Abstract
The construction of a predictive model of an entire eukaryotic cell that describes its dynamic structure from atomic to cellular scales is a grand challenge at the intersection of biology, chemistry, physics, and computer science. Having such a model will open new dimensions in biological research and accelerate healthcare advancements. Developing the necessary experimental and modeling methods presents abundant opportunities for a community effort to realize this goal. Here, we present a vision for creation of a spatiotemporal multi-scale model of the pancreatic β–cell, a relevant target for understanding and modulating the pathogenesis of diabetes.
Keywords: Whole-cell modeling, integrative modeling, structure modeling, modeling of dynamics, multi-scale modeling, data-driven modeling, pancreatic β– cells, diabetes, convergent bioscience
The goals of this perspective are to outline characteristics of a comprehensive whole-cell model, to point out opportunities for community collaboration in its construction, and to highlight several challenges that should be addressed along the way. By bringing attention to specific areas where there are gaps in knowledge or a lack of tools, we hope this piece will serve as a ‘call-to-arms’ for both the β-cell biology and whole-cell modeling fields to work together and address these challenges. It is likely that as the whole-cell modeling field evolves, so will the definition of a comprehensive whole-cell model. We hope to ignite the conversation about and effort towards this grand challenge. We believe that it will take a community effort to prescribe convincingly and in detail how to construct predictive cellular models. Therefore, the focus of the commentary is to shed light on various attributes of comprehensive whole-cell models and the importance of modeling β-cells while at the same time noting the dearth of applicable data. To begin, we review previous efforts in line with whole-cell modeling and what is currently possible.
Previous whole-cell modeling efforts
A whole-cell model describes one or more aspects of the entire cell as a function of its components and relationships among them. Such models increase our understanding of how the cell functions, how it can be modulated, and how it evolved. Recent efforts in whole-cell modeling are wide-ranging and include static visual reconstructions of cellular landscapes, dynamic structural simulations of molecular interactions, systems of mathematical equations recapitulating biochemical reaction pathways, and more. For example, atomic resolution models of cytoplasmic subsections of Mycoplasma genitalium (Feig et al., 2015; Yu et al., 2016) and Escherichia coli (Hasnain et al., 2014; McGuffee and Elcock, 2010) were assembled and used for simulating dynamics via Brownian Dynamics (BD) or Molecular Dynamics (MD), to investigate diffusion and protein stability under crowded cellular conditions. Other efforts focused on assembling 3D cellular landscapes using experimental data, including for example, models of HIV-1 virus and Mycoplasma mycoides using cellPACK (a software tool that assembles large-scale models from molecular components using packing algorithms, www.cellpack.org) (Johnson et al., 2014, 2015), an atomic resolution snapshot of a synaptic bouton using quantitative immunoblotting, mass spectrometry, electron microscopy and super-resolution fluorescence imaging (Wilhelm et al., 2014), and an ultrastructure model of mouse pancreatic β–cell using electron tomography (Noske et al., 2008). Additionally, mathematical models using differential equations and flux balance analysis have been used to construct cellular (e.g. (Karr et al., 2012) and metabolic networks (e.g. (King et al., 2016) of whole-cells to predict phenotype from genotype. Many other platforms for modeling cellular processes using various techniques have been developed over the last two decades. One example is V-Cell, a modeling platform that simulates a variety of molecular mechanisms, including reaction kinetics, membrane transport, and flow, using spatial restraints derived from microscope images (Cowan et al., 2012; Moraru et al., 2008). Another popular cellular modeling platform is M-Cell that also uses spatial 3D cellular models and Monte Carlo methods to simulate reactions and movement of molecules (Stiles et al., 1996). Similarly, the E-Cell platform simulates cell behavior using differential equations and user-defined reaction rules regarding aspects like protein function, regulation of gene-expression, and protein-protein interactions (Tomita et al., 1999). Collectively, these efforts required both an enormous amount of data as well as integrative computational methods. While each of these models offered some degree of insight and represented important milestones in whole-cell modeling, none was able to fully represent the complexity and scope of an entire cell.
A whole-cell model – the ideal
A comprehensive whole-cell model should allow us to address questions from multiple scientific fields, incorporate all available experimental information, and harness the power of a wide variety of computational and database resources. Biologists, chemists, physicists, and many others should be able to use the model to ask a myriad of scientific questions depending on the researcher’s particular interest. For example, biologists could query the effects of a drug on a cell’s expression patterns, chemists could test the stability of a particular compound in a cellular environment, and physicists could examine the relationships between reaction rates in biochemical contexts. For the model to be useful to many disciplines, it should integrate data generated from a wide range of experimental platforms. For instance, in the model, each of the cell’s components that are determined by omics approaches should be connected to their conformational data determined through structural biology approaches. Similarly, subcellular localization data should be determined by microscopy, and so forth. To connect these disparate pieces of information, the model will need to integrate a wide variety of database tools and will also require the incorporation of extensive computational resources to perform simulations and queries. The scope of biological questions accessible through a comprehensive whole-cell model will continue to evolve as the available data and technology evolve.
Attributes of a comprehensive whole-cell model
In our view, a comprehensive model of the cell will have the following attributes:
Complete and multiscale
The model will consist of all cellular components, including individual atoms, small molecules (e.g., water and metabolites), macromolecules (e.g., proteins, nucleic acids, and polysaccharides), complexes (e.g., ribosomes, nuclear pore complex, and proteasome), as well as organelles and cellular compartments (e.g., nucleus, mitochondria, and vesicles). The model will describe the cell at multiple levels of its hierarchical organization, from atoms to cellular compartments.
Space and time
The spatial organization of the cell will be mapped by defining the positions/distributions of its components (Thul et al., 2017) as well as how these positions/distributions change over time, through atomic fluctuations, chemical reactions, diffusion, and active mass transport.
Logical organization
The model will also define the logical organization of the cell (e.g., via molecular networks, mass and information flow), providing a sense of functional relationships among components of the cell; for example, how insulin secretion pathways change and are affected by drug molecules in the environment.
Couple multiple representations
A comprehensive cell model will need to simultaneously include various representations of the cell that researchers have established and published in the past. Examples of some of these representations include:
a static description of the spatial distribution of cellular components derived from various experiments like fluorescent imaging and quantitative proteomics (Johnson et al., 2015; Murphy, 2012, 2016; Wilhelm et al., 2014);
static molecular networks used in systems biology (Fabregat et al. 2017);
ordinary differential equations (ODEs) commonly used for modeling metabolic pathways (Karr et al., 2012);
a flux balance analysis (FBA) for modeling biochemical networks (King et al., 2016);
a description of dynamics by statistical inferencing from spatiotemporal distribution of components and live imaging (Komatsu et al., 2011; Mikuni et al., 2016);
particle diffusion in crowded environments via Brownian Dynamics (Ando and Skolnick, 2011; Długosz and Trylska, 2011); and
atomic fluctuations by MD simulations (Yu et al., 2016).
Each of these representations are informed by different inputs and provide answers to different questions. Therefore, each will be useful to include as a different layer for modeling whole-cells.
Although the representations will be derived from diverse sources of information, they must be in harmony and coupled with each other in the final model. The model of the cell should couple diverse representations, directly or indirectly, such that a change in a model of one cellular component described by one representation will be reflected in all models of all components. For example, a change in concentrations derived from mathematical modeling implies a change in spatial model. Similarly, information about diffusion of complexes and metabolites from Brownian Dynamics simulations should feed back into reaction-diffusion models of functional processes, the structure of a protein complex implies a molecular network involving the constituent proteins (Mosca et al., 2013) and a change in the structure of a subunit in a complex may imply a change in the complex structure.
Integrative
Because of the vast complexities of cellular functions, all relevant information must be used to construct the cellular model. This information includes varied experimental datasets, physical theories, statistical inferences, and/or prior models. Thus, integrative modeling will need to be relied on to convert the diverse input information into the cellular model (Alber et al., 2007; Ward et al., 2013).
Specify uncertainty
Importantly, any model must include a quantification of its uncertainty as consideration of model uncertainty is essential for its interpretation. Model uncertainty depends on the sparseness of the input information and the uncertainty about how input information limits the model (Schneidman-Duhovny et al., 2014). For example, the B-factor used in structural biology, specifies the uncertainty in atomic positions in a model of protein structure (Schneidman-Duhovny et al., 2014; Yuan et al., 2005). Likewise, localization probability densities quantify the uncertainty of an integrative model of macromolecular complex structure (Alber et al., 2007).
Descriptive and predictive
A good model will allow rationalization of existing experimental observations. It will also be quantitatively predictive rather than providing only a table or visualization of experimental observations. Thus, a model will allow testable predictions to be made (i.e., be falsifiable).
Reflect heterogeneity
Even cells of the same type vary in terms of abundance and localization of components (e.g., molecules, complexes, and organelles). Therefore, a whole-cell model will describe the variation among individual cells of the same subtype, among cells of different sub-types (Aguayo-Mazzucato et al., 2017; Dorrell et al., 2016; Gutierrez et al., 2017; Tamura et al., 1992), and among cells from different individuals in the human population. It should also be able to take into account different environments and perturbations. These variations are likely to play significant roles in the progression of disease and drug response, and therefore must be accounted for in a whole-cell model.
Iterative
A model must be capable of continued refinements, reflecting the growth in quantity and quality of input information as well as improvements in computing capacity and modeling methods. A good model will facilitate the identification of the most informative future experiments, and thus expedite the scientific cycle of iterating through experiment and modeling (Sanghvi et al., 2013).
Why model a β–cell?
The human pancreatic β–cell is an attractive target for a multi-scale model of a eukaryotic cell for several reasons. First, a β–cell can be mechanistically simplified as input/output machine, receiving blood glucose as its primary input and subsequently releasing insulin as its primary output, a paradigm that is suitable for coarse-grained model development. Second, there is a wealth of β–cell experimental data available in the literature, especially from electron tomography and transcriptomics (e.g., (Blodgett et al., 2015; Brackeva et al., 2015; Li et al., 2016; Nica et al., 2013; Noske et al., 2008; Pfeifer et al., 2015). Finally, as β–cells are responsible for insulin production, a thorough understanding of their molecular processes would directly benefit researchers focused on diabetes, a disease that affects hundreds of millions of individuals worldwide (Guariguata et al., 2014). This effort will also provide an example for how to construct models of other cell types.
What data are necessary?
To build a comprehensive model that accurately represents the 3D structure, organization, and function of a β–cell, we require:
the basic ultrastructural architecture of the cell, including the overall shape and size of the cell and abundance, location, and membrane topology of all the different organelles,
an exhaustive list of the identities of the β–cell components, including proteins, nucleic acids, sugars, lipids, metabolites, etc., and macromolecular assemblies,
the quantities and dynamic localization of each of the organelles, components and assemblies,
high-resolution structural information of all the components and assemblies from the atomic to the cellular scale,
variability of all structural and compositional features between individual cells in a population, including the plasticity of its architectural organization and variations in the identities, amount, and locations of its components in different cells.
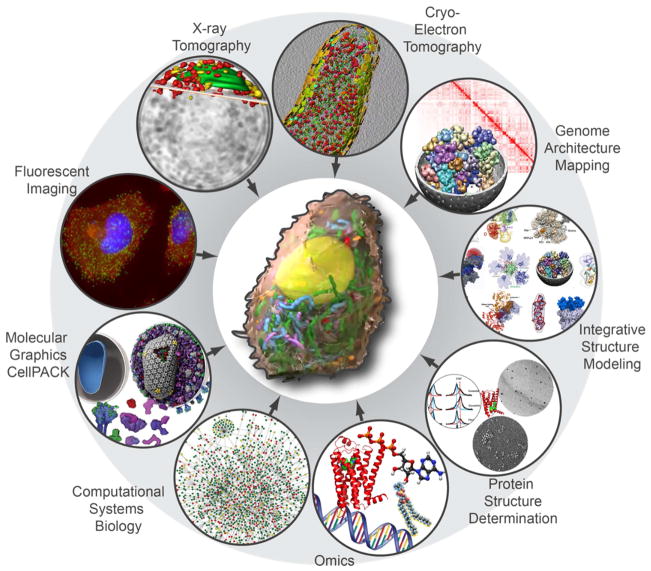
Ideally, data from both single cells and primary tissues will be used to capture both single cell variability as well as the influence of native environment to fully understand cellular function. The required toolbox for acquiring the necessary data is illustrated in Figure 1.
Figure 1. Tools for deriving a spatiotemporal multi-scale model of the human pancreatic β-cell.
Methods of interest: Fluorescent imaging (super-resolution imaging, live imaging), X-ray tomography, cryo-electron tomography, genome architecture mapping (Hi-C maps, fluorescent in-situ hybridization, etc.), integrative structure modeling, protein structure determination (X-ray crystallography, electron microscopy, nuclear magnetic resonance spectroscopy), omics (proteomics, transcriptomics, metabolomics, genomics, lipidomics), computational systems biology, and molecular graphics and packing tools. These methods collectively cover a wide range of scales from atomic to the cellular level. Images were adapted from (Kalhor et al., 2011; Xu et al., 2015) (Jeong et al., 2001; Liu et al., 2012; Sali et al., 2015; Song et al., 2017; Yang et al., 2015)
Collecting the building blocks
The architecture of a β–cell
X-ray and cryo-electron tomography are capable of determining the ultrastructural architecture of β–cells, each at different spatial scales. X-ray tomograms can be used to extract information regarding quantity, location, shape, and size of the nucleus, golgi, mitochondria, and other organelles. At a higher resolution, cryo-electron tomography can elucidate the topology of membranes and compartments within a cell, as well as reveal individual macromolecular complexes. Even the low resolution electron tomograms taken a decade ago by serial sectioning of high-pressure frozen and plastic-embedded cells showed a surprisingly tight packing of insulin vesicles inside islet β–cells (Noske et al., 2008; Pfeifer et al., 2015). But with recent advances including focused ion beam milling, direct detectors, and phase plates, we can increase the resolution and contrast of captured images so that even macromolecules can be visualized (Danev and Baumeister, 2016; Rigort et al., 2012). Computational methods like template-based search (Beck et al., 2009; Nickell et al., 2006) and template-free pattern mining (Xu et al., 2015; Xu et al., 2011) are also under development for the detection of macromolecules in cellular tomograms. With further technological advances and increased resolution, the combination of x-ray and cryo-electron tomographic data along with accurate computational techniques will be key for creating spatial and temporal distribution maps of the organelles and macromolecular complexes of β–cells under various environmental conditions (Beck et al., 2009; Earnest et al., 2017).
Genes and proteins expressed in β–cells
The other requirement for building a 3D cellular model is an exhaustive list of the cell’s components for a given cell state and point in time. This would include both RNA expression levels and measures of protein abundance. Currently, only a limited number of transcriptomics and even fewer proteomics datasets are available for human pancreatic β–cells. Previous analyses were largely performed on model organisms, such as rodents because human sample availability is limited. Studies with human samples were mostly carried out on whole pancreatic islets due to the difficulty to segregate β–cells from islets, limiting their utility in this context.
We compiled and curated the available transcriptomics data for human pancreatic β–cells and compared it with the sole available proteomics dataset of pancreatic β–cells. This assessment showed that the proteomics analysis represents a partial survey of the β–cell proteome. There are only 750 proteins included in the proteomics dataset and 11,606 genes available via transcriptomics corresponding to 11,700 proteins (Supplementary Tables S1 and S2, respectively). Many important proteins are missing from the proteomics data, including G protein-coupled receptors (GPCRs) e.g., glucagon-like peptide 1 receptor (GLP-1R) and other cell membrane proteins that are known to be expressed in human pancreatic β–cells and instrumental in glucose and insulin signaling. Hence, there is a need for global proteomic analyses on human pancreatic β–cells and single cell RNA-seq for a more descriptive analysis and also to capture the variability between individual cells.
Another crucial piece of information is the respective protein copy numbers per cell, as well as the variability of the copy numbers across individual cells. The relative abundance of proteins in a cell population can in principle be extracted from proteomics datasets. However, this information is still lacking due to the limited number of proteomics datasets available for human β-cells. Recently, new techniques have been developed for single cell proteomics (Hughes et al., 2014; Budnik et al., 2017; Su et al., 2017), but they have not yet been widely applied.
A complete model of a cell also requires similar quantitative information about metabolites, lipids, polysaccharides, and other molecules, including their identities and relative concentrations. Partial data is available from omics datasets for rodent cells/islets and human islets (e.g. (Gooding et al., 2016; Huang and Joseph, 2012; Pearson et al., 2016; Roomp et al., 2017; Wallace et al., 2013), but not for isolated human β–cells. A complete list of the components of the cell is necessary to begin to understand what processes occur inside a cell as a result of the interactions and reactions of these components.
Protein structures in human pancreatic β–cells
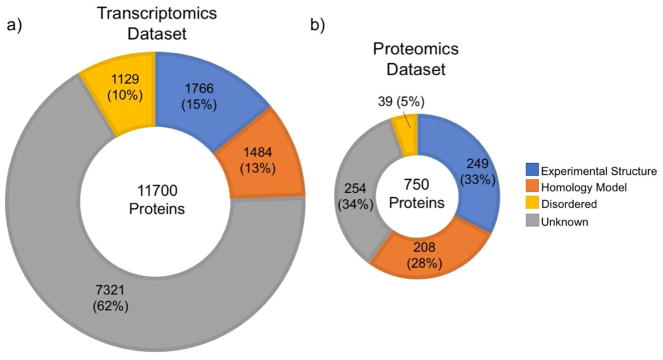
Because our overall focus is on building multiscale 3D models of β–cells, we need not only the list and relative abundance of component proteins, but also their atomic structures. We analyzed how many atomic protein structures are available for our list of 11,700 β–cell proteins. As of March 1, 2018, only ~28% of the proteins had either an experimental structure in the PDB (Berman et al., 2000) (sequence coverage ≥ 75%) or a reliable homology model in SWISS-MODEL (Kiefer et al., 2009; Kopp and Schwede, 2004 (sequence coverage ≥ 75%, normalized-QMEAN ≥ 0.6) (Benkert et al., 2008). 10% of proteins are structurally disordered (fraction of disordered residues ≥ 50%) (Oates et al., 2013), while almost 62% of proteins have no reliable 3D structures (Figure 2). This lack of structural knowledge is even more pronounced when considering protein complexes, which have not yet been exhaustively identified let alone structurally characterized (Im et al., 2016; Mosca et al., 2013).
Figure 2. Pancreatic β-cell protein classes and structural coverage.
a, b) Structural Coverage of β–cell proteins identified in analyses of (a) transcriptomic and (b) proteomics data were categorized according to the amount of available structural information. The categories are Experimental Structures (PDB: sequence coverage ≥ 75%), Reliable Homology Models (SWISS-MODEL: sequence coverage ≥ 75%, normalized-QMEAN ≥ 0.6), Disordered Proteins (D2P2: Disordered residues ≥ 50%), Unknown (no reliable experimental structure, no reliable homology model, fraction of disordered residues < 50%). We used three transcriptomics datasets (Blodgett et al., 2015; Li et al., 2016; Nica et al., 2013) to create a combined list of genes expressed in β–cells. There were approximately 13,400 unique genes (with RPKM/FPKM/TPM values ≥ 1) in all three datasets combined. To improve confidence, we limited the list to genes present in at least two of the three datasets (11,606 genes) for which we could identify 11,700 protein products in UniProtKB (The UniProt Consortium, 2017) (Supplementary Table S1). For the proteomics dataset, we used the only available quantitative proteomics dataset specific to human pancreatic β–cells, which reported only 750 proteins (Brackeva et al., 2015) (Supplementary Table S2). Disordered content information came from the D2P2 database (Oates et al., 2013) and homology models were downloaded from SWISS-MODEL (Kiefer et al., 2009).
The scarcity of omics data as well as the limited number of reliable structures presents a great opportunity for discovery and for a community effort to contribute information about human β–cells under various conditions.
Mathematical models
As mentioned earlier, no one approach or single representation will suffice. A number of modeling approaches and/or models should be coupled together for whole-cell modeling. There are many published mathematical models of functional processes in pancreatic β–cells using ODEs, including modeling of Ca+2 flux (Fridlyand et al., 2003), electrophysiological responses (Riz et al., 2014), and insulin secretory pathway (Fridlyand and Philipson, 2016; Jiang et al., 2007; Topp et al., 2000). However, tuning of these models is required for human datasets as most of these models were constructed using datasets from rodent cell-lines/islets (Babtie and Stumpf, 2017).
Putting the pieces together
Coupling the structural information of cell architecture and experimentally-derived spatial distribution of cell components with mathematical models is an important aspect of the whole-cell model. Platforms like V-Cell (Moraru et al., 2008) and M-Cell (Stiles et al., 1996) already integrate such spatial information with mathematical modeling. V-Cell can simulate various molecular mechanisms using ordinary and/or partial differential equations (ODEs) in the context of compartments, whereas M-Cell can simulate cellular environments via 3D reaction-diffusion using Monte-Carlo methods. Other studies have also considered spatial aspects. For example, Earnest et al. extracted cell geometry from cryo-electron tomography experiments and used stochastic simulations to study the effect of cell structure on reaction network (Earnest et al., 2017). Other studies have relied on fluorescent images to extract ultrastructure and spatial distribution of proteins for cell modeling using V-Cell or M-Cell (Murphy, 2012, 2016). These methods do not represent cellular environments, including proteins, lipids and macromolecular assemblies, as three-dimensional structures. M-Cell represents components as diffusing particles and V-Cell models component concentrations via ODEs. In the future, new versions of these modeling platforms that include 3D representations of components and physical interactions are likely to be applied to larger and more complex systems, such as human β–cells.
Computing resources for whole-cell modeling
One reason why an effort to model the cell is timely is that the required computational capabilities seem to be within reach. As already discussed, it is highly likely that a variety of different types of modeling are needed to construct a model of the cell. Thus, different types of computing architectures will also be required, at least initially. These architectures will include special purpose hardware such as the Anton supercomputer for MD simulations (Miao et al., 2015), Graphics Processing Units (GPUs) for image processing (Zheng et al., 2011), general purpose parallel computer clusters for modeling that requires significant data bandwidth (Alber et al., 2007), and finally cloud computing for modeling with relatively low volume of data transfer among the central repository and individual compute nodes. Large-scale recruitment of personal computers on the Internet, as in Folding@Home (Folding@home, 2017), may also be helpful. Some computing capabilities of each one of these types are already available. As the modeling methods evolve and it becomes more clear what types of modeling are required, it will also be beneficial to construct increasingly specialized and thus efficient computing architectures, perhaps culminating in a special purpose computer for simulating cells. Just as the need to solve the phase problem in crystallography motivated the development of early computer hardware (Kendrew, 1963), we anticipate that the goal of modeling whole-cells will also attract computer hardware and software engineers with a commensurate support from governments, philanthropic foundations, and industry partnerships. It is encouraging that a number of government institutions have already recognized the need for increased computing for biology in general and are investing in it, e.g., the Cloud Credits Models by NIH (Commonfund.nih.gov, 2017) and The European Cloud Initiative (Digital Single Market, 2017).
In addition to computing hardware for modeling, the modeling efforts will also require resources for archiving and disseminating the data and models. It is likely that this functionality will be best achieved through a federated archive of repositories. One such arrangement is envisioned for integrative structures of biological macromolecules and corresponding data for the integrative/hybrid methods initiative of wwPDB (Burley et al., 2017; Sali et al., 2015). The Biostudies resource (McEntyre et al., 2015) at European Bioinformatics Institute (EBI) may provide a convenient portal to the participating archives. Therefore, it is conceivable that the common goal of modeling cells will encourage the communities that generate the data to come together and define appropriate standards for archiving, annotating, validating, and disseminating their data to provide maximal use of the accumulated results.
Value of whole-cell model
Accurate multiscale models will contribute to our understanding of how the cell functions, how it can be modulated, and how it evolved. Models of whole-cells will lead to innovative ways of designing therapeutics, just as structures of individual proteins often facilitate rational, structure-based drug discovery. Drug design efforts have focused at the molecular level on isolated proteins for decades. It is only logical to move towards designing drugs by modeling their effect at the cellular level. Whole-cell models could help in target selection for drug discovery by predicting a potential drug target’s impact on modulating various cell functions. In addition, these models could also provide a basis for rational-based cell therapies. There are a rising number of cell therapy clinical trials, but only one recently approved cell therapy (Golubovskaya et al., 2017). There is therefore scope for models to help improve this outlook. In the case of diabetes, a major area of investigation for improving therapy is focused on generating healthy β–cells in vitro. Thus, studies focused on holistically understanding β–cells are a clear necessity. Efforts towards deriving β–cells from iPSCs and implanting healthy β–cells in patients are active areas of research and provide many challenges (Millman et al., 2016; Rezania et al., 2014). Accurate β–cell models can potentially be used to inform the design of these approaches for therapy (Purcell et al., 2013).
Whole-cell models may also provide new insights about the molecular links from genotype to phenotype and shed light on more complex emergent properties that arise from molecular interactions, such as coordinated insulin release from clusters of β–cells. Multi-scale models will also provide a way to comprehend heterogeneous data from a vast array of complementary experiments and push computational limits so that complex biological questions can be answered. Many of these points have also been considered in other contexts (Betts and Russell, 2007; Carrera and Covert, 2015; Horwitz, 2016; Horwitz and Johnson, 2017; Macklin et al., 2014; Roberts, 2014). One specific outcome of an initial focus on β–cells would be establishment of a platform consisting of the basic pipeline, framework, toolbox, methods, approaches, experimental and computational infrastructure, etc., that could be then extended to other cell types. Beyond that, it’s important to think about how these individual cells integrate into tissues and such complex structures could become an additional target for modeling efforts. To take the long view, development of whole-cell and eventually tissue-scale models will allow for deeper insights into a myriad of health conditions, including diseases like diabetes and cancer, each of which affects hundreds of millions of people worldwide.
Concluding Remarks – a call-to-arms
The scale of many challenges in biology and biomedicine is evolving, prompting corresponding changes towards increased collaboration and communication among scientists. To pursue grand challenges more systematically and strategically, it is becoming increasingly common to adopt a community-wide approach, because these undertakings require both a large effort and varied expertise in diverse fields. Community-wide collaborations are the future of science and are typically structured to allow for rapid sharing of information and tools. Currently, there are several active large, collaborative around the world, including the Cancer Cell Map Initiative (Krogan et al., 2015), Chan Zuckerberg Biohub (https://www.czbiohub.org/), Allen Brain Institute, 4D Nucleome (Dekker et al., 2017), and European Brain Project (Amunts et al., 2016) to name a few. These efforts will develop new technologies and generate data leading to new discoveries, which in turn will change the way we think about grand challenges in biology.
There are many facets of whole-cell modeling that will require a community effort. For example, how these whole-cell models should be built will remain unclear until the field collectively starts working towards this ambitious goal and navigates success and failures. It is insufficient for the modelers to simply work together. Rather the modelers, experimentalists, hardware and software developers and potentially entire institutions must unite in these efforts. This approach will also be useful for developing new computing technologies that will extend the power of whole-cell modeling. As more and more data is collected, the cell types, conditions, and the archiving process can be standardized and streamlined to facilitate these efforts. Additionally, large collaborations will be necessary to collect and integrate disparate data, including transcriptomics, proteomics, cryo-electron tomography, X-ray tomography, live-cell imaging, cryo-electron microscopy, X-ray crystallography, and single particle analysis, under similar conditions for capturing different dimensions of the same state of a cell (Figure 1).
Now is the time to take advantage of the technological advancements in each of these fields to build 3D models of human pancreatic β–cells, thereby providing a method for testing and developing new therapies targeting diabetes. The development of these models will also provide a prototype method to examine and understand other cell types, tissues and the human body.
Supplementary Material
List of the proteins corresponding to genes identified in at least two out of three trancriptomics datasets used in this analysis. Only those genes were considered that have RPKM/FPKM/TPM value > 1. Genes were matched to proteins using UniprotKB (June 2017). Transcriptomics datasets were used from Blodgett et al., 2015; Li et al., 2016; Nica et al., 2013
List of the proteins corresponding to proteomics dataset used in this analysis. Proteins were matched to genes using UniprotKB (June 2017). Proteomics dataset was used from Brackeva et al., 2015
Acknowledgments
This work was supported in part by funding from the Bridge Institute, USC Dornsife College of Letters, Arts and Sciences, Provost Fellowship (J.S.), NIH R01GM096089 (F.A.), and NIH R01 GM083960 and P41 GM109824 (A.S.). The authors would like to thank Angela Walker and Timothy James for feedback on the manuscript, and the entire BISC-599 (2017) class members and faculty guests for discussions on this topic. Authors also thank reviewers and editor for their careful consideration and suggestions to improve the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguayo-Mazzucato C, van Haaren M, Mruk M, Lee TB, Crawford C, Hollister-Lock J, Sullivan BA, Johnson JW, Ebrahimi A, Dreyfuss JM, et al. β cell aging markers have heterogeneous distribution and are induced by insulin resistance. Cell Metab. 2017;25:898–910e5. doi: 10.1016/j.cmet.2017.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- Amunts K, Ebell C, Muller J, Telefont M, Knoll A, Lippert T. The human brain project: creating a european research infrastructure to decode the human brain. Neuron. 2016;92:574–581. doi: 10.1016/j.neuron.2016.10.046. [DOI] [PubMed] [Google Scholar]
- Ando T, Skolnick J. Brownian dynamics simulation of macromolecule diffusion in a protocell. Quantum Bioinform IV (2010) 2011;28:413–426. doi: 10.1142/9789814343763_0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babtie AC, Stumpf MPH. How to deal with parameters for whole-cell modelling. J R Soc Interface. 2017:14. doi: 10.1098/rsif.2017.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Malmström JA, Lange V, Schmidt A, Deutsch EW, Aebersold R. Visual proteomics of the human pathogen Leptospira interrogans. Nat Methods. 2009;6:817–823. doi: 10.1038/nmeth.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert P, Tosatto SCE, Schomburg D. QMEAN: A comprehensive scoring function for model quality assessment. Proteins. 2008;71:261–277. doi: 10.1002/prot.21715. [DOI] [PubMed] [Google Scholar]
- Betts MJ, Russell RB. The hard cell: from proteomics to a whole cell model. FEBS Lett. 2007;581:2870–2876. doi: 10.1016/j.febslet.2007.05.062. [DOI] [PubMed] [Google Scholar]
- Blodgett DM, Nowosielska A, Afik S, Pechhold S, Cura AJ, Kennedy NJ, Kim S, Kucukural A, Davis RJ, Kent SC, et al. Novel Observations From Next-Generation RNA Sequencing of Highly Purified Human Adult and Fetal Islet Cell Subsets. Diabetes. 2015;64:3172–3181. doi: 10.2337/db15-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackeva B, Kramer G, Vissers JPC, Martens GA. Quantitative proteomics of rat and human pancreatic beta cells. Data Brief. 2015;3:234–239. doi: 10.1016/j.dib.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik B, Levy E, Slavov N. Mass-spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. bioRxiv. 2017:102681. doi: 10.1186/s13059-018-1547-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Kurisu G, Markley JL, Nakamura H, Velankar S, Berman HM, Sali A, Schwede T, Trewhella J. PDB-Dev: a Prototype System for Depositing Integrative/Hybrid Structural Models. Structure. 2017;25:1317–1318. doi: 10.1016/j.str.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera J, Covert MW. Why Build Whole-Cell Models? Trends Cell Biol. 2015;25:719–722. doi: 10.1016/j.tcb.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commonfund.nih.gov. [Accessed 20 Nov. 2017];Cloud Credits | NIH Common Fund. [online] 2017 Available at: https://commonfund.nih.gov/bd2k/cloudcredits.
- Cowan AE, Moraru II, Schaff JC, Slepchenko BM, Loew LM. Spatial modeling of cell signaling networks. Methods Cell Biol. 2012;110:195–221. doi: 10.1016/B978-0-12-388403-9.00008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danev R, Baumeister W. Cryo-EM single particle analysis with the Volta phase plate. Elife. 2016:5. doi: 10.7554/eLife.13046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Belmont AS, Guttman M, Leshyk VO, Lis JT, Lomvardas S, Mirny LA, O’Shea CC, Park PJ, Ren B, et al. The 4D nucleome project. Nature. 2017;549:219–226. doi: 10.1038/nature23884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digital Single Market. [Accessed 20 Nov. 2017];The European Cloud Initiative. [online] 2017 Available at: https://ec.europa.eu/digital-single-market/en/%20european-cloud-initiative.
- Dorrell C, Schug J, Canaday PS, Russ HA, Tarlow BD, Grompe MT, Horton T, Hebrok M, Streeter PR, Kaestner KH, et al. Human islets contain four distinct subtypes of β cells. Nat Commun. 2016;7:11756. doi: 10.1038/ncomms11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Długosz M, Trylska J. Diffusion in crowded biological environments: applications of Brownian dynamics. BMC Biophys. 2011;4:3. doi: 10.1186/2046-1682-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnest TM, Watanabe R, Stone JE, Mahamid J, Baumeister W, Villa E, Luthey-Schulten Z. Challenges of Integrating Stochastic Dynamics and Cryo-Electron Tomograms in Whole-Cell Simulations. J Phys Chem B. 2017;121:3871–3881. doi: 10.1021/acs.jpcb.7b00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fda.gov. [Accessed 16 Nov. 2017];FDA approves CAR-T cell therapy to treat adults with certain types of large B-cell lymphoma. [online] 2017 Available at: https://www.fda.gov/newsevents/newsroom/pressannouncements/ucm581216.htm.
- Feig M, Harada R, Mori T, Yu I, Takahashi K, Sugita Y. Complete atomistic model of a bacterial cytoplasm for integrating physics, biochemistry, and systems biology. J Mol Graph Model. 2015;58:1–9. doi: 10.1016/j.jmgm.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folding@home. [Accessed 20 Nov. 2017];Front Page - Folding@home. [online] 2017 Available at: http://folding.stanford.edu/
- Fridlyand LE, Philipson LH. Pancreatic Beta Cell G-Protein Coupled Receptors and Second Messenger Interactions: A Systems Biology Computational Analysis. PLoS ONE. 2016;11:e0152869. doi: 10.1371/journal.pone.0152869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridlyand LE, Tamarina N, Philipson LH. Modeling of Ca2+ flux in pancreatic beta-cells: role of the plasma membrane and intracellular stores. Am J Physiol Endocrinol Metab. 2003;285:E138–54. doi: 10.1152/ajpendo.00194.2002. [DOI] [PubMed] [Google Scholar]
- Golubovskaya V, Berahovich R, Zhou H, Xu S, Harto H, Li L, Chao C-C, Mao MM, Wu L. CD47-CAR-T Cells Effectively Kill Target Cancer Cells and Block Pancreatic Tumor Growth. Cancers (Basel) 2017:9. doi: 10.3390/cancers9100139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding JR, Jensen MV, Newgard CB. Metabolomics applied to the pancreatic islet. Arch Biochem Biophys. 2016;589:120–130. doi: 10.1016/j.abb.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guariguata L, Whiting DR, Hambleton I, Beagley J, Linnenkamp U, Shaw JE. Global estimates of diabetes prevalence for 2013 and projections for 2035. Diabetes Res Clin Pract. 2014;103:137–149. doi: 10.1016/j.diabres.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Gutierrez GD, Gromada J, Sussel L. Heterogeneity of the pancreatic beta cell. Front Genet. 2017;8:22. doi: 10.3389/fgene.2017.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasnain S, McClendon CL, Hsu MT, Jacobson MP, Bandyopadhyay P. A new coarse-grained model for E. coli cytoplasm: accurate calculation of the diffusion coefficient of proteins and observation of anomalous diffusion. PLoS ONE. 2014;9:e106466. doi: 10.1371/journal.pone.0106466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz R. Integrated, multi-scale, spatial-temporal cell biology--A next step in the post genomic era. Methods. 2016;96:3–5. doi: 10.1016/j.ymeth.2015.09.007. [DOI] [PubMed] [Google Scholar]
- Horwitz R, Johnson GT. Whole cell maps chart a course for 21st-century cell biology. Science. 2017;356:806–807. doi: 10.1126/science.aan5955. [DOI] [PubMed] [Google Scholar]
- Huang M, Joseph JW. Metabolomic analysis of pancreatic β-cell insulin release in response to glucose. Islets. 2012;4:210–222. doi: 10.4161/isl.20141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AJ, Spelke DP, Xu Z, Kang CC, Schaffer DV, Herr AE. Single-cell western blotting. Nat Methods. 2014;11:749–755. doi: 10.1038/nmeth.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im W, Liang J, Olson A, Zhou HX, Vajda S, Vakser IA. Challenges in structural approaches to cell modeling. J Mol Biol. 2016;428:2943–2964. doi: 10.1016/j.jmb.2016.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411(6833):41. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Jiang N, Cox RD, Hancock JM. A kinetic core model of the glucose-stimulated insulin secretion network of pancreatic beta cells. Mamm Genome. 2007;18:508–520. doi: 10.1007/s00335-007-9011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GT. [Accessed 21 Nov. 2017];Rapid Visual Inventory & Comparison of Complex 3D Structures. [online] 2017 Available at: https://www.youtube.com/watch?v=Dl1ufW3cj4g.
- Johnson GT, Goodsell DS, Autin L, Forli S, Sanner MF, Olson AJ. 3D molecular models of whole HIV-1 virions generated with cellPACK. Faraday Discuss. 2014;169:23–44. doi: 10.1039/c4fd00017j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GT, Autin L, Al-Alusi M, Goodsell DS, Sanner MF, Olson AJ. cellPACK: a virtual mesoscope to model and visualize structural systems biology. Nat Methods. 2015;12:85–91. doi: 10.1038/nmeth.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2011;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karr JR, Sanghvi JC, Macklin DN, Gutschow MV, Jacobs JM, Bolival B, Assad-Garcia N, Glass JI, Covert MW. A whole-cell computational model predicts phenotype from genotype. Cell. 2012;150:389–401. doi: 10.1016/j.cell.2012.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrew J. Myoglobin and the Structure of Proteins: Crystallographic analysis and data-processing techniques reveal the molecular architecture. Science. 1963;139(3561):1259–1266. doi: 10.1126/science.139.3561.1259. [DOI] [PubMed] [Google Scholar]
- Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–92. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King ZA, Lu J, Dräger A, Miller P, Federowicz S, Lerman JA, Ebrahim A, Palsson BO, Lewis NE. BiGG Models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 2016;44:D515–22. doi: 10.1093/nar/gkv1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu T, Johnsson K, Okuno H, Bito H, Inoue T, Nagano T, Urano Y. Real-Time Measurements of Protein Dynamics Using Fluorescence Activation-Coupled Protein Labeling Method. Journal of the American Chemical Society. 2011;133(17):6745–6751. doi: 10.1021/ja200225m. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Lippman S, Agard DA, Ashworth A, Ideker T. The cancer cell map initiative: defining the hallmark networks of cancer. Mol Cell. 2015;58:690–698. doi: 10.1016/j.molcel.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Klughammer J, Farlik M, Penz T, Spittler A, Barbieux C, Berishvili E, Bock C, Kubicek S. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep. 2016;17:178–187. doi: 10.15252/embr.201540946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Horst R, Katritch V, Stevens RC, Wüthrich K. Biased signaling pathways in β2-adrenergic receptor characterized by 19F-NMR. Science. 2012;335:1106–1110. doi: 10.1126/science.1215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macklin DN, Ruggero NA, Covert MW. The future of whole-cell modeling. Curr Opin Biotechnol. 2014;28:111–115. doi: 10.1016/j.copbio.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEntyre J, Sarkans U, Brazma A. The BioStudies database. Mol Syst Biol. 2015;11:847. doi: 10.15252/msb.20156658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffee SR, Elcock AH. Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput Biol. 2010;6:e1000694. doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Feixas F, Eun C, McCammon JA. Accelerated molecular dynamics simulations of protein folding. J Comput Chem. 2015;36:1536–1549. doi: 10.1002/jcc.23964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikuni T, Nishiyama J, Sun Y, Kamasawa N, Yasuda R. High-Throughput, High-Resolution Mapping of Protein Localization in Mammalian Brain by In Vivo Genome Editing. Cell. 2016;165(7):1803–1817. doi: 10.1016/j.cell.2016.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millman JR, Xie C, Van Dervort A, Gürtler M, Pagliuca FW, Melton DA. Generation of stem cell-derived β-cells from patients with type 1 diabetes. Nat Commun. 2016;7:11463. doi: 10.1038/ncomms11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraru II, Schaff JC, Slepchenko BM, Blinov ML, Morgan F, Lakshminarayana A, Gao F, Li Y, Loew LM. Virtual Cell modelling and simulation software environment. IET Syst Biol. 2008;2:352–362. doi: 10.1049/iet-syb:20080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca R, Céol A, Aloy P. Interactome3D: adding structural details to protein networks. Nat Methods. 2013;10:47–53. doi: 10.1038/nmeth.2289. [DOI] [PubMed] [Google Scholar]
- Murphy RF. CellOrganizer: Image-derived models of subcellular organization and protein distribution. Methods Cell Biol. 2012;110:179–193. doi: 10.1016/B978-0-12-388403-9.00007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RF. Building cell models and simulations from microscope images. Methods. 2016;96:33–39. doi: 10.1016/j.ymeth.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nica AC, Ongen H, Irminger JC, Bosco D, Berney T, Antonarakis SE, Halban PA, Dermitzakis ET. Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome. Genome Res. 2013;23:1554–1562. doi: 10.1101/gr.150706.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickell S, Kofler C, Leis AP, Baumeister W. A visual approach to proteomics. Nat Rev Mol Cell Biol. 2006;7:225–230. doi: 10.1038/nrm1861. [DOI] [PubMed] [Google Scholar]
- Nobelprize.org. [Accessed 21 Nov. 2017];John C. Kendrew - Banquet Speech. [online] 2017 Available at: https://www.nobelprize.org/nobel_prizes/chemistry/laureates/1962/kendrew-speech.html.
- Noske AB, Costin AJ, Morgan GP, Marsh BJ. Expedited approaches to whole cell electron tomography and organelle mark-up in situ in high-pressure frozen pancreatic islets. J Struct Biol. 2008;161:298–313. doi: 10.1016/j.jsb.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates ME, Romero P, Ishida T, Ghalwash M, Mizianty MJ, Xue B, Dosztányi Z, Uversky VN, Obradovic Z, Kurgan L, et al. D2P2: database of disordered protein predictions. Nucleic Acids Res. 2013;41:D508–16. doi: 10.1093/nar/gks1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson GL, Mellett N, Chu KY, Boslem E, Meikle PJ, Biden TJ. A comprehensive lipidomic screen of pancreatic β-cells using mass spectroscopy defines novel features of glucose-stimulated turnover of neutral lipids, sphingolipids and plasmalogens. Mol Metab. 2016;5:404–414. doi: 10.1016/j.molmet.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer CR, Shomorony A, Aronova MA, Zhang G, Cai T, Xu H, Notkins AL, Leapman RD. Quantitative analysis of mouse pancreatic islet architecture by serial block-face SEM. J Struct Biol. 2015;189:44–52. doi: 10.1016/j.jsb.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell O, Jain B, Karr JR, Covert MW, Lu TK. Towards a whole-cell modeling approach for synthetic biology. Chaos. 2013;23:025112. doi: 10.1063/1.4811182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezania A, Bruin JE, Arora P, Rubin A, Batushansky I, Asadi A, O’Dwyer S, Quiskamp N, Mojibian M, Albrecht T, et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol. 2014;32:1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- Rigort A, Bäuerlein FJB, Villa E, Eibauer M, Laugks T, Baumeister W, Plitzko JM. Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography. Proc Natl Acad Sci U S A. 2012;109:4449–4454. doi: 10.1073/pnas.1201333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riz M, Braun M, Pedersen MG. Mathematical modeling of heterogeneous electrophysiological responses in human β-cells. PLoS Comput Biol. 2014;10:e1003389. doi: 10.1371/journal.pcbi.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E. Cellular and molecular structure as a unifying framework for whole-cell modeling. Curr Opin Struct Biol. 2014;25:86–91. doi: 10.1016/j.sbi.2014.01.005. [DOI] [PubMed] [Google Scholar]
- Roomp K, Kristinsson H, Schvartz D, Ubhayasekera K, Sargsyan E, Manukyan L, Chowdhury A, Manell H, Satagopam V, Groebe K, et al. Combined lipidomic and proteomic analysis of isolated human islets exposed to palmitate reveals time-dependent changes in insulin secretion and lipid metabolism. PLoS ONE. 2017;12:e0176391. doi: 10.1371/journal.pone.0176391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Berman HM, Schwede T, Trewhella J, Kleywegt G, Burley SK, Markley J, Nakamura H, Adams P, Bonvin AMJJ, et al. Outcome of the First wwPDB Hybrid/Integrative Methods Task Force Workshop. Structure. 2015;23:1156–1167. doi: 10.1016/j.str.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghvi JC, Regot S, Carrasco S, Karr JR, Gutschow MV, Bolival B, Covert MW. Accelerated discovery via a whole-cell model. Nat Methods. 2013;10:1192–1195. doi: 10.1038/nmeth.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Pellarin R, Sali A. Uncertainty in integrative structural modeling. Curr Opin Struct Biol. 2014;28:96–104. doi: 10.1016/j.sbi.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Yang D, Wang Y, de Graaf C, Zhou Q, Jiang S, Liu K, Cai X, Dai A, Lin G, et al. Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators. Nature. 2017;546:312–315. doi: 10.1038/nature22378. [DOI] [PubMed] [Google Scholar]
- Stiles JR, Van Helden D, Bartol TM, Salpeter EE, Salpeter MM. Miniature endplate current rise times less than 100 microseconds from improved dual recordings can be modeled with passive acetylcholine diffusion from a synaptic vesicle. Proc Natl Acad Sci U S A. 1996;93:5747–5752. doi: 10.1073/pnas.93.12.5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Shi Q, Wei W. Single cell proteomics in biomedicine: High-dimensional data acquisition, visualization, and analysis. Proteomics. 2017:17. doi: 10.1002/pmic.201600267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura H, Matsumoto G, Itakura Y, Terai H, Ikebuchi K, Mitarai T, Isoda K. A case of congenital dyserythropoietic anemia type II associated with hemochromatosis. Intern Med. 1992;31:380–384. doi: 10.2169/internalmedicine.31.380. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM, et al. A subcellular map of the human proteome. Science. 2017:356. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- Tomita M, Hashimoto K, Takahashi K, Shimizu TS, Matsuzaki Y, Miyoshi F, Saito K, Tanida S, Yugi K, Venter JC, et al. E-CELL: software environment for whole-cell simulation. Bioinformatics. 1999;15:72–84. doi: 10.1093/bioinformatics/15.1.72. [DOI] [PubMed] [Google Scholar]
- Topp B, Promislow K, deVries G, Miura RM, Finegood DT. A model of beta-cell mass, insulin, and glucose kinetics: pathways to diabetes. J Theor Biol. 2000;206:605–619. doi: 10.1006/jtbi.2000.2150. [DOI] [PubMed] [Google Scholar]
- Wallace M, Whelan H, Brennan L. Metabolomic analysis of pancreatic beta cells following exposure to high glucose. Biochim Biophys Acta. 2013;1830:2583–2590. doi: 10.1016/j.bbagen.2012.10.025. [DOI] [PubMed] [Google Scholar]
- Ward AB, Sali A, Wilson IA. Biochemistry. Integrative structural biology. Science. 2013;339:913–915. doi: 10.1126/science.1228565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BG, Mandad S, Truckenbrodt S, Kröhnert K, Schäfer C, Rammner B, Koo SJ, Claβen GA, Krauss M, Haucke V, et al. Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science. 2014;344:1023–1028. doi: 10.1126/science.1252884. [DOI] [PubMed] [Google Scholar]
- Xu M, Beck M, Alber F. Template-free detection of macromolecular complexes in cryo electron tomograms. Bioinformatics. 2011;27:i69–76. doi: 10.1093/bioinformatics/btr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Tocheva EI, Chang YW, Jensen GI, Alber F. De novo visual proteomics in single cells through pattern mining. 2015 arXiv preprint arXiv:1512.09347. [Google Scholar]
- Yang L, Yang D, de Graaf C, Moeller A, West GM, Dharmarajan V, Wang C, Siu FY, Song G, Reedtz-Runge S, et al. Conformational states of the full-length glucagon receptor. Nat Commun. 2015;6:7859. doi: 10.1038/ncomms8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I, Mori T, Ando T, Harada R, Jung J, Sugita Y, Feig M. Biomolecular interactions modulate macromolecular structure and dynamics in atomistic model of a bacterial cytoplasm. Elife. 2016:5. doi: 10.7554/eLife.19274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Bailey TL, Teasdale RD. Prediction of protein B-factor profiles. Proteins. 2005;58:905–912. doi: 10.1002/prot.20375. [DOI] [PubMed] [Google Scholar]
- Zheng SQ, Branlund E, Kesthelyi B, Braunfeld MB, Cheng Y, Sedat JW, Agard DA. A distributed multi-GPU system for high speed electron microscopic tomographic reconstruction. Ultramicroscopy. 2011;111:1137–1143. doi: 10.1016/j.ultramic.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of the proteins corresponding to genes identified in at least two out of three trancriptomics datasets used in this analysis. Only those genes were considered that have RPKM/FPKM/TPM value > 1. Genes were matched to proteins using UniprotKB (June 2017). Transcriptomics datasets were used from Blodgett et al., 2015; Li et al., 2016; Nica et al., 2013
List of the proteins corresponding to proteomics dataset used in this analysis. Proteins were matched to genes using UniprotKB (June 2017). Proteomics dataset was used from Brackeva et al., 2015