Abstract
Although high mortality of lung cancer is greatly due to distant metastasis, the mechanism of this metastasis remains unclear. Here, we investigate in lung cancer that SOX30 is sharply under-expressed in metastatic tumors compared with non-metastatic tumors, and suppresses plenty of metastasis related processes or pathways. SOX30 strongly inhibits tumor cell metastasis in vitro and in vivo. Sox30 deficiency promotes lung metastasis in Sox30−/− mice and this uncontrollable lung-metastasis is re-inhibited upon Sox30 re-expression. Mechanistically, SOX30 diminishes Wnt-signaling via directly transcriptional repressing β-catenin or interacting with β-catenin to compete with TCF for binding to β-catenin. The carboxyl-terminus of SOX30 is required for attenuating β-catenin transcriptional activity, whereas the amino-terminus of SOX30 is required for its interaction with β-catenin protein. Enhance of β-catenin attenuates the anti-metastatic role of SOX30. Moreover, Sox30 deficiency promotes tumor metastasis and reduces survival of mice. In addition, nuclear SOX30 expression is closely associated with metastasis and represents a favorable independent prognostic biomarker of lung cancer patients. Altogether, these results highlight an important role and mechanism of SOX30 in lung cancer metastasis, providing a potential therapeutic target for anti-metastasis.
Keywords: Lung cancer, Metastasis, SOX30, Wnt-signaling, Molecular mechanism
Highlights
-
•
SOX30 is closely associated with lung cancer metastasis, and strongly inhibits cancer cell metastasis in vitro and in vivo.
-
•
SOX30 suppresses cancer metastasis via transcriptional repressing β-catenin or competing with TCF for β-catenin binding.
-
•
SOX30 deficiency promotes tumor long-distance metastasis and reduces overall survival of mice and lung cancer patients.
The high mortality of lung cancer is largely due to distant-metastases. However, the mechanism of this metastasis remains unclear. Here, we demonstrate that SOX30 strongly inhibits lung cancer metastasis in vitro and in vivo. As an important metastatic suppressor, SOX30 prevents long-distant metastases and causes fine prognosis inhibiting Wnt-signaling via transcriptional repressing β-catenin or competing with TCF for interaction with β-catenin. This study provides useful information for effective therapies against tumor-metastasis. Considering key role of β-catenin in tumor-metastasis and ineffective treatment using inhibitors against β-catenin, it is a potential choice to suppress Wnt/β-catenin activity via targeting the upstream SOX30.
1. Introduction
Lung cancer represents the most common malignancy and is the leading cause of cancer-related mortality worldwide [1]. Non-small cell lung cancer includes adenocarcinoma (ADC, ~40%), squamous cell carcinoma (SCC, ~30%) and large cell carcinoma (LCC, ~15%), constituting over 80% of lung cancers. The prognosis of lung cancer patients is very poor and 5-year survival rate is <10% [2,3]. The poor prognosis and high mortality of lung cancer patients is mostly attributable to distant-metastasis [[4], [5], [6]]. However, the molecular mechanism leading to metastasis remains elusive, and delineating the mechanism is crucial for developing new and effective therapeutic approaches.
SRY (sex determining region Y)-box family factors (SOXs) contain a highly conserved high mobility group (HMG)-box domain, which is frequently indispensable for SOXs binding to targets or interacting with proteins[[7], [8], [9]]. Mounting evidence indicates that SOX proteins play important roles in a wide variety of embryonic and postnatal developmental processes, such as sex determination [10]; neural [11] and skeletal [12] development; and, especially the definitive endoderm development that gives rise to various organs including liver, pancreas, intestine and lung [[13], [14], [15]]. Moreover, SOXs are altered in a variety of human diseases, highlighting their involvement in these diseases, particularly in malignancies [16]. However, the pathogenetic pathways and specific mechanisms of SOXs in malignancies are poorly understood.
SOX30, as a relatively new SOX gene, has been considered to be involved in spermatogonial differentiation and spermatogenesis [[17], [18], [19], [20]]. For years, the role and molecular mechanism of SOX30 in human cancer are completely unexplored. Only recently, SOX30 has been identified as a preferentially methylated gene and a key player in tumorigenesis [21]. However, the precise roles and mechanisms of SOX30 in tumorigenesis are still largely unexplored.
In this study, we assessed a hitherto unknown role of SOX30 on lung cancer metastasis using clinical association analysis, expression profile of microarray, differentially expressed cell models, nude mice, Sox30-knockout mice and recoverable Sox30-knockout mice models. The molecular mechanism of SOX30 function was further clarified both at the transcriptional level and posttranslational level. Our findings demonstrate that SOX30 is a new metastatic suppressor inhibiting Wnt-signaling via double-track regulation of β-catenin in lung cancer, providing a “fail-safe” mechanism for SOX30 suppressing metastasis.
2. Materials and Methods
2.1. Cell Lines and Patient Samples
The human cell lines (A549, LTEP-a-2 and HEK293), mouse B16-F10 cell line (High lung metastatic model) and mouse LL/2 (Lewis lung carcinoma) cell line were obtained from the Cell Bank of Chinese Academy of Science (CBCAS, Shanghai, China) and the American Type Culture Collection (ATCC, Manassas, VA, USA), cultured in F12K (Sigma, St. Louis, MO, USA)/RPMI-1640 (HyClone, Logan, UT, USA)/DMEM (Sigma)/McCoy's 5A (Invitrogen, Carlsbad, CA, USA) media supplemented with 10% fetal bovine serum (FBS, HyClone), and incubated in 5% CO2 at 37 °C.
A total of 553 lung cancer patients (307 ADC, 241 SCC and 5 LCC) undergone surgical resection between 2004 and 2010 were recruited from Southwest Hospital or Daping Hospital Affiliated to Third Military Medical University in Chongqing, China. The surgical resected specimens were examined macroscopically. The specimens were fixed in 4% Paraformaldehyde Fix Solution and then paraffin-embedded. The specimens were sectioned at 4 μm for H&E staining or IHC. The patients diagnosed as relapse were excluded, and the patients who received preoperative radiation, chemotherapy or biotherapy were also not included. The clinico-pathologic information was retrieved from the patients' electronic medical records. The ages of 307 ADC patients (145 females and 162 males) range from 20 to 84 years and the median age is 58.97 years. The ages of 241 SCC patients (16 females and 225 males) range from 33 to 82 years and the median age is 61.95 years. The ages of 5 LCC patients (5 males) range from 51 to 67 years and the median age is 59.32 years. The tumors were staged according to the TNM staging system of the American Joint Committee on Cancer (AJCC) 7th edition. The study was approved by the ethics committee of Southwest Hospital and Daping Hospital. All experiments were performed in accordance with the approved guidelines of the Third Military Medical University, and informed consent was signed by all of the patients.
2.2. RNA Extraction and Gene Expression Microarray
Total RNA was extracted and purified by RNeasy mini kit (QIAGEN, GmBH, Hilden, Germany) according to the manufacturer's protocol. Expression profiles were generated using Agilent expression arrays (4 × 44 K, Agilent, Mississauga, ON, USA). After RNA amplification and labeling, each slide was hybridized with 1.65 μg labeled sample. Slides were washed in staining dishes after 17 h hybridization and scanned using an Agilent Microarray Scanner (Agilent Technologies, Santa Clara, CA, USA). Data were extracted with the Feature Extraction software 10.7 (Agilent Technologies) and normalized by Quantile algorithm, Gene Spring Software 11.0 (Agilent Technologies). After applying a filter compared SOX30 with vector control, 747 up-regulated and 834 down-regulated genes (fold change > 2 or <1/2).were identified.
2.3. Gene Ontology (GO) and KEGG Pathway Analysis
GO and KEGG pathway analyses were performed using Cytoscape V2.7 (http://cytoscape.org/) with the ClueGo V1.3. ClueGo determines the distribution of the target list across the GO terms and pathways. An adjusted p-value, 0.05 indicated a statistically significant deviation from the expected distribution, and the corresponding GO terms and pathways were enriched in target genes.
2.4. Network Analysis
The human protein-protein interaction networks (PPINs) were downloaded from the Human Protein Reference Database. The igraph package of the statistical language R was applied for functional profiling. The network visualization and Cytoscape were used to find the putative target genes. The Markov cluster algorithm was used to identify highly connected modules within the global networks.
2.5. Hierarchical Clustering
Cluster version 3.0 and Java TreeView version 1.1.6 were used to perform Hierarchical clusteringt. With the use of clustering algorithms, the samples and probes were grouped based on similarities in the expression profiles. The probe set was filtered based on the standard deviation to exclude the probes of least variance. The average linkage and median centering were the chosen parameters. The unsupervised and supervised clustering was used. In the unsupervised method, all genes were included, whereas in supervised, significant genes were involved input.
2.6. Sox30-Null and Inducible Sox30-Null Mouse Models
Sox30-null mice were generated by the Model Animal Research Center at Nanjing University (the strategy as Supplemental Fig. S1). Briefly, BAC clones contain targeted gene were purchased. Targeted gene including homologous arms was retrieved from the BAC vector, then 1st LoxP-SA-IRES-GFP-NEO-STOP-PPS-2nd LoxP were introduced between Exon1 and Exon2 by homologous recombination in E.coli. The targeting vector was confirmed by PCR, enzyme digestion and sequencing. The targeting vector was linearized with AsiSI and electroporated into C57BL/6 ES cells. The recombinants were selected for their G418 and ganciclovir (Ganc) resistance. The targeted ES cells were screened by PCR and southern blot. The positive clones were chosen for microinjection and then get chimeras. Cross the chimeras with C57BL/6 mice to test if any heterozygous produced.
To restore Sox30 expression, we generated mice expressing Cre recombinase by breeding Sox30 heterozygous (Sox30+/−) mice with B6·Cg-Tg (CAG-cre/Esr1) 5Amc/JNju tool mice (No: 004682, The Jackson Laboratory), to produce Sox30-KI and Cre double positive heterozygote mice. Tamoxifen-inducible Cre-mediated recombination result in deletion of the floxed sequences in the offspring of these mice.
The genotype of the offspring was identified by PCR. All mouse experiments were carried out with the permission of the Institutional Animal Care and Use Committee of Third Military Medical University, China.
2.7. RNA and Protein Extraction
Total RNA was extracted using TRIzol Reagent (Invitrogen), and approximately 5 μg total RNA was treated with DNase I (Sigma) to eliminate the genomic DNA contamination. Complementary DNAs were synthesized using the GoScript™ Reverse Transcription System (Promega) according to the instruction and were stored at −20 °C. Cells or tissues were lysed in lysis buffer (Beyotime, Shanghai, China) containing complete protease inhibitor (Roche, Mannheim, Germany) to get total protein.
The cytoplasmic and nuclear extracts of lung cancerous and peri-cancerous pair tissues were obtained using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce, Rockford, lL, USA). Briefly, approximately 40 mg clinical specimens were cut into small pieces and washed in PBS. The tissues were then homogenized with homogenizer in 400 μl cold CER I. The cold CER II (22 μl) was next added, and the supernatant (cytoplasmic extract) was transferred into a clean tube. The pellet was suspended in 200 μl cold NER and the supernatant was transferred into another clean tube as the nuclear extract.
2.8. RT-PCR and RT-qPCR
RT-PCR (Reverse transcription polymerase chain reaction) and RT-qPCR (quantitative real-time PCR) were performed as previously described [21]. Primers are listed in Supplemental Table S1.
2.9. Tissue Microarray Generation and Immunohistochemical (IHC) Analysis
The tissue microarray generation, IHC analysis and quantitatively defined expression levels are described in a previous study [22]. Briefly, positive percentage of staining was classified into 5 categories: 0 score (<10% positive cells), 1 score (10% to 25% positive cells), 2 score (26% to 50% positive cells), 3 score (51% to 75% positive cells) and 4 score (>75% positive cells). Intensity of staining was graded into four groups: 0 score (negative staining), 1 score (weak staining), 2 score (moderate staining) and 3 score (strong staining). All core biopsies were independently reviewed by two blinded pathologists. The expression levels of SOX30 were calculated by the product of category for the positive percentage and grade for the intensity of staining (the range of the calculation was therefore 0–12) [23,24]. IHC staining was performed using the antibody against SOX30 (1100, Santa Cruz Biotechnology, Heidelberg, Germany).
2.10. Western Blotting (WB) Analysis
WB analysis was performed as previously described [21]. The following primary antibodies were used: SOX30 rabbit polyclonal antibody (1:800; Santa Cruz Biotechnology, sc-20104), β-catenin mouse monoclonal antibody (1:800; Santa Cruz Biotechnology, sc-7963), p-β-catenin mouse monoclonal antibody (1:600; Santa Cruz Biotechnology, sc-57535), TCF4 goat monoclonal antibody (1:600; Santa Cruz Biotechnology, SC-8631), MYC rabbit polyclonal affinity purified antibody (1:500; Cell Signaling Technology, Boston, MA, USA, #13987), CCND1 mouse monoclonal antibody (1:500; Santa Cruz Biotechnology, sc-450), MMP7 goat polyclonal antibody (1:600; Santa Cruz Biotechnology, sc-8832), HNF6 mouse monoclonal antibody (1:800; Santa Cruz Biotechnology, sc-376167), and FN1 rabbit polyclonal antibody (1500; Abzoom, Dallas, TX, USA, AM2784). Secondary antibodies were horseradish peroxidase (HRP)-conjugated.
2.11. Construction of Plasmids and Stable Cells
The construction of plasmids and stable cells were performed as previously described [21].
2.12. Wound Healing Assay
A549 and LTEP-a-2 cells stably transfected with SOX30 or control vector were cultured in 6-well plates at 5 × 105 cells/well. When the cells grew up to 90% confluence, three scratch wounds were made across each well using a P-200 pipette tip (Axygen, Union City, CA, USA), and the cells were then incubated at 5% CO2 and 37 °C in F12K/RPMI-1640 media containing 0.1% FBS. Images of the wound closure areas were obtained at 0, 24, 48 and 72 h after scratch wound. The experiment was performed in triplicate wells at least twice.
2.13. Transwell Assay
Transwell assays were performed in 6-well plates (8 μm, Corning, Acton, MA, USA) without or with matrigel (BD Bioscience, San Jose, CA, USA) to assess the cell migration and invasion, respectively. A549 and LTEP-a-2 cells stably transfected with SOX30 or control vector, and A549-SOX30 stable cells stably transfected with miRNAs or negative control were suspended at 2 × 105 cells/ml in serum-free medium, and seeded in the upper well of the chamber. The lower well of the chamber contained media supplemented with 10% FBS. The cells that migrated to the lower chamber after 12 h (migration) or 18 h (invasion) were stained with 0.1% crystal violet and counted under an inverted microscope. All experiments were performed in triplicate and repeated thrice.
2.14. Metastasis Assay in Nude Mice
A549 cells stably transfected with SOX30 or empty vector in PBS were injected into tail vein (1 × 106 cells/mouse) of 6-week-old Balb/c nude mice. The metastasis was observed at 8 weeks after injection. All of the animal experiments conformed to the Guide for Care and Use of Laboratory Animals, and were approved by the Institutional Committee of Laboratory Animal Experimentation at the Third Military Medical University in China.
2.15. Lung-Metastasis Assay in Sox30-Null Mice
Mouse wild-type B16-F10 (B16) melanoma cells or LL/2 cells were injected into tail vein of about 8 weeks old C57/Bl6 Sox30+/+, Sox30+/− and Sox30−/− mice at a concentration of 5 × 105cells/mouse or 1 × 106cells/mouse. Mice were sacrificed at 21 days after B16 injection and at 30 days after LL/2 injection, and lung metastasis was quantified. The animal experiments conformed to the Guide for Care and Use of Laboratory Animals and were approved by the Institutional Committee of Laboratory Animal Experimentation at the Third Military Medical University in China.
2.16. Metastasis Assay in Inducible Sox30-Null Mice
Tamoxifen (tam, 10 mg/ml, Sigma) was administered via i.p. to induce Cre-mediated Sox30 expression in half (another half of Sox30−/− mice were injected with solvent via i.p. following the same steps) of the Sox30−/− mice at a week post the B16 or LL/2 injection by tail vein. The mice were injected once a day for 5 days with 1 mg tam/mice. The mice were sacrificed at 21 days after B16 injection or at 30 days after LL/2 injection, and metastasis was evaluated in lung tissues. All of the animal experiments conformed to the Guide for Care and Use of Laboratory Animals and were approved by the Institutional Committee of Laboratory Animal Experimentation at the Third Military Medical University in China.
2.17. The β-Catenin/TCF Transcriptional Activity and Luciferase Reporter Assays
A549 and HEK293T cells were transfected with TOP-Flash/FOP-Flash plasmids containing wild-type and mutated TCF/LEF binding sites, respectively. The TOP-Flash/FOP-Flash plasmids were purchased from Addgene (12456 and 12457, Cambridge, MA, USA). Transfections were performed using ViaFect™ Transfection Reagent (Promega). Luciferase reporter assays were analyzed by the fluorescence microplate reader measurement system Varioskan LUX (Thermo Fisher, Waltham, MA, USA) using a Dual-luciferase reporter kit (Promega) as previously described [21]. A Renilla reniformis luciferase reporter was used as an internal control. Luciferase activities were measured at 36 h post-transfection. Each experiment was repeated thrice.
2.18. Chromatin-Immunoprecipitation Assay
Chromatin-immunoprecipitation (ChIP) assays were analyzed using a ChIP assay kit (Cell Signaling Technology, #9004) according to the manufacturer's protocol. Briefly, 4 × 106 A549 and HEK293 cells were fixed in a final concentration of 1% formaldehyde, digested with micrococcal nuclease, chromatin immunoprecipitated after analysis of chromatin digestion, eluted of chromatin and purified DNA. The immunoprecipitated and input DNA were used as templates for RT-qPCR analysis using the primers listed in Supplemental Table S1.
2.19. Electrophoretic Mobility Shift Assay
Electrophoretic mobility shift assay (EMSA) was performed using a LightShift® Chemiluminescent EMSA Kit (Pierce, 20148) to detect DNA-protein interaction according to the manufacturer's instruction. Briefly, biotin 5′ end-labeled DNA probes containing putative binding sites for SOX30 without or with 100-fold unlabeled DNA probes (an oligonucleotide competitor) were incubated with the nuclear extracts prepared using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce) from A549 cells expressing empty vector or SOX30. The DNA-protein complex was subjected to 8% polyacrylamide gel electrophoresis and then transferred onto nylon membrane (Pierce). The membrane was immediately cross-linked with a hand-held UV lamp equipped with 254 nm bulbs for 10 min at a distance of about 0.5 cm, and was then detected by chemiluminescence. The probe sequences are listed in Supplementary Table S1.
2.20. Site-Directed Mutagenesis Assay
The SOX30 binding sites in the β-catenin promoter and SOX30 HMG-box constructs were mutated using a QuikChange Lightning Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) according to the instruction. Briefly, mutagenic primers were designed, and the mutant strand was synthesized by RT-PCR. The amplification product was digested by Dpn I, and then the Dpn I-treated DNA was transformed into XL10-Gold Ultracompetent cells. The mutations were validated by sequencing. The primers used in the site-directed mutagenesis were listed in Supplemental Table S1.
2.21. Co-Immunoprecipitation (Co-IP) Assay
Total extracts of A549 and HEK293 cells or lung tissues from mice with or without SOX30 expression were lysed with IP lysis buffer (Pierce). The co-IP analyses were performed using a Co-Immunoprecipitation Kit (Pierce, 26149) according to the manufacturer's protocol. Briefly, the experimental steps: pre-clear lysate using the control agarose resin, Co-IP, elution of Co-IP, resin regeneration and preparation for SDS-PAGE analysis were carried out in turn. Subsequent WB analyses were performed as described above. The experiment was repeated thrice.
2.22. Fluorescence Resonance Energy Transfer (FRET) Assay
The HEK293 and A549 cells were plated in 12-well culture plates or special small dishes (Nest Biotechnology Co. LTD), and co-transfected with pEYFP-SOX30 (SOX30 fused to yellow fluorescent protein)/pEYFP-Vector and pECFP-Catenin (β-catenin fused to cyan fluorescent protein). FRET analyses were performed as previously reported [25,26]. The transfected cells were prepared into suspension 48 h after transfection, and FRET was analyzed by the fluorescence microplate reader measurement system Varioskan LUX (Thermo Fisher) in NUNC 384-well black bottom plates (Thermo Fisher). The cells were also fixed 48 h after transfection, and FRET analysis was determined by LSM800 confocal microscope (Zeiss, Jena, Germany). The experiment was carried out thrice in triplicate wells.
2.23. Structural Prediction
A structure of SOX30 was obtained from the Robetta server which is an automated tool for protein structure prediction. As the confidence of matching to a known structure was low, the de novo Rosetta fragment insertion method was used for SOX30 construction. The complex of the β-catenin with TCF4 was used high-resolution crystal structure (2gl7) from PDB dataset. The structures in protein-protein interactions of SOX30 and β-catenin were achieved by Z-dock server (http://zdock.umassmed.edu/) with the best top 10 model selected from the overlapped results.
2.24. Statistical Analysis
Statistical analyses were performed using SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 6 software (La Jolla, CA, USA). The data were performed at least triplicate and expressed as the mean ± standard error of the mean (SEM). The statistical comparisons were analyzed using Chi-square test, Student's t-test (only two groups) or One-way ANOVA (three or four groups). Survival analyses were calculated by log-rank test Kaplan-Meier and Cox regression methods. The expression was categorized as high or low according to the median score. The p-values of <0.05 (two-sided) were considered statistically significant.
3. Results
3.1. SOX30 Is Tightly Associated with Metastasis of Lung Cancer Patients
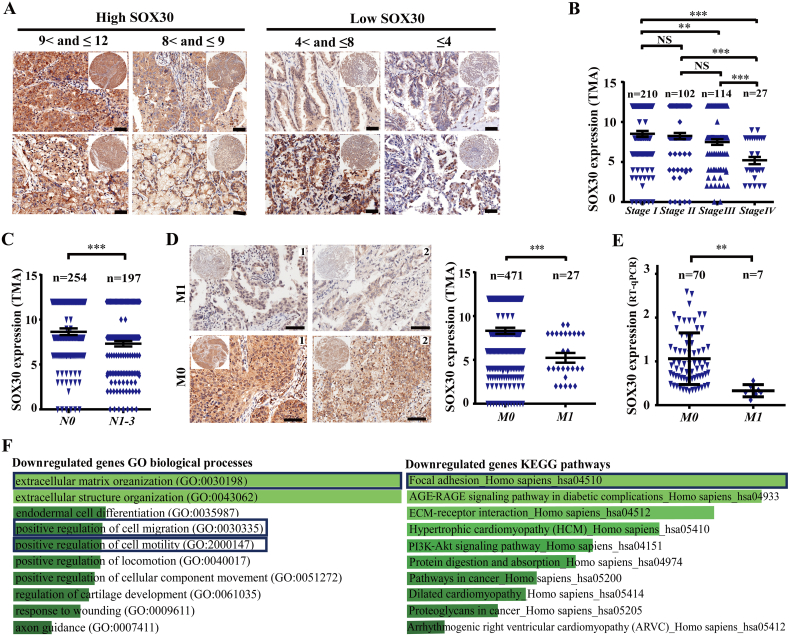
To examine SOX30 expression, the lung biopsy specimens of 550 lung cancer patients were probed using immunohistochemistry (IHC). Based on the product of positive percentage and intensity of staining, the patients were classified into two groups: SOX30 high (scores 8< and ≤12) and SOX30 low (scores ≤ 8) (Fig. 1A). When analyzing SOX30 expression with clinicopathologic characteristics of these patients, SOX30 was closely correlated with clinical stages (p = 0.001) and long-distance metastasis (p = 0.019) (Supplemental Table S2). The percentage of SOX30 high-expression samples was decreased in patients from clinical early stages (I + II, 31.41%, 98/312) to advanced stages (IV + III, 17.02%, 24/141) groups (Supplemental Table S2). SOX30 expression was significantly reduced from clinical stage I, II, III to IV groups (p < 0.0001, Fig. 1B). Moreover, SOX30 expression was obviously decreased in the patients of lymph nodal metastasis (N1–3) group compared with no lymph nodal metastasis (N0) group (p = 0.0002, Fig. 1C). When separating the specimens into two groups based on the clinical diagnosis of metastasis, the percentage of SOX30 high-expression samples was significantly lower in the group of distant metastasis (7.41%, 2/27) than that of non-metastasis (28.03%, 132/471) (p = 0.019) (Supplemental Table S2). SOX30 expression was much low in the group of metastasis compared to that of non-metastasis (Fig. 1D), which was further confirmed in an independent cohort at mRNA level (Fig. 1E). These data reveal that SOX30 is closely associated with metastasis of lung cancer patients.
Fig. 1.
SOX30 is associated with tumor-metastasis of patients and metastatic processes/pathways. (A) Images of SOX30 expression are presented at different score levels. SOX30 low expression group contains the ≤8 score patients. SOX30 high expression group contains the 8< and ≤12 score patients. Scale bars, 20 μm. (B) SOX30 expression was significantly lower in lung cancer patients at advanced-stage than that at early-stage. The p value was measured with Student's t-tests. NS, nonstatistical significance; **, p < 0.01; ***, p < 0.001. (C) SOX30 expression was obviously lower in lung cancer patients with lymph node metastasis (N1–3) than that without lymph node metastasis (N0). The p value was measured with Student's t-tests. ***, p < 0.001. (D) SOX30 expression was significantly lower in lung cancer patients with distant-metastasis (M1) than that without metastasis (M0). The p value was measured with Student's t-tests. ***, p < 0.001. Scale bars, 20 μm. (E) SOX30 expression levels were analyzed in non-metastatic and metastatic cancers in an independent cohort. The p value was measured with Student's t-tests. **, p < 0.01. (F) The top 10 categories of the GO biological processes and the KEGG pathways associated with down-regulated genes lists (SOX30/vector). The blue boxes represent the biological processes or pathways correlated with tumor metastasis.
To further identify the association of SOX30 with metastasis, we examined the RNA array profiles of differential expression mRNAs in SOX30 and vector expressed A549 cells. Based on these differentially expressed mRNAs, Gene ontology (GO) and KEGG enrichment was performed. Among the top 10 biological processes or pathways of down-regulated genes, many biological processes or pathways were correlated with tumor metastasis (Fig. 1F), indicating a potentially prominent role of SOX30 on metastasis.
3.2. SOX30 Suppresses Cancer Cell Migration and Invasion In Vitro
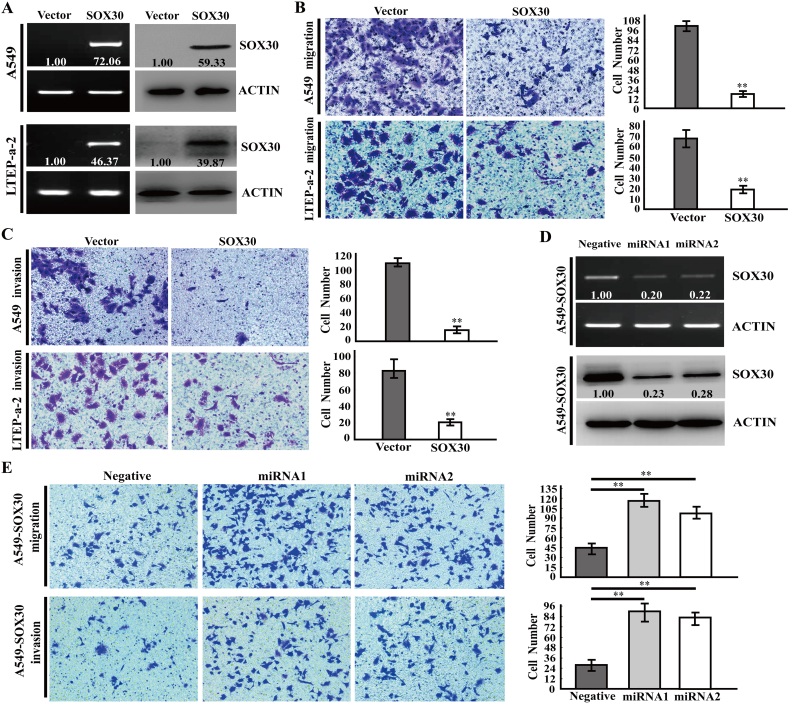
To investigate the role of SOX30 on cancer cell migration and invasion, we generated gain-of-function in human adenocarcinoma A549 and LTEP-a-2 cells by transfecting SOX30 or empty vector plasmids (Fig. 2A and Supplemental Fig. S2A), and determined cell migration and invasion by wound healing scratch and transwell assays. The cancer cell migration ability was decreased in SOX30-transfected cells compared with empty vector-transfected cells (Fig. 2B and Supplemental Fig. S2B). The cancer cell invasion ability was clearly reduced in SOX30-transfected cells (Fig. 2C). To further confirm the roles of SOX30, the effect of SOX30 knockdown on cancer cell migration and invasion in a stable transfectants (A549-SOX30) with micro RNA (miRNA) was investigated. In contrast with the results of SOX30 overexpression, knockdown of SOX30 increased cancer cell migration and invasion compared with negative control (Fig. 2D and E). These data suggest that SOX30 sharply suppresses cancer cell migration and invasion.
Fig. 2.
SOX30 inhibits cancer cell migration and invasion. (A) Ectopic expression of SOX30 in A549 and LTEP-a-2 cell lines was demonstrated by RT-PCR and WB. The relative intensities of the proteins versus internal control (ACTIN) are shown. ACTIN represents β-actin. (B) The migrated cells were analyzed by transwell assays without matrigel in stable SOX30 or vector A549 and LTEP-a-2 cells. The cells in the lower chamber were fixed, stained and determined by the average count of five random microscopic fields. Error bars indicate SEM. The p value was measured with Student's t-tests. **, p < 0.01. (C) The invaded cells were analyzed by transwell assays with matrigel in stable SOX30 or vector A549 and LTEP-a-2 cells. The cells in the lower chamber were fixed, stained and determined by the average count of five random microscopic fields. Error bars indicate SEM. The p value was measured with Student's t-tests. **, p < 0.01. (D) SOX30 knockdown was confirmed by RT-PCR and WB in A549-SOX30 stable cells. ACTIN represents β-actin. (E) Migration and invasion analyses of the migrated or invaded cells were performed in A549-SOX30 stable cells transfected with miRNAs or negative control. The p value was measured with Student's t-tests. **, p < 0.01.
3.3. SOX30 Inhibits Tumor Metastasis in Nude Mice and Sox30-Knockout Mice
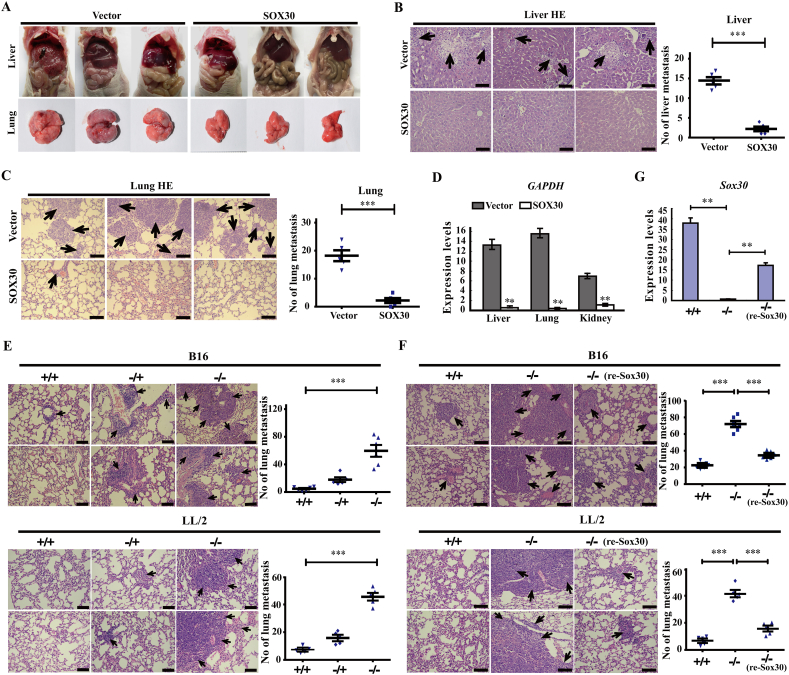
To confirm the functional role of SOX30 in vivo, the metastasis of tumor cells (A549 transfectants) were examined in nude mice. The nude mice of SOX30 over-expressing group were demonstrated significantly decreased lung and liver metastatic tumors compared with that of empty vector group by tissue observation and HE staining (Fig. 3A-C). The inhibitory liver and lung metastatic tumors of SOX30 were further confirmed by detecting human-specific GAPDH levels to quantify metastatic human cancer cells (Fig. 3D). In addition, SOX30 over-expression also inhibited kidney metastatic tumors (Fig. 3D). To further evaluate SOX30 inhibitory function on metastasis, we created a Sox30-null C57BL/6 mouse model by homologous recombination. The lung cancer cell lines (A549 and LTEP-a-2) were first injected into wild-type (Sox30+/+), heterozygous (Sox30+/−) and homozygous (Sox30−/−) mice by tail vein to produce metastasis models. However, the injections of the two lung cancer cell lines did not work in these mice. B16-F10 (B16) mouse melanoma cell line, a classical model with very high lung metastatic potential [27,28], and Lewis lung carcinoma (LL/2) cell line widely used as a model for lung-metastasis were then selected and injected into Sox30+/+, Sox30+/− and Sox30−/− mice through tail vein to produce lung metastasis models. The Sox30−/− and Sox30+/− mice were significantly increased in lung colonization of B16 or LL/2 cells as compared with Sox30+/+ mice after injection (Fig. 3E, Supplemental Fig. S3A and B). These results show that SOX30 strongly inhibits tumor cell metastasis in vivo.
Fig. 3.
SOX30 represses tumor metastasis in vivo. (A–C) Liver and lung metastases of A549 cells were determined by tissue observation and HE staining and quantitative analyses of metastases were measured in liver and lung tissues. Arrows indicate the metastasis loci. Scale bar, 50 μm. HE, haematoxylin and eosin. The p value was measured with Student's t-tests. ***, p < 0.001. (D) Liver, lung and kidney metastases were measured at 8 weeks after implantation of A549 cells by detecting human-specific gene expression using RT-qPCR. Human-specific GAPDH levels were used to quantify metastatic human cancer cells. The p value was measured with Student's t-tests. **, p < 0.01. (E) Lung metastases were evaluated by HE staining in Sox30+/+, Sox30+/− and Sox30−/− mice injected with B16 cells or LL/2 cells by tail-vein. The numbers of metastasis were determined at 21 days after B16 or 30 days after LL/2 injection. Arrows indicate the metastasis loci. Scale bar, 50 μm. The p value was measured with One-way ANOVA. ***, p < 0.001. (F) The uncontrolled lung metastasis of B16 cells or LL/2 cells due to Sox30 loss can be re-inhibited upon Sox30 re-expression in an inducible system. Arrows indicate the metastasis loci. Scale bar, 50 μm. −/− (re-Sox30), Sox30 re-expression Sox30−/− mice. The p value was measured with Student's t-tests. ***, p < 0.001. (G) Sox30 expression was restored in lung tissues of Sox30−/− mice by tamoxifen treatment in the inducible system. The p value was measured with Student's t-tests. **, p < 0.01.
3.4. The Wild Lung-Metastasis in Sox30−/− Mice is Re-Inhibited upon Sox30 Re-Expression
To further determine whether the uncontrolled lung-metastasis of B16 melanoma or LL/2 cells in Sox30−/− mice was restorable when re-expression of Sox30, we used an inducible system to manipulate Sox30 expression and determined the metastasis of B16 or LL/2 cells. Sox30 expression was restored via removing the insertion cassette with cre–recombinase [Sox30−/− (re-Sox30)] induced by tamoxifen (tam) in half of the Sox30−/− mice at a week after B16 or LL/2 injection. A significant decrease in lung colonization of B16 or LL/2 cells was found in the Sox30−/− (re-Sox30) animals compared with the control Sox30−/− mice at 21 days after B16 injection or 30 days after LL/2 injection (Fig. 3F, G and Supplemental Fig. S3C), indicating that wild lung-metastasis caused by Sox30-deletion was re-inhibited upon Sox30 re-expression. The result demonstrates that SOX30 exerts a critical inhibitory role on tumor cell metastasis.
3.5. SOX30 Dramatically Attenuates Wnt-Signaling Pathway
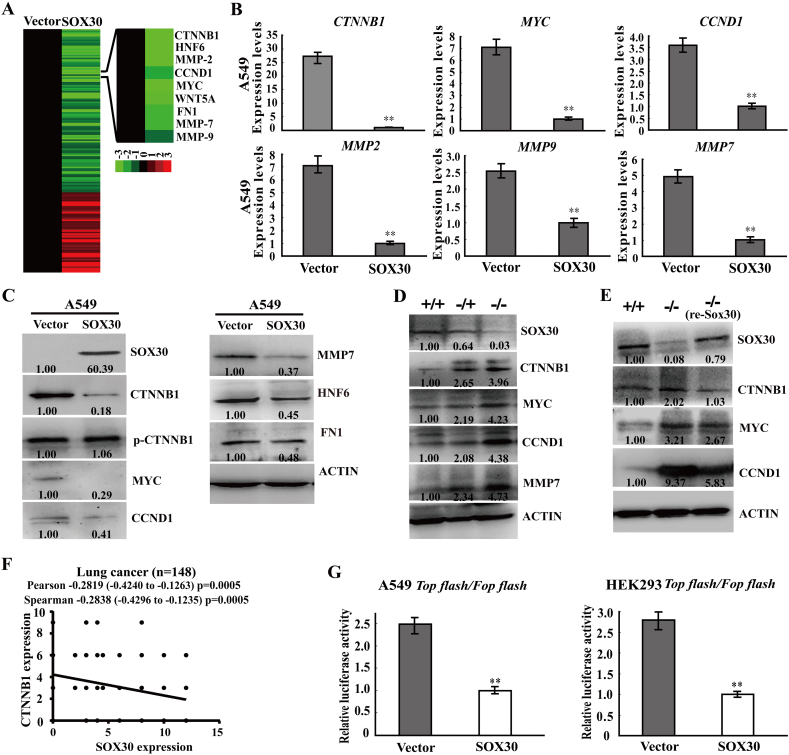
To explore the mechanism of SOX30 anti-metastatic function, we built a global SOX30-regulated networks using potential targets from RNA array profiles, and identified an important module: Wnt-signaling regulated gene network (Supplemental Fig. S4). The expressions of Wnt-signaling genes, such as β-catenin (CTNNB1), MYC, CCND1, WNT5A, MMP2, MMP7, MMP9, JUN, DKK1, FOSL1 and NKD2 associated with cell migration and invasion, were down-regulated in SOX30 transfectants (Fig. 4A, Supplemental Fig. S5A and Supplemental Table S3). These genes were indeed notly down-regulated in tumor cells when SOX30 overexpression (Fig. 4B, C and Supplemental Fig. S5B). Several downstream probes, including CXXC4, DAAM1 and PLCB1, which have been found upregulated by SOX30 in microarray dataset were also indeed up-regulated when SOX30 overexpression (Supplemental Fig. S5B). In addition, a clear inverse correlation in expression was noted between SOX30 and Wnt-signaling targets in TCGA database (Supplemental Fig. S5C and D). Deletion of Sox30 resulted in enhanced expressions of β-catenin, MYC, CCND1 and MMP7 in Sox30-null mouse model (Fig. 4D). In the inducible system, β-catenin, MYC and CCND1 expression were expectedly re-inhibited when restoring Sox30 expression in Sox30−/− (re-Sox30) mice (Fig. 4E). In human clinical samples, SOX30 expression was negatively correlated with β-catenin expression in lung cancer patients (p = 0.0005) (Fig. 4F). To further ensure that SOX30 definitely suppresses Wnt-signaling activity, TOPflash/FOPflash reporter assays were performed in A549 cell line and HEK293 cell line (a star cell line for mechanism study). Ectopic expression of SOX30 dramatically repressed relative Wnt-signaling transcriptional activity compared with empty vector control (Fig. 4G). These data demonstrate that the anti-metastatic role of SOX30 is mediated by suppressing Wnt-signaling pathway.
Fig. 4.
SOX30 attenuates Wnt-signaling by down-regulation of related genes. (A) Hierarchical clustering of potential targets of SOX30 from RNA array profiles was made in stably transfected A549 cells. The mRNAs were clustered according to the profiles of two-fold differential expression between SOX30 and vector groups (SOX30/Vector). (B) Ectopic expression of SOX30 significantly represses the endogenous mRNA expression of β-catenin (CTNNB1), MYC, CCND1, MMP2, MMP9 and MMP7 by RT-qPCR. The relative intensities of the proteins versus internal control are shown. ACTIN was used as an internal control. Error bars indicate SEM. The p value was measured with Student's t-tests. **, p < 0.01. (C) Ectopic expression of SOX30 suppresses the endogenous protein expression of β-catenin, MYC, CCND1, MMP7, HNF6 and FN1 by WB. ACTIN was used as an internal control. (D) Loss of SOX30 activates the protein expression of β-catenin, MYC, CCND1 and MMP7 in lung tissues from Sox30-null mouse model. The relative intensities of the proteins versus internal control are shown. ACTIN was used as an internal control. (E) The functional recovery of Sox30-null mice on metastasis when re-expression of Sox30 is associated with Wnt-signaling. The β-catenin, MYC and CCND1 were re-inhibited when restoring Sox30 expression in Sox30−/− (re-Sox30) mice. The relative intensities of the proteins versus internal control (ACTIN) are shown. (F) SOX30 expression is negatively associated with β-catenin expression in lung cancer patients. The p value was measured with Pearson and Spearman correlations. CTNNB1, β-catenin. (G) SOX30 decreased TOP-flash (contains wild-type LEF/TCF-binding sites) reporter activation in A549 and HEK293 cells. TOP-flash or FOP-flash (contains the mutant LEF/TCF-binding sites) plasmid was co-transfected with SOX30 or empty vector and PRL-TK plasmids into the cells to determine the transcriptional activity of Wnt/β-catenin signaling. Error bars indicate SEM. The p value was measured with Student's t-tests. **, p < 0.01.
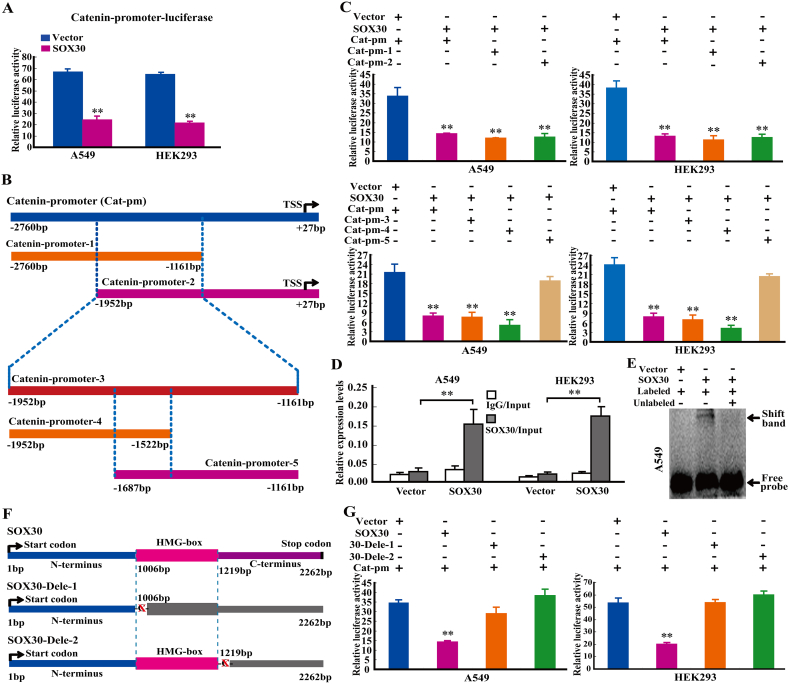
3.6. SOX30 Represses Wnt-Signaling by Direct Binding to β-Catenin Promoter
To elucidate the molecular mechanism of SOX30 suppressing Wnt-signaling, we analyzed whether SOX30 could regulate β-catenin (the master gene of Wnt-signaling) at transcriptional level. Bioinformatics analysis revealed that β-catenin promoter contains consensus binding sites for SOX30 (Supplemental Fig. S6A and B). Luciferase reporter assays in A549 and HEK293 cell lines indicated that SOX30 over-expression significantly attenuated the activity of β-catenin promoter (−2760 to +27 bp) (Fig. 5A), suggesting that SOX30 antagonizes Wnt-signaling by binding to β-catenin promoter and inhibits its transcription. To identify SOX30-binding site(s) within β-catenin promoter, two different regions of β-catenin promoter (Catenin-promoter-1: -2760 to -1161 bp and Catenin-promoter-2:-1952 to +27 bp) were constructed and analyzed by luciferase reporter assays. SOX30 still attenuated the activity of both Catenin-promoter-1 and Catenin-promoter-2, suggesting that the overlap region (-1952 to -1161 bp) of Catenin-promoter-1 and Catenin-promoter-2 was required for SOX30-binding (Fig. 5B and C). Then this overlap region (-1952 to -1161 bp) within β-catenin promoter was constructed as Catenin-promoter-3 and further segmented into Catenin-promoter-4: -1952 to -1522 bp and Catenin-promoter-5: -1687 to -1161 bp, and analyzed by luciferase reporter assays. SOX30 still attenuated the activity of both Catenin-promoter-3 and Catenin-promoter-4, but failed to attenuate the activity of Catenin-promoter-5, insinuating that the specific region (-1952 to-1687) of Catenin-promoter-4 was required for SOX30-binding (Fig. 5B and C). This key region of β-catenin promoter for SOX30 binding was further confirmed by ChIP-qPCR assay and EMSA (Fig. 5D and E). These findings demonstrate that β-catenin is a direct target of SOX30.
Fig. 5.
SOX30 represses Wnt/β-catenin signaling by directly binding to β-catenin promoter. (A) SOX30 targets β-catenin as detected by luciferase reporter assay. A luciferase reporter linked with full-length native promoter of β-catenin (Catenin-promoter) was used for the luciferase reporter assay in A549 and HEK293 cells. Results were normalized with internal controls and presented as averages with SEM from three experiments. The p value was measured with Student's t-tests. **, p < 0.01. (B) Different regions of β-catenin promoter (Cat-pm-1, -2, -3 -4 and -5) were constructed into luciferase reporter vector. The region in the dotted line is the overlap region of Cat-pm-1 and -2 or Cat-pm-4 and -5. TSS represents transcription start site. (C) The effects of SOX30 on different partial regions of β-catenin promoter activity were analyzed by luciferase reporter assays. The SOX30-binding region in β-catenin promoter was identified at −1952 to −1687 bp. The p value was measured with Student's t-tests. **, p < 0.01. (D) ChIP-qPCR was performed to identify β-catenin as a direct binding target of SOX30. **, p < 0.01. (E) EMSA was performed to identify β-catenin as a direct binding target of SOX30. The lanes from left to right are vector+ Biotin-labeled probe, SOX30+ Biotin-labeled probe and SOX30+ Biotin-labeled probe+100-fold unlabeled probe. (F) The constructs with or without HMG-box and/or C-terminal domain of SOX30 were built. SOX30-Dele-1 is the construct without HMG-box and C-terminal regions. SOX30-Dele-2 is the construct without C-terminal region. (G) The effects of SOX30 with or without HMG-box and/or C-terminal domains on β-catenin promoter activity were determined. The p value was measured with Student's t-tests. **, p < 0.01.
3.7. SOX30 Binds to β-Catenin Promoter Independent on the HMG-Box
To determine whether the HMG-box (the DNA-binding domain of Sox genes) is required for SOX30 binding to β-catenin promoter, the deletions of HMG-box domain (336-406AA) and/or carboxyl-terminal region (C-terminus, 407-753AA) were generated by site-directed mutagenesis (Fig. 5F and Supplemental Fig. S6C). SOX30 failed to attenuate the activity of β-catenin promoter when the HMG-box and C-terminus region were both deleted and it still failed to attenuate β-catenin promoter activity when the C-terminus region was deleted (Fig. 5G). These data indicate that the C-terminus of SOX30 is required for attenuating activity of β-catenin promoter independent on the HMG-box.
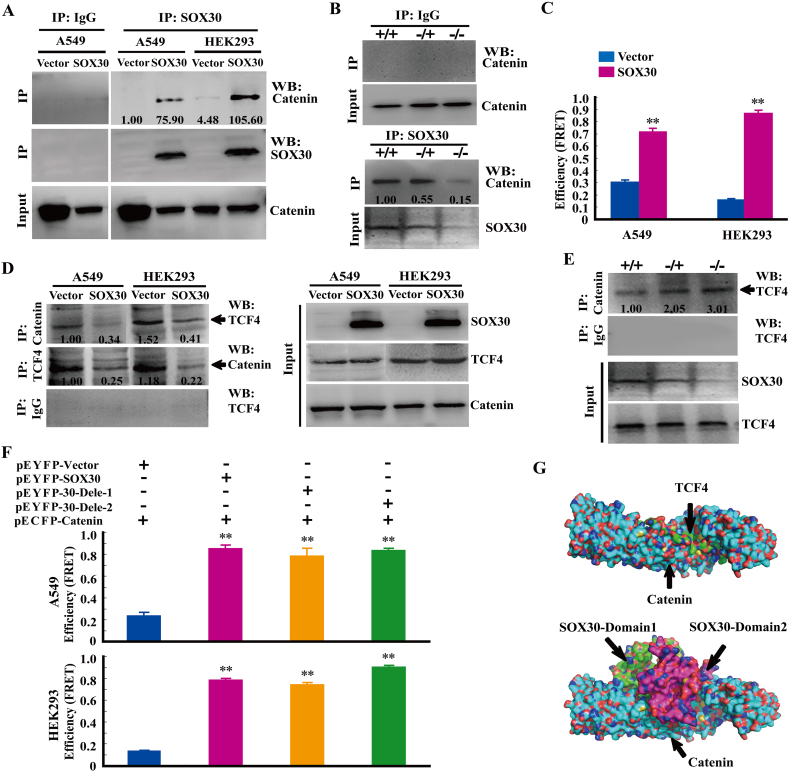
3.8. SOX30 Diminishes Wnt/β-Catenin-Dependent Transcription by Direct Interacting with β-Catenin Protein
Previous studies demonstrate that Wnt/β-catenin signaling activity frequently depends on posttranslational regulation of β-catenin levels, and some SOX members may bind to β-catenin protein, suggesting that SOX30-mediated repression of Wnt/β-catenin signaling may involve in a protein interaction. To test this hypothesis, co-IP assays were performed in A549 cell line, HEK293 cell line and lung tissues of Sox30-null mice. β-catenin can be co-immunoprecipitated with SOX30 in A549 and HEK293 cells, and lung tissues of animal models (Fig. 6A and B). Furthermore, FRET assays analyzed using Varioskan LUX revealed that efficient energy transfer between ECFP and EYFP occurred only between pEYFP-SOX30 and pECFP-Catenin, not between pEYFP-SOX30 and pECFP-Vector in A549 and HEK293 cell lines (Fig. 6C), which was further confirmed by FRET assays analyzed using LSM800 confocal microscope in HEK293 cell line (Supplemental Fig. S7A and B). Additionally, structural prediction showed that Z-dock binding of SOX30 identified an interaction surface on β-catenin (Supplemental Fig. S7C). These data reveal a direct interaction between SOX30 and β-catenin proteins.
Fig. 6.
SOX30 antagonizes Wnt/β-catenin signaling via directly interact with β-catenin for competitive binding to β-catenin with TCF. (A, B) Co-IP was performed on whole lysates from cells with or without SOX30 or from lung tissues of Sox30-null mouse model using SOX30 antibody. The relative intensities of the blots (IP) versus internal control (input) are shown. (C) The interaction of SOX30 with β-catenin was measured by FRET assay. FRET efficiency of A549 and HEK-293 cells co-transfected with full-length β-catenin labeled with CFP and full-length SOX30 labeled with YFP were presented. The p value was measured with Student's t-tests. **, p < 0.01. (D) The transfected cell lysates were subjected to IP using β-catenin or TCF4 antibodies followed by WB with TCF4 or β-catenin antibodies. Reciprocal IP was performed using β-catenin or control IgG followed by WB with the TCF4 antibody. The relative intensities of the blots (IP) versus internal control (input) are shown. (E) The lung lysates form Sox30+/+, Sox30+/− and Sox30−/− mice were subjected to IP using β-catenin or IgG antibody. Reciprocal IP was performed using β-catenin antibody or control IgG followed by WB with TCF4 antibody. The relative intensities of the blots (IP) versus internal control (input) are shown. (F) The N-terminus of SOX30 was required for interacting with β-catenin protein by FRET. The pEYFP-30-Dele-1 was the construct without the HMG-box and C-terminus regions. The pEYFP-30-Dele-2 was the construct without the C-terminus region. The p value was measured with Student's t-tests. **, p < 0.01. (G) Z-dock binding of TCF4 or SOX30 docking fragment (domain 1 and domain 2) identified an interaction surface on β-catenin. The β-catenin was represented in light blue; TCF4 or SOX30-domain1 in green; SOX30-domain 2 in pink. The atom of oxygen was colored red and the nitrogen was represented with blue.
3.9. SOX30 Directly Interacts with β-Catenin to Compete with TCF for Binding to β-Catenin
As the level of p-β-catenin, the key event of β-catenin protein degradation, seemed to be no obvious change in SOX30 transfected cells (Fig. 4C), SOX30 may interact with β-catenin to compete with TCF for binding to β-catenin not to affect β-catenin stability. To validate this hypothesis, competitive co-IP assays were performed in A549 and HEK293 cell lines using β-catenin or TCF4 (a well-known binding partner of β-catenin in Wnt/β-catenin signaling) antibodies. Decreased levels of endogenous TCF4 protein were detected in β-catenin precipitates upon SOX30 over-expression and vice versa (detect β-catenin in TCF4 precipitate, Fig. 6D). Luciferase reporter activity was consistently decreased in the presence of SOX30 when overexpressing β-catenin using TOPflash/FOPflash reporter assays (Supplemental Fig. S7D). Accordingly, increased levels of endogenous TCF4 protein were detected in β-catenin precipitates upon Sox30-null in lung tissues of Sox30−/− mice (Fig. 6E). These results demonstrate that SOX30 directly interacts with β-catenin to compete with TCF for binding to β-catenin.
3.10. The N-Terminus of SOX30 is Required for Interacting with β-Catenin Protein
To determine crucial region of SOX30 required for interacting with β-catenin, the deletions of HMG-box and/or C-terminus were generated, and the interacting activity of SOX30 with β-catenin was then evaluated by FRET using Varioskan LUX. SOX30 can interact with β-catenin when C-terminus region was deleted, and it still can interact with β-catenin when HMG-box and C-terminus regions were both deleted (Fig. 6F). These results reveal that amino-terminal region (N-terminus, 1-335AA) of SOX30 is required for interacting with β-catenin. As the crystal structure of β-catenin/Tcf4 complex described, Tcf4 can specifically recognize β-catenin using two distinct sites of interaction between the proteins [29,30]. The docking results of SOX30 with β-catenin from protein-protein interaction exhibited that the N-terminal (domain 1 and domain 2) of SOX30 was bonded from competition with TCF4 in the similar region of β-catenin (Fig. 6G). The data suggest that the N-terminus of SOX30 is required for interacting with β-catenin.
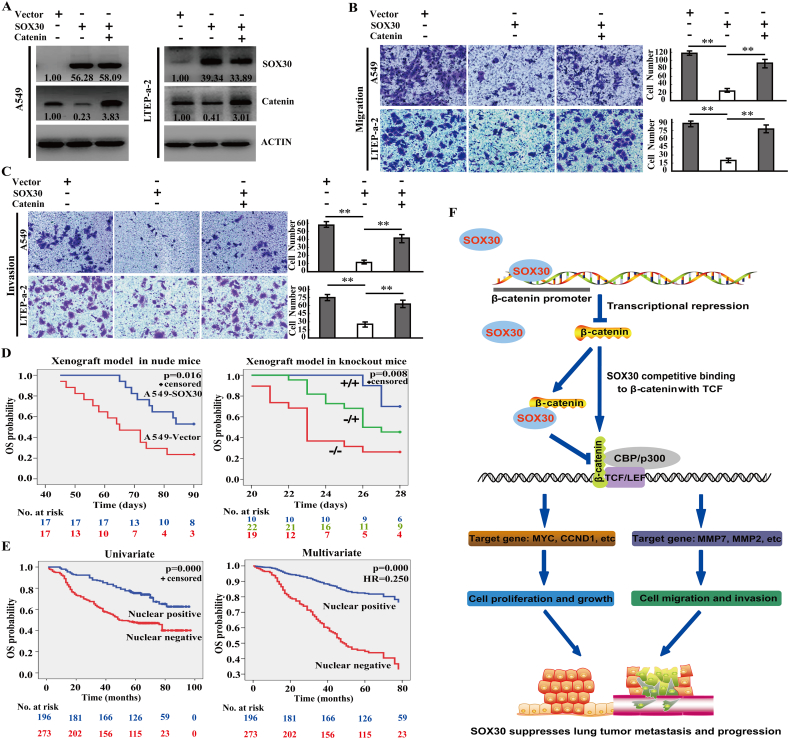
3.11. Restoration of β-Catenin Attenuates the Anti-Metastatic Role of SOX30
To define whether β-catenin is required for the anti-metastatic role of SOX30, we enforced the expression of β-catenin using its expression plasmid when over-expression of SOX30 in A549 and LTEP-a-2 cell lines, and determined the recovery of tumor cell migration and invasion (Fig. 7A). Enhance of β-catenin when over-expression of SOX30 diminished the anti-metastatic role of SOX30 in both A549 and LTEP-a-2 cells (Fig. 7B and C). These data illustrate that β-catenin is required for SOX30 as a tumor metastasis suppressor.
Fig. 7.
SOX30 inhibits tumor metastasis and improves survival of patients by regulating β-catenin. (A) The expression of SOX30 and β-catenin was analyzed by WB at 48 h transfected with empty-vector, SOX30 and co-transfected with SOX30 and β-catenin. The relative intensities of the proteins versus internal control (ACTIN) are shown. (B, C) Analysis of cell migration and invasion in A549 and LTEP-a-2 cells transfected with empty-vector, SOX30 and co-transfected with SOX30 and β-catenin. The migrated or invaded cell numbers were quantitatively analyzed by the average count of five random microscopic fields. The p value was measured with Student's t-tests. **, p < 0.01. (D) Kaplan-Meier curves indicating survival of A549-SOX30 nude mice compared with A549-Vector nude mice after injection A549 cells into tail vein or of Sox30+/+, Sox30+/− and Sox30−/− mice after injection B16 cells into tail vein. Statistical significance was determined by the log rank test. Sox30−/− (re-Sox30) mice are the Sox30 re-expression Sox30−/− mice. (E) Survival analyses of nuclear SOX30 expression on OS of 469 lung cancer patients split into positive and negative groups. Survival analyses were evaluated by Kaplan-Meier survival curve and multivariate Cox regression. HR represents hazard ratio. (F) A schematic illustration of how SOX30 regulates tumor metastasis in lung cancer.
3.12. SOX30 Improves Survival of Mice with Metastasis in Nude and Knockout Mice Models
To determine the prognostic value of SOX30 in animal models, we established xenograft tumor in nude mice and knockout mice, and evaluated the long-term consequence. Thirty-four nude mice were monitored for changes in survival after injected A549-SOX30 or -vector stable transfectants into tail vein. The A549-SOX30 mice had a significantly prolonged survival compared with A549-vector mice (p = 0.016, Fig. 7D). About 52.94% (9/17) of A549-SOX30 mice lived at the 90 days after injection, whereas only 23.53% (4/17) of A549-vector mice survived at that time. To evaluate the long-term consequence of Sox30 deletion, a cohort of Sox30+/+ (n = 10), Sox30+/− (n = 22) and Sox30−/− (n = 19) mice were monitored for changes in survival after injection of B16 cells by tail vein. The Sox30−/− and Sox30+/− mice showed a significant decrease in survival compared with wild-type littermates (p = 0.008, Fig. 7D). Up to 70% (7/10) of the Sox30+/+ mice lived for >28 days, whereas only 45.5% (10/22) of the Sox30+/− and 26.3% (5/19) of the Sox30−/− mice survived on 28 days after injection of B16 cells. These results demonstrate that SOX30 is a strong prognostic candidate.
3.13. Nuclear SOX30 Expression is Associated with Metastasis and Represents a Favorable Independent Prognostic Biomarker
As SOX30 is a transcriptional factor, we first determined the levels of SOX30 in nucleus and cytoplasm in pairs of lung cancerous and peri-cancerous tissues by nucleus/cytoplasm extraction method, and found that the nuclear SOX30 expression was much higher in peri-cancerous tissues than that in cancerous tissues (Supplemental Fig. S8A). In addition, the negative correlation of SOX30 and β-catenin expression was also observed in nucleus and cytoplasm extraction of the pair clinical specimens, respectively (Supplemental Fig. S8A). We then quantitated the nuclear staining of SOX30 in clinical specimens (Supplemental Fig. S8B), and analyzed its correlations with disease outcome, clinical stages and other clincopathological parameters in the lung cancer patient cohort. Nuclear SOX30 expression was strongly associated with clinical stages (p = 0.001) and long-distance metastasis (p = 0.000) (Supplemental Table S4). The percentage of SOX30 positive nuclear expression samples was decreased in patients from clinical early stages (I + II, 43.59%, 136/312) to advanced stages (IV + III, 31.91%, 45/141) groups (Supplemental Table S4). Furthermore, the percentage of SOX30 positive nuclear expression samples was sharply lower in the group of distant metastasis (3.70%, 1/27) than that of non-metastasis (42.25.03%, 199/471) groups (Supplemental Table S4). The correlation between nuclear SOX30 expression and overall survival (OS) of the lung cancer patients was further evaluated. Nuclear SOX30 expression was clearly associated with the OS of the patients by Kaplan-Meier (p = 0.000) and Cox-Regression (HR = 0.250, p = 0.000) analyses (Fig. 7E and Supplemental Table S5), and the patients with positive nuclear expression of SOX30 lived much longer than that with negative nuclear expression. These data reveal that nuclear SOX30 expression is strongly associated with metastasis, and is a favorable independent prognostic marker of lung cancer patients.
4. Discussion
Distant-metastasis greatly contributes to lung cancer poor survival, and it is urgently necessary to understand the mechanisms of this metastasis. SOX30 has been recently identified as a novel key player in lung cancer [21,22]. To further explore precise role and mechanism of SOX30 in lung cancer, we performed clinical association analyses and expression profiling of microarray, and revealed a possible role of SOX30 on tumor metastasis. Gain and loss of function studies demonstrated that SOX30 strongly inhibited cancer cell migration and invasion in vitro, and suppressed tumor cell metastasis in vivo. These data indicate that SOX30 functions as a strong metastatic suppressor.
To further determine the inhibitory role of SOX30 on metastasis, we created an induced Sox30 knockout C57BL/6 mouse model. Due to lung cancer cell lines did not work in Sox30+/+ Sox30−/+ and Sox30−/− mice by injecting into tail vein to produce metastasis models. B16-F10 mouse melanoma cell line was used to produce lung metastasis models. The Sox30−/− and Sox30+/− mice were significantly increased in lung colonization of B16 cells as compared with Sox30+/+ mice after injection. Furthermore, the wild lung-metastasis in Sox30−/− mice was clearly re-inhibited upon Sox30 re-expression. These results show that SOX30 expression in mice strongly reduces the metastatic ability of B16 cells to lung tissue. Although B16 melanoma cell line is not a lung cancer cell line, the model looking at metastasis of B16 to lung still reveals the inclusion that SOX30 can inhibit tumor cell metastasis. Moreover, we then generated the orthotopic graft model of mouse Lewis Lung Carcinoma (LL/2) cell line, a more suitable cell line for the functional roles of SOX30 in vivo, in the SOX30-null C57BL/6 model. The data of LL/2 lung colonization was consistent with the results using a B16 melanoma mouse model. These data show that SOX30 is indeed a critical inhibitor of tumor metastasis in lung cancer.
To explore the mechanism of SOX30 function, we built a global SOX30-regulated networks, and Wnt-signaling was identified as an important mediator of SOX30 repressing metastasis. Wnt-signaling plays key roles in a remarkable variety of cellular processes, including cell survival and metastasis [[31], [32], [33]]. This signaling activity frequently depends on the regulation of β-catenin protein [[34], [35], [36]]. Previous studies have demonstrated that Sox proteins appear to modulate β-catenin activity mainly via regulating protein stability of β-catenin independent on DNA-binding [35,[37], [38], [39]]. But in our present study, we found the metastatic suppressor SOX30 as a critical regulator of Wnt/β-catenin signaling via direct binding to β-catenin promoter and repressing its transcription. Generally, the HMG-box of Sox proteins is essential for transcriptional regulating targets. However, the C-terminal region not HMG-box of SOX30 is required for binding to β-catenin promoter, implying the presence of new transcriptional repression domain in the C-terminal region. Further studies are needed to elucidate the new functional domain of SOX30.
Some SOX proteins are shown to interact with β-catenin physically [[35], [36], [37], [38]]. To test interaction between SOX30 and β-catenin proteins, co-IP and FRET assays were performed. SOX30 repressed Wnt/β-catenin signaling also via a direct protein-protein interaction with β-catenin, which was confirmed in the Sox30-null mouse model. There are two most probable mechanisms for SOX30 repressing Wnt/β-catenin signaling via interacting with β-catenin: regulating β-catenin stability or competing with TCF for β-catenin binding. Based on the possibility for SOX30 competition with TCF in the similar region of β-catenin by structural prediction and no obvious difference of p-β-catenin level between SOX30 and empty vector transfected cells, we prefer to believe the competition model. The following competitive co-IP assays demonstrate that SOX30 indeed competes with TCF4 for binding to β-catenin.
Previous studies have suggested a few SOX proteins binding to β-catenin in the region that overlaps with the sites where TCF proteins bind, but few evidences exist for this competition model [[36], [37], [38], [39], [40]]. In our present study, we provided direct evidences both in vitro and in vivo to support the competition model of SOX30 with TCF for β-catenin binding. Further studies revealed that the N-terminus is required for SOX30 interacting with β-catenin protein, and the docking results also demonstrated that the N-terminal of SOX30 was bonded from competition with TCF4 in the similar region of β-catenin.
SOX30 acts as a metastatic suppressor via inhibiting β-catenin in lung cancer, and we then wanted to know the nucleus and cytoplasm expressions of SOX30 and β-catenin in pairs of lung cancerous and peri-cancerous tissues. The negative correlation of SOX30 and β-catenin expression was also found in nucleus and cytoplasm extraction of the pair clinical specimens, respectively, which further verifies the conclusion that SOX30 indeed inhibits β-catenin expression in lung cancer. Moreover, the nuclear SOX30 expression was observed much higher in peri-cancerous tissues than that in cancerous tissues, which suggests that SOX30 is little expressed in nucleus of cancerous tissues and it may do not work in cancerous tissues as a transcriptional factor. To determine the key role of nuclear SOX30 expression in lung cancer, we analyzed its correlations with disease outcome and other clincopathological parameters in the patient cohort. The patients with negative nuclear SOX30 expression were preferential the ones at advanced clinical stages or with long-distance metastasis. Moreover, the patients with positive nuclear expression of SOX30 live much longer than these with negative nuclear expression. These results confirm that SOX30 is indeed a key metastatic inhibitor in lung cancer.
Our previous studies have indicated that SOX30 is silenced by methylation and genetic alterations are very rare [18,21]. These data suggest that the silence of SOX30 in lung cancer patients is caused by DNA methylation not by genetic alterations unlike other key tumor suppressors such as TP53. Given that DNA methylation is reversible and SOX30 acts important role on anti-tumor metastasis, SOX30 could be used to develop a new therapy by demethylation for lung cancer disease. The latest technologies for targeted DNA methylation editing using CRISPR/Cas9-based approaches [41,42] will be useful for developing this new therapy. In addition, considering the key role of β-catenin in tumor metastasis and ineffective treatment using inhibitors against β-catenin, it is a potential choice to suppress Wnt/β-catenin activity via targeting the upstream SOX30.
5. Conclusions
In conclusion, SOX30 is an important metastatic suppressor inhibiting Wnt-signaling via transcriptional repressing β-catenin dependent on C-terminus or via competing with TCF for β-catenin binding dependent on N-terminus (Fig. 7F), providing a “fail-safe” mechanism to ensure SOX30 suppressing tumor-metastasis. The metastasis-inhibitory role and mechanism of SOX30 provides effective therapies against tumor metastasis in lung cancer.
Funding Sources
This work was supported by the National Natural Science Foundation of China (No. 81502551 and 81172714) and the Postdoctoral Science Foundation special funded project of Chongqing (No. Xm2014122).
Conflicts of Interest
The authors declare no competing financial interests.
Author Contributions
Conception and design: F. Han, J. Cao and J-Y. Liu; Development of methodology: F. Han, W-B. Liu, X-Y, Shi, J-T. Yang, X. Jiang and L.Yin; Acquisition of data: X. Zhang, J-J, Li, and Y. Dong; Analysis and interpretation of data: F. Han, W-B. Liu, Z-M. Li and X-Y. Shi; Writing, review, and/or revision of the manuscript: F. Han, J. Cao J-Y. Liu and C-S. Huang; Administrative, technical, or material support: F. Han, W-B. Liu, X-Y. Shi, J-T. Yang, X. Jiang, L.Yin, X. Zhang, J-J. Li and C-S. Huang; Study supervision: F. Han, J. Cao and J-Y. Liu.
Acknowledgements
The authors would like to thank all patients involved in this study and thank AJE (American Journal Experts) for language editing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.04.026.
Contributor Information
Jia Cao, Email: caojia1962@126.com.
Jin-yi Liu, Email: jinyiliutmmu@163.com.
Appendix A. Supplementary data
Supplementary material
References
- 1.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Azzoli C.G., Baker S., Jr., Temin S., Pao W., Aliff T., Brahmer J. American Society of Clinical Oncology clinical practice guideline update on chemotherapy for stage IV non-small-cell lung cancer. J. Clin. Oncol. 2009;27:6251–6266. doi: 10.1200/JCO.2009.23.5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jemal A., Siegel R., Xu J., Ward E. Cancer statistics, 2010. CA Cancer J. Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 4.Parkin D.M., Bray F., Ferlay J., Pisani P. Global cancer statistics, 2002. CA Cancer J. Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 5.Steeg P.S. Tumor metastasis: mechanistic insights and clinical challenges. Nat. Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 6.Wan L., Pantel K., Kang Y. Tumor metastasis: moving new biological insights into the clinic. Nat. Med. 2013;19:1450–1464. doi: 10.1038/nm.3391. [DOI] [PubMed] [Google Scholar]
- 7.Gubbay J., Collignon J., Koopman P., Capel B., Economou A., Münsterberg A. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- 8.Chew L.J., Gallo V. The yin and Yang of sox proteins: activation and repression in development and disease. J. Neurosci. Res. 2009;87:3277–3287. doi: 10.1002/jnr.22128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Man C.H., Fung T.K., Wan H., Cher C.Y., Fan A., Ng N. Suppression of SOX7 by DNA methylation and its tumor suppressor function in acute myeloid leukemia. Blood. 2015;125:3928–3936. doi: 10.1182/blood-2014-06-580993. [DOI] [PubMed] [Google Scholar]
- 10.Sinclair A.H., Berta P., Palmer M.S., Hawkins J.R., Griffiths B.L., Smith M.J. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 11.Lai T., Jabaudon D., Molyneaux B.J., Azim E., Arlotta P., Menezes J.R. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57:232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 12.Wagner T., Wirth J., Meyer J., Zabel B., Held M., Zimmer J. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 13.Kanai-Azuma M., Kanai Y., Gad J.M., Tajima Y., Taya C., Kurohmaru M. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development. 2002;129:2367–2379. doi: 10.1242/dev.129.10.2367. [DOI] [PubMed] [Google Scholar]
- 14.Park K.S., Wells J.M., Zorn A.M., Wert S.E., Whitsett J.A. Sox17 influences the differentiation of respiratory epithelial cells. Dev. Biol. 2006;294:192–202. doi: 10.1016/j.ydbio.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 15.Shimoda M., Kanai-Azuma M., Hara K., Miyazaki S., Kanai Y., Monden M. Sox17 plays a substantial role in late-stage differentiation of the extraembryonic endoderm in vitro. J. Cell Sci. 2007;120:3859–3869. doi: 10.1242/jcs.007856. [DOI] [PubMed] [Google Scholar]
- 16.Castillo S.D., Sanchez-Cespedes M. The SOX family of genes in cancer development: biological relevance and opportunities for therapy. Expert Opin. Ther. Targets. 2012;16:903–919. doi: 10.1517/14728222.2012.709239. [DOI] [PubMed] [Google Scholar]
- 17.Ballow D., Meistrich M.L., Matzuk M., Rajkovic A. Sohlh1 is essential for spermatogonial differentiation. Dev. Biol. 2006;294:161–167. doi: 10.1016/j.ydbio.2006.02.027. [DOI] [PubMed] [Google Scholar]
- 18.Han F., Dong Y., Liu W., Ma X., Shi R., Chen H. Epigenetic regulation of Sox30 is associated with testis development in mice. PLoS One. 2014;9 doi: 10.1371/journal.pone.0097203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han F., Wang Z., Wu F., Liu Z., Huang B., Wang D. Characterization, phylogeny, alternative splicing and expression of Sox30 gene. BMC Mol. Biol. 2010;11:98. doi: 10.1186/1471-2199-11-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osaki E., Nishina Y., Inazawa J., Copeland N.G., Gilbert D.J., Jenkins N.A. Identification of a novel Sry-related gene and its germ cell-specific expression. Nucleic Acids Res. 1999;27:2503–2510. doi: 10.1093/nar/27.12.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han F., Liu W., Jiang X., Shi X., Yin L., Ao L. SOX30, a novel epigenetic silenced tumor suppressor, promotes tumor cell apoptosis by transcriptional activating p53 in lung cancer. Oncogene. 2015;34:4391–4402. doi: 10.1038/onc.2014.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han F., Liu W., Xiao H., Dong Y., Sun L., Mao C. High expression of SOX30 is associated with favorable survival in human lung adenocarcinoma. Sci. Rep. 2015;5 doi: 10.1038/srep13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai D., Zhang J., Li T., Hang R., Liu Y., Tian Y. The ATPase hCINAP regulates 18S rRNA processing and is essential for embryogenesis and tumour growth. Nat. Commun. 2016;7 doi: 10.1038/ncomms12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xing C., Lu X.X., Guo P.D., Shen T., Zhang S., He X.S. Ubiquitin-specific protease 4-mediated deubiquitination and stabilization of PRL-3 is required for potentiating colorectal oncogenesis. Cancer Res. 2016;76:83–95. doi: 10.1158/0008-5472.CAN-14-3595. [DOI] [PubMed] [Google Scholar]
- 25.Gau D., Ding Z., Baty C., Roy P. Fluorescence resonance energy transfer (FRET) -based detection of profilin-VASP interaction. Cell. Mol. Bioeng. 2011;4:1–8. doi: 10.1007/s12195-010-0133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muntau A.C., Roscher A.A., Kunau W.H., Dodt G. The interaction between human PEX3 and PEX19 characterized by fluorescence resonance energy transfer (FRET) analysis. Eur. J. Cell Biol. 2003;82:333–342. doi: 10.1078/0171-9335-00325. [DOI] [PubMed] [Google Scholar]
- 27.Raz A., Bucana C., McLellan W., Fidler I.J. Distribution of membrane anionic sites on B16 melanoma variants with differing lung colonising potential. Nature. 1980;284:363–364. doi: 10.1038/284363a0. [DOI] [PubMed] [Google Scholar]
- 28.Valles S.L., Benlloch M., Rodriguez M.L., Mena S., Pellicer J.A., Asensi M. Stress hormones promote growth of B16-F10 melanoma metastases: an interleukin 6- and glutathione-dependent mechanism. J. Transl. Med. 2013;11:72. doi: 10.1186/1479-5876-11-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poy F., Lepourcelet M., Shivdasani R.A., Eck M.J. Structure of a human Tcf4-beta-catenin complex. Nat. Struct. Biol. 2001;8:1053–1057. doi: 10.1038/nsb720. [DOI] [PubMed] [Google Scholar]
- 30.Sampietro J., Dahlberg C.L., Cho U.S., Hinds T.R., Kimelman D., Xu W. Crystal structure of a beta-catenin/BCL9/Tcf4 complex. Mol. Cell. 2006;24:293–300. doi: 10.1016/j.molcel.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Inestrosa N.C., Arenas E. Emerging roles of Wnts in the adult nervous system. Nat. Rev. Neurosci. 2010;11:77–86. doi: 10.1038/nrn2755. [DOI] [PubMed] [Google Scholar]
- 32.Logan C.Y., Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 33.Moon R.T., Kohn A.D., De Ferrari G.V., Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat. Rev. Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 34.Jamieson C., Sharma M., Henderson B.R. Wnt signaling from membrane to nucleus: β-catenin caught in a loop. Int. J. Biochem. Cell Biol. 2012;44:847–850. doi: 10.1016/j.biocel.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 35.MacDonald B.T., Tamai K., He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valenta T., Hausmann G., Basler K. The many faces and functions of β-catenin. EMBO J. 2012;31:2714–2736. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akiyama H., Lyons J.P., Mori-Akiyama Y., Yang X., Zhang R., Zhang Z. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 2004;18:1072–1087. doi: 10.1101/gad.1171104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iguchi H., Urashima Y., Inagaki Y., Ikeda Y., Okamura M., Tanaka T. SOX6 suppresses cyclin D1 promoter activity by interacting with beta-catenin and histone deacetylase 1, and its down-regulation induces pancreatic beta-cell proliferation. J. Biol. Chem. 2007;282:19052–19061. doi: 10.1074/jbc.M700460200. [DOI] [PubMed] [Google Scholar]
- 39.Mansukhani A. Sox2 induction by FGF and FGFR2 activating mutations inhibits Wnt signaling and osteoblast differentiation. J. Cell Biol. 2005;168:1065–1076. doi: 10.1083/jcb.200409182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zorn A.M., Barish G.D., Williams B.O., Lavender P., Klymkowsky M.W., Varmus H.E. Regulation of Wnt signaling by sox proteins: XSox17 alpha/beta and XSox3 physically interact with beta-catenin. Mol. Cell. 1999;4:487–498. doi: 10.1016/s1097-2765(00)80200-2. [DOI] [PubMed] [Google Scholar]
- 41.Liu X.S., Wu H., Ji X., Stelzer Y., Wu X., Czauderna S. Editing DNA methylation in the mammalian genome. Cell. 2016;167:233–247. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vojta A., Dobrinić P., Tadić V., Bočkor L., Korać P., Julg B. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016;44:5615–5628. doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material