Abstract
Background
Hypoxia is associated with a poor prognosis in prostate cancer. This work aimed to derive and validate a hypoxia-related mRNA signature for localized prostate cancer.
Method
Hypoxia genes were identified in vitro via RNA-sequencing and combined with in vivo gene co-expression analysis to generate a signature. The signature was independently validated in eleven prostate cancer cohorts and a bladder cancer phase III randomized trial of radiotherapy alone or with carbogen and nicotinamide (CON).
Results
A 28-gene signature was derived. Patients with high signature scores had poorer biochemical recurrence free survivals in six of eight independent cohorts of prostatectomy-treated patients (Log rank test P < .05), with borderline significances achieved in the other two (P < .1). The signature also predicted biochemical recurrence in patients receiving post-prostatectomy radiotherapy (n = 130, P = .007) or definitive radiotherapy alone (n = 248, P = .035). Lastly, the signature predicted metastasis events in a pooled cohort (n = 631, P = .002). Prognostic significance remained after adjusting for clinic-pathological factors and commercially available prognostic signatures. The signature predicted benefit from hypoxia-modifying therapy in bladder cancer patients (intervention-by-signature interaction test P = .0026), where carbogen and nicotinamide was associated with improved survival only in hypoxic tumours.
Conclusion
A 28-gene hypoxia signature has strong and independent prognostic value for prostate cancer patients.
Keywords: Gene expression signature, Hypoxia, Prognostic biomarker, Prostate cancer, Radiotherapy
Highlights
-
•
We identified genes regulated by hypoxia in prostate cancer cell lines.
-
•
In a cohort of primary tumours, the hypoxia regulated genes were reduced to a prognostic signature.
-
•
The mRNA abundance signature was prognostic of biochemical recurrence/metastatic recurrence in 11 independent cohorts.
Hypoxia, i.e. insufficient supply of oxygen, is an important micro-environmental factor in solid tumours, including prostate cancer. In this work, we identified genes whose abundance were affected by induced hypoxia (1% oxygen concentration) from prostate cancer cell lines. The hypoxia-regulated genes were then refined into a signature, i.e. a biomarker consisting of multiple genes, based on their ability to predict patient outcome after radical prostatectomy. The gene signature predicted the risk of developing biochemical and metastatic recurrence in >1000 patients with primary tumours receiving varying treatments. The prognostic value was independent from existing clinic-pathological factors.
1. Introduction
Ninety percent of prostate cancer (PCa) patients are diagnosed with localized carcinoma, which have a highly variable course of disease progression. Hypoxia is a common micro-environmental feature in most solid tumours, which leads to changes in transcriptomic profiles, a higher potential to metastasise and resistance to radiotherapy [1]. Localized prostate cancer has marked and heterogeneous hypoxia [2], and hypoxia is an adverse prognostic feature [[3], [4], [5]]. Combining hypoxia-targeting treatment with radiotherapy was shown to improve local control of tumours and survival of patients in head and neck and bladder cancers [[6], [7], [8], [9]]. There is good evidence from the literature that the most hypoxic tumours benefit the most from hypoxia-modifying therapy. However, there is no clinically validated method of selecting prostate cancer patients who would benefit from hypoxia modifying treatment.
Hypoxia gene signatures were successfully derived for multiple tumour sites including head and neck, bladder, soft tissue sarcoma and cervical cancers, which were not only independently prognostic but also predictive of benefit from hypoxia-modifying therapy in head and neck and bladder cancers [[10], [11], [12], [13], [14], [15], [16], [17], [18]]. Work in prostate cancer showed some prognostic significance for signatures derived in other tumour types [3] or associated with the hypoxia marker pimonidazole [15].
We previously showed that hypoxia gene signatures are better if tumour site specific [19]. The aim of this study was to generate and validate a prostate cancer-specific transcriptomic signature. In vitro analysis of genes regulated by hypoxia was combined with in vivo analysis of a gene co-expression network and patient survival using data from a retrospective cohort. Following signature derivation, validation was performed in multiple independent cohorts.
2. Methods
2.1. Patient Cohorts
This study included patients with prostate carcinoma from twelve cohorts, including the cancer genome atlas project (TCGA) [20], GSE54460 [21], GSE21032 [22], CPC-GENE [23], Cambridge [24,25], six retrieved from the Decipher GRID™ prostate cancer database (NCT02609269) [[26], [27], [28], [29], [30], [31]], and one from Belfast [32]. A summary of the cohorts and procedures for pre-processing of transcriptomic data are provided in Supplementary Table 1 and Supplementary Methods. Patient cohort characteristics are summarised in Supplementary Table 2. The TCGA cohort was used as the training cohort. In addition, bladder cancer patients enrolled in a randomized trial of hypoxia-modifying therapy were available in the BCON cohort [6,18]. Informed consent protocols were approved by local Institutional Review Boards.
2.2. Endpoints and Statistical Analysis
Biochemical recurrence (BCR) free survival was the clinical endpoint of this study in all but one cohort. Distant metastasis (DMET) free survival was the clinical endpoint of the other cohort. Patient follow up data were censored at 5-year. The signature derived from this work assigned one hypoxia signature score for each individual tumour and in each cohort patients were stratified into high vs. low hypoxia based on the median cohort signature score. Survival estimates were performed using the Kaplan-Meier method. The Log-rank test was used to test the null hypothesis of equality of survival distributions. Hazard ratios (HR) and 95% confidence intervals (CI) were obtained using the Cox proportional hazard model. In case-cohort studies (originally designed to sample the adverse pathology population to estimate risk of metastasis after radical prostatectomy), randomly sampled sub-cohorts were used in order to reduce over-estimation of events in evaluation of the BCR endpoint. Odds ratios (OR) were obtained using logistic regression models. Association between hypoxia signature score and Gleason group, tumour stage was estimated using simple linear regression. All P-values were two sided and statistical significance was set as 0.05.
2.3. Prostate Cancer Hypoxia Signature Development
A network-based methodology was applied for generation of a prostate cancer-specific hypoxia signature, hypothesising that in vitro hypoxia regulated genes co-expressing with each other in vivo collectively indicate tumour hypoxia. Briefly, genes up- and down-regulated by hypoxia (1% O2, 24 h) were identified with RNA sequencing in four PCa cell lines (PNT2-C2, LNCaP, DU-145, and PC-3). The four cell lines were chosen as they were derived from human tissues and are widely used as in vitro PCa models [33]. A gene co-expression network was constructed in a TCGA training cohort and a putative gene module enriched with in vitro hypoxia genes was identified. Genes within the putative module were ranked by their connectivity and added iteratively into the signature. The final signature was selected based on prognostic significance. The signature scores are a continuous variable, which were then binarised into high and low categories using the median value for each cohort. The hypoxia signature derived here was then independently validated in the other eleven PCa cohorts. In multivariable analysis, the hypoxia gene signature was adjusted for standard clinic-pathological factors and a genomic classifier Decipher [27]. More details were given in Supplementary Methods.
2.4. Comparison with Literature Transcriptomic and Genomic Signatures
Seven hypoxia [10,11,[14], [15], [16], [17]] and eleven prostate cancer transcriptomic signatures [[34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44]]were curated from the literature and their prognostic significances were evaluated for comparison (Supplementary methods). Furthermore, the potential benefit of combining the de novo hypoxia signature with a prognostic 31-loci DNA classifier [45] was also investigated by entering both into a Cox model.
3. Results
3.1. In Vitro Hypoxia Responsive Genes
Genes up or down regulated under hypoxia in four PCa cell lines were identified. 84, 306 and 848 genes were differentially expressed in greater than four, three and two cell lines, respectively. A seed gene database was constructed consisting of the 848 genes differentially expressed in two or more cell lines (Supplementary Table 3). Eight cell cycle and metabolism pathways were enriched in the hypoxia seed gene dataset (Supplementary Table 4). For many of the above identified genes, their regulation under hypoxia was well documented [46,47].
3.2. In Vivo Prostate Cancer-specific Hypoxia Gene Module
A PCa gene co-expression network was built from pre-treatment tumour biopsies in the TCGA cohort. The network, containing 1856 genes (including 113 seed genes) of good variability was partitioned into 34 gene modules. One module of 66 genes was identified most hypoxia-related as 33% of its member genes (22/66, P = 4.85 ∗ 10−12, Supplementary Fig. 1) were seed genes. Another 24 genes from the 66-gene module were hypoxia regulated in one cell line (Supplementary Fig. 2). The final signature, containing 28 genes from the 66-gene module, was derived by evaluating prognostic significance in cross validation. The signature gene coefficients were determined from a Cox survival model (Supplementary Table 5). Patients with high signature scores were associated with significantly poorer outcome (log rank P = 1.9 ∗ 10−8, Supplementary Fig. 3).
3.3. 28-Gene Signature Is Prognostic of BCR in PCa Cohorts Receiving Radical Prostatectomy
The high correlation among the 28 hypoxia signature genes was well preserved in the independent validation cohorts (Supplementary Fig. 4), suggesting the good reproducibility of the signature. The 28 signature genes also clustered tumours in independent validation cohorts into two clear groups (Supplementary Figs. 5 and 6).
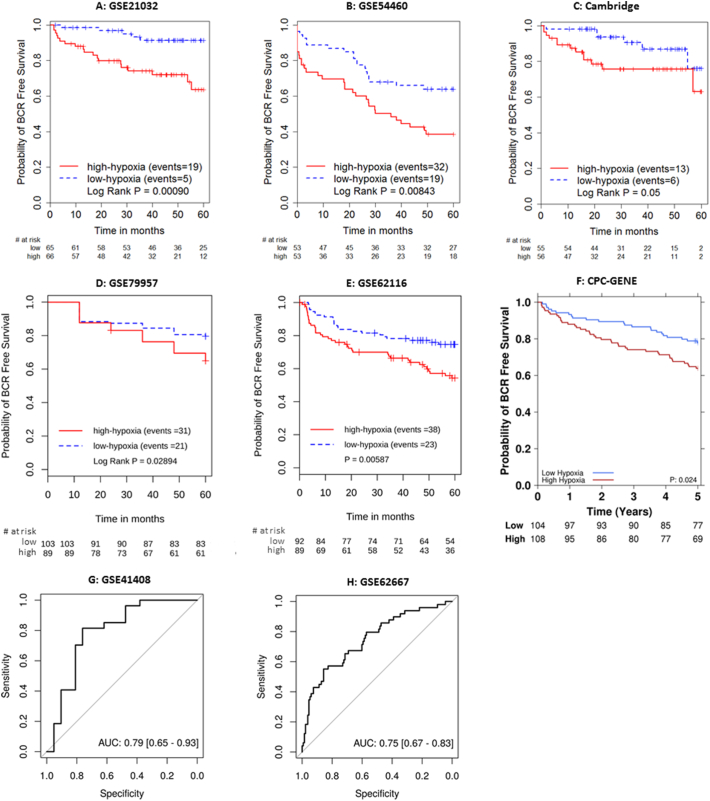
The 28-gene signature and all coefficients were frozen and applied to independent validation cohorts. The prognostic value of the 28-gene hypoxia signature was first validated in the GSE21032 (fresh frozen), GSE54460 (FFPE) and Cambridge (fresh frozen) prostatectomy cohorts. In both GSE21032 and GSE54460, tumours with high 28-gene signature scores were associated with significantly poorer 5-year BCR in the GSE21032 (n = 131, HR 4.59, 95% CI 1.71–12.32, P = .0025, Fig. 1A) and GSE54460 (n = 106, HR 2.12, 95% CI 1.20–3.74, P = .0098, Fig. 1B) cohorts. In the Cambridge cohort, borderline prognostic significance was observed with the default median cut-off signature score (n = 111, HR 2.54, 95% CI 0.96–6.69, P = .06, Fig. 1C), which improved when an upper quartile split was used (HR 4.54, 95% CI 1.82–11.36, P = .0011). A continuous 28-gene signature score also achieved prognostic significance (P = .047) in the Cambridge cohort.
Fig. 1.
Kaplan-Meier or receiver operating characteristic plots for independent validation of the 28-gene hypoxia signature in prostatectomy-treated cohorts.
In other five prostatectomy-treated cohorts, tumours with high 28-gene signature scores were again associated with significantly poorer outcome (GSE79957: n = 260, HR 1.60, P = .055; GSE62116: n = 235, HR 2.04 P = .007; GSE41408: n = 48, OR 7.60, P = .0022; GSE62667: n = 182, OR 5.90, P = .000036; and CPC-GENE: n = 212, HR 1.80, P = .026. Fig. 1D to H).
Multivariable analysis was performed adjusting the hypoxia signature with clinicopathological covariates and the Decipher genomic classifier, wherever possible (Table 1). Statistical significance was retained in three cohorts (GSE21032: HR 3.51, P = .021; GSE54460: HR 1.84, P = .048; and CPC-GENE: HR 1.81, P = .021), and was borderline in another two cohorts (GSE62116: HR 1.36, P = .059; and GSE41408: OR 2.89, P = .081).
Table 1.
Multivariable analysis of the 28-gene hypoxia signature in independent cohorts of PCa patients receiving radical prostatectomy.
| Study | Variable | Multivariable analysis | |
|---|---|---|---|
| Cohort and case-cohort studies | HR (95% CI) | P-value | |
| GSE21032 | Hypoxia | 3.51 (1.21–10.15) | 0.021 |
| Gleason Group 2 VS 1 | 0.69 (0.20–2.47) | 0.57 | |
| Gleason Group 3 VS 1 | 4.12 (1.32–12.91) | 0.015 | |
| Gleason Group ≥ 4 VS 1 | 2.45 (0.72–8.31) | 0.15 | |
| Log2 pre-treatment PSA | 1.04 (0.64–1.69) | 0.88 | |
| SVI | 3.86 (1.25–11.92) | 0.019 | |
| SM | 1.44 (0.55–3.74) | 0.46 | |
| ECE | 2.58 (0.70–9.52) | 0.155 | |
| GSE54460 | Hypoxia | 1.84 (1.00–3.39) | 0.048 |
| Gleason Group 2 VS 1 | 1.49 (0.42–5.34) | 0.54 | |
| Gleason Group 3 VS 1 | 2.77 (0.76–10.13) | 0.123 | |
| Gleason Group 4 VS 1 | 2.21 (0.45–10.74) | 0.33 | |
| Gleason Group 5 VS 1 | 2.33 (0.50–10.83) | 0.28 | |
| Log2 pre-treatment PSA | 1.74 (1.21–2.50) | 0.0029 | |
| Clinical T Stage 2 VS 1 | 0.47 (0.22–1.01) | 0.052 | |
| Clinical T Stage ≥ 3 VS 1 | 0.62 (0.23–1.62) | 0.33 | |
| SM | 2.47 (1.35–4.55) | 0.0035 | |
| Cambridge | Hypoxia | 1.53 (0.54–4.30) | 0.42 |
| Gleason Group 2 VS 1 | 1.84 (0.20–17.28) | 0.59 | |
| Gleason Group 3 VS 1 | 8.27 (0.79–86.99) | 0.078 | |
| Gleason Group 4 VS 1 | 15.66 (1.36–181.00) | 0.028 | |
| Gleason Group 5 VS 1 | 490.3 (11.7–20,542.2) | 0.0011 | |
| Log2 pre-treatment PSA | 0.54 (0.22–1.37) | 0.19 | |
| SM | 1.32 (0.46–3.76) | 0.61 | |
| ECE | 1.42 (0.42–4.72) | 0.57 | |
| CPC-GENE | Hypoxia | 1.81 (1.02–3.21) | 0.021 |
| Gleason Group 2 VS 1 | 0.85 (0.35–2.05) | 0.68 | |
| Gleason Group 3 VS 1 | 0.82 (0.31–2.20) | 0.65 | |
| Gleason Group ≥ 4 VS 1 | 3.00 (0.27–33.85) | 0.310 | |
| Pre-treatment PSA | 1.04 (0.98–1.11) | 0.152 | |
| Clinical T Stage 2 VS 1 | 1.80 (1.03–3.13) | 0.018 | |
| GSE62116 | Hypoxia | 1.36 (0.99–1.88) | 0.059 |
| Decipher | 1.15 (0.86–1.53) | 0.36 | |
| Gleason Group 3 VS ≤ 2 | 2.21 (1.10–4.40) | 0.026 | |
| Gleason Group 4 VS ≤ 2 | 1.29 (0.57–2.93) | 0.54 | |
| Gleason Group 5 VS ≤ 2 | 1.13 (0.53–2.44) | 0.75 | |
| Log2 pre-treatment PSA | 1.21 (0.96–1.52) | 0.10 | |
| SVI | 0.87 (0.46–1.66) | 0.68 | |
| SM | 1.67 (0.95–2.95) | 0.076 | |
| ECE | 1.83 (1.01–3.31) | 0.046 | |
| LNI | 0.73 (0.31–1.74) | 0.48 | |
| GSE79957 | Hypoxia | 0.93 (0.58–1.48) | 0.76 |
| Decipher | 1.58 (1.11–2.23) | 0.010 | |
| Gleason Group 3 VS ≤ 2 | 0.84 (0.33–2.17) | 0.72 | |
| Gleason Group 4 VS ≤ 2 | 1.32 (0.46–3.82) | 0.61 | |
| Gleason Group 5 VS ≤ 2 | 2.46 (1.20–5.03) | 0.014 | |
| Log2 pre-treatment PSA | 1.25 (0.94–1.66) | 0.122 | |
| SVI | 1.57 (0.82–3.03) | 0.180 | |
| SM | 1.96 (0.99–3.88) | 0.052 | |
| ECE | 2.18 (0.85–5.60) | 0.104 | |
| LNI | 4.71 (2.43–9.14) | <0.001 | |
| Case control studies | OR (95% CI) | P-value | |
| GSE62667 | Hypoxia | 1.19 (0.62–2.30) | 0.61 |
| Decipher | 2.26 (1.28–4.23) | 0.0069 | |
| Gleason Group 3 VS ≤ 2 | 1.77 (0.47–6.38) | 0.39 | |
| Gleason Group 4 VS ≤ 2 | 2.03 (0.43–8.98) | 0.36 | |
| Gleason Group 5 VS ≤ 2 | 8.68 (1.94–42.91) | 0.0057 | |
| Log2 pre-treatment PSA | 1.56 (0.97–2.57) | 0.071 | |
| SVI | 2.70 (0.81–8.99) | 0.103 | |
| SM | 2.20 (0.80–6.39) | 0.133 | |
| ECE | 3.99 (1.01–20.55) | 0.067 | |
| GSE41408 | Hypoxia | 2.89 (0.94–10.79) | 0.081 |
| Decipher | 0.41 (0.089–1.66) | 0.22 | |
| Gleason Group 3 VS ≤ 2 | 1 (0-NA) | 0.99 | |
| Gleason Group ≥ 4 VS ≤ 2 | 3.94 (0.29–67.75) | 0.31 | |
| Log2 pre-treatment PSA | 3.02 (1.29–9.55) | 0.031 | |
PSA: prostate specific antigen; SVI: seminal vesicle invasion; SM: surgical margin; ECE: extracapsular extension; LNI: lymph node invasion.
3.4. 28-Gene Signature is a Strong Independent Prognostic Factor in PCa Cohorts Receiving Radiotherapy
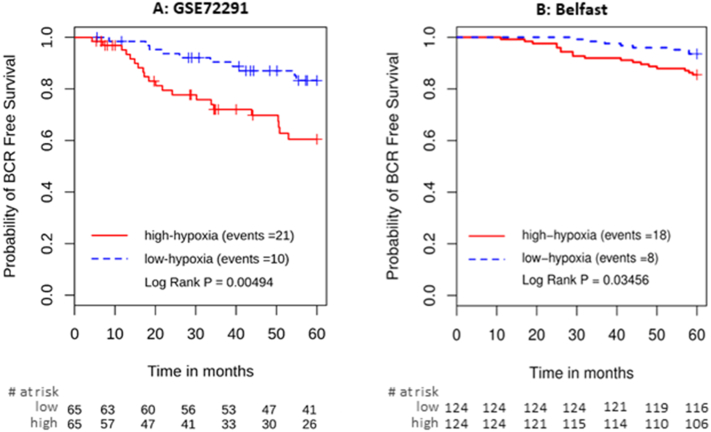
The 28-gene signature predicted biochemical recurrence in the GSE72291 cohort treated with post-operative radiotherapy (n = 130, HR 2.81, 95% CI 1.33–6.00, P = .007, Fig. 2A). In multivariable analysis (Table 2), the hypoxia signature remained a strong prognostic factor (HR = 2.17, P = .014) alongside Decipher (HR 2.24, P = .0061), Gleason group 5 (HR 8.50, P = .0012), and pre-treatment PSA level (HR 1.74, P = .0042). The hypoxia signature was also prognostic in the Belfast cohort of intermediate/high-risk patients who underwent definitive radiotherapy (n = 248, log rank P = .035, Fig. 2B). Multivariable analysis was not performed due to the low event rate.
Fig. 2.
Kaplan-Meier plots for independent validation of the 28-gene hypoxia signature in A) post-prostatectomy radiotherapy cohort; B) radiotherapy-treated cohort.
Table 2.
Multivariable analysis of the 28-gene signature in predicting biochemical recurrence in patients receiving radical prostatectomy and adjuvant radiotherapy.
| Study | Variable | Multivariable analysis | |
|---|---|---|---|
| GSE72291 | Hypoxia | 2.17 (1.17–4.01) | 0.014 |
| Decipher | 2.24 (1.26–4.00) | 0.0061 | |
| Gleason Group 3 VS ≤ 2 | 2.49 (0.72–8.56) | 0.148 | |
| Gleason Group 4 VS ≤ 2 | 2.34 (0.67–8.21) | 0.183 | |
| Gleason Group 5 VS ≤ 2 | 8.50 (2.34–30.92) | 0.0012 | |
| Log2 pre-treatment PSA | 1.74 (1.19–2.55) | 0.0042 | |
| SVI | 1.21 (0.51–2.85) | 0.66 | |
| SM | 0.60 (0.25–1.49) | 0.27 | |
| ECE | 3.15 (0.89–11.17) | 0.075 | |
PSA: prostate specific antigen; SVI: seminal vesicle invasion; SM: surgical margin; ECE: extracapsular extension; LNI: lymph node invasion.
3.5. 28-Gene Signature Predicts Metastatic Outcome
The 28-gene signature was also evaluated in a pooled cohort of 631 PCa patients from four studies with metastatic outcome [[29], [30], [31],48]. The signature predicted distant metastasis events with a HR of 2.41 (95% CI 1.38–4.19, P = .002). In multivariable analysis, the hypoxia signature remained significant (HR 2.57, 95% CI 1.38–4.77, P = .003) alongside Decipher, seminal vesicle invasion and lymph node invasion (Table 3).
Table 3.
Multivariable analysis of the 28-gene signature in predicting metastatic outcome.
| Study | Variable | Multivariable analysis | |
|---|---|---|---|
| Pooled cohort | Hypoxia | 2.57 (1.38–4.77) | 0.003 |
| Decipher | 1.22 (1.06–1.41) | 0.007 | |
| Gleason Group 2 VS 1 | 1.13 (0.14–9.30) | 0.907 | |
| Gleason Group 3 VS 1 | 3.50 (0.43–28.26) | 0.239 | |
| Gleason Group 4 VS 1 | 3.67 (0.45–30.06) | 0.226 | |
| Gleason Group 5 VS 1 | 5.83 (0.74–45.96) | 0.094 | |
| Log2 pre-treatment PSA | 1.02 (0.79–1.33) | 0.864 | |
| AGE | 0.99 (0.95–1.03) | 0.522 | |
| SVI | 2.01 (1.15–3.51) | 0.014 | |
| SM | 1.45 (0.84–2.51) | 0.18 | |
| ECE | 1.39 (0.70–2.75) | 0.346 | |
| LNI | 1.95 (1.04–3.67) | 0.037 | |
PSA: prostate specific antigen; SVI: seminal vesicle invasion; SM: surgical margin; ECE: extracapsular extension; LNI: lymph node invasion.
3.6. 28-Gene Signature Is Associated With Gleason Score and Tumour Stage
A positive and significant correlation was seen between the 28-gene signature score and pathological Gleason score in five of the six examined cohorts (Supplementary Fig. 7). Hypoxia score increased significantly with tumour stage in four of the six cohorts (Supplementary Fig. 8), but was not associated with pre-treatment PSA (Supplementary Fig. 9). In GSE21032, the hypoxia signature scores were significantly higher in metastatic compared with primary tissue (P = .007, Supplementary Fig. 10).
3.7. Prediction of Benefit from Hypoxia Modification of Radiotherapy
The 28-gene prostate signature was further validated in the BCON phase III randomized trial of radiotherapy alone or with carbogen and nicotinamide (CON) for bladder cancer patients. Tumours were stratified as high or low from median, first, and third quartiles of cohort 28-gene signature scores. When the first quartile was used, high hypoxic tumours were associated with a non-significant trend of poor prognosis in patients treated with radiotherapy only (overall survival, n = 75, HR 1.68, P = .17, Supplementary Fig. 11). BCON tumours stratified as high hypoxic had improved survival with the addition of CON (n = 113, HR 0.54, 95% CI 0.32–0.91, P = .021). For low-hypoxic tumours, giving CON with RT was associated with poorer survival (n = 38, HR 2.49, 95% CI 1.06–5.89, P = .037, Supplementary Fig. 12). A signature-intervention interaction test confirmed the predictive value of the signature for both binary hypoxic status (P = .0026) and continuous hypoxia score (P = .037). CON was shown to have a detrimental effect on the 25% BCON patients with low 28-gene signature scores. As one given treatment would only benefit a subset of patients and might even produce adverse effect on others, this emphasized the importance of identifying reliable predictive biomarker.
3.8. Comparison with Literature Gene Signatures
The prognostic value of published hypoxia and PCa transcriptomic signatures were compared in both GSE21032 and GSE54460 (Supplementary Tables 6 and 7). One PCa hypoxia signature [15] reached (borderline) prognostic significance in both cohorts. The Toustrup signature [16], derived for head and neck cancer, was prognostic in both cohorts. Our 28-gene signature scores correlated moderately with Toustrup signature scores (GSE21032: Pearson r = 0.33, P = .00011; GSE54460: Pearson r = 0.51, P = 2.32 ∗ 10−8, Supplementary Fig. 13). The 28-gene signature had higher prognostic significance and was the only significant factor when both signatures were entered into a Cox proportional hazards regression model (Supplementary Table 8).
3.9. Association with Copy Number Alteration Burden and Genomic Classifier
Recent studies suggested global copy number alteration (CNA) burden as a prognostic marker of PCa patients [22,49]. A very moderate correlation was found between global CNA burden, defined as the percentage of the tumour genome affected by CNA, and hypoxia signature scores for both TCGA (Pearson correlation 0.36, Supplementary Fig. 14) and GSE21032 (Pearson correlation 0.40, Supplementary Fig. 14). Hypoxia signature (binary or continuous) and CNA burden (continuous) both retained significance when entered together into a Cox model in both cohorts (Supplementary Tables 9 and 10).
A 100-loci prognostic classifier for localized PCa [3] was recently identified and reduced to a 31-loci signature [45]. Both reflect genomic instability and were shown to have high prognostic value in >500 prostate cancer patients. We examined if adding the 28-gene signature and genomic classifier could further improve prognostication for PCa patients. In the CPC-GENE cohort [23], the 28-gene signature (log rank P = .023, Fig. 1F) achieved similar prognostic significance compared with the 31-loci genomic instability classifier (log rank P = .042, Supplementary Fig. 15). Combining the 31-loci genomic classifier with the 28-gene signature improved prognostication (log rank P = .0035, Supplementary Fig. 16). Using patients with low 28-gene signature and low 31-loci signature as a reference, patients with high 28-gene signature and high 31-loci signature had a significantly poorer outcome (HR 2.75, 95% CI 1.42–5.31, P = .0026). Patients with only positive 28-gene signature (P = .58) or 31-loci signature (P = .41) had similar survival as the double negative group, respectively (Supplementary Table 11). The above analyses suggested that improved prognostication can be achieved by combining the 28-gene signature and indicators of genome instability, consistent with the previous observation from Lalonde et al. [3].
4. Discussion
A 28-gene signature was derived and shown to be independently prognostic in eleven cohorts of low- to high-risk prostate cancer patients with localized disease. Ten genes (ATF3, BHLHE40, SLC2A3, EGR1, KLF10, FOSL2, CYR61, SLC2A14, KLF6, and TIPARP) in the 28-gene signature were previously shown to be hypoxia regulated in different tumour sites (Supplementary Table 12). The study showed that the Toustrup hypoxia signature [16], prognostic and predictive of hypoxia-modifying therapy for head and neck cancer, was prognostic in prostate cancer patients. The Ragnum signature [15], the only prostate cancer-specific hypoxia signature in the literature, had (borderline) prognostic significance for biochemical recurrence. A moderate correlation was found between the Toustrup 15-gene and our 28-gene hypoxia signatures score. The derived 28-gene signature could be combined with a recently published genomic classifier to identify a group of high-risk PCa patients with very poor prognosis. The 28-gene hypoxia signature has translational significance. The prognostic value of the signature was independently validated in seven cohorts of biopsies preserved in FFPE blocks, a routine technique for preserving tissues in hospitals. Degradation and chemical modification of nucleic acids in FFPE samples significantly reduces RNA quality. [50] The prognostic significance of the signature was also confirmed using three major high-throughput expression profiling platforms, i.e. Affymetrix array, Illumina BeadArray and RNA-seq. The results of the current study clearly indicate the robustness of the signature for routine clinical application.
One major limitation for the derived 28-gene signature is the lack of benchmarking against gold-standard hypoxia measurement with polographic needle oxygen electrodes. Prognostic information was used in the signature derivation procedure, therefore the final signature could reflect other poor prognostic parameters beyond hypoxia. Use of needle electrode presents great technical challenge, where to date the Toustrup [16] is the only gene expression biomarker that reflected oxygen partial pressure in human tissue. Ragum [15] derived a prostate hypoxia signature based on pimonidazole staining, another approach for assessing tumour hypoxia.
In this study, 1% oxygen concentration was chosen for in vitro experiment as it is widely used in the literature and protein expression of HIF, a major regulator of transcriptional responses to hypoxia, stabilizes in cell lines around this oxygen concentration. One limitation is that certain transcriptional program, for example unfolded protein response genes [51] which are generally activated in oxygen level ≤ 0.2%, might be missed. Furthermore, previous research suggested that prostate tumours were profoundly hypoxic with a median oxygen concentration of ~0.5% [52]. Therefore, the signature derived here could be further refined in future studies by adding genes regulated at even lower oxygen concentration.
Another limitation is that the 28-gene signature was prospectively validated in retrospective cohorts, where no standardisation could be achieved in terms of tissue handling, RNA extraction, the amount of tumour material, etc. However, the prognostic significance was clearly validated, showing a good level of signature robustness. Other limitations include the inability to validate the signature in a prostate cancer phase III trial involving hypoxia modification with both gene expression data and patient outcome. The 28-gene prostate signature was predictive of hypoxia modification therapy for bladder cancer – the only hypoxia modification trial available with full gene expression data. Use of a median score as a cut-off might not be optimal but was chosen as a pre-determined threshold to avoid bias and to provide balanced groups. Also, a median cut-off was used for our head and neck and bladder signatures. Different thresholds were used for different hypoxia signatures and arrays [16,53], and there is no consensus on the ideal method to define tumours as hypoxic and no methodological study exists to date assessing and comparing the performance of the different methods. In a prospective clinical trial the first ~50 patient samples could be used to generate a median threshold in newer FFPE blocks for classification.
The 28-gene signature might be further reduced while achieving comparable or slightly worse but acceptable prognostic value. Identifying the minimum set of genes for a signature is a very open research problem. The enormous number of unique combinations of 28 genes definitely opens the door for potential reduction of the signature.
In conclusion, a hypoxia gene signature for primary prostate tumours was derived. Employed as a potential tool for personalized medicine, the signature identifies that more hypoxic tumours have poorer outcome. The signature warrants final qualification in a prospective 28-gene signature-driven randomized trial of hypoxia-modifying therapy.
Conflictof Interest
MT, NE, MA, and ED are employees of GenomeDx Biosciences. RJK received research grant and royalties from GenomeDX.
Author Contribution
Bioinformatics: LY, MT, NE, VB, WCC, SH, AMBM, DM, MA.
Cell line work: DR, BASB, NT.
Result interpretation: LY, DR, MT, NE, VB, WCC, SH, FMB, RBD, AC, CMLW.
Provide data access: AR, ES, RBD, RJK, EK, PJH, SJ, SJF, ADL, DEN, RGB, PCB, ED.
Manuscript preparation: LY, MT, WCC, MA, FMB, PCB, AC, CMLW.
Design of methodology: LY, FMB, AC, CMLW.
Project supervision: DEN, RGB, PCB, ED, AC, CMLW.
All authors approved the final version of the manuscript.
Acknowledgement
Belfast-Manchester Movember Centre of Excellence (CE013_2-004), funded in partnership with Prostate Cancer UK (PG14 008 TR2). Cancer Research UK Major Centre funding. Cancer Research UK Manchester Institute Cancer Research Molecular Biology Core Facility and the University of Manchester Clinical Immune and Molecular Monitoring Laboratory for use of Good Clinical Practice facilities. Medical Research Council of the UK (grant G0801525). Cancer Research UK (grant C2094/A11365). Experimental Cancer Medicine Centre funding. CIHR New Investigator Award, TFRI New Investigator Award, CIHR Graduate Fellowship, Prostate Cancer Canada and the Movember Foundation (Grant RS2014-01). Cancer Research UK (grant 23969) and Oxford MRC/CRUK Institute. The work was supported by researchers at the NIHR Manchester Biomedical Research Centre. The funding sources have no roles in writing or decision or submit it for publication.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.04.019.
Appendix A. Supplementary Data
Supplementary material
References
- 1.Hill R.P., Bristow R.G., Fyles A., Koritzinsky M., Milosevic M., Wouters B.G. Hypoxia and predicting radiation response. Semin Radiat Oncol. 2015;25:260–272. doi: 10.1016/j.semradonc.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Parker C., Milosevic M., Toi A., Sweet J., Panzarella T., Bristow R. Polarographic electrode study of tumor oxygenation in clinically localized prostate cancer. Int J Radiat Oncol Biol Phys. 2004;58:750–757. doi: 10.1016/S0360-3016(03)01621-3. [DOI] [PubMed] [Google Scholar]
- 3.Lalonde E., Ishkanian A.S., Sykes J., Fraser M., Ross-Adams H., Erho N. Tumour genomic and microenvironmental heterogeneity for integrated prediction of 5-year biochemical recurrence of prostate cancer: a retrospective cohort study. Lancet Oncol. 2014;15:1521–1532. doi: 10.1016/S1470-2045(14)71021-6. [DOI] [PubMed] [Google Scholar]
- 4.Milosevic M., Warde P., Ménard C., Chung P., Toi A., Ishkanian A. Tumor hypoxia predicts biochemical failure following radiotherapy for clinically localized prostate cancer. Clin Cancer Res. 2012;18:2108–2114. doi: 10.1158/1078-0432.CCR-11-2711. [DOI] [PubMed] [Google Scholar]
- 5.Turaka A., Buyyounouski M.K., Hanlon A.L., Horwitz E.M., Greenberg R.E., Movsas B. Hypoxic prostate/Muscle P<span class="small">o</span><sub>2</sub> ratio predicts for outcome in patients with localized prostate cancer: long-term results. Int J Radiat Oncol Biol Phys. 2012;82:e433–e439. doi: 10.1016/j.ijrobp.2011.05.037. [DOI] [PubMed] [Google Scholar]
- 6.Hoskin P.J., Rojas A.M., Bentzen S.M., Saunders M.I. Radiotherapy with concurrent carbogen and nicotinamide in bladder carcinoma. J Clin Oncol. 2010;28:4912–4918. doi: 10.1200/JCO.2010.28.4950. [DOI] [PubMed] [Google Scholar]
- 7.Janssens G.O., Rademakers S.E., Terhaard C.H., Doornaert P.A., Bijl H.P., Ende P.V.D. Accelerated radiotherapy with carbogen and nicotinamide for laryngeal cancer: results of a phase III randomized trial. J Clin Oncol. 2012;30:1777–1783. doi: 10.1200/JCO.2011.35.9315. [DOI] [PubMed] [Google Scholar]
- 8.Overgaard J. Hypoxic modification of radiotherapy in squamous cell carcinoma of the head and neck – a systematic review and meta-analysis. Radiother Oncol. 2011;100:22–32. doi: 10.1016/j.radonc.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Overgaard J., Sand Hansen H., Overgaard M., Bastholt L., Berthelsen A., Specht L. A randomized double-blind phase III study of nimorazole as a hypoxic radiosensitizer of primary radiotherapy in supraglottic larynx and pharynx carcinoma. Results of the Danish Head and Neck Cancer Study (DAHANCA) Protocol 5-85. Radiother Oncol. 1998;46:135–146. doi: 10.1016/s0167-8140(97)00220-x. [DOI] [PubMed] [Google Scholar]
- 10.Betts G.N.J., Eustace A., Patiar S., Valentine H.R., Irlam J., Ramachandran A. Prospective technical validation and assessment of intra-tumour heterogeneity of a low density array hypoxia gene profile in head and neck squamous cell carcinoma. Eur J Cancer. 2013;49:156–165. doi: 10.1016/j.ejca.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 11.Buffa F.M., Harris A.L., West C.M., Miller C.J. Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. Br J Cancer. 2010;102:428–435. doi: 10.1038/sj.bjc.6605450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eustace A., Mani N., Span P.N., Irlam J.J., Taylor J., Betts G.N.J. A 26-gene hypoxia signature predicts benefit from hypoxia-modifying therapy in laryngeal cancer but not bladder cancer. Clin Cancer Res. 2013;19:4879–4888. doi: 10.1158/1078-0432.CCR-13-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fjeldbo C.S., Julin C.H., Lando M., Forsberg M.F., Aarnes E.-K., Alsner J. Integrative analysis of DCE-MRI and gene expression profiles in construction of a gene classifier for assessment of hypoxia-related risk of chemoradiotherapy failure in cervical cancer. Clin Cancer Res. 2016;22:4067–4076. doi: 10.1158/1078-0432.CCR-15-2322. [DOI] [PubMed] [Google Scholar]
- 14.Lendahl U., Lee K.L., Yang H., Poellinger L. Generating specificity and diversity in the transcriptional response to hypoxia. Nat Rev Genet. 2009;10:821–832. doi: 10.1038/nrg2665. [DOI] [PubMed] [Google Scholar]
- 15.Ragnum H.B., Vlatkovic L., Lie A.K., Axcrona K., Julin C.H., Frikstad K.M. The tumour hypoxia marker pimonidazole reflects a transcriptional programme associated with aggressive prostate cancer. Br J Cancer. 2015;112:382–390. doi: 10.1038/bjc.2014.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toustrup K., Sørensen B.S., Nordsmark M., Busk M., Wiuf C., Alsner J. Development of a hypoxia gene expression classifier with predictive impact for hypoxic modification of radiotherapy in head and neck cancer. Cancer Res. 2011;71:5923–5931. doi: 10.1158/0008-5472.CAN-11-1182. [DOI] [PubMed] [Google Scholar]
- 17.Winter S.C., Buffa F.M., Silva P., Miller C., Valentine H.R., Turley H. Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res. 2007;67:3441–3449. doi: 10.1158/0008-5472.CAN-06-3322. [DOI] [PubMed] [Google Scholar]
- 18.Yang L., Taylor J., Eustace A., Irlam J., Denley H., Hoskin P.J. A gene signature for selecting benefit from hypoxia modification of radiotherapy for high risk bladder cancer patients. Clin Cancer Res. 2017;23:4761–4768. doi: 10.1158/1078-0432.CCR-17-0038. [DOI] [PubMed] [Google Scholar]
- 19.Harris B.H.L., Barberis A., West C.M.L., Buffa F.M. Gene expression signatures as biomarkers of tumour hypoxia. Clin Oncol. 2015;27:547–560. doi: 10.1016/j.clon.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Abeshouse A., Ahn J., Akbani R., Ally A., Amin S., Andry Christopher D. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long Q., Xu J., Osunkoya A.O., Sannigrahi S., Johnson B.A., Zhou W. Global transcriptome analysis of formalin-fixed prostate cancer specimens identifies biomarkers of disease recurrence. Cancer Res. 2014;74:3228–3237. doi: 10.1158/0008-5472.CAN-13-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor B.S., Schultz N., Hieronymus H., Gopalan A., Xiao Y., Carver B.S. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fraser M., Sabelnykova V.Y., Yamaguchi T.N., Heisler L.E., Livingstone J., Huang V. Genomic hallmarks of localized, non-indolent prostate cancer. Nature. 2017;541:359–364. doi: 10.1038/nature20788. [DOI] [PubMed] [Google Scholar]
- 24.Dunning M.J., Vowler S.L., Lalonde E., Ross-Adams H., Boutros P., Mills I.G. Mining human prostate cancer datasets: the camcApp shiny app. EBioMedicine. 2017;17:5–6. doi: 10.1016/j.ebiom.2017.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross-Adams H., Lamb A.D., Dunning M.J., Halim S., Lindberg J., Massie C.M. Integration of copy number and transcriptomics provides risk stratification in prostate cancer: a discovery and validation cohort study. EBioMedicine. 2015;2:1133–1144. doi: 10.1016/j.ebiom.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boormans J.L., Korsten H., Ziel-van der Made A.J.C., van Leenders G.J.L.H., de Vos C.V., Jenster G. Identification of TDRD1 as a direct target gene of ERG in primary prostate cancer. Int J Cancer. 2013;133:335–345. doi: 10.1002/ijc.28025. [DOI] [PubMed] [Google Scholar]
- 27.Erho N., Crisan A., Vergara I.A., Mitra A.P., Ghadessi M., Buerki C. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8 doi: 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freedland S.J., Choeurng V., Howard L., De Hoedt A., Du Plessis M., Yousefi K. Utilization of a genomic classifier for prediction of metastasis following salvage radiation therapy after radical prostatectomy. Eur Urol. 2016;70:588–596. doi: 10.1016/j.eururo.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 29.Karnes R.J., Bergstralh E.J., Davicioni E., Ghadessi M., Buerki C., Mitra A.P. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol. 2013;190:2047–2053. doi: 10.1016/j.juro.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross A.E., Johnson M.H., Yousefi K., Davicioni E., Netto G.J., Marchionni L. Tissue-based genomics augments post-prostatectomy risk stratification in a natural history cohort of intermediate- and high-risk men. Eur Urol. 2016;69:157–165. doi: 10.1016/j.eururo.2015.05.042. [DOI] [PubMed] [Google Scholar]
- 31.Zhao S.G., Evans J.R., Kothari V., Sun G., Larm A., Mondine V. The landscape of prognostic outlier genes in high-risk prostate cancer. Clin Cancer Res. 2016;22:1777–1786. doi: 10.1158/1078-0432.CCR-15-1250. [DOI] [PubMed] [Google Scholar]
- 32.Jain S., Lyons C., Walker S.M., Mcquaid S., Hynes S., Mitchell D.M. A metastatic biology gene expression assay to predict the risk of distant metastases in patients with localized prostate cancer treated with primary radical treatment. J Clin Oncol. 2017;35:11. [Google Scholar]
- 33.Cunningham D., You Z. 2015. In Vitro and In Vivo Model Systems Used in Prostate Cancer Research; p. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bibikova M., Chudin E., Arsanjani A., Zhou L., Garcia E.W., Modder J. Expression signatures that correlated with Gleason score and relapse in prostate cancer. Genomics. 2007;89:666–672. doi: 10.1016/j.ygeno.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 35.Bismar T.A., Demichelis F., Riva A., Kim R., Varambally S., He L. Defining aggressive prostate cancer using a 12-gene model. Neoplasia (New York, NY) 2006;8:59–68. doi: 10.1593/neo.05664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuzick J., Swanson G.P., Fisher G., Brothman A.R., Berney D.M., Reid J.E. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12:245–255. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glinsky G.V., Glinskii A.B., Stephenson A.J., Hoffman R.M., Gerald W.L. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest. 2004;113:913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irshad S., Bansal M., Castillo-Martin M., Zheng T., Aytes A., Wenske S. A molecular signature predictive of indolent prostate cancer. Sci Transl Med. 2013;5 doi: 10.1126/scitranslmed.3006408. (202ra122-202ra122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knezevic D., Goddard A.D., Natraj N., Cherbavaz D.B., Clark-Langone K.M., Snable J. Analytical validation of the Oncotype DX prostate cancer assay – a clinical RT-PCR assay optimized for prostate needle biopsies. BMC Genomics. 2013;14:690. doi: 10.1186/1471-2164-14-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long Q., Johnson B.A., Osunkoya A.O., Lai Y.-H., Zhou W., Abramovitz M. Protein-coding and MicroRNA biomarkers of recurrence of prostate cancer following radical prostatectomy. Am J Pathol. 2011;179:46–54. doi: 10.1016/j.ajpath.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh D., Febbo P.G., Ross K., Jackson D.G., Manola J., Ladd C. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 42.Stephenson A.J., Smith A., Kattan M.W., Satagopan J., Reuter V.E., Scardino P.T. Integration of gene expression profiling and clinical variables to predict prostate carcinoma recurrence after radical prostatectomy. Cancer. 2005;104:290–298. doi: 10.1002/cncr.21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu C.-L., Schroeder B.E., Ma X.-J., Cutie C.J., Wu S., Salunga R. Development and validation of a 32-gene prognostic index for prostate cancer progression. Proc Natl Acad Sci. 2013;110:6121–6126. doi: 10.1073/pnas.1215870110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu J., Yu J., Rhodes D.R., Tomlins S.A., Cao X., Chen G. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res. 2007;67:10657–10663. doi: 10.1158/0008-5472.CAN-07-2498. [DOI] [PubMed] [Google Scholar]
- 45.Lalonde, E., Alkallas, R., Chua, M. L. K., Fraser, M., Haider, S., Meng, A., Zheng, J., Yao, C. Q., Picard, V., Orain, M., Hovington, H., Murgic, J., Berlin, A., Lacombe, L., Bergeron, A., Fradet, Y., Têtu, B., Lindberg, J., Egevad, L., Grönberg, H., Ross-Adams, H., Lamb, A. D., Halim, S., Dunning, M. J., Neal, D. E., Pintilie, M., VAN DER Kwast, T., Bristow, R. G. & Boutros, P. C. 2017. Translating a prognostic DNA genomic classifier into the clinic: retrospective validation in 563 localized prostate tumors. Eur Urol [DOI] [PubMed]
- 46.Gardner L.B., Li Q., Park M.S., Flanagan W.M., Semenza G.L., Dang C.V. Hypoxia inhibits G1/S transition through regulation of p27 expression. J Biol Chem. 2001;276:7919–7926. doi: 10.1074/jbc.M010189200. [DOI] [PubMed] [Google Scholar]
- 47.Guillaumond F., Leca J., Olivares O., Lavaut M.-N., Vidal N., Berthezène P. Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proc Natl Acad Sci. 2013;110:3919–3924. doi: 10.1073/pnas.1219555110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spratt D.E., Yousefi K., Deheshi S., Ross A.E., Den R.B., Schaeffer E.M. Individual patient-level meta-analysis of the performance of the decipher genomic classifier in high-risk men after prostatectomy to predict development of metastatic disease. J Clin Oncol. 2017;35:1991–1998. doi: 10.1200/JCO.2016.70.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hieronymus H., Schultz N., Gopalan A., Carver B.S., Chang M.T., Xiao Y. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci U S A. 2014;111:11139–11144. doi: 10.1073/pnas.1411446111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall J.S., Taylor J., Valentine H.R., Irlam J.J., Eustace A., Hoskin P.J. Enhanced stability of microRNA expression facilitates classification of FFPE tumour samples exhibiting near total mRNA degradation. Br J Cancer. 2012;107:684–694. doi: 10.1038/bjc.2012.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mujcic H., Rzymski T., Rouschop K.M.A., Koritzinsky M., Milani M., Harris A.L. Hypoxic activation of the unfolded protein response (UPR) induces expression of the metastasis-associated gene LAMP3. Radiother Oncol. 2009;92:450–459. doi: 10.1016/j.radonc.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 52.Mckeown S.R. Defining normoxia, physoxia and hypoxia in tumours—implications for treatment response. Br J Radiol. 2014;87 doi: 10.1259/bjr.20130676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chi J.-T., Wang Z., Nuyten D.S.A., Rodriguez E.H., Schaner M.E., Salim A. Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers. PLoS Med. 2006;3 doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material