HLA-DQ binding islet autoantigens in type 1 diabetes. Diabetes 2014;63:237–247 [DOI] [PubMed] [Google Scholar]
- 18.McGinty JW, Chow IT, Greenbaum C, Odegard J, Kwok WW, James EA. Recognition of posttranslationally modified GAD65 epitopes in subjects with type 1 diabetes. Diabetes 2014;63:3033–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rondas D, Crèvecoeur I, D’Hertog W, et al. Citrullinated glucose-regulated protein 78 is an autoantigen in type 1 diabetes. Diabetes 2015;64:573–586 [DOI] [PubMed] [Google Scholar]
- 20.Delong T, Wiles TA, Baker RL, et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science 2016;351:711–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kracht MJ, van Lummel M, Nikolic T, et al. Autoimmunity against a defective ribosomal insulin gene product in type 1 diabetes. Nat Med 2017;23:501–507 [DOI] [PubMed] [Google Scholar]
- 22.Acevedo-Calado M, James EA, Morran MP, et al. Identification of unique antigenic determinants in the amino terminus of IA-2 (ICA512) in childhood and adult autoimmune diabetes: new biomarker development. Diabetes Care 2017;40:561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Babon JA, DeNicola ME, Blodgett DM, et al. Analysis of self-antigen specificity of islet-infiltrating T cells from human donors with type 1 diabetes. Nat Med 2016;22:1482–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwok WW, Domeier ML, Raymond FC, Byers P, Nepom GT. Allele-specific motifs characterize HLA-DQ interactions with a diabetes-associated peptide derived from glutamic acid decarboxylase. J Immunol 1996;156:2171–2177 [PubMed] [Google Scholar]
- 25.Chang KY, Unanue ER. Prediction of HLA-DQ8beta cell peptidome using a computational program and its relationship to autoreactive T cells. Int Immunol 2009;21:705–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chow IT, Yang J, Gates TJ, et al. Assessment of CD4+ T cell responses to glutamic acid decarboxylase 65 using DQ8 tetramers reveals a pathogenic role of GAD65 121-140 and GAD65 250-266 in T1D development. PLoS One 2014;9:e112882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Chow IT, Sosinowski T, et al. Autoreactive T cells specific for insulin B:11-23 recognize a low-affinity peptide register in human subjects with autoimmune diabetes. Proc Natl Acad Sci U S A 2014;111:14840–14845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwok WW, Roti M, Delong JH, et al. Direct ex vivo analysis of allergen-specific CD4+ T cells. J Allergy Clin Immunol 2010;125:1407–1409.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipson KL, Ghosh R, Urano F. The role of IRE1alpha in the degradation of insulin mRNA in pancreatic beta-cells. PLoS One 2008;3:e1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neal MD, Sodhi CP, Jia H, et al. Toll-like receptor 4 is expressed on intestinal stem cells and regulates their proliferation and apoptosis via the p53 up-regulated modulator of apoptosis. J Biol Chem 2012;287:37296–37308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Lesort M, Guttmann RP, Johnson GV. Modulation of the in situ activity of tissue transglutaminase by calcium and GTP. J Biol Chem 1998;273:2288–2295 [DOI] [PubMed] [Google Scholar]
- 32.Geiger R, Duhen T, Lanzavecchia A, Sallusto F. Human naive and memory CD4+ T cell repertoires specific for naturally processed antigens analyzed using libraries of amplified T cells. J Exp Med 2009;206:1525–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Godkin A, Friede T, Davenport M, et al. Use of eluted peptide sequence data to identify the binding characteristics of peptides to the insulin-dependent diabetes susceptibility allele HLA-DQ8 (DQ 3.2). Int Immunol 1997;9:905–911 [DOI] [PubMed] [Google Scholar]
- 34.Kampstra AS, van Heemst J, Moustakas AK, Papadopoulos GK, Huizinga TW, Toes RE. The increased ability to present citrullinated peptides is not unique to HLA-SE molecules: arginine-to-citrulline conversion also enhances peptide affinity for HLA-DQ molecules. Arthritis Res Ther 2016;18:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arif S, Tree TI, Astill TP, et al. Autoreactive T cell responses show proinflammatory polarization in diabetes but a regulatory phenotype in health. J Clin Invest 2004;113:451–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Araki E, Oyadomari S, Mori M. Endoplasmic reticulum stress and diabetes mellitus. Intern Med 2003;42:7–14 [DOI] [PubMed] [Google Scholar]
- 37.Lipson KL, Fonseca SG, Urano F. Endoplasmic reticulum stress-induced apoptosis and auto-immunity in diabetes. Curr Mol Med 2006;6:71–77 [DOI] [PubMed] [Google Scholar]
- 38.Wu J, Kaufman RJ. From acute ER stress to physiological roles of the unfolded protein response. Cell Death Differ 2006;13:374–384 [DOI] [PubMed] [Google Scholar]
- 39.Fonseca SG, Lipson KL, Urano F. Endoplasmic reticulum stress signaling in pancreatic beta-cells. Antioxid Redox Signal 2007;9:2335–2344 [DOI] [PubMed] [Google Scholar]
- 40.Volchuk A, Ron D. The endoplasmic reticulum stress response in the pancreatic β-cell. Diabetes Obes Metab 2010;12(Suppl. 2):48–57 [DOI] [PubMed] [Google Scholar]
- 41.Scheuner D, Kaufman RJ. The unfolded protein response: a pathway that links insulin demand with beta-cell failure and diabetes. Endocr Rev 2008;29:317–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oyadomari S, Takeda K, Takiguchi M, et al. Nitric oxide-induced apoptosis in pancreatic beta cells is mediated by the endoplasmic reticulum stress pathway. Proc Natl Acad Sci U S A 2001;98:10845–10850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brozzi F, Eizirik DL. ER stress and the decline and fall of pancreatic beta cells in type 1 diabetes. Ups J Med Sci 2016;121:133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marré ML, Profozich JL, Coneybeer JT, et al. Inherent ER stress in pancreatic islet β cells causes self-recognition by autoreactive T cells in type 1 diabetes. J Autoimmun 2016;72:33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Halvorsen TL, Leibowitz G, Levine F. Telomerase activity is sufficient to allow transformed cells to escape from crisis. Mol Cell Biol 1999;19:1864–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marré ML, James EA, Piganelli JD. β cell ER stress and the implications for immunogenicity in type 1 diabetes. Front Cell Dev Biol 2015;3:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morran MP, Casu A, Arena VC, et al. Humoral autoimmunity against the extracellular domain of the neuroendocrine autoantigen IA-2 heightens the risk of type 1 diabetes. Endocrinology 2010;151:2528–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spanier JA, Sahli NL, Wilson JC, et al. Increased effector memory insulin-specific CD4+ T cells correlate with insulin autoantibodies in patients with recent-onset type 1 diabetes. Diabetes 2017;66:3051–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.