Abstract
The ordered assembly of amyloidogenic proteins causes a wide spectrum of common neurodegenerative diseases, including Alzheimer’s and Parkinson’s diseases. These diseases share common features with prion diseases, in which misfolded proteins can self-replicate and transmit disease across different hosts. Deciphering the molecular mechanisms that underlie the amplification of aggregates is fundamental for understanding how pathological deposits can spread through the brain and drive disease. Here, we used single-molecule microscopy to study the assembly and replication of tau at the single aggregate level. We found that tau aggregates have an intrinsic ability to amplify by filament fragmentation, and determined the doubling times for this replication process by kinetic modeling. We then simulated the spreading time for aggregates through the brain and found this to be in good agreement with both the observed time frame for spreading of pathological tau deposits in Alzheimer’s disease and in experimental models of tauopathies. With this work we begin to understand the physical parameters that govern the spreading rates of tau and other amyloids through the human brain.
Keywords: Prion propagation, Alzheimer’s disease, kinetic modeling, single-molecule microscopy, tau, biophysics
For many years, cell autonomous mechanisms were believed to account for human neurodegenerative diseases. However, mounting evidence suggests that the protein assemblies, which are involved in various neurodegenerative diseases, including aggregated tau, are capable of self-sustaining amplification.1 The latter is believed to enable the propagation of pathological assemblies along connected brain cells, resulting in progression from restricted areas to large numbers of brain regions. This phenomenon is commonly referred to as prion-like spreading. According to the prion hypothesis, upon entry into the cytoplasm of cells, tau assemblies can seed the aggregation of native monomers, thereby initiating the formation of additional aggregates, which can be released and spread to neighboring cells.2,3 Synthetic tau filaments made from recombinant protein as well as filamentous material extracted from tau mouse models or Alzheimer’s disease (AD) brains have been shown to act as seeds in various model systems and initiate the spreading of tau pathology.4
On a molecular level, to achieve propagation of protein aggregates through the brain, an amplification of seeds is required. This requires secondary aggregation processes, by which new seeds are produced through the fragmentation of filaments and/or new assemblies form on the surface of existing filaments.5 For prion diseases, amplification appears to occur predominantly through fragmentation.6,7 Thus, an inverse correlation between the stability of prions and their infectivity has been described.7–9 The mechanisms of amplification of tau seeds are less well studied. In experimental studies of tau transmission or propagation in cells or animal models, seed material is often amplified artificially by sonication before administration to the model system to increase the efficiency of templated seeding. Here, we set out to quantify the intrinsic ability of tau to amplify in order to gain insights into the key mechanisms governing prion-like spreading of tau in vivo. For this purpose, we characterized the aggregation kinetics of unlabeled full-length wild-type and mutant P301S tau using the recently developed single-molecule technique SAVE (Single Aggregate Visualization by Enhancement) imaging.10
Results and Discussion
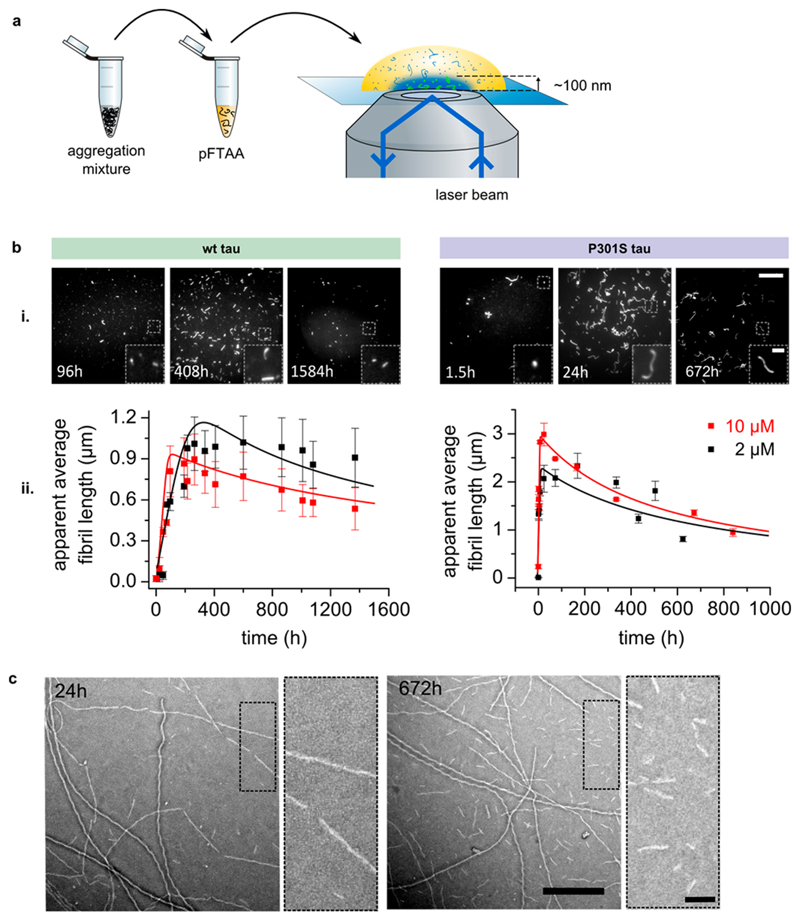
As the kinetic rates for the elongation and fragmentation of amyloid proteins can be derived from the length distributions of filaments during aggregation,11,12 we employed SAVE imaging to follow the evolution of fibrillar tau aggregates. In more detail, wild-type and P301S tau (both 0N4R isoform) were aggregated in vitro using heparin as an inducer; at regular time points, aliquots of these aggregation reactions were mixed with the luminescent conjugated oligothiophene pFTAA13 and imaged on a total internal reflection fluorescence (TIRF) microscope (see scheme in Figure 1a). As pFTAA has a high affinity for tau aggregates14 and becomes highly fluorescent upon binding, we could readily detect the evolution of single tau fibrils after the induction of aggregation using this approach (see Figure 1b-i for representative images). It should be noted that our method was not able to detect small fibrillar species (<100 nm), either due to the reduced number of dye binding sites or reduced binding of small aggregates to the glass surface, and therefore fewer species were visible on our images at the early and late time-points.
Figure 1.
Tau aggregation is a two stage process of elongation and fragmentation. (a) Observing the aggregation of full length tau by SAVE imaging. Wild-type and P301S tau (0N4R) were incubated under aggregating conditions for up to 2 months. At regular time points, aliquots of the reaction mixtures were stained with pFTAA, adsorbed onto a glass cover slide and imaged on a TIRF microscope. (b) (i) Representative images of fibrils during midelongation phase, at maximum length and at the end point. Scale bar: 15 μm. Inset: 3-fold magnification of boxed areas. Scale bar: 2.5 μm. (ii) The apparent average length of fibrils was analyzed and plotted as a function of time. To account for the diffraction limited resolution, the “apparent” average length was plotted whereby diffraction limited spots are counted as fibrils with a length of 1 pixel (204 nm). N = 3 (3 different batches of protein, each in triplicates). Error bars: SEM. Solid lines: fit to fragmentation model. (c) Representative electron micrographs of the 2 μM P301S aggregation reaction after 24 h and after 672 h. Scale bar: 500 nm. Dotted areas were magnified and shown on the right. Scale bar: 100 nm.
In order to obtain quantitative mechanistic insights from our SAVE images, we extracted the average lengths of fibrillar aggregates present at each time point and plotted these as a function of aggregation time. Aggregates smaller than the diffraction limit were counted as fibrils with the length of 1 pixel (204 nm). This revealed two stages of aggregation for both wild-type and mutant tau: first a relatively short stage (<200 h for the wild-type protein and <24 h for the P301S mutant) dominated by an increase of the average fibril length by elongation, and a long second phase during which the average fibril length decreased (see Figure 1b-ii). Wild-type tau aggregated slowly into straight fibrils with a maximum apparent length of 900 to 1,000 nm. By contrast, mutant P301S tau elongated markedly faster into long fibrils with curly appearance, reaching an apparent average length of 2,000 to 3,000 nm. In the late stage of aggregation, fibrils from both protein variants decreased in length over time and this effect was more apparent for P301S tau than for wild-type tau. At 10 μM aggregation concentration, wild-type fibrils decreased in size from around 900 nm at maximum to around 570 nm at the last measurement and mutant fibrils decreased from around 3000 nm to around 1300 nm. A comparison of the size histograms of P301S tau fibrils at the end of the elongation phase (24 h/1 day) with late fibrils (672 h/28 days) showed a redistribution of long fibrils toward small fibrils with a length less than 400 nm (Supporting Figure 1a). To corroborate this finding and to get an estimate of the fragment lengths by a nondiffraction limited method, transmission electron microscopy (TEM) was performed on P301S tau aggregates after 1 day and 28 days of aggregation (see representative electron micrographs in Figure 1c). The length histograms obtained from these images confirmed an increase of small fragments with a length of less than 100 nm after 28 days (Supporting Figure 1b). Taken together, this data shows that tau undergoes slow spontaneous fibril fragmentation, leading to a buildup of small fibrillar fragments over long periods of time. No proteolysis of tau was observed over the course of aggregation studied here, indicating that the protein stayed intact (see Supporting Figure 2).
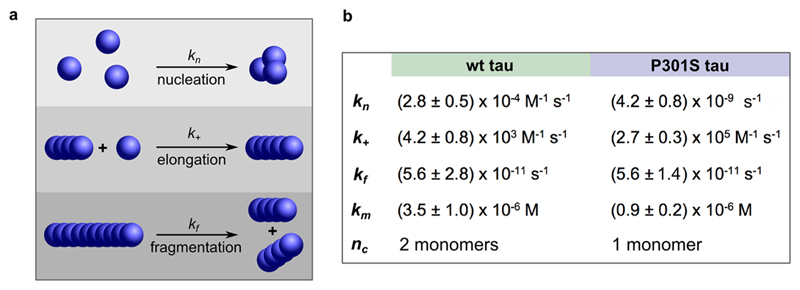
Next, the experimentally determined average fibril lengths were fitted to a kinetic model of filamentous aggregation. Only a kinetic model describing aggregation as a nucleation-elongation mechanism with fibril fragmentation15,16 (solid lines in Figure 1b-ii; kinetic model Figure 2a) was able to fully reproduce the data (see Supporting Information for details). By contrast, a model in which the fibrils self-replicate via secondary nucleation and do not fragment was unable to reproduce the data (see supporting Figure 3). The rate constants obtained from the fits are shown in Figure 2b. Both the elongation and fragmentation kinetics of tau determine the efficiency by which new fibrillar seeds are formed from existing aggregates. Here, we found that P301S tau has a 64 times faster elongation rate compared to wild type tau, possibly due to P301S tau existing in a more favorable conformation for addition to fibril ends than wild-type tau. The fragmentation rates of both protein variants are the same within experimental error. The primary nucleation rate constant and the critical nucleus size do not influence the amplification rate of existing aggregates and are therefore not relevant for aggregate spreading in vivo.
Figure 2.
Kinetic model. (a) Schematic of microscopic steps during tau aggregation and corresponding rate constants. (b) Rate constants obtained from fits. Km is the critical concentration for saturated elongation; nc is the critical nucleus size. It is assumed that only the nucleation step is heparin dependent. Errors are allowed relative deviation.
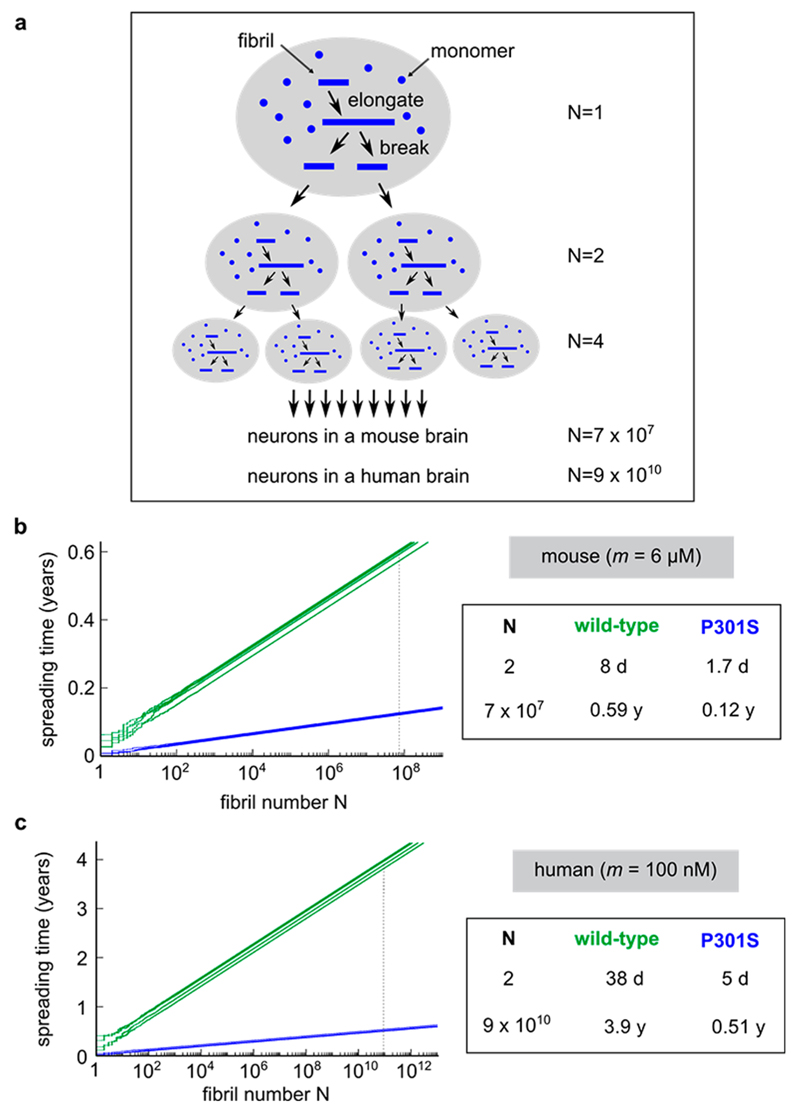
Importantly, it was shown that the phenotype of different yeast prion strains can be predicted by a simple analytical model which takes into account the availability of monomeric protein, the rate of cell division, and the elongation and fragmentation rates of the different prion strains.8 Here, we employ a similar model based on the experimentally determined rates for tau fibril growth and fragmentation to obtain estimates for the rate of prion-like replication of tau in vivo (see Figure 3a), since recombinant tau fibrils similar to the ones studied here have previously been shown to seed endogenous tau in recipient cells3,17–20 and in PS19 mice (transgenic for 1N4R P301S tau).21 If one assumes that the elongation and fragmentation rates are unaltered in vivo, we can derive the doubling time of tau aggregates from stochastic simulations of fibril growth and fragmentation. Notably, these processes are dependent on the availability of free monomer which can be added onto fibril ends during elongation. In the htau P301S mouse model overexpressing human P301S tau, we estimate the cytosolic human tau concentration to be ≈6 μM, assuming negligible binding of human tau to microtubules in the presence of mouse tau (see Supporting Figure 4). As shown in our simulations, fibril amplification is efficient at this concentration of free monomer (Figure 3b). On average, the doubling time t2 for a tau seed is given by
| (1) |
where m is the concentration of monomer, k+ is the elongation rate, kf is the fragmentation rate, and Km is the critical concentration for saturated elongation.15 Under these conditions, we obtain a doubling time of 8 days for wild-type tau and 1.7 days for P301S tau. In order to put these doubling times into the context of tau propagation in a mouse brain, containing around 70 million neurons,22 we calculated the time it would take to obtain one aggregate per cell (i.e., 70 million aggregates). According to our simple model, this would take around 7 months for wild-type tau and 1.5 months for P301S tau, values which are within an order of magnitude agreement of the experimentally observed life span of transgenic mice.23
Figure 3.
Simulations of tau fibril replication. (a) Hypothetical mechanism for tau fibril amplification and spreading in tissues. (b,c) Simulations of stochastic fibril amplification for wild-type (green) and P301S tau (blue) aggregates (b) in the htau P301S mouse model or (c) in humans. On the left, representative simulations are shown for each condition; average amplification times of tau aggregates are shown on the right. For each condition, 100 independent simulations were performed to obtain a reliable amplification time.
In human neuronal cells, the cytosolic concentration of monomeric tau is markedly lower as the protein is expressed at endogenous levels and binds with nanomolar affinity to microtubules.24,25 Therefore, when we lowered the cytosolic tau concentration in our simulations to 100 nM, the doubling times for wild-type and P301S tau aggregates increased distinctly to 38 days and 5 days, respectively (see Figure 3c). Based on these doubling times and the larger number of cells in a human brain (~90 billion cells26), we obtain estimates of approximately 4 years for the spreading of wild-type tau through the brain or approximately 6 months for the frontotemporal dementia (FTD) associated mutant P301S tau. By comparison, the age of onset for sporadic AD is around 65,27 while early onset FTD in individuals with the P301S mutation starts in their 20s or 30s.28,29 The time between diagnosis and death is similar in both cases with typically 8–10 years.
It is important to note that the spreading of aggregates in vivo is dependent on many factors such as the efficiency of uptake, the rate of aggregate clearance, the release of new seeds from the cytosol, and so forth. These and other factors likely reduce the spreading efficiency, i.e., the fraction of aggregates which successfully enter a cell and amplify. Thus, seeding efficiencies in vitro, where recombinant aggregates are added to cultured cells, are usually much lower than 100%.17,30 However, due to the exponential nature of fibril replication, our model is relatively insensitive to changes in spreading efficiencies, with estimated spreading times remaining within an order of magnitude of a human life span even at very low spreading efficiencies such as 0.0001% (see Supporting Table 1).
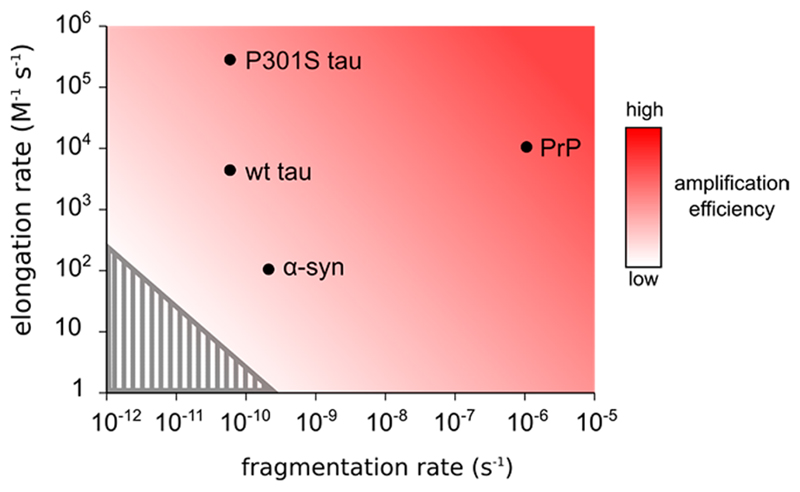
We have recently also measured the elongation and fragmentation rate for murine PrP and human α-synuclein31 under similar experimental conditions (see Supporting Table 2), and these are plotted in Figure 4 for comparison. Since the doubling time depends on the product of the elongation and fragmentation rate, it is possible to have a slow fragmentation rate but a fast elongation rate for efficient aggregate amplification as is the case for P301S tau. Murine PrP has high values of both rate constants and is therefore predicted to amplify and hence spread much faster than the other proteins. Both wild-type tau and α-synuclein are predicted to amplify more slowly and appear to have doubling times close to the limit for the disease to occur in the human lifetime (Figure 4, gray area). Intriguingly, these values seem to correlate with the time of disease progression in the respective diseases, with the most common prion disease Creutzfeldt–Jakob disease having the shortest survival time of a few months to 2 years,32 followed by tauopathies such as AD and FTD with a survival time of around 10 years27–29 and Parkinson’s disease (the most common α-synucleinopathy) where disease worsens over long periods of time (10–20 years).33
Figure 4.
Elongation and fragmentation rates for murine PrP, tau, and α-synuclein. Elongation and fragmentation rates of several disease associated proteins. The conditions used to determine these rate constants are shown in Supporting Table 2. The shaded area represents the minimum product of rate constants that would yield 90 billion aggregates in 100 years at a protein concentration of 100 nM.
Based on our findings, we suggest that the exponential amplification of aggregates by fragmentation is the basis for prion-like spreading in tauopathies such as AD and could play an important role in other neurodegenerative conditions. The exponential phase may be preceded by an initial lag phase during which the first tau aggregates are formed. Subsequently, a long silent period would follow, during which tau pathology slowly spreads but remains undetectable by common staining protocols. This hypothesis is supported by the recent finding that abnormal tau is indeed present in subcortical regions in a large fraction of young asymptomatic individuals.34,35 Furthermore, brain tissue derived from mice transgenic for P301S tau shows seeding activity long before pathological tau deposits can be detected by immunohistological staining,36,37 suggesting that seeds are present well before they can be detected by common staining protocols. Similarly, in tissue from human AD subjects at early Braak stages, seeding activity was observed including cortical regions which lack overt neurofibrillary pathology.38 If correct, this makes it very hard to study this process directly due to the long doubling times and modeling enables extrapolation from experiments performed under conditions where seeding is more favorable and can hence be more easily measured. While not fully quantitative, our simple model provides insights into the key factors that will likely affect spreading rates. Higher expression of tau in specific cells would lead to faster doubling times in these cells while conversely efficient removal of aggregates by the cell would extend the doubling time. These factors in combination could provide a simple explanation for why certain cells are more susceptible to aggregate formation. Furthermore, different tau aggregate strains may have different fragmentation or elongation rates and thus different doubling times as is the case for prion strains,8 which could account for the varying rates of disease progression in different tauopathies and patients.27–29 It is worth noting that tau aggregates present in cells will continue to grow and fragment with time resulting in a loss of function and several aggregates being present in a cell, which could be toxic to cells and ultimately contribute to neuronal death. Cell division will reduce the number of aggregates, offering a possible explanation for why spreading occurs via connected neurons since they do not divide.
In summary, we have quantified the growth and amplification processes of full length tau and found that it can sustain a slow prion-like spreading mechanism. Further, we have developed a simple model that is able to make semiquantitative predictions about the replication of tau aggregates in the brain, which are in good agreement with experimental observations. Finally, extending the doubling time by reducing the rate of elongation or fragmentation may be a valid strategy for the treatment of tauopathies.
Methods
Tau Purification
Human 0N4R tau and human 0N4R P301S tau were expressed as previously described.19,29 Induced cells were cultured for 3–4 h at 37 °C, then harvested by centrifugation, and finally resuspended in buffer A (50 mM MES, 0.1 mM PMSF, 2 mM TCEP, pH 6.5), containing one EDTA-free complete protease inhibitor cocktail tablet per liter of culture. The cells were lysed by sonication (1 × 1.5 min, 5 s on, 10 s off, 40% amplitude), and the soluble fraction was cleared from cell debris by centrifugation at 4 °C, 15 000g (JA-20 rotor, Beckman Coulter). The cleared lysate was then filtered through a 0.45 μM syringe driven filter and loaded onto a HiTrap Capto S column (5 mL, GE Healthcare) previously equilibrated with buffer A. After any unbound proteins were removed from the column by washing with 10 column volumes of buffer A, tau was eluted using a linear gradient from 0 to 50% buffer B (buffer A + 1 M NaCl) over 10 column volumes. Fractions containing tau protein were then pooled and precipitated with 20% ammonium sulfate at 4 °C for 1h and the precipitate collected by centrifugation at 4 °C, 15,000 g (JA-20 rotor, Beckman Coulter). This pellet was either stored at –20 °C overnight or used directly for the next purification step. Tau was further purified by size-exclusion chromatography on a Superdex 200 Increase 10/300 GL size exclusion column (GE Healthcare Life Sciences), previously equilibrated with 1x SSPE and 1 mM TCEP. The fractions collected were analyzed by SDS-PAGE, and those showing the purest bands corresponding to tau were pooled. The protein concentration was determined by measuring its absorbance at 280 nm on a Nanodrop 2000 using an extinction coefficient of 7450 M−1 cm−1 and by BCA assay. Aliquots were frozen in liquid nitrogen and stored at −80 °C.
TIRF Microscope
The samples were imaged using a home-built total internal reflection fluorescence microscope. The total internal reflection mode restricts the fluorescence illumination to within 200 nm from the sample slide. A 488 nm laser (Cobolt MLD) was aligned and directed parallel to the optical axis at the edge of a 1.49 NA TIRF objective (APON60XO TIRF, Olympus), mounted on an inverted Nikon Eclipse Ti microscope. Fluorescence was collected by the same objective and separated from the returning TIRF beam by a dichroic mirror (Di01-R405/488/532/635, Semrock), and passed through an emission filters (FF03-525/50-25, Semrock). The control of the hardware was performed using custom-written scripts (bean-shell) for MicroManager (NIH). The images were recorded on an EMCCD camera (Evolve 512 Delta, Photometrics) operating in frame transfer mode (EMGain of 4.4 e−/ADU and 250 ADU/photon). Each pixel was 204 nm in length. Images were recorded for 30 frames with an exposure time of 50 ms and averaged using ImageJ (NIH) software.
Sample Preparation for TIRF Imaging
Borosilicate glass coverslips (VWR international, Ø 50 mm) were cleaned using an argon plasma cleaner (PDC-002, Harrick Plasma) for 30 min to remove any fluorescent residues. Multiwell slide chambers (CultureWell chambered cover glass 50 well, Grace Bio-Laboratories) were separated from the original cover glass and affixed to the cleaned cover slides. To stain aggregates for imaging, samples were diluted into 30 nM pFTAA to a final protein concentration of 50 nM tau. Then 10 μL of each sample was adsorbed to the cover slides for 15 min before imaging.
Analyzing Number and Length of Aggregates
To analyze the number and length of tau aggregates, a Hessian based filament detection algorithm was employed as described by Salji et al.39
Electron Microscopy
Samples were placed on glow-discharged 400 meshed Formvar/carbon film-coated copper grids (Sigma-Aldrich) for 3 min and stained with uranyl acetate. Images were taken on a Philips Spirit transmission electron microscope at a magnification of 16 000.
Model
In order to explain the concentration dependent aggregation of tau proteins, we propose a three-step mechanism: nucleation, saturated elongation and fragmentation, as shown in Figure 2a. Here, we need to take the saturation effect into consideration, since for wild-type tau and P301S tau the maximal average fibril length under 2 μM and 10 μM conditions varies by a factor of less than 1.4 times, in contrast to the prediction of 2.2 times for simple first order elongation.40 This is a clear sign that monomer saturation needs to be included.
Accordingly, the following kinetic equation can be used to quantify the time dependent fiber length distribution of tau fibrils,
| (2) |
where [Fj](t) (j ≥ nc) is the concentration of tau fibrils constituted by j monomers, m(t) is the concentration of free tau monomers, and is the total concentration of tau proteins in the system. kn, ke, and kf are the rate constants governing the primary nucleation, elongation, and fragmentation of tau fibrils, respectively. nc is the critical nucleus size for primary nucleation, and Km is the critical concentration for saturated elongation. With respect to [Fj](t), we can further introduce as the number P and mass concentration M of tau fibrils which are observable in our single molecule measurement. The smallest observable aggregate was defined as an aggregate with a length of 100 nm, corresponding to nb = 340 monomers, calculated from the mass-per length for full length tau fibrils of around 160 kDa/nm.41
Determining the Spreading Time Based on Stochastic-Deterministic Combined Simulations
In order to assess the potential of tau to propagate in a prion-like manner, i.e., by cycles of seeded growth and amplification, and spread over the whole mouse/human brain under normal physiological conditions, numerical investigations were performed on the basis of the elongation-fragmentation mechanism with kinetic rates given in eq 3,
| (3) |
with m0 as the initial monomer concentration. Instead of direct stochastic simulations, which cannot reach a sufficiently large number of fibrils within an adequate computational time (more than 500 years on a personal computer), we adopted an alternative efficient algorithm by combining stochastic and deterministic simulations. In detail, starting from a single tau fibril of an initial length L0 = 1000 monomers (≅300 nm) in a cell-like volume (10 μm)3, stochastic simulations using a Gillespie algorithm implementation were first performed. Then we switched to the deterministic ODE solver and continued the calculation from the end point of stochastic simulations, once there were enough fibrils generated in the system (fibril mass exceeds 10 million monomers, corresponding to ~1000 fibrils, in our simulation). For each case, 100 independent simulations were performed to obtain a reliable spreading time.
Determining Mouse Tau Concentration by ELISA
Whole brains from P301S tau transgenic mice or wild-type C57BL/6 mice were snap frozen, suspended in 10x brain mass of A68 buffer (10 mM Tris-HCl, pH 7.4, 0.8 M NaCl, 1 mM EGTA, and 10% sucrose) and homogenized using a hand-held homogenizer (Millipore). Debris was pelleted by centrifugation at 21 000g for 20 min at 4 °C. The concentration of tau in homogenates was determined using a sandwich ELISA approach with HT7 (Life Technologies) as capture antibody and BR13342 as detection antibody. A standard curve was obtained using recombinant human P301S tau. To calculate the average brain concentration of tau, it was assumed that brains had a density of 1.1 g/mL.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschemneuro.8b00094.
Detailed description of the experimental procedures, data analysis methods, and kinetic modeling; figures illustrating fibril length distributions of P301S tau aggregates by TIRFM and TEM, SDS-PAGE analysis of tau to show absence of degradation during aggregation, misfits to a model which includes secondary nucleation, concentration measurements of P301S tau in P301S Tg mouse brains by ELISA; tables showing hypothetical tau spreading times at different spreading efficiencies and experimental conditions used for mPrP, α-synuclein, and tau aggregations from which data was derived for Figure 5 (PDF)
Acknowledgments
The authors would like to thank Dr. Mathew Horrocks for help with the image acquisition, Dr. Therése Klingstedt for providing pFTAA, and Jason Sang, Dr. Rohan Ranasinghe, and David Wirthensohn for helpful discussions.
Funding
D.K. acknowledges funding from the ERC (Grant Number 669237) and the Royal Society. F.K. acknowledges funding from the Augustus Newman foundation and the ERC. L.H. was supported by the Tsinghua University Initiative Scientific Research Programme (Grants 20151080424) and the program of China Scholarships Council (CSC). W.A.M. is supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (grant number 206248/Z/17/Z). T.C.T.M. acknowledges funding from Peterhouse College Cambridge and the Swiss National Science Foundation. G.M. was supported by Sidney Sussex College Cambridge. M.G. is an Honorary Professor in the Department of Clinical Neurosciences of the University of Cambridge. His work is supported by the UK Medical Research Council (MC_U105184291) and the European Union (Joint Programme-Neurodegeneration Research, JPND-REfrAME, and Horizon 2020 IMPRiND).
Abbreviations
- AD
Alzheimer’s disease
- ELISA
enzyme-linked immunosorbent assay
- FTD
frontotemporal dementia
- pFTAA
pentameric formyl thiophene acetic acid
- SAVE
Single Aggregate Visualization by Enhancement
- TEM
transmission electron microscopy
- TIRF(M)
total internal reflection fluorescence (microscopy)
Footnotes
ORCID
Franziska Kundel: 0000-0001-5013-0004
Georg Meisl: 0000-0002-6562-7715
Tuomas J. P. Knowles: 0000-0002-7879-0140
Author Contributions
D.K. jointly conceived the study with M.G. and T.J.P.K. F.K., W.A.M., B.F., and N.E. performed the experiments and analyzed the data. L.H., T.C.T.M., and G.M. conducted the kinetics fits and simulations. F.K., D.K., and M.G. wrote the manuscript. All authors discussed the results and implications and commented on the manuscript at all stages. All authors have given approval to the final version of the manuscript.
Notes
The authors declare no competing financial interest.
References
- (1).Goedert M. Alzheimer’s and Parkinson’s diseases: The prion concept in relation to assembled Aβ, tau, and α-synuclein. Science. 2015;349(6248):1255555. doi: 10.1126/science.1255555. [DOI] [PubMed] [Google Scholar]
- (2).Clavaguera F, et al. Transmission and spreading of tauopathy in transgenic mouse brain. Nat Cell Biol. 2009;11(7):909–913. doi: 10.1038/ncb1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Frost B, Jacks RL, Diamond MI. Propagation of tau misfolding from the outside to the inside of a cell. J Biol Chem. 2009;284(19):12845–12852. doi: 10.1074/jbc.M808759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Goedert M, Eisenberg DS, Crowther RA. Propagation of tau aggregates and neurodegeneration. Annu Rev Neurosci. 2017;40:189–210. doi: 10.1146/annurev-neuro-072116-031153. [DOI] [PubMed] [Google Scholar]
- (5).Knowles TPJ, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15(6):384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- (6).Bett C, et al. Biochemical properties of highly neuroinvasive prion strains. PLoS Pathog. 2012;8(2):e1002522. doi: 10.1371/journal.ppat.1002522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Prusiner SB. Biology and genetics of prions causing neurodegeneration. Annu Rev Genet. 2013;47:601–623. doi: 10.1146/annurev-genet-110711-155524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442(7102):585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- (9).Derdowski A, Sindi SS, Klaips CL, DiSalvo S, Serio TR. A size threshold limits prion transmission and establishes phenotypic diversity. Science. 2010;330(6004):680–683. doi: 10.1126/science.1197785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Horrocks MH, et al. Single-molecule imaging of individual amyloid protein aggregates in human biofluids. ACS Chem Neurosci. 2016;7(3):399–406. doi: 10.1021/acschemneuro.5b00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Michaels TCT, et al. The length distribution of frangible biofilaments. J Chem Phys. 2015;143(16):164901. doi: 10.1063/1.4933230. [DOI] [PubMed] [Google Scholar]
- (12).Xue W-F, Radford SE. An imaging and systems modeling approach to fibril breakage enables prediction of amyloid behavior. Biophys J. 2013;105(12):2811–2819. doi: 10.1016/j.bpj.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Klingstedt T, et al. Synthesis of a library of oligothiophenes and their utilization as fluorescent ligands for spectral assignment of protein aggregates. Org Biomol Chem. 2011;9(24):8356–8370. doi: 10.1039/c1ob05637a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Brelstaff J, et al. The fluorescent pentameric oligothiophene pFTAA identifies filamentous tau in live neurons cultured from adult P301S tau mice. Front Neurosci. 2015;9:184. doi: 10.3389/fnins.2015.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Knowles TPJ, et al. An analytical solution to the kinetics of breakable filament assembly. Science. 2009;326(5959):1533–1537. doi: 10.1126/science.1178250. [DOI] [PubMed] [Google Scholar]
- (16).Hong L, Yong W-A. Simple moment-closure model for the self-assembly of breakable amyloid filaments. Biophys J. 2013;104(3):533–540. doi: 10.1016/j.bpj.2012.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Guo JL, Lee VM-Y. Seeding of normal tau by pathological tau conformers drives pathogenesis of Alzheimer-like tangles. J Biol Chem. 2011;286(17):15317–15331. doi: 10.1074/jbc.M110.209296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Nonaka T, Watanabe ST, Iwatsubo T, Hasegawa M. Seeded aggregation and toxicity of α-synuclein and tau - Cellular models of neurodegenerative diseases. J Biol Chem. 2010;285(45):34885–34898. doi: 10.1074/jbc.M110.148460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Falcon B, et al. Conformation determines the seeding potencies of native and recombinant tau aggregates. J Biol Chem. 2015;290(2):1049–1065. doi: 10.1074/jbc.M114.589309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Clavaguera F, et al. “Prion-like” templated misfolding in tauopathies. Brain Pathol. 2013;23(3):342–349. doi: 10.1111/bpa.12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Iba M, et al. Synthetic tau fibrils mediate transmission of neurofibrillary tangles in a transgenic mouse model of Alzheimer’s-like tauopathy. J Neurosci. 2013;33(3):1024–1037. doi: 10.1523/JNEUROSCI.2642-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Herculano-Houzel S, Mota B, Lent R. Cellular scaling rules for rodent brains. Proc Natl Acad Sci U S A. 2006;103(32):12138–12143. doi: 10.1073/pnas.0604911103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Allen B, et al. Abundant tau filaments and nonapoptotic neurodegeneration in transgenic mice expressing human P301S tau protein. J Neurosci. 2002;22(21):9340–9351. doi: 10.1523/JNEUROSCI.22-21-09340.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ackmann M, Wiech H, Mandelkow E. Nonsaturable binding indicates clustering of tau on the microtubule surface in a paired helical filament-like conformation. J Biol Chem. 2000;275(39):30335–30343. doi: 10.1074/jbc.M002590200. [DOI] [PubMed] [Google Scholar]
- (25).Makrides V, Massie MR, Feinstein SC, Lew J. Evidence for two distinct binding sites for tau on microtubules. Proc Natl Acad Sci U S A. 2004;101(17):6746–6751. doi: 10.1073/pnas.0400992101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Azevedo FAC, et al. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J Comp Neurol. 2009;513(5):532–541. doi: 10.1002/cne.21974. [DOI] [PubMed] [Google Scholar]
- (27).Bird TD. Alzheimer Disease Overview. In: Adam MP, Ardinger HH, Pagon RA, et al., editors. GeneReviews. University of Washington; Seattle, WA: 1993. [accessed November 17, 2017]. available at: http://www.ncbi.nlm.nih.gov/books/NBK1161/ [PubMed] [Google Scholar]
- (28).Sperfeld AD, et al. FTDP-17: an early-onset phenotype with parkinsonism and epileptic seizures caused by a novel mutation. Ann Neurol. 1999;46(5):708–715. doi: 10.1002/1531-8249(199911)46:5<708::aid-ana5>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- (29).Bugiani O, et al. Frontotemporal dementia and corticobasal degeneration in a family with a P301S mutation in tau. J Neuropathol Exp Neurol. 1999;58(6):667–677. doi: 10.1097/00005072-199906000-00011. [DOI] [PubMed] [Google Scholar]
- (30).McEwan WA, et al. Cytosolic Fc receptor TRIM21 inhibits seeded tau aggregation. Proc Natl Acad Sci U S A. 2017;114(3):574–579. doi: 10.1073/pnas.1607215114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Sang JC, et al. Direct observation of protein aggregate replication in vitro to model prion and prion-like diseases. submitted.
- (32).Pocchiari M, et al. Predictors of survival in sporadic Creutzfeldt–Jakob disease and other human transmissible spongiform encephalopathies. Brain. 2004;127(10):2348–2359. doi: 10.1093/brain/awh249. [DOI] [PubMed] [Google Scholar]
- (33).Kalia LV, Lang AE. Parkinson’s disease. Lancet. 2015;386(9996):896–912. doi: 10.1016/S0140-6736(14)61393-3. [DOI] [PubMed] [Google Scholar]
- (34).Braak H, Del Tredici K. The pathological process underlying Alzheimer’s disease in individuals under thirty. Acta Neuropathol. 2011;121(2):171–181. doi: 10.1007/s00401-010-0789-4. [DOI] [PubMed] [Google Scholar]
- (35).Elobeid A, Soininen H, Alafuzoff I. Hyper-phosphorylated tau in young and middle-aged subjects. Acta Neuropathol. 2012;123(1):97–104. doi: 10.1007/s00401-011-0906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Holmes BB, et al. Proteopathic tau seeding predicts tauopathy in vivo. Proc Natl Acad Sci U S A. 2014;111(41):E4376–E4385. doi: 10.1073/pnas.1411649111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Kaufman SK, Thomas TL, Del Tredici K, Braak H, Diamond MI. Characterization of tau prion seeding activity and strains from formaldehyde-fixed tissue. Acta Neuropathol Commun. 2017;5(1):41. doi: 10.1186/s40478-017-0442-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Furman JL, et al. Widespread tau seeding activity at early Braak stages. Acta Neuropathol. 2017;133(1):91–100. doi: 10.1007/s00401-016-1644-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Salji CJ, et al. The JCMT Gould Belt Survey: properties of star-forming filaments in Orion A North. Mon Not R Astron Soc. 2015;449(2):1782–1796. [Google Scholar]
- (40).Hong L, Qi X, Zhang Y. Dissecting the kinetic process of amyloid fiber formation through asymptotic analysis. J Phys Chem B. 2012;116(23):6611–6617. doi: 10.1021/jp205702u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).von Bergen M, et al. The Core of Tau - Paired Helical Filaments Studied by Scanning Transmission Electron Microscopy and Limited Proteolysis. Biochemistry. 2006;45(20):6446–6457. doi: 10.1021/bi052530j. [DOI] [PubMed] [Google Scholar]
- (42).Goedert M, Spillantini MG, Jakes R, Rutherford D, Crowther RA. Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron. 1989;3(4):519–526. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.