Abstract
Objective
A comprehensive search on electronic databases was conducted to identify all eligible studies of TNF-α polymorphisms and knee osteoarthritis (OA).
Methods
Eight studies on TNF-α -308 G > A and three on TNF-α -238G > A polymorphism were identified.
Results
Overall, the pooled ORs indicated that neither TNF-α -238G > A nor -238G > A polymorphism was associated with knee OA risk. Similarly, in the stratified analysis by ethnicity, no significant association was found.
Conclusion
This meta-analysis results inconsistent with the previous meta-analyses showed that the TNF-α -308 G > A and −238G > A polymorphisms may not be associated with the susceptibility to knee OA.
Keywords: Osteoarthritis, Knee, Tumor necrosis factor-alpha, Polymorphism, Meta-analysis
1. Introduction
Osteoarthritis (OA) is a progressive and irreversible degenerative condition, that affecting synovial joints, most frequently the knees, hips, hands, and spine. 1, 2, 3 OA, one of the most common causes of disability, affects more than 100 million people worldwide.4,5 Radiological studies revealed that more than 20 million people in the US suffer from knee OA.5 A wide array of factors including biomechanical, biochemical, and genetic factors predisposition are known to be involved in the list of causative agents of OA. 1,6,7 Researchers continue to search for immunological and genetic clues OA. Patients with OA have high levels of TNF-α in the synovial fluid and it plays an important role in inflammation and joint destruction that are hallmarks of OA.8
TNF-α has an extremely broad spectrum of biological activities.9 It is a cytokine that plays an important role in acute inflammation and immune responses.10 It is stimulates cytokine production, which enhances the expression of adhesion molecules and activation of neutrophils.11 Increasing amount of evidence from clinical studies shows that the blood levels of TNF-α correlate with development and progression of different diseases.12
The TNF-α gene is located on chromosome 6p21.3, within the class III region of HLA, contains four exons, and spans approximately 3 kbp.13 Several single nucleotide polymorphisms have been identified in the promoter region of human TNF-α gene14. Among these, two common polymorphism in the promoter including −308 G > A and −238G > A has been studied intensively. To date, a few studies based on different ethnicities have reported conflicting evidence regarding the association of −308 G > A and −238G > A polymorphisms in TNF-α gene with risk of knee OA. Thus, to summarize more reliable and large-scale evidence on whether TNF-α-308 G > A and −238G > A polymorphisms are associated with knee OA susceptibility we have performed a meta-analysis.
2. Materials and methods
2.1. Case-control study
2.1.1. Study population
The study protocol was approved by the Ethics Committee and all participants signed written informed consent. From January 2014 to September 2017, a total of 110 patients with radiographically confirmed knee OA and, 120 age and sex control subjects who had no symptoms or signs of OA or related disease were recruited to this case-control study. Most of the patients were residing in the central region of Iran and though some of participants were from north western of Iran. The diagnosis of knee OA was based on the criteria of the American College of Rheumatology, which included primary OA with any symptoms and radiographic signs of OA according to the Kellgren–Lawrence (KL) grading system.15
2.1.2. Genotyping
Genomic DNA from each participant was isolated from 4 ml EDTA anti-coagulated whole blood using Genomic DNA Mini Kit (Qiagen Inc., Hilden, Germany) based on the instructions of the protocol. Extracted DNA was labeled and suspended in Tris buffer and stored at −20 °C until used. The TNF-α-308 G > A and −238G > A polymorphisms were determined using polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP-PCR) as described by before. Primers forward (5′-AGGCAATAGGTTTTGAGGGCCAT-3′) and reverse (5′-TCCTCCCTGCTCCGATTCCG-3′) were used to amplify the 107-bp DNA fragment of the TNFα−308 A > G polymorphism. In addition, to identify the TNF-α -238G > A polymorphism, the following primers were used: forward (5′-AGAAGACCCCCCTCGGAACC-3′) and reverse (5′-ATCTGGAGGAAGCGGTAGTG-3′). All PCR was carried out in 50 μl containing 0.1 μg of DNA, 5 μl of 10x buffer, 5 μl of 50 mM MgCl2, 1 μl of 10 mM dNTPs, 5 μM of each primers, 2.5U Taq DNA polymerase. PCR conditions were 5 min for initial denaturation at 95 °C; 35 cycles at 95 °C for 1 min for denaturation, 30 s at 65 °C for annealing and 30 s at 72 °C for extension, followed by 5 min at 72 °C for final extension. The PCR products for TNF-α −308 G > A and −238G > A polymorphisms was digested with enzyme NcoI (Frementase, Vilnius, Lithuania) and MspI (Roche diagnostics, Swiss) restriction enzymes for 3 h at 37 °C, respectively. The products were electrophoresed on a 3% agarose gel.
2.1.3. Statistical analysis
The distribution of the genotype frequencies of TNF-α −308 G > A and −238G > A polymorphisms for patients and healthy control were compared using the chi-squared test. Moreover, the Odds ratio (OR) with 95% confidence interval (CI) for knee OA susceptibility was also calculated. The distribution of the genotypes in the control population was tested for Hardy–Weinberg equilibrium (HWE) using a goodness-of-fit Chi-square test. All statistical tests were two-sided and were performed with SPSS software version 29 (Chicago Illinois). All comparisons were considered to be statistically significant at P < 0.05.
2.2. Meta-analysis
2.2.1. Search strategy
We performed a systematic literature search in PubMed, Embase, ISI Web of Science, Cochrane Library, and Chinese National Knowledge Infrastructure until April 2018. Searching tasks were independently performed by 2 researchers. The search strategy involved the combination of the following keywords: (osteoarthritis OR knee OR knee OA) AND (tumor necrosis factor-alpha OR TNF-α OR TNFSF2 OR Cachectin) AND (−308 G > A OR rs1800629) AND (−238G > A OR rs361525) AND (genetic OR polymorphism OR variant OR mutation). There was no restriction on time period, sample size, population, language, or type of report. The reference lists of relevant articles were also manually searched to acquire additional eligible original articles and to supplement the yield of initial search in the databases.
2.2.2. Selection criteria
Studies were considered eligible if they satisfied the following criteria: (1) using a case–control or cohort study design; (2) evaluating the association of TNF-α −308 G > A and −238G > A polymorphisms and susceptibility to knee OA; (3) providing explicit genotypes and allele frequencies in each group or presenting sufficient information for odds ratio (OR) and 95% confidence interval (CI) calculation; (4) full-text article. We excluded the article if not meet the inclusion criteria. Studies were excluded if they met the following criteria: (1) it is not an original study; (2) unpublished data, reviews, lectures, editorials or correspondence letters; (3) it is not the study of the association of TNF-α −308 G > A and −238G > A polymorphisms with knee OA; (4) it does not have control data; (5) studies in which family members had been studied because their analysis are based on linkage considerations; (6) it does not provide reusable data. For studies with overlapping data, we selected the most recently published study with the largest sample size.
2.2.3. Data extraction
By using a standardized form the data were checked and extracted by two investigators independently.
The following data were collected from each study: first author, year of publication, country origin, ethnicity, total number of cases and controls, the frequencies of genotypes, minor allele frequencies (MAFs), P-value for Hardy–Weinberg equilibrium (HWE). In case of disagreement (in the data extraction), consensus was resolved through consensus, or a third author would assess these articles. Disagreements were resolved by discussion among all authors until consensus was reached.
2.2.4. Statistical analysis
All meta-analyses were conducted using Comprehensive Meta-Analysis (CMA) software version 2.0 (Stata Corp., College Station, TX, USA) and a P value below 0.05 was considered statistically significant. The association of TNF-α −308 G > A and −238G > A polymorphisms with knee OA was assessed by determining the pooled ORs and 95% CIs under the allele model (G vs. A), the homozygote model (GG vs. AA), the heterozygote model (AG vs. GG), the dominant model (GG vs. GA + AA), and the recessive model (GG + GA vs. AA). The significance of the pooled OR was determined by a Z-test and P < 0.05 was considered statistically significant. We used the Cochran’s Q-statistic (considered statistically significant if P < 0.10) to evaluate potential heterogeneity between studies.16,17 Additionally, between-study heterogeneity quantified using the I2 value, Venice criteria for the I2 test included: “I2 < 25% represents no heterogeneity, I2 = 25–50% represents moderate heterogeneity, I2 = 50–75% represents large heterogeneity, and I2 > 75% represents extreme heterogeneity’’. Thus, if results were not heterogeneous, the pooled ORs were calculated by the fixed-effect model (the Mantel–Haenszel method)18; otherwise, a random-effect model (the DerSimonian and Laird method)19 was used. To explore the sources of heterogeneity, we conducted subgroup analysis by ethnicity, source of controls, and genotyping methods. For each study, deviations from the Hardy-Weinberg equilibrium (HWE) in the healthy controls were tested examined using the x2 test (or Fisher exact test).20 One-way sensitivity analyses were performed by iteratively removing one study at a time to assess the stability of the meta-analysis results. Begg’s funnel plots and Egger’s linear regression test were used to assess publication bias.21,22 If publication bias existed, the “trim and fill” method was used to estimate the number of missing studies and to adjust the pooled result.
3. Results
3.1. Case-control study
The frequencies of the polymorphic genotypes between cases and controls are shown in Table 1.The genotype frequencies were in HWE for both TNF-α −308 G > A and −238G > A polymorphisms (Table 2). There was no significant difference in the distribution of genotypes between the cases and controls for both TNF-α −308 G > A and −238G > A polymorphisms (P > 0.05). However, percentage of the TNF-α −308 A allele was significantly higher in control compared to knee OA cases (p = 0.001).
Table 1.
Comparisons of genotypes and alleles frequencies of TNF-α −308 G > A and -238G > A polymorphisms in melanoma cases and controls.
| Polymorphism | Cases (n = 110) | Control (n = 120) | OR (95% CI) | p-value |
|---|---|---|---|---|
| TNF-α-308 G > A | ||||
| Genotypes | ||||
| GG | 79 (71.8) | 85 (70.8) | 1.00 | |
| AG | 30 (27.2) | 33 (27.5) | 0.989 (0.553–1.766) | 0.969 |
| AA | 1 (0.9) | 2 (1.6) | 0.541 (0.048–6.054) | 0.618 |
| Allele | ||||
| G | 203 (92.3) | 194 (80.8) | 1.00 | |
| A | 17 (7.7) | 46 (19.2) | 0.353 (0.196–0.637) | 0.001 |
| TNF-α -238G > A | ||||
| Genotypes | ||||
| GG | 91 (82.7) | 105 (87.5) | 1.00 | |
| AG | 19 (17.3) | 14 (11.7) | 1.581 (0.750–3.330) | 0.228 |
| AA | 0 (0.0) | 1 (0.8) | 0.360 (0.015–8.942) | 0.533 |
| Allele | ||||
| G | 201 (91.4) | 224 (93.3) | 1.00 | |
| A | 19 (8.6) | 16 (6.7) | 1.323 (0.663–2.643) | 0.427 |
OR: Odds Ratio; CI: Confidence Interval.
Table 2.
Characteristics of the individual studies included in the meta-analysis.
| First Author | Country (Ethnicity) |
SOC | Genotyping Method |
Case/Control | Cases |
Control |
MAFs | HWE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotypes |
Alleles |
Genotypes |
Alleles |
|||||||||||||
| TNF-α −308 G > A | GG | AG | AA | G | A | GG | AG | AA | G | A | ||||||
| Moos 200023 | Germany(Caucasian) | HB | RFLP-PCR | 55/240 | 36 | 18 | 1 | 90 | 20 | 166 | 74 | 0 | 406 | 74 | 0.154 | 0.004 |
| Sezgin 200824 | Turkey(Caucasian) | HB | NS | 151/84 | 121 | 26 | 4 | 268 | 34 | 72 | 12 | 0 | 156 | 12 | 0.071 | 0.480 |
| Han 201225 | Korea(Asian) | PB | RFLP-PCR | 301/291 | 79 | 188 | 34 | 346 | 256 | 258 | 33 | 0 | 549 | 33 | 0.056 | 0.305 |
| Ji 201326 | China(Asian) | PB | TaqMan | 200/305 | 143 | 50 | 7 | 336 | 64 | 253 | 50 | 2 | 556 | 54 | 0.088 | 0.781 |
| Munoz-Valle 201427 | Mexico(Mixed) | PB | RFLP-PCR | 50/100 | 44 | 6 | 0 | 94 | 6 | 93 | 7 | 0 | 193 | 7 | 0.035 | 0.716 |
| Vnukov 201628 | Russia(Caucasian) | NS | NS | 117/94 | 84 | 31 | 2 | 199 | 35 | 65 | 26 | 3 | 156 | 32 | 0.170 | 0.839 |
| Abdel Galil 201729 | Egypt(African) | NS | TaqMan | 210/210 | 180 | 25 | 2 | 386 | 34 | 115 | 82 | 13 | 314 | 106 | 0.257 | 0.748 |
| Sobhan 2018 | Iran(Asian) | PB | RFLP-PCR | 110/120 | 79 | 30 | 1 | 203 | 17 | 85 | 33 | 2 | 194 | 46 | 0.191 | 0.909 |
| TNF-α -238 G > A | GG | GA | AA | G | A | GG | GA | AA | G | A | ||||||
| Ji 201326 | China(Asian) | PB | TaqMan | 200/305 | 186 | 14 | 0 | 386 | 14 | 282 | 23 | 0 | 587 | 23 | 0.037 | 0.493 |
| Munoz-Valle 201427 | Mexico(Mixed) | PB | RFLP-PCR | 50/100 | 47 | 3 | 0 | 97 | 3 | 89 | 11 | 0 | 189 | 11 | 0.055 | 0.560 |
| Sobhan 2018 | Iran(Asian) | PB | RFLP-PCR | 110/120 | 91 | 19 | 0 | 201 | 19 | 105 | 14 | 1 | 224 | 16 | 0.066 | 0.493 |
SOC: Source of Control; HB: Hospital Based; PB: Population Based; NS: Not Stated; PCR-RFLP: Polymerase Chain Reaction-Restriction Fragment Length Polymorphism; MAF: Minor Allele Frequency; HWE: Hardy–Weinberg Equilibrium.
3.2. Meta-analysis
3.2.1. Characteristics of the studies
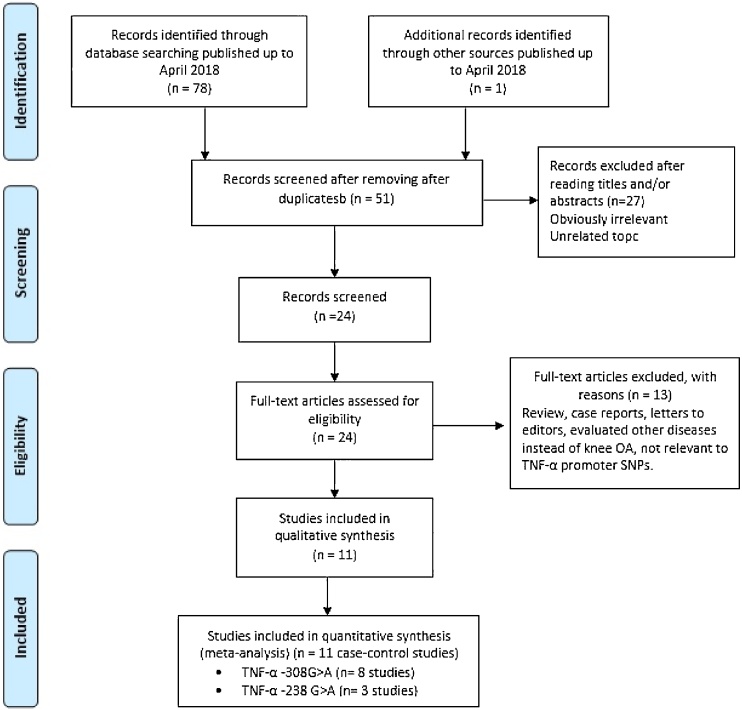
A flow diagram summarizing the literature review process and reasons for exclusion is presented in Fig. 1. A total of eleven studies in eight publications, including eight case-control studies23, 24, 25, 26, 27, 28, 29 for TNF-α −308 G > A polymorphism (1194 cases and 1444 controls) and case-control three studies26,27 for TNF-α -238G > A (360 cases and 525 controls) were selected for this meta-analysis. Table 2 summarizes the characteristics of the studies included in our analysis. All of these eleven case-control studies provided sufficient data to calculate the association of TNF-α −308 G > A and -238G > A polymorphisms with knee OA. These studies were reported from 2000 to 2018, and geographically, the studies were conducted Germany, turkey, Korea, china, Mexico, Russia, Egypt and Iran. Two genotyping methods were applied in the present case-control studies such as PCR-RFLP and TaqMan. All studies showed that the distribution of genotypes in the control group was in agreement with the HWE except for one case-control study23 for TNF-α −308 G > A.
Fig. 1.
Flow diagram of the study selection process.
3.2.2. Quantitative data synthesis
3.2.2.1. TNF-α −308 G > A polymorphism
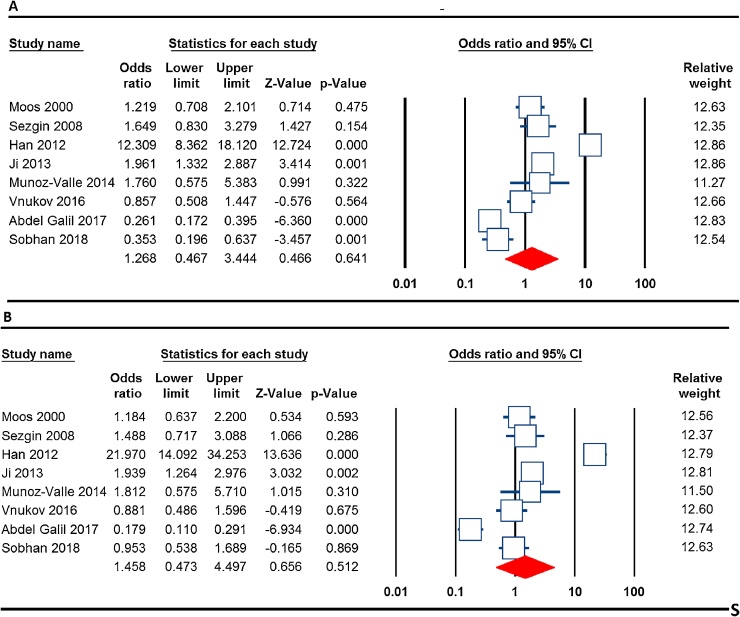
Table 3 listed the main results of the meta-analysis of TNF-α −308 G > A polymorphism and knee OA risk. When all the eligible studies were pooled into the meta-analysis of TNF-α −308 G > A polymorphism, no evidence of a significant between TNF-α −308 G > A polymorphism and knee OA risk in all the five genetic models, i.e., allele (A vs. G: OR = 1.268, 95% CI 0.467–3.444, p = 0.641, Fig. 2A), homozygote (AA vs. GG: OR = 2.302, 95% CI 0.329–16.091, p = 0.401), heterozygote (AC vs. GG: OR = 1.406, 95% CI 0.482–4.096, p = 0.533), dominant (AA + AG vs. GG: OR = 1.458, 95% CI 0.473–4.497, p = 0.512, Fig. 2B) and recessive model (AA vs. AG + GG: OR = 2.232, 95% CI 0.439–11.33, p = 0.334). The studies were further stratified on the basis of ethnicity backgrounds. When stratified by ethnicity, a significant association between TNF-α −308 G > A polymorphism and knee OA did not detected among Asians and Caucasians (Table 3).
Table 3.
The meta-analysis of TNF-α -308 G > A and -238 G > A polymorphisms with knee OA risk.
| Subgroup | Genetic Model | Type of Model | Heterogeneity |
Odds Ratio |
Publication Bias |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| I2 (%) | PH | OR | 95% CI | Ztest | POR | PBeggs | PEggers | |||
| TNF-α -308 G > A | ||||||||||
| Overall | A vs. G | Random | 96.70 | ≤0.001 | 1.268 | 0.467–3.444 | 0.466 | 0.641 | 0.901 | 0.602 |
| AA vs. GG | Random | 82.39 | ≤0.001 | 2.302 | 0.329–16.091 | 0.840 | 0.401 | 0.367 | 0.215 | |
| AG vs. GG | Random | 96.31 | ≤0.001 | 1.406 | 0.482–4.096 | 0.624 | 0.533 | 0.901 | 0.570 | |
| AA + AG vs. GG | Random | 96.82 | ≤0.001 | 1.458 | 0.473–4.497 | 0.656 | 0.512 | 0.901 | 0.635 | |
| AA vs. AG+ GG | Random | 74.71 | 0.001 | 2.232 | 0.439–11.33 | 0.968 | 0.334 | 0.229 | 0.168 | |
| By Ethnicity | ||||||||||
| Caucasian | A vs. G | Fixed | 13.29 | 0.316 | 1.136 | 0.816–1.581 | 0.756 | 0.449 | 0.296 | 0.321 |
| AA vs. GG | Fixed | 48.02 | 0.146 | 1.616 | 0.401–6.510 | 0.675 | 0.500 | 0.296 | 0.043 | |
| AG vs. GG | Fixed | 0.00 | 0.785 | 1.080 | 0.740–1.576 | 0.397 | 0.691 | 0.296 | 0.354 | |
| AA + AG vs. GG | Fixed | 0.00 | 0.538 | 1.120 | 0.773–1.621 | 0.598 | 0.550 | 0.296 | 0.290 | |
| AA vs. AG+ GG | Fixed | 46.16 | 0.156 | 1.606 | 0.400–6.442 | 0.668 | 0.504 | 0.296 | 0.049 | |
| Asian | A vs. G | Random | 98.11 | ≤0.001 | 2.066 | 0.321–13.300 | 0.764 | 0.445 | 1.000 | 0.440 |
| AA vs. GG | Random | 85.37 | 0.001 | 6.219 | 0.211–183.489 | 1.058 | 0.290 | 1.000 | 0.904 | |
| AG vs. GG | Random | 97.52 | ≤0.001 | 3.199 | 0.538–19.018 | 1.279 | 0.201 | 1.000 | 0.632 | |
| AA + AG vs. GG | Random | 97.81 | ≤0.001 | 3.455 | 0.536–22.25 | 1.304 | 0.192 | 1.000 | 0.674 | |
| AA vs. AG+ GG | Random | 70.92 | 0.032 | 5.559 | 0.509–60.68 | 1.407 | 0.160 | 1.000 | 0.884 | |
| TNF-α -238 G > A | ||||||||||
| Overall | A vs. G | Fixed | 0.00 | 0.452 | 1.009 | 0.641–1.587 | 0.038 | 0.970 | 1.000 | 0.411 |
| AA vs. GG | Fixed | 0.00 | 1.000 | 0.384 | 0.015–9.550 | −0.583 | 0.560 | NA | NA | |
| AG vs. GG | Fixed | 14.54 | 0.310 | 1.060 | 0.661–1.710 | 0.243 | 0.808 | 1.000 | 0.545 | |
| AA + AG vs. GG | Fixed | 0.796 | 0.365 | 1.036 | 0.648–1.657 | 0.148 | 0.882 | 1.000 | 0.497 | |
| AA vs. AG+ GG | Fixed | 0.00 | 1.000 | 0.360 | 0.015–8.942 | −0.623 | 0.533 | NA | NA | |
NA: Not Applicable.
Fig. 2.
Forest plots of the association between TNF-α −308 G > A polymorphism with knee OA. (A) the allele model (A vs. G); and (B) the dominant model (AA + AG vs. GG).
3.2.2.2. TNF-α -238G > A polymorphism
Table 3 also listed the main results of the meta-analysis of TNF-α -238G > A polymorphism and knee OA risk. When all the eligible studies were pooled into the meta-analysis of TNF-α -238G > A polymorphism, no evidence of a significant between TNF-α -238G > A polymorphism and knee OA risk in all the five genetic models, i.e., allele (A vs. G: OR = 1.009, 95% CI 0.641–1.587, p = 0.970), homozygote (AA vs. GG: OR = 0.384, 95% CI 0.015–9.550, p = 0.560), heterozygote (AC vs. GG: OR = 1.060, 95% CI 0.661–1.710, p = 0.808), dominant (AA + AG vs. GG: OR = 1.036, 95% CI 0.648–1.657, p = 0.882) and recessive model (AA vs. AG + GG: OR = 0.360, 95% CI 0.015–8.942, p = 0.533).
3.2.3. Between-Study heterogeneity
The heterogeneity was significant for TNF-α −308 G > A polymorphism in all five genetic models, but not for TNF-α -238G > A, which suggested that the TNF-α −308 G > A polymorphism was source of between-study heterogeneity. Therefore, to explore the sources of heterogeneity, we performed further subgroup analyses by ethnicity and source of controls respectively. As a result, ethnicity was found to contribute to substantial heterogeneity (Table 3).
3.2.4. Sensitivity analyses
We have performed the sensitivity analyses to determine whether modification of the inclusion criteria of this meta-analysis affected the results. The sensitivity analyses was conducted by sequential omission of each individual studies one at a time or by omitting studies in which the genotypes distributions in the controls significantly deviated from the HWE, did not materially alter the pooled ORs for TNF-α −308 G > A and −238G > A polymorphism in overall, indicating that the results was stable. However, after excluding Sobhan et al. study in stratified analysis among Asians, statistically significant association was observed for TNF-α −308 G > A polymorphism under the recessive model (AA vs. AG + GG: OR = 15.60, 95% CI 01.267–192.113, p = 0.032).
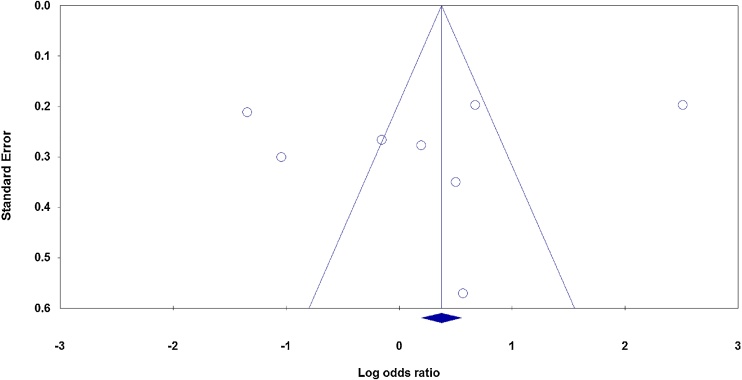
3.2.5. Publication bias
No obvious visual asymmetry was observed in Begg’s funnel plots (Fig. 3), and the results of Egger’s test also failed to reveal any evidence of publication bias among studies for both TNF-α −308 G > A and -238G > A polymorphisms (all P < 0.05).
Fig. 3.
Funnel plot analysis to detect publication bias for TNF-α −308 G > A polymorphism under the allele model.
4. Discussion
Given the pivotal role of TNF-α in the in the imbalance between anabolic and catabolic processes of knee OA, the previous studies have reported the TNF-α promoter −308 G > A and −238G > A polymorphism as a marker of knee OA.25,26,29 Although several case-control studies have discussed the TNF-α promoter region polymorphism and susceptibility to knee OA, but they did not include all of the eligible studies. Therefore, we performed the meta-analysis to investigate the association between the promoter region polymorphisms of TNF-α gene and knee OA risk.
To the best of our knowledge, this is the first meta-analysis assessing the association between TNF-α −238G > A polymorphism and knee OA. Previously, Abdel Galil et al. reported there is a significant association between TNF-α −308 G > A polymorphism and individual susceptibility to and severity of early-onset knee OA in the Egyptian females. The study by Ji et al. also found the allele A of TNF-α −308 may increase the risk for OA, whereas TNF-α -238G > A polymorphism do not play a role in knee OA in the Han Chinese population. In another study, Han et al. suggest close relationships between TNF-α −308 G > A and -238G > A polymorphisms and individual susceptibility to OA in the Korean population. Muñoz-Valle et al. reported that the TNFα levels were increased in GG versus GA genotypes in both TNF-α −308 G > A and -238G > A polymorphisms. However, they have suggest that the TNF-α −308 G > A and −238G > A polymorphisms are not associated with the risk of knee OA. However, our data revealed no significant association of TNF-α −308 G > A and −238G > A polymorphisms with knee OA risk. We suggest that the number and sample size of the selected studies is small and not sufficient enough to get a conclusive result.
In 2014, Kou et al., in a meta-analysis of seven studies, with 983 OA cases and 1355 controls, discussed the association between TNF-α −308 G > A and knee OA.30 Inconsistent with our meta-analysis they have found a significant association between TNF-α −308 G > A and knee OA risk under the allele model (OR = 2.30, 95% CI = 1.08–4.90) and the recessive genetic model analysis (OR = 11.08, 95% CI = 4.75-25.86, p < 0.001). However, we found that they wrongly included two studies by Romero et al. and Cheng et al. in their meta-analysis, which reported on the same or overlapping data from Munoz-Valle et al. 27 and Ji et al. 26 studies, respectively. Hence, it may significantly affect their total conclusions. Therefore, compared with Kou et al. meta-analyses, our meta-analysis involved a larger number of studies and provided a more comprehensive and reliable results.
In our analysis, there was evidence of heterogeneity between studies. It may be due to some factors, including the diversity in design, sample sizes, gender distribution, difference of ethnicity, the selection of methods, definition of cases, and sample sizes.31, 32, 33 Therefore, we conducted the subgroup analysis by ethnicity, control of sources and genotyping method trying to clarify the sources of heterogeneity. The results showed that Asian populations contribute to substantial between-study heterogeneity in the current meta-analysis. Notably, after removing the study by Sobhan et al. study in stratified analysis among Asians, statistically significant association was found for TNF-α −308 G > A polymorphism under the recessive model (AA vs. AG+GG: OR = 15.60, 95% CI 01.267–192.113, p = 0.032). This finding indicates that the Asian populations might have been genetically heterogeneous, with differences in terms of lifestyle and environment (e.g., East Asians vs. Iranian).
To our knowledge, this is the most accurate and first meta-analysis on TNF-α −308 G > A and −238G > A polymorphisms association with knee OA susceptibility. However, several limitations should be taken into consideration when explaining the results. First, the current meta-analysis was limited by the small sample size on both TNF-α −308 G > A and −238G > A polymorphisms, which needs further investigations. Second, only one of included studies was conducted in African and Latinos populations, thus we could not determine the role of TNF-α −308 G > A and −238 G > A polymorphisms in African and Latinos populations through the current meta-analyses. Third, due to the limited studies, we did not perform meta-analysis for association of the combined TNF-α −308 G > A and −238G > A polymorphisms and knee OA risk. Forth, given that we included only published studies in the current meta-analysis, publication bias may be present, although our results of publication bias showed no significance. Fifth, there was a significant between-study heterogeneity on TNF-α −308 G > A polymorphism under all five genetic models, and as such, results may be distorted. Different ethnicity and different sources of controls may contribute to the between-study heterogeneity. Finally, gene-gene and gene-environment interactions may have influenced the current meta-analysis findings, as knee OA is a multifactorial complex disorder mainly caused by genetic and environmental factors. However, no appropriate information was available to test this.
5. Conclusion
The current meta-analysis results indicate that the TNF-α −308 G > A and −238G > A polymorphisms might not be a risk factor for knee OA. Due to the limited studies, these findings are not robust and could be due to chance, and more epidemiologic studies with large sample that allow stratification for other gene-gene and gene-environment should also be performed in future analyses.
Conflicts of interest
The authors declare that they have no conflict of interest.
Acknowledgements
Not applicable.
References
- 1.Yazdi M.M., Jamalaldini M.H., Sobhan M.R. Association of ESRα gene Pvu II T & C, XbaI A & G and BtgI G & A polymorphisms with knee osteoarthritis susceptibility: a systematic review and meta-analysis based on 22 case-control studies. Arch Bone Jt Surg. 2017;5(6) [PMC free article] [PubMed] [Google Scholar]
- 2.Sobhan M.R., Mehdinejad M., Jamaladini M.H., Mazaheri M., Zare-Shehneh M., Neamatzadeh H. Association between aspartic acid repeat polymorphism of the asporin gene and risk of knee osteoarthritis: a systematic review and meta-analysis. Acta Orthop Traumatol Turc. 2017;51(5) doi: 10.1016/j.aott.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sobhan M.R., Mahdinezhad-Yazdi M., Jafari M., Mazaheri M., Neamatzadeh H., Daliri K. Association of ESRα XbaI A&G, PvuII T&C and ESRβ AlwNI T&C polymorphisms with the risk of adolescent idiopathic scoliosis: a systematic review and genetic meta-analysis. Rev Bras Ortop (English Ed) 2018;(March) doi: 10.1016/j.rboe.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woolf A.D., Pfleger B. Burden of major musculoskeletal conditions. Bull World Health Organ. 2003;81(9):646–656. http://www.ncbi.nlm.nih.gov/pubmed/14710506 (Accessed 10 April 2018) [PMC free article] [PubMed] [Google Scholar]
- 5.Bhatia D., Bejarano T., Novo M. Current interventions in the management of knee osteoarthritis. J Pharm Bioallied Sci. 2013;5(1):30–38. doi: 10.4103/0975-7406.106561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhai G., Hart D.J., Kato B.S., MacGregor A., Spector T.D. Genetic influence on the progression of radiographic knee osteoarthritis: a longitudinal twin study. Osteoarthr Cartil. 2007;15(2):222–225. doi: 10.1016/j.joca.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Wallace I.J., Worthington S., Felson D.T. Knee osteoarthritis has doubled in prevalence since the mid-20th century. Proc Natl Acad Sci U S A. 2017;114(35):9332–9336. doi: 10.1073/pnas.1703856114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Özler K., Aktaş E., Atay Ç, Yılmaz B., Arıkan M., Güngör Ş. Serum and knee synovial fluid matrixmetalloproteinase-13 and tumor necrosis factor-alpha levels in patients with late stage osteoarthritis. Acta Orthop Traumatol Turc. 2016;50(6):670–673. doi: 10.1016/j.aott.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Vasanthi P., Nalini G., Rajasekhar G. Role of tumor necrosis factor-alpha in rheumatoid arthritis: a review. APLAR J Rheumatol. 2007;10(4):270–274. [Google Scholar]
- 10.Parameswaran N., Patial S. Tumor necrosis factor-α signaling in macrophages. Crit Rev Eukaryot Gene Expr. 2010;20(2):87–103. doi: 10.1615/critreveukargeneexpr.v20.i2.10. http://www.ncbi.nlm.nih.gov/pubmed/21133840 . (Accessed 12 April 2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee C.-W., Lin C.-C., Luo S.-F. Tumor necrosis factor-alpha enhances neutrophil adhesiveness: induction of vascular cell adhesion molecule-1 via activation of Akt and CaM kinase II and modifications of histone acetyltransferase and histone deacetylase 4 in human tracheal smooth muscle cells. Mol Pharmacol. 2008;73(5):1454–1464. doi: 10.1124/mol.107.038091. [DOI] [PubMed] [Google Scholar]
- 12.Zhou J., Kawai T., Yu Q. Pathogenic role of endogenous in the development of Sjögren’s-like sialadenitis and secretory dysfunction in non-obese diabetic mice. Lab Invest. 2017;97(4):458–467. doi: 10.1038/labinvest.2016.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan Z., Lei-Butters D., Engelhardt J.F., Leno G.H. Indexing TNF-alpha gene expression using a gene-targeted reporter cell line. BMC Biol. 2009;7(8) doi: 10.1186/1741-7007-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mo Y.-Y., Zeng X.-T., Weng H., Cen Y., Zhao Q., Wen X. Association between tumor necrosis factor-alpha G-308A polymorphism and dental peri-implant disease risk. Medicine (Baltimore) 2016;95(35) doi: 10.1097/MD.0000000000004425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.KELLGREN J.H., LAWRENCE J.S. Radiological assessment of osteo-arthrosis. Ann Rheum Dis. 1957;16(4):494–502. doi: 10.1136/ard.16.4.494. http://www.ncbi.nlm.nih.gov/pubmed/13498604 . (Accessed 12 April 2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khoram-Abadi K.M., Forat-Yazdi M., Kheirandish S. DNMT3B -149 C&T and -579 G&T polymorphisms and risk of gastric and colorectal cancer: a meta-analysis. Asian Pacific J Cancer Prev. 2016;17(6) [PubMed] [Google Scholar]
- 17.Nedooshan J.J., Kargar S., Neamatzadeh H., Haghighi F., Abadi R.D.M., Seddighi N. Lack of association of the fat mass and obesity associated (FTO) gene rs9939609 polymorphism with breast cancer risk: a systematic review and meta-analysis based on case – control studies. Asian Pac J Cancer Prev. 2017;18(4) doi: 10.22034/APJCP.2017.18.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mantel N., Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. http://www.ncbi.nlm.nih.gov/pubmed/13655060. (Accessed 16 March 2018) [PubMed] [Google Scholar]
- 19.DerSimonian R., Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. http://www.ncbi.nlm.nih.gov/pubmed/3802833 . (Accessed 16 March 2018) [DOI] [PubMed] [Google Scholar]
- 20.Sadeghiyeh T., Biouki F.H., Mazaheri M., Zare-Shehneh M., Neamatzadeh H., Poursharif Z. Association between Catechol-O-Methyltransferase Val158Met (158G/A) polymorphism and suicide susceptibility: a meta-analysis. J Res Health Sci. 2017;17(2) [PubMed] [Google Scholar]
- 21.Begg C.B., Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. http://www.ncbi.nlm.nih.gov/pubmed/7786990 . (Accessed 16 March 2018) [PubMed] [Google Scholar]
- 22.Egger M., Davey Smith G., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. http://www.ncbi.nlm.nih.gov/pubmed/9310563 . (Accessed 16 March 2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moos V., Rudwaleit M., Herzog V., Höhlig K., Sieper J., Müller B. Association of genotypes affecting the expression of interleukin-1β or interleukin-1 receptor antagonist with osteoarthritis. Arthritis Rheum. 2000;43(11):2417–2422. doi: 10.1002/1529-0131(200011)43:11<2417::AID-ANR7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 24.Sezgin M., Barlas I.O., Ankarali H.C. Tumour necrosis factor alpha -308G/A gene polymorphism: lack of association with knee osteoarthritis in a Turkish population. Clin Exp Rheumatol. 2008;26(5):763–768. http://www.ncbi.nlm.nih.gov/pubmed/19032806 . (Accessed 9 April 2018) [PubMed] [Google Scholar]
- 25.Han L., Song J.H., Yoon J.H. TNF-α and TNF-β polymorphisms are associated with susceptibility to osteoarthritis in a Korean population. Korean J Pathol. 2012;46(1):30. doi: 10.4132/KoreanJPathol.2012.46.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji B., Shi J., Cheng X. Association analysis of two candidate polymorphisms in the tumour necrosis factor-α gene with osteoarthritis in a Chinese population. Int Orthop. 2013;37(10):2061–2063. doi: 10.1007/s00264-013-1931-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muñoz-Valle J.F., Oregón-Romero E., Rangel-Villalobos H. High expression of TNF alpha is associated with −308 and −238 TNF alpha polymorphisms in knee osteoarthritis. Clin Exp Med. 2014;14(1):61–67. doi: 10.1007/s10238-012-0216-3. [DOI] [PubMed] [Google Scholar]
- 28.Vnukov V.V., Krolevets I.V., Panina S.B. Gene polymorphism and levels of pro-inflammatory cytokines in plasma and synovial fluid of patients with posttraumatic knee osteoarthritis. Adv Gerontol. 2016;29(1):52–58. http://www.ncbi.nlm.nih.gov/pubmed/28423246 . (Accessed 9 April 2018) [PubMed] [Google Scholar]
- 29.Abdel Galil S.M., Ezzeldin N., Fawzy F., El-Boshy M. The single-nucleotide polymorphism (SNP) of tumor necrosis factor α −308G/A gene is associated with early-onset primary knee osteoarthritis in an Egyptian female population. Clin Rheumatol. 2017;36(11):2525–2530. doi: 10.1007/s10067-017-3727-1. [DOI] [PubMed] [Google Scholar]
- 30.Kou S., Wu Y. Meta-analysis of tumor necrosis factor alpha -308 polymorphism and knee osteoarthritis risk. BMC Musculoskelet Disord. 2014;15(1):373. doi: 10.1186/1471-2474-15-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jafari Nedooshan J., Kargar S., Neamatzadeh H., Haghighi F., Dehghani Mohammad Abadi R., Seddighi N. Lack of association of the fat mass and obesity associated (FTO) Gene rs9939609 polymorphism with breast cancer risk: a systematic review and meta-analysis based on case – control studies. Asian Pac J Cancer Prev. 2017;18(4):1031–1037. doi: 10.22034/APJCP.2017.18.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jafari Nedooshan J., Forat Yazdi M., Neamatzadeh H., Zare Shehneh M., Kargar S., Seddighi N. Genetic association of XRCC1 Gene rs1799782, rs25487 and rs25489 polymorphisms with risk of thyroid cancer: a systematic review and meta-analysis. Asian Pac J Cancer Prev. 2017;18(1):263–270. doi: 10.22034/APJCP.2017.18.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Namazi A., Forat-Yazdi M., Jafari M. Association of Interleukin-10-1082 a/G (Rs1800896) polymorphism with susceptibility to gastric cancer: meta-analysis of 6,101 cases and 8,557 controls. Arq Gastroenterol. 2018;55(1):33–40. doi: 10.1590/S0004-2803.201800000-18. [DOI] [PubMed] [Google Scholar]