Abstract
Enhancements in the diagnostic capabilities using host biomarkers are currently much needed where sensitivity and specificity issues plague the diagnosis of Hand, Foot and Mouth Disease (HFMD) in pediatrics clinical samples. We investigated miRNome profiles of HFMD saliva samples against healthy children and developed miRNA-based diagnosis models. Our 6-miRNA scoring model predicted HFMD with an overall accuracy of 85.11% in the training set and 92.86% in the blinded test set of Singapore cohort. Blinded evaluation of the model in Taiwan HFMD cases resulted in 77.08% accuracy with the 6-miRNA model and 68.75% with the 4-miRNA model. The strongest predictor of HFMD in all of the panels, hsa-miR-221 was found to be consistently and significantly downregulated in all of our HFMD cohorts. This is the first study to prove that HFMD infection could be diagnosed by circulating miRNAs in patient's saliva. Moreover, this study also serves as a stepping stone towards the future development of other infectious disease diagnosis workflows using novel biomarkers.
Keywords: miRNA, Biomarker, Saliva, HFMD, Machine learning
Highlights
-
•
Targeted salivary miRNome profiling was conducted between Hand Foot and Mouth disease patients and healthy individuals.
-
•
HFMD diagnosis models were established with training data from Singapore HFMD cohort using the support vector machine.
-
•
The models were also validated blindly using the testing data from Singapore HFMD cohort and the entire Taiwan HFMD cohort.
Using saliva as the medium for diagnosis of human diseases has been a long dream for doctors and patients. In this research article, we developed a rapid test for detecting hand foot and mouth disease using molecules known as miRNA in saliva. We created a mathematical model to detect a specific pattern of miRNA response of the HFMD infection to identify HFMD infected patients. Our model can accurately distinguish HFMD patients from the healthy person by 92.86% in the blinded testing set of Singapore HFMD cohort.
1. Introduction
Hand, foot, and mouth disease (HFMD) is a widespread endemic viral infection which afflicts millions of infants and children yearly in the Western Pacific regions caused by the human enterovirus species A (HEV-A) from the genus Enterovirus. HFMD is customarily self-resolving, characterized by fever and papulovesicular, sometimes maculopapular, rash on the palms, soles, elbows, and trunk along with mouth ulcers [1]. However, in a modest proportion of cases, EV71-associated HFMD can rapidly advance into severe neurological complications such as encephalitis and acute flaccid paralysis [2]. These complications may in turn swiftly progress to cardiopulmonary failure and mortality [3]. Even though neurological complications have been largely associated with EV71 [4], CA16 also has also been reported to cause similar aggravating conditions [5]. The various complications that could arise from enterovirus infections strongly necessitate a rapid and accurate identification of enterovirus such that efficient isolation of infected patients could be carried out to prevent further transmission.
HFMD is transmitted either via fecal-oral or droplet route and is currently diagnosed by via clinical symptoms and additional laboratory testing is mostly deemed unnecessary for mild cases [6]. Nevertheless, such practices may lead to misdiagnosis due to the lack of a robust and definitive screening test and aggravate transmission of HFMD in atypical and mild cases. In addition, there is currently no cure for HFMD [7]. Treatment options are confined to alleviation of physical symptoms and supportive management [7]. Therefore, development of novel and rapid diagnostic methods, spanning a range of enteroviruses is critical when there is a risk of neurological complication leading to fatality [8]. The golden criterion of laboratory confirmation of HFMD is the identification of enterovirus isolates from clinical samples such as throat, stool or skin vesicle swab [9]. Enteroviruses could be isolated in human muscle rhabdomyosarcoma (RD) cells or African green monkey kidney (Vero) cells and could subsequently be confirmed using reverse-transcription polymerase chain reaction (PCR) of viral RNA, indirect immunofluorescence and viral microneutralization assays [9]. However, the abovementioned approaches are rather laborious, lengthy and time-consuming [10]. Although rapid diagnostic methods utilizing modern molecular methods such as quantitative real-time PCR (qRT-PCR) were recently developed to address those issues [4], the sensitivity and specificity of such assays need further refinements due to diverse genetic differences between serotypes and genotypes of enteroviruses [11].
MicroRNAs (miRNA) are single-stranded RNA molecules of approximately 22 nucleotides that negatively regulate gene expression either via degradation of its target mRNA through various mechanism such as Dicer cleavage or by repressing translation machinery [12]. Having a partial complementarity to its target mRNA, miRNA uniquely regulates hundreds of cellular gene expression hence making it a conceivable indicator of the state of a cell [13]. A number of miRNA-based diagnostic tests utilizing serum of HFMD patients were recently developed [13,30] (Cui et al.). Moreover, miRNA is also known to be readily isolated from exosomes which are cell-secreted vesicles in human saliva. As saliva collection is significantly less invasive than other specimens such as skin vesicle, rectal swab and blood, a saliva-based diagnostic test could be especially beneficial and convenient for HFMD that chiefly affects children. In addition, salivary miRNA has remarkable stability and resistance to cellular and physical degradation [14] thereby conferring it as a potential clinical biomarker. Here, we described a salivary miRNA qPCR analysis which could identify HFMD patients with near 90% accuracy in blinded model evaluation of the “Singapore Cohort”.
2. Materials and Methods
2.1. Patient Samples and Infection
Total of 35 HFMD suspected throat swab and saliva clinical samples were obtained from Kandang Kerbau (KK) Women's and Children's Hospital from August 2012 to February 2016. The collection was under the approval of centralized institutional review board (CIRB) of Singhealth under CIRB number 2012/448/E. Twenty-four HFMD samples in “Taiwan Cohort” were collected under IRB 104-3836B which was approved by the Research Ethics Board of Chang Gung Memorial Hospital in Taiwan. Presence of enteroviruses in patient samples were confirmed using previously established pan-entero PCR reactions in saliva samples. Healthy saliva samples were collected from multiple child care centers which participated in saliva collection drive under National University of Singapore Ethical review board approval number B-14-273.
2.2. miRNA Extraction and Reverse Transcription
miRNA was extracted from 50ul of saliva using biofluid extraction kit (Exiqon, Inc.) with 1 μg of MS2 carrier RNA (Roche, Ltd.) and eluted in 30 μl of water. 7ul of RNA is used to reverse transcribed previously extracted miRNA using universal cDNA synthesis kit following manufacture protocol (Exiqon, Inc.).
2.3. Primary Screen Using miRNA qPCR Panel
Pools of saliva (described in the result section) were screened for dysregulated miRNA primarily using serum plasma focused miRCURY LNA™ microRNA PCR (Exiqon, Inc.). The panel is chosen as miRnome of saliva was previously found to be significantly overlapping with those from serum/plasma [12]. cDNA synthesized were diluted 40 folds in water and 2 μl is added to each well of qPCR plates. Quantitative real-time PCR reactions were carried out according to manufacture protocol using ExiLENT SYBR® Green master mix (Exiqon, Inc.).
2.4. Individual qPCR Assays for Validation
Validation was carried out using individual qRT-PCR assays using selected 8 miRNAs along with 1 normalizer RNA. Saliva were extracted, reverse transcribed and amplified as described above. Melting curve of each reaction is analyzed to ensure specific amplification of targets and only samples with normalizer having CT value of <30 were taken into account for further analysis.
2.5. Statistical Analysis
During the primary screening, data normalization was done in two steps. Firstly, inter-plate calibration was carried out across all plates using inter-plate calibrator present in each panel across all plates to minimize run to run variations. Second step of normalization involved determining reference gene. Raw CT values were analyzed for the most stably present miRNA with CT value below 30 using NormFinder algorithm [15]. Significantly deregulated miRNAs were determined using GenEx qPCR analysis software (http://genex.gene-quantification.info). Raw CT values from validation studies were normalized using the selected normalizer and normalized CT values were further analyzed with R software [16]. Individual miRNA performance was determined with “easyROC” [17]. Support vector machine with radial classification analysis was performed to build predictive models with R software in “caret” package [18]. Statistical significances of risk score differences between HFMD and Healthy groups were calculated using non-parametric Mann-Whitney test using Prism (GraphPad Software, Inc.).
3. Results
3.1. Patient Information and Study Design
Saliva samples used in this study were collected from multiple patient cohorts (Fig. 1). HFMD patients from both “Singapore Cohort” (n = 35) and “Taiwan Cohort” (n = 24) were hospitalized for symptomatic HFMD at the time of sample collection and collected saliva samples were diagnosed as enterovirus positive by using adapted pan-entero PCR protocol as described previously [19]. We also collected healthy samples (considering confounding factors such as age, gender and race) from 24 children in Singapore whose saliva samples were tested negative for HFMD by pan-entero PCR. Details on patient characteristics are summarized in Table 1.
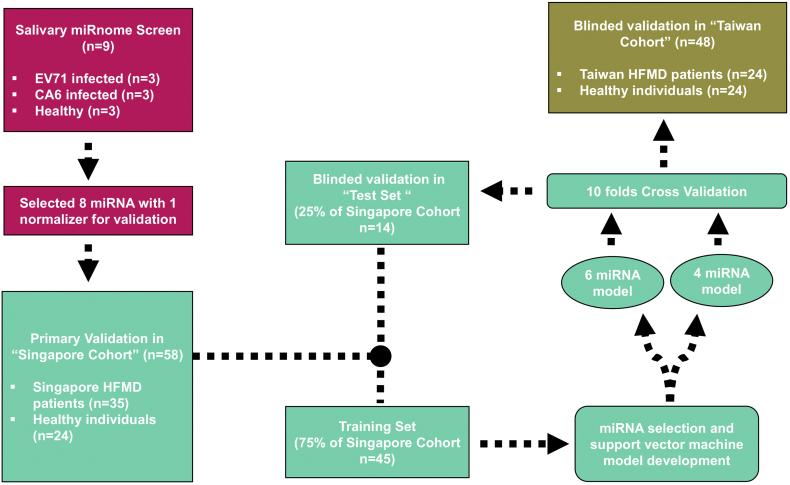
Fig. 1.
Overall study design and patient cohorts involved in model development and validations. Screening of potential miRNA biomarkers for HFMD infection was performed on 3 EV71, 3 CA6 and 3 healthy pooled saliva samples. The 8 putative miRNA predictors from primary screen was subsequently used to form diagnostic models using the training set which includes 75% of the “Singapore Cohort” with support vector machines. Cross validation was carried out using k-fold validation method for 10 folds and respective performances of the two models were determined. Blinded validation of the two models was carried out using the test set which includes 25% of the “Singapore Cohort’ and the whole “Taiwan Cohort”.
Table 1.
Pathological findings of HFMD patients in “Singapore and Taiwan Cohort”.
| Categories | Singapore cohort (n = 35) | Taiwan Cohort (n = 24) |
|---|---|---|
| Age (<1 year old) | 8/35 | 7/24 |
| Age (1–5 years old) | 24/35 | 13/24 |
| Age (>5 years) | 3/35 | 4/24 |
| Duration of hospitalization (<3 days) |
6/35 | 3/24 |
| Duration of hospitalization (3–5 days) |
24/35 | 18/24 |
| Duration of hospitalization (>5 days) |
5/35 | 3/24 |
| Fever | 30/35 | 22/24 |
| Breathlessness | 1/35 | 0/24 |
| Vomiting | 15/35 | 3/24 |
| Rash | 24/35 | 15/24 |
| Oral ulcer | 5/35 | 19/24 |
3.2. Differential miRNA Expression of HFMD Patients in the Screening Population
Differential salivary miRNA expression between HFMD and healthy samples were profiled using Exiqon miRNA qPCR panel. The primary screen was carried out by identification of dysregulated miRNAs in pooled EV71 and CA6 patient saliva samples against the healthy group (n = 3 each). Pooled samples were spiked with synthetic miRNA, uniSP6 which was later used to ensure absence of PCR inhibitors in each pool. To reduce plate to plate variation, inter-plate calibration was carried out using pre-defined inter-plate calibrators from the manufacturer. miRNA expressions normalization was carried out by selecting stably expressing miRNA with least variance and student t-test was used to determine significantly (p < 0.05) dysregulated miRNA across different pools.
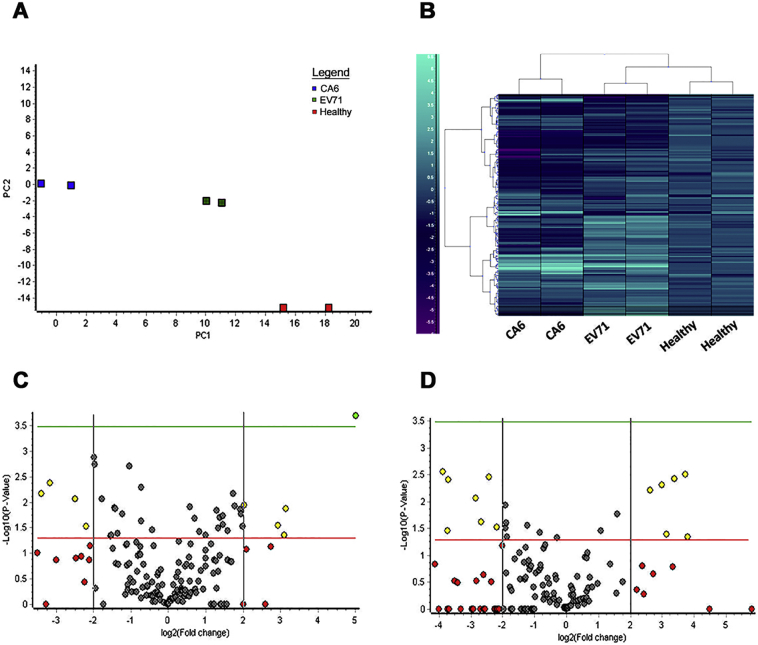
A total of 179 miRNAs were analyzed and the primary screen classified a subset of miRNA to be significantly regulated in HFMD saliva respect to the healthy control pool. We found 23 significantly expressed miRNA between EV71 against the healthy controls pool and 10 between CA6 against healthy controls pool with overlap of 7 miRNAs using p-value of <0.05 and absolute 4-fold change difference (Fig. S1). After dimension reduction with the principle component analysis, miRNA expression between independent repeats of screening arrays were found to be closely correlated (Fig. 2A) and one-way hierarchical clustering analysis exhibited clear clusters with differential pattern of molecular miRNA signatures between healthy controls and enterovirus patients (Fig. 2B). Gene set enrichment analysis was also carried out to classify over represented pathways using KEEG and identified a number of biological process involved in viral infection such as endocytosis, cytoskeleton regulation, and MAPK signaling pathways (Table S1). Later, significantly dysregulated 8 miRNA signatures with 1 normalizer (Table S2) from the primary screen were then selected to validate in “Singapore Cohort” and “Taiwan Cohort” in individual patients using qPCR with specific primers.
Fig. 2.
Salivary miRNA expression of HFMD in the salivary miRnome screen. (A) PCA plot based on miRNA expression values across pools (n = 3 each). (B) Heat map showing differentially expressed miRNA gene expression pattern across different samples (n = 3). Volcano plot was used to illustrate distribution of significant and non-significant miRNA in Singapore HFMD Cohort 1 screen in C. Healthy Vs EV71 (n = 6) and D. Healthy Vs CA6 (n = 6). Red colour represent population with absolute 4-fold change while significant miRNAs were represented in yellow or green while red is non-significant. Student t-test (non-parametric) was performed and HITs were determined using absolute fold change of >4-fold and p-value < 0.05.
3.3. Diagnosis Performance of Individual miRNAs, Feature Selection and the Model Development
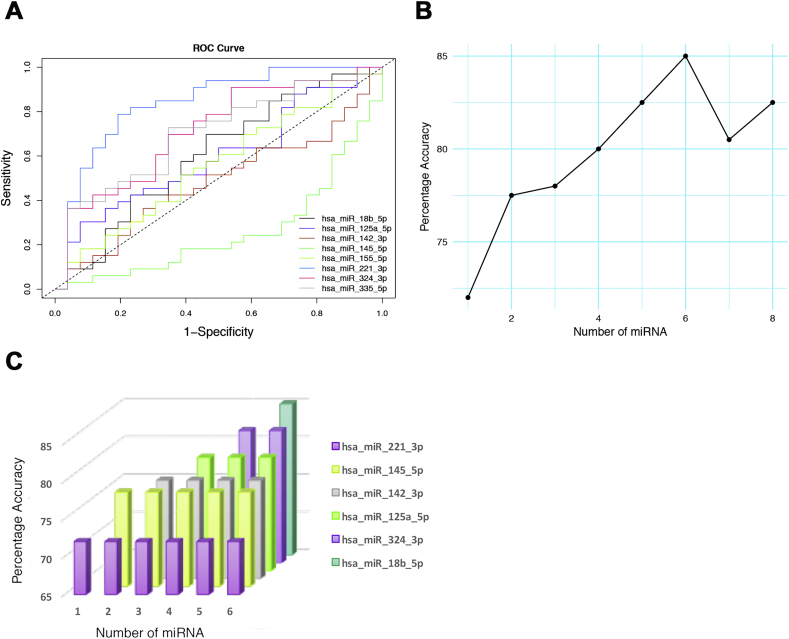
After determining the expression levels of the 8-miRNA signatures in the “Singapore Cohort”, receiver operating characteristics analysis was employed to evaluate respective sensitivity and specificity of individual miRNAs at a given threshold by using “caret” and “ROC” package implemented in R software. While expression levels of the majority of miRNAs exhibited positive association with the disease status of HFMD (Fig. 3A), hsa-miR-221-3p unveiled the highest accuracy in diagnosing HFMD with the positive predictive value of 83.90% and negative predictive value of 75.00% (Supplementary). hsa-miR-18b-5p, on the contrary, was the least effective in HFMD identification with the positive predictive value of 63.00% and negative predictive value of 50.00%.
Fig. 3.
miRNA selection and performance tuning. A. Logistic regression of hit miRNAs. ROC curve was used to display sensitivity and specificity of individual miRNA in HFMD diagnosis with the entire “Singapore Cohort”. B. Overall accuracy of the diagnosis model resolved with increasing number of miRNA classifiers using support vector machine model in the training set of the “Singapore Cohort”. C. Importance of individual miRNA in the 6-miRNA model. Accuracy for each model was calculated using “caret” package in R software. “ggplot2” library was used for illustration using R software.
Subsequently, to determine if the diagnosis accuracy could be further improved by integrating the expression of multiple miRNAs, the support vector machine algorithm was implemented for miRNA expression combination with the radial kernel in “caret” package in R software (R script for the model training and validations are shown in detail in the supplementary method section). It is of note that expression levels of certain miRNA pairs were highly correlated to one other (data not shown) and therefore to reduce redundancy in the model, we determined the best performing miRNAs and combinations to be included in the final diagnosis model. For evaluation, the “Singapore Cohort” was randomly divided into the training set and the test set with the constraint such that both training and test set contained the same fraction of HFMD patients. The training set comprised 75% of the entire “Singapore Cohort” and was then applied to train the HFMD diagnosis model with all possible numbers of miRNAs and combinations. Finally, the model converged at peak accuracy of 85.00% with 6 miRNAs (Fig. 3B). Although the diagnosis model with 6-miRNA resulted with the highest diagnosis accuracy of 85.00%, 4-miRNA model also rendered an acceptable accuracy of 80.00% and was therefore selected for further validations (Fig. 3B).
3.4. Performance Evaluation of the Diagnosis Model with the 10-Fold Cross-Validation in the Training Set and Blinded Assessment in the Test Set
Since the training dataset was relatively limited in size, in order to avoid overfitting, we leveraged the 10-fold cross-validation method to fairly evaluate the performance of the former 6-miRNA and 4-miRNA diagnosis models. Instead of including a separate validation set, 10-fold cross-validation method equally divided the training dataset into 10 parts; trained the diagnosis model with 9 parts of the dataset, tested the model performance on the remaining one part of the dataset and repeated the process for 10 iterations [20]. The final performance of the model in the training set was determined by averaging the accuracy, sensitivity and specificity from each round (Table 2). As expected, the 6-miRNA model was slightly more effective in the diagnosis of HFMD than the 4-miRNA counterpart with about 4% increase in accuracy.
Table 2.
Performance of predictive models in HFMD discrimination. 4 and 6 miRNA predictor models were constructed with miRNA expressions on the training set of the “Singapore Cohort”. The two models were evaluated on the test set of the “Singapore Cohort” and the “Taiwan Cohort”. The overall accuracy (ACC) is shown together with sensitivity (SEN, the probability to accurately predict HFMD patient as “HFMD”) and specificity (SPE, the probability to predict healthy individuals as “Healthy”). Respective accuracy, sensitivity and specificity were calculated using package “crossval” implemented in R. k-fold cross-validation was carried out using package “caret” and was repeated for 10 folds.
| Set | ACC % | SEN % | SPE % |
|---|---|---|---|
| “Singapore Cohort” (6 miRNA predictors with the training set) | 85.11 | 88.89 | 82.76 |
| “Singapore Cohort” (6 miRNA predictors with the test set) | 92.86 | 100.00 | 88.89 |
| “Singapore Cohort” (4 miRNA predictors with the training set) | 80.85 | 83.33 | 79.31 |
| “Singapore Cohort” (4 miRNA predictors with the test set) | 91.67 | 100.00 | 87.50 |
| “Taiwan Cohort” (6 miRNA predictors) | 77.08 | 78.26 | 76.00 |
| “Taiwan Cohort” (4 miRNA predictors) | 68.75 | 68.00 | 69.57 |
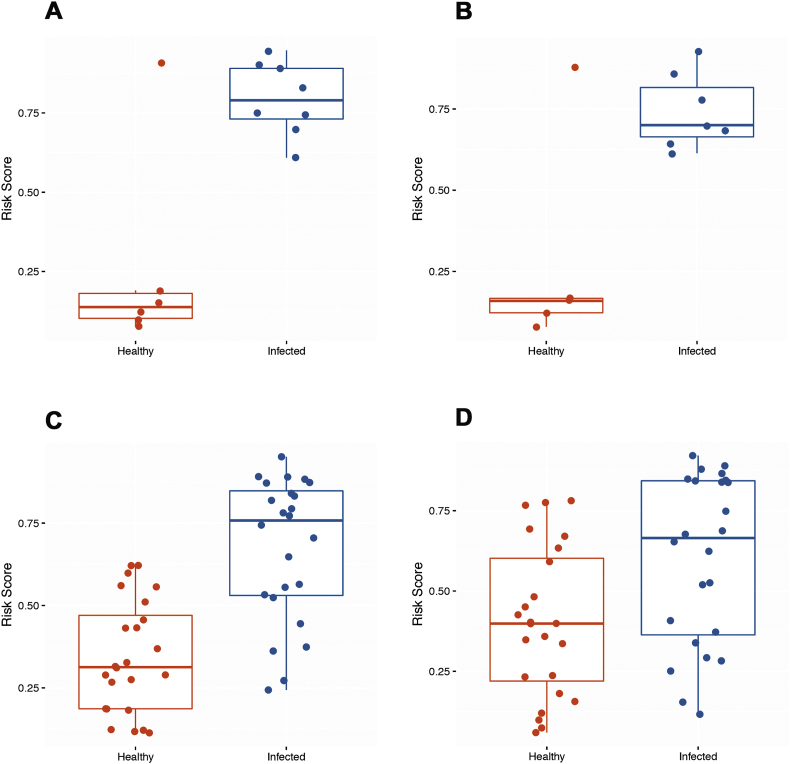
Additionally, the fit of the model was also assessed in blinded fashion for the difference between estimated and true infection status with the test dataset which constituted 25% of the “Singapore Cohort”. Surprisingly, both 6 and 4 miRNAs models performed excellently with the test set data in classifying blinded HFMD cases with respective accuracy of 92.86% and 91.67% (Table 2). Such high accuracies were a result of well separated risk scores in blinded test set data (Fig. 4A and B) which reflected the appropriate complexity in our models, avoiding overfitting with a fine balance between variance and bias.
Fig. 4.
Risk score of healthy and HFMD patients in blinded validation. Risk index of HFMD was obtained for (A) the 6-miRNA model and (B) the 4-miRNA model in testing set of “Singapore Cohort”. The two models were also validated in the “Taiwan Cohort” using (C) the 6-miRNA model and (D) the 4-miRNA model. Circles denote data points with triplicate readings. Box plot was constructed using ggplot2 library in R software.
3.5. Blinded Model Evaluation with HFMD Patients from Different Geographical Origin
It is of note that our patient samples were obtained solely from a single hospital throughout three years of our study and hence it is possible that our miRNA signatures previously validated were prone to bias from geographical, racial or other unknown factors. In order to avoid inherent study bias, we assessed the performance of the HFMD diagnosis model to the independent Taiwan HFMD Cohort which included 24 HFMD patients from Taiwan and 24 healthy individuals. HFMD patient samples from Taiwan were obtained in a different time period than the samples from Singapore. Interestingly, despite the initial model development was performed using the “Singapore Cohort”, the HFMD diagnosis model was still remarkably accurate in discriminating HFMD in Taiwan, having 77.08% overall accuracy with the 6-miRNA model and 68.75% in the 4-miRNA model (Table 2). We also analyzed the risk score of HFMD in “Taiwan Cohort” and found that the mean risk score of HFMD patients was observed to be significantly higher than healthy controls in both 8 (Fig. 4C) and 4 miRNA models (Fig. 4D) with p-value <.0001.
4. Discussion
Developing a robust HFMD diagnostic kit is conventionally hampered by many factors. Firstly, HFMD is known to be caused by a wide range of enteroviruses such as EV71, CA16, CA6 and in some cases, by Echoviruses. Moreover, intra-serotype variabilities of viral gene sequences are also commonly observed with enteroviruses. These reasons challenge the development of an appropriate screening assay of sufficient sensitivity and specificity. These reasons effectively render developing an assay, which could detect all strains of HFMD causing enteroviruses, while retaining a decent specificity and sensitivity greatly challenging. Our novel diagnostic test attempts to address those issues by identifying general miRNA signature responses triggered by HFMD to circumvent the above-mentioned issues.
A number of miRNAs purposed as HFMD diagnostic markers were reported recently (Cui et al., [21]). Many of these tests utilized serum miRNA in symptomatic HFMD patients and notably those infected with a specific strain, EV71. Although the miRNA present in serum and saliva are known to be highly similar [12], we did not observe a significant overlap of signatures between our studies and other previous reports profiling serum of HFMD patients. To our knowledge, this study is the first to attempt saliva-based miRNA analysis in HFMD patients whereas the previous studies were focused on serum markers (Cui et al., [22]). Out of the validated 6 and 4 miRNAs from our study, hsa-miR-221-3p was found to be the most important miRNA in both panels. In addition, Wang et al. reported that, hsa-miR-221-3p was also significantly downregulated in severe EV71 cases [22]. Although all the rest of the miRNAs identified in our study were different from those previously published, such findings were expected since our study utilized saliva instead of serum.
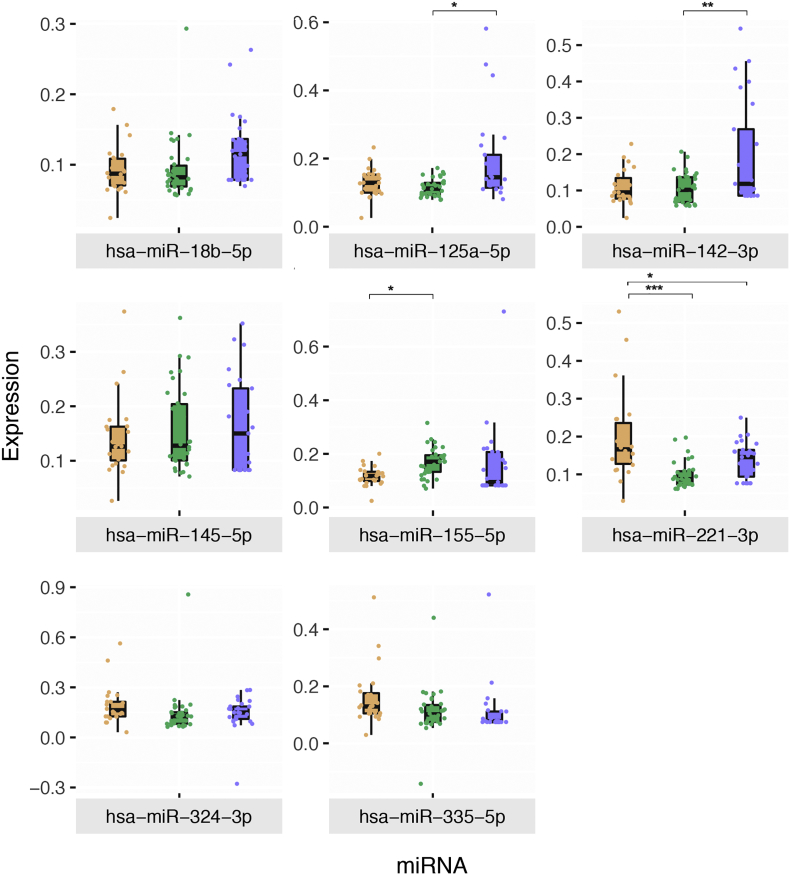
A salivary miRNA-based HFMD diagnostic test is more desirable in the clinical context for the following reasons: (1) it requires only 50ul of saliva, (2) collection of saliva is non-invasive and (3) complete profiling of miRNA from extraction to data analysis can be performed within 4 h which is considerably faster than current methods of traditional pan-entero PCR which requires gel electrophoresis or virus isolation which requires days to complete. A rather less important yet interesting question is whether our signature miRNA studied were released from the infected tissues or from the surrounding uninfected ones as an elicited immune response against the viral infection. Entire miRNA panel identified in this study is known to be present in the serum and plasma of human blood samples [23,24]. hsa-miR-221-3p is known to be upregulated in many types of cancer [25,26] and the function of the miRNA is regarded to be positive regulator of apoptosis through inhibiting ARF4 protein [27]. It is possible that significant downregulation of hsa-miR-221-3p in both of our HFMD cohorts (Fig. 5) seems to suggest an indication of the specific viral pathogenesis mechanism which inhibits the apoptosis of infected cell which might allow further replication of the enterovirus [28]. Interestingly, we found that most of our miRNA predictors were generally upregulated during HFMD. In this study, hsa-miR-324-3p was found to be significantly upregulated in saliva samples (Fig. 5). hsa-miR-324-3p expression levels were not known to alter in many dysregulated cellular processes such as infections and cancers. However, interestingly, we also found that EV71, CA16 and CA6 contains has-miR-324-3p target sites in their respective genomes predicted by ViTa software (http://vita.mbc.nctu.edu.tw) (Supplementary Results). Therefore, we postulated that increase in hsa-miR-324-3p expression level was a possible large spectrum specific antiviral response against enteroviruses, although further in vitro studies are warranted to cement the phenomena. Although diagnosis accuracy of the 6-miRNA model in blinded evaluation of the “Taiwan Cohort” was significantly a notch under at 77.08% compared to the 92.86% of the “Singapore Cohort”; it is understandable that miRNA responses between different geographical region could slightly differ due to genetics and epigenetics differences.
Fig. 5.
Differential expression of hit miRNAs in the Singapore and Taiwan cohorts. Boxplots of miRNA expressions in healthy (orange), “Singapore HFMD” (green) and “Taiwan HFMD” (blue) cohorts. Stars indicate significance between HFMD and healthy. (***P < 10−3, **P < 10−2, **P < 0.05, FDR-adjusted non-parametric ANOVA with Dunnett's multiple comparison test using 95% CI).
We envisioned that a systematic comparison of the viral load and our miRNA predictor levels in a larger HFMD cohort will be needed to fine tune our detection algorithm in the future. We consider the next critical step in the HFMD diagnosis test formulation is to validate the identified miRNA biomarkers in sub-clinical HFMD cases. Since a vast majority of HFMD cases are sub-clinical, yet responsible for silent transmission of HFMD [29], we believe routine HFMD test using a robust molecular based detection method in high-risk areas such as schools in endemic countries will be beneficial to deter disease transmission. We agree that validation of the biomarkers with other viral and bacterial diseases in which the physical symptoms overlap with HFMD is also necessary to prevent false-positive diagnoses. Besides, to validate the identified biomarkers as truly circulating miRNAs in exosomes, targeted enrichment of exosomes, followed by diagnostic miRNA expression profiling will be performed in the future. Due to the limited scope of the study, we did not cross compare the expression of identified diagnostic miRNAs in other biofluid samples rather than saliva. We firmly believe that it is worth expanding the study scope in the future to validate our findings in multiple non-invasive samples such as serum, urine and sweat to adequately appreciate the miRNA landscape in other biofluids and decipher their interplay in enterovirus pathogenesis. Additionally, we also did not explore the expression of identified miRNA biomarkers in severe HFMD cases as our cohort consists exclusively of mild HFMD cases; it will be intriguing to examine the expression changes of identified miRNA signatures between mild and severe HFMD cases which may potentially open doors for severity prognosis. Nevertheless, given the cross-validated and blindly evaluated data, it can be safely assumed that the 6-miRNA composition is unlikely to be altered for the HFMD diagnosis model. We believe our saliva test have the potential to further develop into a point of care device where general public and school could use to routinely monitor HFMD to prevent disease transmission.
Acknowledgments
Acknowledgements
We will like to thank Kirill Eremenko, Hadelin de Ponteves and SuperDataScience Team for the R scripts deposited in UDEMY which were used as backbone codes in the model development of this study.
Fundings
J.J.H.C. was supported by the MINDEF DRIP Grant [R571-000-210-232]; and the CBRG grant [CBRG13nov02] from National Medical Research Council, Ministry of Health, Singapore). R.W. was supported by grants from the Ministry of Science and Technology, Taiwan [MOST-106-2320-B-182-007]; and Chang Gung Memorial Hospital Research Fund [CMRPD1G0021, CMRPD1A0193, CMRPD1A0553, CMRPD1F0281-2 and CMPRD1E0411-3].
Conflict of Interest Statement
“The authors have declared that no conflict of interest exists.”
Author Contributions
NM and SVPD carried out experiments, and NM analyzed the data and drafted the manuscript; AAW and CJC performed sample collection; RYLW and JJHC gave specialized expertise, samples and reagents. JJHC, RYLW, NWHT and CYC planned the study, supervised, and edited the manuscript, along with other co-authors. RYLW and JJHC contributed equally to this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.05.006.
Contributor Information
Nyo Min, Email: micnm@nus.edu.sg.
Previtha Dawn Sakthi Vale, Email: a0131641@u.nus.edu.
Anng Anng Wong, Email: wong.anng.anng@kkh.com.sg.
Natalie Woon Hui Tan, Email: natalie.tan.w.h@singhealth.com.sg.
Chia Yin Chong, Email: chong.chia.yin@singhealth.com.sg.
Robert Y.L. Wang, Email: yuwang@mail.cgu.edu.tw.
Justin Jang Hann Chu, Email: miccjh@nus.edu.sg.
Appendix A. Supplementary data
Supplementary material
References
- 1.Wang S.-M., Liu C.-C. Update of enterovirus 71 infection: epidemiology, pathogenesis and vaccine. Expert Rev Anti Infect Ther. 2014;12:447–456. doi: 10.1586/14787210.2014.895666. [DOI] [PubMed] [Google Scholar]
- 2.Lin C.Y., Wang M.C., Zeng X.P., Kuang S.Z., Lin X.B. Epidemiologic features of hand-foot-mouth disease in Haikou city from 2008 to 2015. Zhonghua Liu Xing Bing Xue Za Zhi. 2016;37:1615–1618. doi: 10.3760/cma.j.issn.0254-6450.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 3.Solomon T., Lewthwaite P., Perera D., Cardosa M.J., Mcminn P., Ooi M.H. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect Dis. 2010;10:778–790. doi: 10.1016/S1473-3099(10)70194-8. [DOI] [PubMed] [Google Scholar]
- 4.Lin T.-Y., Twu S.-J., Ho M.-S., Chang L.-Y., Lee C.-Y. Enterovirus 71 outbreaks, Taiwan: occurrence and recognition. Emerg. Infect Dis. 2003;9:291–293. doi: 10.3201/eid0903.020285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LI L.J. Review of hand, foot and mouth disease. Front Med China. 2010;4:139–146. [Google Scholar]
- 6.Li W., Zhang X., Chen X., Cheng Y.P., Wu Y.D., Shu Q. Epidemiology of childhood enterovirus infections in Hangzhou, China. Virol J. 2015;12:58. doi: 10.1186/s12985-015-0294-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunaseelan S., Chu J.J.H. Identifying novel antiviral targets against enterovirus 71: where are we? Futur Virol. 2017;12:171–191. [Google Scholar]
- 8.Chan K.P., Goh K.T., Chong C.Y., Teo E.S., Lau G., Ling A.E. Epidemic hand, foot and mouth disease caused by human enterovirus 71, Singapore. Emerg Infect Dis. 2003;9:78–85. doi: 10.3201/eid1301.020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez-Ruiz M. Human rhabdomyosarcoma cells for rapid detection of enteroviruses by shell-vial assay. J Med Microbiol. 2003;52:789–791. doi: 10.1099/jmm.0.05237-0. [DOI] [PubMed] [Google Scholar]
- 10.Ooi M.H., Wong S.C., Lewthwaite P., Cardosa M.J., Solomon T. Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol. 2010;9:1097–1105. doi: 10.1016/S1474-4422(10)70209-X. [DOI] [PubMed] [Google Scholar]
- 11.Liang Z., Mao Q., Gao F., Wang J. Progress on the research and development of human enterovirus 71 (EV71) vaccines. Front Med. 2012;7:111–121. doi: 10.1007/s11684-012-0237-z. [DOI] [PubMed] [Google Scholar]
- 12.Weber J.A., Baxter D.H., Zhang S., Huang D.Y., How Huang K., Jen Lee M. The MicroRNA Spectrum in 12 body fluids. Clin Chem. 2010;56:1733. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cookson V.J., Bentley M.A., Hogan B.V., Horgan K., Hayward B.E., Hazelwood L.D. Circulating microRNA profiles reflect the presence of breast tumours but not the profiles of microRNAs within the tumours. Cell Oncol (Dordr) 2012;35:301–308. doi: 10.1007/s13402-012-0089-1. [DOI] [PubMed] [Google Scholar]
- 14.Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen C.L., Jensen J.L., Orntoft T.F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 16.Team R.C. R: A language and environment for statistical computing. R foundation for statistical computing. 2017. https://www.R-project.org/ (Vienna, Austria)
- 17.Karaagaoglu A.E., Goksuluk D., Korkmaz S., Zararsiz G. easyROC: an interactive web-tool for ROC curve analysis using R language environment. R I Dent J. 2016;8:213–230. [Google Scholar]
- 18.Thao N.T., Ngoc N.T., Tu P.V., Thuy T.T., Cardosa M.J., McMinn P.C. Development of a multiplex polymerase chain reaction assay for simultaneous identification of human enterovirus 71 and coxsackievirus A16. J Virol Methods. 2010;170:134–139. doi: 10.1016/j.jviromet.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardosa M.J., Perera D., Brown B.A., Cheon D., Chan H.M., Chan K.P. Molecular epidemiology of human enterovirus 71 strains and recent outbreaks in the Asia-Pacific region: comparative analysis of the VP1 and VP4 genes. Emerg Infect Dis. 2003;9:461–468. doi: 10.3201/eid0904.020395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lever J., Krzywinski M., Altman N. Model selection and overfitting. Nat Methods. 2016;13:703. [Google Scholar]
- 21.Jia H.-L., He C.-H., Wang Z.-Y., Xu Y.-F., Yin G.-Q., Mao L.-J. MicroRNA expression profile in exosome discriminates extremely severe infections from mild infections for hand, foot and mouth disease. BMC Infect Dis. 2014;14:506. doi: 10.1186/1471-2334-14-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang R.Y., Weng K.F., Huang Y.C., Chen C.J. Elevated expression of circulating miR876-5p is a specific response to severe EV71 infections. Sci Rep. 2016;6:24149. doi: 10.1038/srep24149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arroyo J.D., Jr Chevillet, Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F. 2011. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cookson V.J., Fau Bentley Ma, Hogan B.V., FAU Hogan B.V., Horgan K., Fau Horgan K. 2012. Circulating microRNA profiles reflect the presence of breast tumours but not the profiles of microRNAs within the tumours. [DOI] [PubMed] [Google Scholar]
- 25.Calin G.A., Ferracin M., Cimmino A., DI Leva G., Shimizu M., Wojcik S.E. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 26.Szafranska A.E., Davison T.S., John J., Cannon T., Sipos B., Maghnouj A. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 27.Wu Q., Ren X., Zhang Y., Fu X., Li Y., Peng Y. MiR-221-3p targets ARF4 and inhibits the proliferation and migration of epithelial ovarian cancer cells. Biochem Biophys Res Commun. 2017;497:1162–1170. doi: 10.1016/j.bbrc.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 28.James E.R., Green D.R. Infection and the origins of apoptosis. Cell Death Differ. 2002;9:355–357. doi: 10.1038/sj.cdd.4400986. [DOI] [PubMed] [Google Scholar]
- 29.Chang L.Y., King C.C., Hsu K.H., Ning H.C., Tsao K.C., Li C.C. Risk factors of enterovirus 71 infection and associated hand, foot, and mouth disease/herpangina in children during an epidemic in Taiwan. Pediatrics. 2002;e88:109. doi: 10.1542/peds.109.6.e88. [DOI] [PubMed] [Google Scholar]
- 30.Cui, L., Qi Y Li, H., Ge, Y., Zhao, K., Qi, X., Guo, X., Shi, Z., Zhou, M., Zhu, B., Guo, Y., Li, J., Stratton, C. W., Tang, Y.-W., Wang, H. Serum microRNA expression profile distinguishes enterovirus 71 and coxsackievirus 16 infections in patients with hand-foot-and-mouth disease. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material