Abstract
Following cell entry, the RNA genome of HIV-1 is reverse transcribed into double-stranded DNA that ultimately integrates into the host cell genome to establish the provirus. These early phases of infection are notably vulnerable to suppression by a collection of cellular anti-viral effectors, called restriction or resistance factors. The host anti-viral protein APOBEC3G (A3G) antagonizes the early steps of HIV-1 infection through the combined effects of inhibiting viral cDNA production, and cytidine-to-uridine driven hypermutation of this cDNA. In seeking to address the underlying molecular mechanism for inhibited cDNA synthesis, we developed a deep sequencing strategy to characterize nascent reverse transcription products and their precise 3'-termini in HIV-1 infected T cells. Our results demonstrate site- and sequence-independent interference with reverse transcription, which requires A3G’s specific interaction with reverse transcriptase (RT) itself. This approach also established, contrary to current ideas, that cellular uracil base excision repair (UBER) enzymes target and cleave A3G-edited uridine-containing viral cDNA. Together, these findings yield further insights into the regulatory interplay between RT, A3G and the cellular DNA repair machinery, and identify the suppression of HIV-1 RT by a directly interacting host protein as a new cell-mediated anti-viral mechanism.
Introduction
Prominent among the anti-retroviral restriction factors1,2 are the APOBEC3 (A3) proteins, a family of cytidine deaminases with DNA editing capability3,4. Humans encode seven A3 proteins of which APOBEC3G (A3G), A3F, A3D and some allelic forms of A3H are packaged into HIV-1 virions and inhibit infection5–10. During wild type HIV-1 infection, A3G and other A3 proteins are largely disabled by the virus-encoded Vif protein, which promotes proteasomal degradation through recruitment to a CUL5 ubiquitin ligase complex11–13. Vif is therefore essential for productive HIV-1 replication in its natural cell targets, CD4-positive T-cells14.
A3 proteins dramatically alter the nascent DNA products of reverse transcription in ensuing rounds of infection: these effects are most pronounced for A3G, which displays the strongest anti-viral phenotype6,15. Not only is the viral complementary DNA (cDNA) littered with numerous cytidine-to-uridine mutations16–18, but the accumulation of cDNA is also severely reduced compared to infection in the absence of A3 proteins14,17,19–22.
Figure 1a illustrates three potential, non-mutually exclusive mechanisms for A3G anti-viral action. 1) Hypermutation: the post-synthetic deamination of cytidine to uridine in minus strand reverse transcripts registers as plus strand guanosine-to-adenosine mutations. At least 10% of guanosines can be mutated14, which severely compromises viral sequence integrity and is typically lethal. 2) cDNA fragmentation and degradation: nascent cDNA is hypermutated as in the first model but with the uracils subsequently recognized by uracil-DNA glycosylases (UDGs) in the uracil base excision repair (UBER) pathway, leading to base excision, recognition by abasic site endonucleases (APE1 or APE2), DNA cleavage and, potentially, degradation. 3) Deamination-independent inhibition of reverse transcription: A3G interferes with the biosynthetic capability of the reverse transcriptase (RT) enzyme, reducing the production of cDNA.
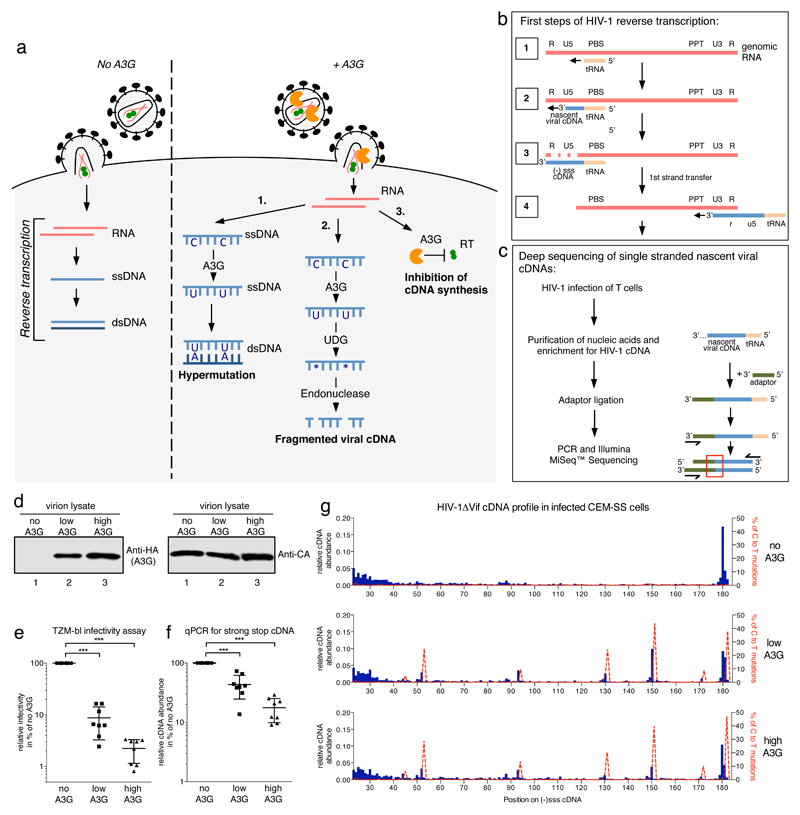
Figure 1. Effects of A3G on profiles of nascent HIV-1 cDNA products in infected T cells.
a) Early steps of the HIV-1 life cycle illustrating three proposed anti-retroviral mechanisms for A3G that are deaminase-dependent (pathways 1 and 2) or -independent (pathway 3). b) Diagram of HIV-1 reverse transcription. The first full intermediate, (-)sss cDNA, is completed in step 3. PBS: primer binding site, PPT: polypurine tract. c) Basic steps of sequencing library preparation. During infection, HIV-1 produces nascent viral cDNAs of increasing length (see step 2 in b). Sequencing reads reveal precise 3’-termini at the points of adaptor-viral DNA ligation (red box). d) Immunoblot analysis of HIV-1 virion lysates from one of six independent virus preparations. ‘Low’ or ‘High’ A3G refers to producer cell transfection ratios of 1:10 or 1:4, respectively (A3G expression plasmid to NL4.3/ΔVif). e) Single-cycle virion infectivity measured by β-galactosidase activity in challenged TZM-bl reporter cells. f) Quantitative PCR measuring cDNA abundance in CEM-SS cells at 4 h post-infection. For e) and f) the individual data points with their mean and standard deviation of eight independent infections from six virus preparations are shown. *** indicates p-value of <0.0001 in an unpaired, two-tailed t-test with Welch’s correction performed in GraphPad Prism®. g) Numbers of unique sequencing reads ending at each nt of the HIV-1NL4.3 (-)sss cDNA were divided by the total read number (Supplementary Figure 4b) within each sample to show the relative abundance of cDNAs for each length between nt positions 23 and 182. Shown in dashed red lines are the percentages of reads carrying C to T/U mutations at that position (scale on the right y-axis). See Method section for analysis details. One representative experiment out of three independent repeats is shown.
While A3-mediated hypermutation is well documented3,4, the two latter mechanisms for reducing cDNA accumulation remain controversial and poorly understood. Whether UDG-dependent processing of A3G-edited viral DNA occurs is unresolved. Although reducing levels of the UDG, UNG2, has been shown to diminish A3G anti-viral activity23,24, other groups have found that UDGs are dispensable for A3G function and the reduction of cDNA levels25–27. A3G’s deaminase-independent activity was originally noted when enzymatically-deficient A3G mutant proteins were shown to exhibit a degree of anti-viral function and to suppress cDNA production21,28.
A common observation from studies of the deaminase-independent effects of A3G is the increasing magnitude of inhibition over the course of reverse transcription and integration, which implies features of processivity. In addition to proposals that A3G inhibits specific steps, such as DNA strand transfers and integration27,29, we have therefore suggested more general interference with RT-catalyzed nucleic acid polymerization14,20. Conceptually, this could occur via a “roadblock” mechanism, in which A3G binds the template genomic RNA and sterically hinders RT translocation, or via direct interference with the biosynthetic capability of RT (or a combination of both). The first hypothesis is supported experimentally by in vitro reconstituted primer extension assays where A3G induces pausing/arrest at specific sites in the U5-R sequence22 and by nucleic acid binding studies using purified A3G30, whereas A3G’s interaction with RT may lend support to the second31.
Results
High throughput sequencing strategy for determining 3’-termini of HIV-1 reverse transcripts
We reasoned that defining populations of nascent viral cDNAs produced during HIV-1 infection would shed light upon the mechanism(s) of A3G-mediated viral inhibition. Here, we developed an Illumina MiSeq™ based deep sequencing method that enables unbiased mapping of the 3’-termini of reverse transcription products at single nucleotide resolution in infected T cells. This protocol focuses on early cDNA species, namely the minus strand strong stop ((-)sss) intermediate (comprising the U5 and R regions of the 5’-long terminal repeat) and early first strand transfer products32,33(Figure 1b). The strategy for library creation is depicted in Figure 1c (Supplementary Figure 1 and online methods). Briefly, T cells are infected with HIV-1, nucleic acids extracted and then enriched for viral DNA using capture oligonucleotides. The open 3’-termini of single stranded cDNA are ligated to a single stranded DNA adaptor (Supplementary Figure 1c), and PCR-amplified for sequencing. Barcoding is used to discard non-unique reads.
Profiles of cDNA intermediates in T cells infected with HIV-1ΔVif in the presence or absence of A3G
To investigate A3G’s effects on reverse transcription, HIV-1ΔVif virions were produced without, or with low or high amounts of packaged A3G (Figure 1d). Both infectivity, measured by challenging TZM-bl reporter cells (Figure 1e), and cDNA accumulation in CEM-SS cells at 4 h post infection, measured by qPCR of total DNA extracts (Figure 1f), inversely correlated with the level of packaged A3G. The suppression of cDNA production was characteristically less pronounced than the inhibition of infectivity20,29,34, presumably because the latter reflects the cumulative effects of deaminase-dependent and -independent activities.
The DNA extracts were then used to prepare libraries for sequencing (Figure 1c). We first analyzed cDNA 3’-termini: unique reads for each (-)sss cDNA 3’-end were counted and divided by total read number, which ranged between 26500 (high A3G) and 81000 (no A3G) (Supplementary Figure 4b), to obtain relative distributions of cDNA lengths. The top panel in Figure 1g depicts the profile for HIV-1ΔVif without A3G, with the most abundant species being the 180 nt (-)sss cDNA (>17% in this experiment). The rest of the profile was notably flat and evenly distributed, with some uplift in the abundance of shorter cDNAs. This pattern differs substantially from what is seen in reconstituted primer extension assays where DNA synthesis pauses at specific sites (Supplementary Figure 3a)35–37.
The presence of A3G resulted in the appearance of five prominent peaks, each representing a specific 3’-terminus (Figure 1g, middle and bottom panels). Importantly, these sites do not match A3G-induced pause sites seen in reconstituted reactions containing purified A3G (Supplementary Figure 3a, lanes 6 to 9)22. Instead, these sites lie one nt 5’ to cytidine-to-uridine mutations identified in longer DNA reads (dashed red lines), and occurring at consensus A3G editing sites5,16,38,39. These peaks also featured in first strand transfer sequences, but were absent when catalytically inactive A3G mutant proteins were tested (Supplementary Figures 4a and 5)28. These observations suggested that A3G editing hotspots serve as cDNA cleavage sites.
Target cell UBER enzymes mediate the detection and cleavage of A3G-edited HIV-1 cDNA
To test whether UDGs, specifically UNG240,41, mediate the processing of uridine-containing cDNA, leading to endonucleolytic cleavage, we utilized the bacteriophage uracil-DNA glycosylase inhibitor (UGI)42. CEM-SS target cells and HEK293T producer cells stably expressing codon optimized UGI (hUGI)25 were generated, the latter being required because UNG2 is packaged into virions43,44. Enzymatic analyses of cell lysates confirmed efficient UDG suppression (Supplementary Figure 6). HIV-1ΔVif was produced in control or hUGI expressing HEK293T cells in the absence or presence of A3G (Figure 2a). In bulk measurements of virus infectivity and cDNA abundance (Figures 2b and 2c), no differences were attributable to hUGI, irrespective of expression in producer, target or both cultures, reconfirming that UNG2 inhibition does not impact A3G anti-viral function25–27. Sequencing analysis revealed that UNG2 inhibition in target cells mitigated A3G’s induction of foreshortened cDNAs, with the overall profiles of 3’-termini resembling those seen without A3G (Figure 2d; and Supplementary Figure 7).
Figure 2. Consequences of UDG inhibition on A3G antiviral phenotype and cDNA profiles.
a) Immunoblot analysis of HIV-1 virion lysates showing increasing amounts of packaged A3G_HA at constant CA levels for virions produced in the presence or absence of a codon optimized (humanized) uracil-DNA glycosylase inhibitor (hUGI). ‘Low’ or ‘High’ A3G refers to a producer cell transfection ratios of 1:10 or 1:1, respectively (A3G expression plasmid to NL4.3/ΔVif). One of three independent sets of virus preparations used for b) and c) is shown. b) Virion infectivity was evaluated by challenging TZM-bl cells and measurement of β-galactosidase activity. c) The abundance of (-)sss containing cDNA in CEM-SS cells at 4 h post-infection was measured by quantitative PCR. For b) and c) each viral preparation was used to infect TZM-bl or CEM-SS target cells with or without hUGI, black dots and grey squares respectively. The individual data points with their mean and standard deviation for three independent viral preparations and infections are shown. d) Sequencing reads from a MiSeq™ library run were analyzed and presented as in Figure 1g. The labeling to the right indicates whether the HEK293T producer cells (Prod) and/or the CEM-SS target (Target) cells expressed hUGI. No A3G indicates the absence of A3G in producer cells and high A3G refers to relative A3G content in the producer cells. Sequencing data are derived from one representative experiment out of two independent repeats.
We therefore conclude: first, that target cell UNG2 can detect A3G-edited cDNAs leading to uracil removal and subsequent cleavage (Figure 1a, pathway 2); second, that UBER-mediated cDNA fragmentation does not lead to their complete degradation, implying that a deamination-independent mechanism likely underlies the inhibition of cDNA accumulation by A3G; and, third, that A3G does not induce site-specific RT pausing, thus arguing against the aforementioned “roadblock” mechanism for RT inhibition.
Interaction between A3G and the HIV-1 RT heterodimer
The 3’-termini profiles seen in the absence of A3G, in its presence but with UNG2 activity abrogated, or with catalytically inactive A3G mutants are very similar to each other, yet levels of cDNA production differ substantially. One model to reconcile these observations is for A3G to affect RT processivity through a mechanism that lacks template sequence specificity. This would yield similarly reduced nucleotide addition efficiency at all positions, regardless of RT location on the template RNA. We hypothesized that this could be achieved through A3G interacting with RT and inhibiting its enzymatic capability.
To evaluate this model, we initially used lysates from transfected HEK293T cells to confirm that A3G co-immunoprecipitates with the HIV-1 RT heterodimer (p51 and p66 subunits) (Figure 3a)31. Since A3G and RT are RNA binding proteins, we employed several techniques to ensure that this interaction is not simply bridged by RNA. First, co-immunoprecipiation assays were performed in the presence of ribonucleases (Figure 3b); although either a heterogeneous RNase mixture (upper panel) or RNase A (lower panel) noticeably reduced the amount of recovered A3G, the effect was saturable with A3G pull-down still remaining clear at high RNase levels.
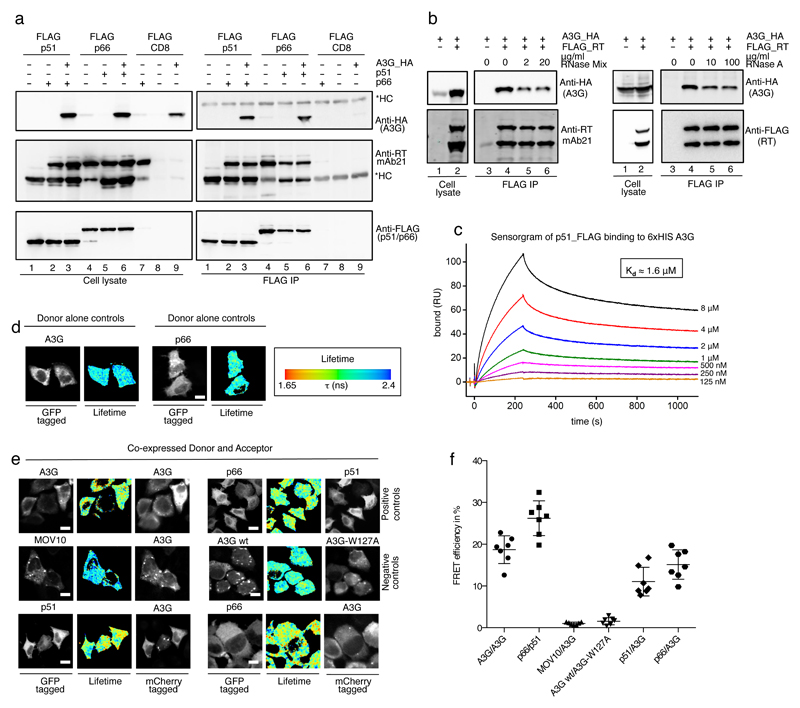
Figure 3. Interaction of A3G with HIV-1 reverse transcriptase.
Co-immunoprecipiation analysis of A3G_HA binding to FLAG tagged HIV-1 RT. Transfected HEK293T cell lysates were subjected to anti-FLAG immunoprecipiation, recovered proteins were detected with anti-HA (for A3G), anti-RT or anti-FLAG antibodies. CD8_FLAG served as an irrelevant protein control. One representative experiment of three repeats is shown. *HC: immunoglobulin heavy chain b) RNase resistance of the A3G-RT complex. Shown are anti-FLAG immunoprecipitations after the bead bound proteins had been subjected to RNase A or RNase Mix treatment, at the indicated concentrations, followed by washing and immunoblotting. One representative experiment of three repeats is shown. Samples without RT_FLAG carry CD8_FLAG as an irrelevant tagged protein control. c) Surface plasmon resonance analysis of purified A3G and p51 on a Biacore T-200 instrument. Association and dissociation curves of p51_FLAG to immobilized A3G_6xHis at the indicated concentrations are shown. The sensorgram indicates specific binding between the two components, and the responses gave good fits to a single interaction binding model with a Kd of ~1.6 μM. d)-f) Measurements of FRET efficiency using FLIM in HeLa cells expressing GFP and mCherry fusion proteins. Representative images with GFP fluorescence from multiphoton laser scanning microscopy (left panel) and corresponding wide field CCD camera images of mCherry fluorescence (right panels (e only)) are shown. The center panels represent pseudo-colored images of GFP lifetime (τ) (blue/green, normal/longer GFP lifetime; yellow/red, shorter GFP lifetime indicating FRET). d)Control images demonstrating normal GFP lifetime in the absence of mCherry acceptor. White scale bars represent 10 μm. e) Co-expression of indicated GFP and mCherry fusion proteins and the fluorescence lifetime according to the scale in d) indicating the presence or absence of FRET. f) Dot plot of FRET efficiencies with their mean and standard deviation from n=7 cells each.
We next carried out surface plasmon resonance (SPR) assays using purified p51 and A3G, both stripped of nucleic acids (Supplementary Figure 8). Measurements provided association and dissociation curves that gave acceptable fits to a single interaction model (Figure 3c), suggesting that the majority of complexes were homogeneous in nature. The association rate constant (kon) was calculated to 1.4 x 103 M-1s-1, and the dissociation rate constant (koff) to 2.2 x 10-3 s-1, yielding a dissociation constant (Kd) of ~1.6 μM; a value close to that calculated using rotational anisotropy (2.3 μM)45. Curve fitting indicated a maximal binding (Bmax) of ~200 RU of p51. Since the A3G ligand surface density is ~200 RU this is consistent with a 1:1 binding stoichiometry for the A3G-p51 complex. Taken together, these findings indicate that A3G and RT interact directly and that RNA may help to stabilize the complex.
Using cell-based FRET assays to measure the A3G-RT interaction
To address further the direct or indirect nature of the A3G-RT interaction, we turned to fluorescence resonance energy transfer (FRET) assays as they circumvent any need for protein purification or enrichment. Specifically, we employed green fluorescent protein (GFP, the donor) and mCherry (the acceptor) fusion proteins, co-expressed in HeLa cells, together with fluorescence lifetime imaging microscopy (FLIM) for the FRET readout. Here, the average lifetime (τ) of the donor is reduced when in a FRET state owing to the proximity of the acceptor (online methods)46. FRET is shown both as representative pseudocolored images with blue/green indicating longer τ than yellow/red colors (Figure 3d and e), and as efficiency percentages (Figure 3f).
Cells were transfected with vector pairs expressing the indicated fusion proteins, fixed and analyzed after 24 h. As expected, A3G_GFP localized to P-bodies, stress granules as well as the cytosol, while the RT subunits were more evenly distributed across the cytoplasm (Figure 3d and e). The oligomerization of A3G47–49 and hetero-dimerization of RT subunits provided positive controls, each visualized as yellow/orange coloration (Figure 3e, top row; and Figure 3f). The RNA helicase MOV10, which co-localizes to P-bodies with A3G, but is not thought to interact with A3G directly since co-immunoprecipitation depends on RNA bridging50 served as a negative control. No change in donor τ was observed, consistent with the lack of a direct A3G-MOV10 interaction. The low FRET efficiency displayed between A3G_GFP and the oligomerization-deficient W127A_mCherry mutant51 provided further assay validation(Figure 3e, second row). In clear contrast, co-expression of A3G_GFP with p51_mCherry or p66_mCherry yielded marked reductions in τ, demonstrating very close proximity between RT subunits and A3G (Figure 3e, third row), consistent with a direct protein-protein interaction.
A3G and RT interact within intact HIV-1 virions
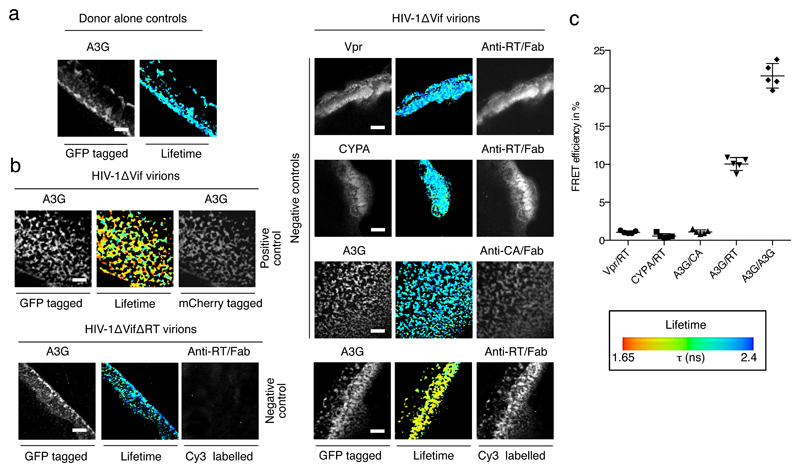
We also adapted our FRET-FLIM system for cell-free, bulk HIV-1 particles (Figure 4). A3G_GFP was packaged into virions by co-expression with HIV-1ΔVif, and these were purified and immobilized on coverslips, before immuno-staining and FRET analysis. Rather than using protein fusions that could interfere with viral assembly or particle, RT was detected using a Cy3-labelled anti-RT Fab fragment. A3G_GFP exhibits a normal GFP lifetime in the absence of a fluorescent acceptor (Figure 4a) and a positive control of co-packaged A3G_GFP and A3G_mCherry demonstrates the suitability of this assay for detecting protein-protein interactions within virions (Figure 4b, left panel, top row; and Figure 4c). A substantial increase in FRET efficiency was evident when RT was detected with labeled Fab (Figure 4b, right panel, bottom row; and Figure 4c), which was not seen with other packaged proteins as acceptors (Vpr_GFP or cyclophilin A_GFP), the antibody-mediated detection of Capsid protein (p24Gag) (Figure 4b, top right panels; and Figure 4c), or for virions engineered to lack RT (Figure 4b, left panel, bottom row). These data demonstrate that A3G and RT interact in viral particles, and support the view that A3G is positioned at the site of reverse transcription.
Figure 4. A3G interaction with HIV-1 RT in virions.
Suspensions of HIV-1 virions with packaged A3G_GFP, GFP_Vpr, GFP_CYPA or A3G_GFP and A3G_mCherry were immobilized on coverslips, fixed and stained with Cy3 labeled anti-RT or anti-CA Fab fragments. a) and b) Representative images show clusters of HIV-1 virions immobilized on fibronectin streaks with green fluorescence (left panel), red fluorescence (Cy3 or mCherry as indicated, right panel) and GFP lifetime as pseudo-colored images according to the indicated scale (as in Figure 3). White scale bars represent 10 μm. a) A3G_GFP demonstrates normal lifetime when packaged into HIV-1 virions. b) FRET is detected for the positive control of A3G_GFP and A3G_mCherry (upper left panel) and between A3G_GFP and Cy3 stained RT (lower right panel), but not between Vpr and RT, CYPA and RT, or A3G and CA (upper right panels). The absence of a signal for red fluorescence with HIV-1ΔRT virions confirmed the specificity of the anti-RT Fab fragments (lower left panel). c) Quantification of FRET efficiencies for n=5 areas. Individual measurements with their mean and standard deviation are shown.
Regions of A3G that interact with HIV-1 RT
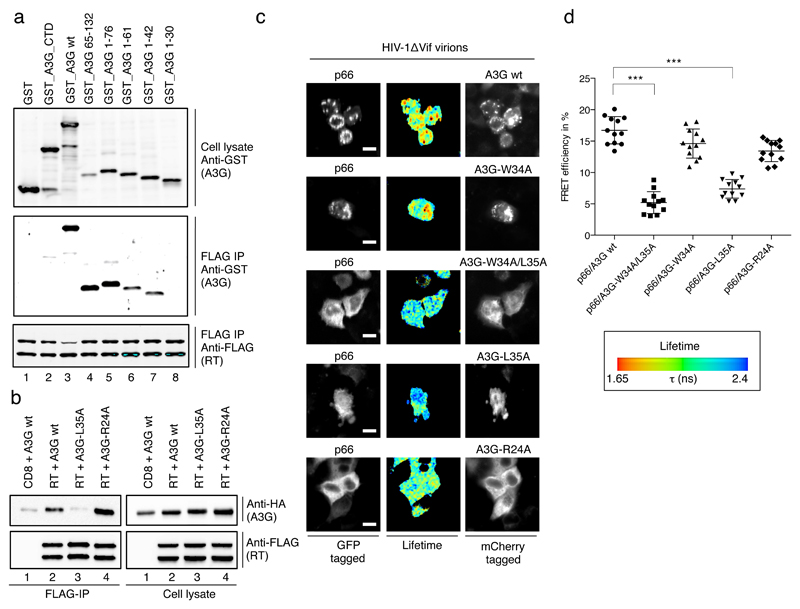
Regions of A3G that interact with RT were mapped by co-expression of RT with a set of A3G-glutathione-S-transferase (GST) fusions and co-immunoprecipitation (Figure 5a). Consistent with earlier results31, full length A3G and a fragment spanning residues 65 to 132 interacted with RT (lanes 3 and 4). Interestingly, the N-terminal 76 amino acid fragment of A3G also bound RT (lane 5), with further truncations revealing dependency on the residues between positions 30 and 42 (lanes 7 and 8). Thus, RT interacts with two discrete and non-overlapping regions of A3G.
Figure 5. Mapping of A3G-RT interaction sites on A3G protein.
a) Anti FLAG immunoprecipitation of p51_FLAG and p66_FLAG co-expressed with GST or GST_A3G fusion proteins, recovered proteins were detected with anti-GST (for A3G) or anti-FLAG antibodies as indicated. A3G truncations are indicated and numbers refer to amino acid positions in A3G. b) Co-immunoprecipitation analysis of wild type or mutant A3G with HIV-1 p51_FLAG and p66_FLAG, recovered proteins were detected with anti-HA (for A3G) or anti-FLAG antibodies. One representative out of three experiments is shown. c) FRET-FLIM analysis of wild type or mutant A3G with the p66 subunit of HIV-1 RT. Representative images show green fluorescence (GFP, left panel) and red fluorescence (mCherry, right panel) and GFP lifetime as pseudo-colored images according to the indicated scale (as in Fig 5). White scale bars represent 10 μm. d) Dot plots showing individual FRET efficiencies with their mean and one standard deviation from n=12 cells each. *** indicates p-value of <0.0001 in an unpaired, two-tailed t-test performed in GraphPad Prism®.
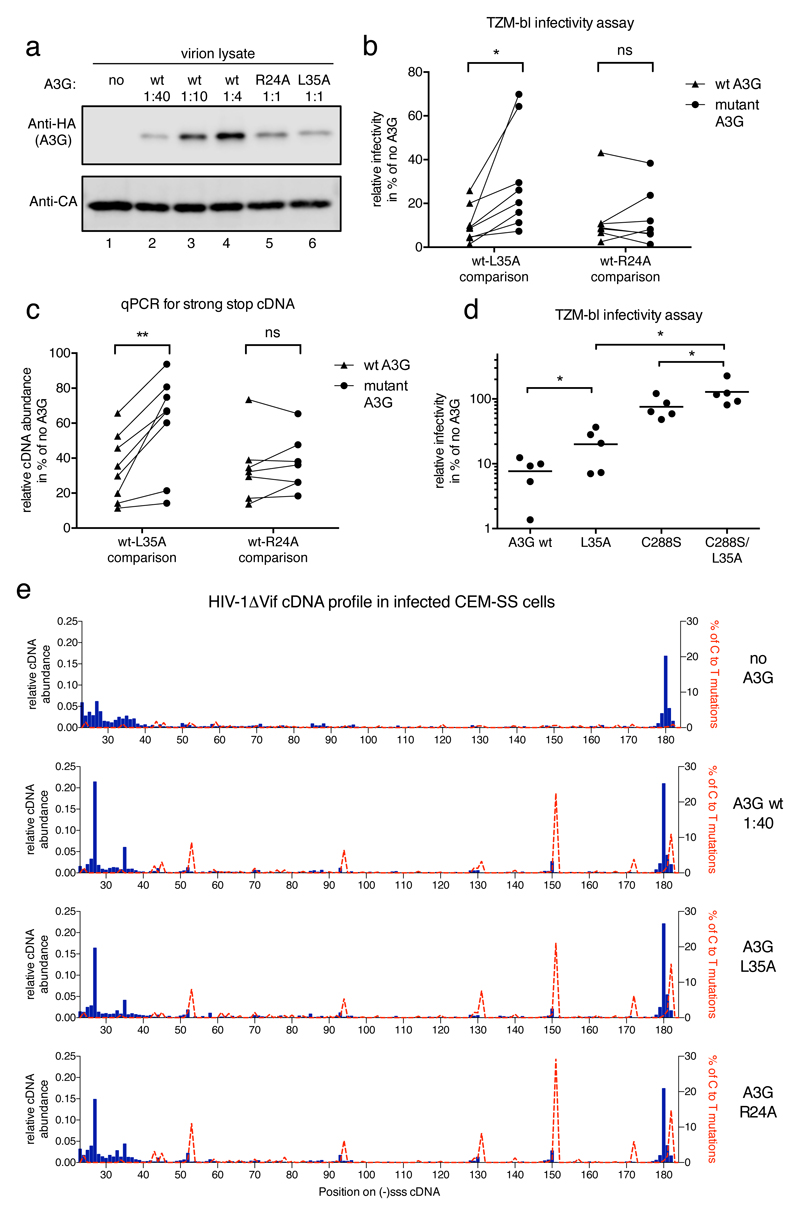
Double alanine scanning mutagenesis between residues 30 and 42 in the context of full length A3G showed that a W34A/L35A double mutant, and subsequently the single L35A mutant, were impaired for interacting with RT (Figure 5b). These mapping data were confirmed in intact cells, as the substitution of leucine at position 35 with alanine (W34A/35A and L35A mutants) significantly reduced FRET efficiency (Figure 5c, row 3 and 4 and Figure 5d). As a control, the R24A mutation in A3G, which disrupts a positively charged patch at the A3G dimer interface52 and inhibits RNA binding51 (Supplementary Figure 3c) was shown not to alter binding to RT (Figure 5b, c and d), thereby reinforcing the conclusion that the A3G-RT interaction is RNA independent. Importantly, like R24A51, the L35A protein maintains full enzymatic activity as measured in an E. coli-based DNA editing assay (Supplementary Figure 9), indicating that these substitutions do not cause global structural defects or misfolding.
Inhibition of the RT interaction diminishes A3G anti-viral activity
Derivation of a mutation in A3G that interferes with RT binding but maintains deaminase function allowed us to evaluate directly the importance of the A3G-RT interaction for anti-viral activity. The L35A protein, similar to R24A51, is not packaged into viruses as efficiently as wild type A3G; however, this defect is partial and is overcome by increasing protein expression in virus producer cells.
For these experiments, we used titrations of wild type A3G in producer cells to yield viruses with a range of packaged A3G levels (Figure 6a; Supplementary Figure 10a). These stocks were then used to infect TZM-bl cells to determine overall infectivity (Supplementary Figure 10b), and CEM-SS cells to measure cDNA production (Supplementary Figure 10c); fitted curves were generated that related infectivity or cDNA levels to amounts of packaged A3G (Supplementary Figure 10d and 10e).
Figure 6. Phenotypes of packaged L35A and R24A A3G mutant proteins on viral infectivity and cDNA profiles.
a) Immunoblot analysis of HIV-1 virions showing relative amounts of packaged wild type or mutant A3G_HA at constant CA levels. Ratios refer to the amounts of transfected A3G expression plasmid to proviral plasmid during virus production. b) A3G-L35A, but not A3G-R24A, displays diminished HIV-1 inhibitory activity. A3G packaging was quantified by immunoblot density measurements and the different wild type A3G packaging levels were plotted over measured infectivity. The extent of infection inhibition exerted by the wild type protein at the empirically determined level of packaged mutant protein was then extrapolated (see Supplementary Fig 10). Inhibition levels, in % relative to the no A3G control, of wild type A3G (triangles) and L35A or R24A (circles) in eight (L35A) or seven (R24A) independent experiments are shown. A paired, two tailed student t test was performed in GraphPad Prism® and * indicates p<0.05 (p=0.0223), ns: not significant c) As in b), but with (-)sss cDNA abundance measured by qPCR in cells 4 h post-infection for virions carrying wild type or mutant A3G. ** indicates p<0.005 (p=0.0028). d) Relative infectivity of n=5 independent virus preparations carrying the indicated wild type or mutant A3G at equal, ‘high’ levels as shown in the representative immunoblot in Supplementary Fig 10g. A paired, two tailed student t test was performed in GraphPad Prism®. * indicates p<0.05 (p=0.0397 for A3G wt – L35A; p=0.0297 for C288S – C288S/L35A and p=0.0137 for L35A – C288S/L35A). e) Sequencing reads from a MiSeq™ library run were analyzed and presented as in Fig 1g. Labels to the right indicate the presence or absence of A3G proteins in virions.
Viruses carrying the L35A or R24A mutants were prepared in parallel and analyzed for packaging, infectivity and cDNA production. By extrapolation along the fitted curves, we compared their effects on infectivity (Figure 6b) and reverse transcription (Figure 6c) with those of wild type A3G at matched packaging levels. Over eight experiments, L35A was significantly less anti-viral than A3G, and this was coupled with increased cDNA production. Critically, R24A displayed equivalent anti-viral activity to wild type A3G at comparable levels of encapsidation, thus phenotypically distinguishing itself from L35A. Moreover, the finding that R24A still suppresses reverse transcription (Figure 6c), while binding RNA less well (Supplementary Figure 3)51, is again consistent with RT suppression not being mediated by the “roadblock” mechanism.
To test whether the remaining anti-viral activity of L35A is deaminase dependent, we combined this change with a mutation, C288S, that ablates deamination but incompletely prevents anti-viral activity (Figure 6d; and Supplementary Figures 5, 10g and 10h)27,28. Importantly, the double mutant (C288S/L35A) displayed a total loss of anti-viral function even at high expression levels (Figure 6d), underscoring the importance of both attributes for full A3G activity.
Lastly, we analyzed the cDNA profiles in CEM-SS cells challenged with HIV-1ΔVif lacking A3G or containing similar levels of wild type A3G, R24A or L35A (Figure 6a and e). The three profiles obtained with A3G-bearing viruses were essentially identical, confirming the editing competency of these mutant proteins and the absence of any site-specific pausing that correlated with inhibited cDNA synthesis. We therefore conclude that A3G regulates HIV-1 RT through a direct interaction that impedes enzymatic capability and suppresses the production of DNA evenly and without sequence or site specificity (Figure 1a, pathway 3).
Discussion
Here, we present a custom method for monitoring the progression of HIV-1 reverse transcription at single nucleotide resolution in infected cells. Our findings have relevance for understanding not only A3-mediated restriction, but also fundamental aspects of reverse transcription. In particular, much of the current knowledge of reverse transcription has been obtained using in vitro reconstituted systems32,33,53,54 (e.g., Supplementary Figure 3). In agreement with such studies, we demonstrate a marked accumulation of a predominant (-)sss species (e.g., Figure 1g)35,55,56, yet defining its precise 3’-terminus has previously been elusive. The abundant (-)sss cDNA we detect has a single 3’-cytidine (nt position 180), and we presume this to be derived from the CapG1-form of HIV-1 RNA. This is consistent with the preferential dimerization and packaging of CapG1 RNA that has a single guanosine at its 5’-terminus, rather than the CapG2- or CapG3-forms that contain two or three 5’-guanosines57,58.
In contrast to the distinctive (-)sss product seen in living cells and reconstituted systems, the foreshortened cDNAs that are detected in reconstitution experiments are negligible in infected cells (Figure 1g; Supplementary Figure 3). Their formation has been ascribed to RT pausing/arrest at homopolymeric motifs or RNA secondary structures35–37,59. These reproducible disparities may reflect: variations in the enzymatic capabilities of HIV-1 RT in the microenvironment of reverse transcription complexes in infected cells versus in reconstituted reactions; differences in the formation of viral genomic RNA secondary and tertiary structures; or the presence or relative abundance of viral (or host) proteins such as the HIV-1 Nucleocapsid protein at the site of reverse transcription.
Not surprisingly, experimental system dependent differences extend to the effects of A3G on reverse transcription. Studies with reconstituted assays had indicated A3G-induced pausing of RT at specific sites22 (Supplementary Figure 3a), which helped formulate the “roadblock” model. Critically, our cell-based DNA sequencing approach provided no evidence for localized A3G-induced RT pausing, with the prominent A3G-dependent cDNA peaks being caused by UBER activity (Figure 2). These findings evoke a sequence- and site-independent mechanism for the suppression of RT by A3G, and imply: first, that there are differences in RT’s behavior in the context of HIV-1 infection; and, second, that the A3G-induced pausing that is seen in reconstituted reactions reflects an assay-specific epiphenomenon.
Intrinsic to our model for A3G function is its negative regulatory interaction with RT, which, based on a series of corroborating biochemical, biophysical and cell-based FRET assays (Figures 3 to 5)31, we consider to be dependent on direct binding. Purposeful interference with this interaction through replacement of leucine-35 resulted in a significant loss of anti-viral activity (Figure 6b and c), with the remaining anti-viral activity of L35A being attributable to its deaminase function since the C288S/L35A double mutant displayed a complete loss of anti-viral phenotype (Figure 6d).
Our sequencing approach also allowed us to examine the effects of UBER on viral cDNA following exposure to A3G. Although, we reveal UNG2-mediated uridine excision from deaminated cDNA, our measurements show ensuing endonucleolytic cleavage is far from complete since substantial proportions of cytidine-to-uridine edited cDNAs remained intact. (Figures 1, 2 and 6). Appreciating this inefficiency helps to reconcile the apparent discrepancy between the predicted degradative fate of deaminated cDNAs (Figure 1a, mechanism 2) and the lack of persuasive evidence for UDG involvement in A3G function. Nevertheless, despite the absence of an evident anti-viral effect in single round infections (Figure 2b), inefficient UBER recognition of edited cDNA could still play a role in the interplay between HIV-1 and infected cells, for instance through sensing of aberrant cDNA fragments as a pathogen-associated molecular pattern1,60. How HIV-1 may circumvent this (aside from Vif induced A3 destruction in virus producing cells) is a matter of conjecture, but we note that HIV-1 Vpr and its interacting CUL4 ubiquitin ligase induces UNG2 degradation23,44,61.
Uracilation of HIV-1 cDNA can also arise from the misincorporation of dUTP in place of dTTP during reverse transcription62, especially in non-dividing cells such as macrophages that have high dUTP:dTTP ratios63,64. UNG2-mediated recognition of the resulting uracilated viral cDNA suppresses infection, with restriction being most evident at the level of DNA integration65–68. Interestingly, and in contrast to observations made with A3G-induced uracilation (Figure 2, Supplementary Figure 7)25–27, UGI-mediated suppression of UNG2 in the face of misincorporation-driven uracilation provokes viral rescue65,66. The basis for this dichotomy remains to be determined, but could relate to variations in the extent of uracilation, or cell type dependent differences in the efficiency of UBER-mediated cDNA recognition or the fate/processing of uracilated cDNA.
Reverse transcription is a vulnerable step in the HIV-1 life cycle: it is highly susceptible to pharmacological inhibition, and is also suppressed by A3G and other host cell restriction factors. TRIM5α destabilizes post-entry viral capsids which compromises the timely onset/completion of reverse transcription69–71, while the deoxynucleotide triphosphohydrolase SAMHD1 interferes with reverse transcription by depleting cellular dNTP pools2. In contrast to these indirect mechanisms, we demonstrate that RT is inhibited by A3G by a distinct mechanism, namely via a direct protein interaction that impedes biosynthetic activity. Establishing the concept that HIV-1 RT can be negatively regulated in this manner raises the possibility that additional cellular factors may modulate RT72.
Future directions for this work include determining: how A3G binding to RT interferes with enzymatic function, how leucine-35 (a residue that is highly conserved within the A3 family) participates in the RT interaction, and which regions of RT bind to A3G. Elucidating the macromolecular structure of the A3G-RT complex will naturally be a major advance, and can assist in resolving the heretofore elusive structure of full-length A3G73. Finally, by revealing that a non-virally encoded interacting partner can regulate HIV-1 RT, we highlight a previously unrecognized vulnerability in the enzyme that may have the potential to be targeted therapeutically.
Methods
Plasmid constructs
The expression vector for Vif-deficient HIV-1NL4.3 has been described previously1. Wild type and mutant A3G proteins carrying the carboxy-terminal 3xHA tag were expressed using pCMV4_HA2,3. Site-directed mutagenesis was carried out based on the QuikChange protocol (Stratagene) using Pfu polymerase (Stratagene). Transfer plasmids for the generation of recombinant baculovirus were generated as follows: wild type or mutant A3G sequences were PCR amplified and cloned into pCGTHCFFLT7 with a carboxy-terminal 6xHis tag sequence using XbaI and AgeI restriction sites. The tagged A3G insert was subsequently PCR amplified and inserted into the pVL1392 transfer vector (BD Biosciences) using EcoRI and XmaI restriction sites. Expression vectors for HIV-1 RT p51 and p66 were created as follows: a codon optimized gag-pol plasmid (pCO Gag-pol,4) was used as the PCR template, and. fragments were subcloned into pCAGGS using XmaI/NotI (p51) or EcoRI/XmaI (p66). The reverse primers were designed with or without a 1xFLAG tag coding sequence. Expression vectors encoding truncated A3G proteins with amino-terminal GST tags were generated by PCR amplification and subcloned into pCAGGS_GST with EcoRI/XhoI. Fluorescent protein fusions were expressed from plasmid constructs carrying the respective cDNA sequence from the aforementioned vectors and subcloned in pEGFP_N1 (Clontech) and pmCherry_N1 (Clontech) vectors. The cDNA for the (human) codon optimized uracil-DNA glycosylase inhibitor (hUGI) gene was a gift from Michael Emerman5 and was subcloned into the MLV-based transfer vector pCMS286. MLV packaging and VSV-G expression vectors have been described7.
Cell lines and stable cell lines
HEK293T and HeLa cells were obtained from ATCC. TZM-bl cells were obtained through the NIH AIDS Reagents Repository Program (ARRP). Cells were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen, UK) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. CEM-SS cells, also from ARRP, were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Invitrogen, UK) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. All cells were tested and found to be negative for mycoplasma contamination. hUGI expressing HEK293T, CEM-SS and TZM-bl cells were created using standard MLV-based transduction vectors. HEK293T cells were co-transfected with expression plasmids for VSV-G, an MLV packaging plasmid and pCMS28 encoding hUGI. Transduced cells were selected with 1μg/ml puromycin.
Virion production
HEK293T cells were seeded in 10 cm tissue culture plates and co-transfected with 10 μg pNL4.3/ΔVif and 0.25 to 10 μg pCMV4_A3G_HA wild type or mutant expression vectors using polyethylenimine (PEI, Molecular Biosciences). Total DNA levels were kept constant with empty pCMV4-HA vector. At 48 h, supernatants were harvested, DNAse (RQ1 RNase free DNAse (Promega)) treated for 1 h at 37°C and viruses then purified through a 20% (w/v) sucrose cushion at 28,000 x g for 75 min at 4°C. Viruses were resuspended in PBS, and quantified according to p24Gag content using an enzyme-linked immunosorbent assay (ELISA; Perkin-Elmer).
TZM-bl reporter assay
TZM-bl reporter assays were utilized to measure single cycle HIV-1 infectivity. Virus inputs equivalent to 10 or 20 ng p24Gag were used to challenge 0.5 x 105 cells in 48-well format. At 48 h post infection, whole-cell lysates were assayed for the induction of β-galactosidase expression using a Galacto-Star system (Applied Biosystems).
Infections
CEM-SS T cells were seeded at 1 x 106 cells per 0.5 ml of medium in a 12-well format. Virus equivalent to 75 ng p24Gag was added and the cells were spin-infected at 2,000 x g for 2 h at 30°C. After centrifugation, cells were left to recover for 1 h in the incubator, before 2 washes with PBS to remove the inoculum. Cells were maintained in fresh media for 1 h before being harvested by centrifugation, and pellets either frozen at -80°C or immediately processed for DNA extraction.
DNA extraction and quantitative real time PCR
Whole cell DNA was isolated using the DNAeasy Blood and Tissue Kit (Qiagen) according to the manufacturer’s protocol. DNA was eluted with 200 μl H2O. 17.5 μl were mixed with 2 μl Cutsmart buffer (NEB) and 0.5 μl DnpI (NEB) (to eliminate any remaining transfected plasmid) for 1 h at 37°C before analysis by quantitative PCR. The remaining nucleic acid fraction was reserved for library preparation. Early reverse transcription products were detected using primers that amplify the region between nucleotides 500 and 635 of the provirus: oHC64 (5’-taactagggaacccactgc-3’) and oHC65 (5’-gctagagattttccacactg-3’) and probe oHC66 (5’-FAM-acacaacagacgggcacacacta-TAMRA-3’). Reactions were performed in duplicate, in TaqMan Universal PCR master mix (no AmpErase), using 0.9 pmol of each primer per μl and 0.25 pmol probe per μl. After 10 min at 95°C, reactions were cycled through 15 s at 95°C, followed by 1 min at 60°C for 40 repeats, carried out on an ABI Prism model 7900HT (Applied Biosystems). pNL4.3/ΔVif, was diluted in herring sperm DNA to generate a standard curve.
Statistical analysis
Where indicated a paired or unpaired, two-tailed t-test was carried out using the t-test function in GraphPad Prism® 6.0.
MiSeq library preparation
Enrichment for HIV-1 specific sequences
Whole cell DNA from infected cells was isolated as above and HIV-1 specific DNA was enriched over the genomic DNA. Biotinylated oligonucleotides complementary to the HIV-1 sequence (within the tRNA primer and within the first 22 nt of the strong stop sequence, see exact sequences below) were annealed to the cDNA sample and a pulldown with magnetic strepdavidin beads was performed: for each sample, 100 μl strepdavidin beads (Promega, Magnasphere (Z5481) were washed (using a magnet) with 100 μl of bind and wash buffer (BW: 5 mM Tris pH 7.5, 0.5 mM EDTA, 1 M NaCl) then blocked for 10 min in casein solution at room temperature, and again washed once in BW buffer. Beads were resuspended in BW buffer containing 50 pmol 5'-cagtgtggaaaatctctagcag-biotin-3’, 5'-cagtggcgcccgaaca-biotin-3’, and 5'-biotin-cagtgtggaaaatctctagcagtggcgcccgaacagggac-biotin-3’. The mixture was left for 30 min at room temperature while rotating. Beads were then washed twice in 1xTEN buffer (10 mM Tris HCl pH 8.0, 1 mM EDTA, 100 mM NaCl) and then resuspended in 10 μl TEN buffer. This was added to 170 μl DNA and 90 μl 3xTEN. Samples were denatured at 92°C for 2 min and incubated at 52°C for 1 h. Finally, beads were washed once with 1xTEN buffer and resuspended in 50 μl H2O. To elute the DNA, samples were heated to 92°C for 2 min, put on the magnet and the supernatant removed quickly.
Adaptor ligation
The barcoded adaptor carries a 5’ phosphate group (PHO) and a 3’ three carbon chain (C3) spacer (SpC3) on to the 3’ hydroxyl group (5’-PHO-tgaagagcctagtcgctgttcannnnnnctgcccatagagagatcggaagagcacacgtct-SpC3-3’) (Integrated DNA Technologies or MWG Eurofins) and was self annealed in T4 DNA ligase buffer by heating to 92°C followed by slow cooling to 16°C (2% slope on Eppendorf PCR machine). Ligation reactions were set up in 60 μl total volume, with 6 μl T4 DNA ligase buffer, 24 μl 50% PEG-8000 (Sigma), 6 μl 5 M betaine (Sigma), 4 μl pre-annealed adaptor (400 pmol total), 1.2 μl T4 DNA ligase (NEB, 2,000,000 units/ml) and 18.8 μl DNA sample. Reactions were incubated at 16°C overnight. As controls, instead of DNA samples, HTP control oligos (listed in Supplemental Figure 2) at 100 pmol/μl were mixed at equimolar ratios and then diluted 1 in 62,500 before being ligated as above.
Adaptor removal and size selection
Ligations were denatured by adding 30 μl gel loading buffer II (Ambion, formamide buffer) and heating for 2 min at 94°C before being put on ice. 6% TBE urea gels (precast, Life Technologies) were prerun in 1xTBE buffer for ~15 min at 250 V, and each sample was loaded into three adjacent wells. Gels were run for 20 min at 250 V before gel strips containing samples were immersed in 1xTBE and SYBR Gold (Thermo Fisher). To avoid cross-contamination between samples during staining and gel cutting, a maximum of two samples were run per gel and gels were cut for separate staining. After 5 min, DNA was visualized on a visible blue light LED transilluminator (Dark Reader, Clare Chemicals), gels were cut right above the adaptor and the smear between the adaptor and the well was divided into three even pieces. Each piece was crushed and 1 ml urea gel extraction buffer (0.5 M NH4CH3CO2, 1 mM EDTA, 0.2% SDS) was added. Tubes were rotated for 3 to 5 h and the suspensions were transferred into SpinX columns (0.2 μm, Acetate; Corning Costar) with a round whatman filter to prevent membrane clogging. Columns were centrifuged at 14 000 x g for 1 min. The elutions were combined with 3 μg polyA carrier RNA, 1 μl glycogen, and 0.7 ml isopropanol. Samples were frozen overnight, centrifuged for 30 min at 14 000 x g and DNA pellets were washed with 80% EtOH. Any remaining EtOH was evaporated at 55°C. Dried pellets were resuspended in 20 μl H2O.
PCR and preparation for MiSeq™ run
PCR reactions contained 20 μl Accuprime Supermix I (Invitrogen), 18 μl DNA, 1 μl of 10 μM MP1.0+22HIV, a sequence modified multiplexing PCR primer 1.0 (MP1.0 (Illumina)) (5’-aatgatacggcgaccaccgagatctacactctttccctacacgacgctcttccgatctcactgctagagattttccacactg-3’) from MWG Eurofins and 1 μl NEB index primer (NEBNext for Illumina Multiplex Oligo Kit). For each of the three gel pieces from the same sample, separate PCR reactions, but the same index primer was used. A 25 cycle PCR was performed at 55°C annealing and 68°C extension. Half of each PCR reaction was pooled together and purified with AmpureXP beads (1.8x ratio) according to the manufacturer’s instructions. Concentrations were determined using a Qubit Fluorometer (Invitrogen) and the size range was determined by Tapestation (Agilent) measurements. Typical tapestation gel images showed material to be between roughly 150 bp to 500 bp long with two wide peaks at about 160 bp and 350 bp. On initial library preparation optimization, TopoTA cloning and individual sequencing of 10 or more colonies was performed to check inserts. The final library concentration was adjusted to 4 nM.
MiSeq™ library runs and data analysis
Libraries were run on an Illumina MiSeq™ Benchtop Sequencer using the MiSeq™ Reagent Kit v3 (150 cycle) and the ‘Generate FASTQ’ workflow at University College London (UCL) Institute for Neurology Next Generation Sequencing Facility or at the King’s College London Genomics Centre. Read lengths were set at 26 bases for Read1 and 142 or 125 bases for Read2. The two libraries from which data are shown in this article had passfilter percentages (PF%) of 93.68% and 93.11% with quality scores (AVG % Q30) of 89.05% and 82.45%, respectively.
The main information extracted from each read were: first, the last nucleotide of the HIV-1 sequence adjacent to the fixed adaptor sequence, which was ligated to the viral cDNAs (see Figure 1c and Supplementary Figure 1b). This represents the open 3’-terminus of the viral cDNA at time of harvest and, second, the base variation of all bases, in particular C to T mutations. For this purpose FASTQ files were subjected to in house analysis. Adaptors were trimmed and sequences that were duplicated (including the barcode) were removed as PCR artifacts. The remaining sequences were aligned to the HIV-1 sequence using Bowtie (http://bowtie-bio.sourceforge.net/index.shtml), allowing a maximum of 3 base mismatches, and the position of 3’-termini for each read was determined from the alignment position. Mutation rates from the template sequence for each base were also calculated. Where required, a linear length-dependent correction factor was calculated from synthesized oligos control library (see Supplementary Figure 2) and applied to the dataset to correct for differences in sequencing efficiency of longer products.
Output files for each indexed sample were designed to yield the number of total unique reads for each nucleotide position in the (-)sss product of the HIV-1NL4.3 proviral sequence. The template that was used spanned the strong stop and first strand transfer sequences up to the polypurine tract (U5-R-U3-PPT; 635 bases total). Of note, only sequences of a minimal length of 22 nt are accounted for in this library due to primer design. Accordingly, there were 613 possible termini for the cDNA sequences in total, though our main analysis focuses on the 160 nt up to base 182, the strong stop site. For base substitution analysis, parse results presented the number of each of the four possible base calls for each nucleotide position. Percentages could therefore be calculated from the total coverage of each base. Notably, coverage for each base itself is not constant for two reasons: one, the 3’-termini of individual reads vary, thereby coverage ends at varying sites; and, two, Read2 sequence length of 142 or 125 (depending on the library) may not reach the 5’ end of strong stop, depending on the starting position.
Relative abundance of cDNA along the (-)sss sequence in the main figures was calculated by dividing the number of total reads for each nt position by the number of total reads up to nt 182 (for total reads see Supplementary Fig 4b). The sole exception is Supplementary Figure 4a, which shows profiles beyond first strand transfer, where the read number was divided by the total read count in the entire sample. All figures displaying cDNA profiles (Fig 1g, 2d, 6e, and Supplementary Fig 2a, 4a, 5 and 7) show the relative abundance of HIV-1 cDNA molecules for each length between nt positions 23 and 182 of the HIV-1NL4.3 (-)sss product (in blue histogram bars, scale on the left y-axis). All positions with cytosine bases in the HIV-1NL4.3 (-)sss sequence were analyzed for the presence of cytosine versus thymine/uracil bases as described above; shown in dashed red lines is the percentage of reads, which carried C to T/U mutations at the indicated position (scale on the right y-axis). Labels to the right of the graphs describe the virions used for infection.
Co-immunoprecipitation assays
HEK293T cells were transfected in 6 well format using PEI. At 36 h, cells were scraped in ice-cold PBS, pelleted (2 min, 500 x g) and lysed in DMEM (Gibco) + 0.5% IGEPAL CA 630 (Sigma) plus protease inhibitor cocktail (Roche). After 10 min on ice, cell lysates were sonicated for 10 s, and clarified by centrifugation at 1000 x g for 10 min. Magnetic protein G Dynabeads (Invitrogen) were washed twice in PBS using a magnetic stand and then pre-incubated with FLAG_M2 antibody (Sigma). The charged magnetic beads were washed once, distributed to the different cell lysates and rocked at 4°C for 2 h. Beads were washed four times with DMEM + 0.5% IGEPAL CA 630 and proteins then eluted with protein loading buffer (according to Laemmli), resolved by SDS PAGE and analyzed by standard immunoblot analysis. Antibodies used include anti-HA-HRP (3F10, Roche), unconjugated anti-HA (3F10, Roche, )anti-FLAG M2-HRP (Sigma), anti-RT (mAb21, NIH AIDS Reagents Repository Program (ARRP)), anti-p24Gag (mouse monoclonal, 24-28) and anti-GST-HRP (RPN1236, Sigma). Horseradish peroxidase-conjugated primary or secondary antibodies were visualized by enhanced chemiluminescence (Pierce) and unconjugated primary antibodies were detected by infrared IRDye-conjugated secondary antibodies (LI-COR Biosciences) and LI-COR infrared imaging technology (LI-COR UK Ltd.) All blots in their un-cropped versions and including molecular weight markers are presented in Supplementary Fig 11.
For co-immunoprecipitation experiments with FLAG_RT and GST_A3G truncations, 50 mM NaCl was added to DMEM + 0.5% IGEPAL CA 630 during washes for more stringency. Additionally, proteins were eluted after the 2 h incubation by competition with 3xFLAG peptide (150 μg/ml) for 30 min instead of direct elution by protein loading buffer.
For co-immunoprecipitation coupled with RNase treatment, all starting materials were doubled and the reactions were carried out as above until the 2 h incubation. Magnetic beads were washed twice in DMEM + 0.5% IGEPAL CA 630 and reactions were split in three parts with one part left untreated and RNaseA (Sigma) or RNase Mix (Roche) added to the two remaining reactions at concentrations up to 100 μg/ml or 20 μg/ml, respectively. All reactions were incubated at room temperature for 30 min while rocking and then washed two more times in DMEM + 0.5% IGEPAL CA 630 before elution in protein loading buffer.
Surface plasmon resonance
Purification of p51 for surface plasmon resonance experiments
HIV-1 p51 RT was purified from 20 10 cm dishes of HEK293T cells transfected with p51_FLAG using anti-FLAG magnetic affinity resin (Sigma). Briefly, at 48 h after transfection, cells were lysed in 50 mM Tris HCL 7.6, 150 mM NaCl, 0.5% Triton, 1 mM EDTA supplemented with 50 μg/ml RNase A (Sigma) and protease inhibitor (Roche), and incubated for 10 min on ice. Lysates were sonicated, clarified by centrifugation, and incubated with anti-FLAG M2 magnetic beads (Sigma) for 3 h. The beads were washed extensively with lysis buffer and eluted three times with lysis buffer containing 150 μg/ml 3xFLAG peptide (Sigma). Elutions were combined and dialyzed against PBS + 0.01% IGEPAL CA 630 (Sigma) to remove the peptides. Samples were concentrated to 10 μM as determined by Bradford assay, and homogeneity was judged by Coomassie staining (see Supplemental Figure 8).
Surface plasmon resonance experiments
p51_FLAG was diluted to 8 μM before preparing a 2-fold dilution series down to 0.16 μM. A3G was purified as below, dialyzed against PBS + 0.01% IGEPAL CA 630 and used at a final concentration of 10 nM. Interaction analyses were performed on a Biacore T200 instrument (GE Healthcare). Binding surfaces for 6xHis tagged A3G were created by immobilizing an anti-His antibody (Biacore/GE Healthcare) onto a Series S CM5 sensor chip surface (Biacore/GE Healthcare) by amine coupling. The capture of the A3G ligand was carried out at low surface density (~200 RU) to minimize potential A3G-A3G interactions and ensure monomeric interactions with p51. The p51 analyte was injected at different concentrations, each in duplicate, in running buffer (PBS + 0.01% IGEPAL CA 630) at 15 μl/min for 4 min followed by a 15 min dissociation time. Standard double referencing data subtraction methods were applied9.
Fluorescence resonance energy transfer (FRET) detected by fluorescence lifetime imaging microscopy (FLIM)
The interaction between A3G and HIV-1 RT subunits was measured by measuring FRET between fluorescent protein tags on the respective proteins of interest. Energy transfer between fluorescent proteins quickly loses its efficiency as it decreases to the sixth power of distance and is limited to a distance of less than 10 nm. The detection of FRET suggests proximity of two fluorophores on the scale of Angstrom10. The efficiency of FRET was determined by FLIM where fluorescent lifetime refers to the period of time a fluorescent molecule stays in an excited state and emitting a photon. FRET leads to a decrease in the fluorescence lifetime of the donor molecule that can be very accurately measured using time-correlated single photon counting. Contrary to other spectral methods of measuring FRET, such as sensitized emission FRET (SE-FRET or ratiometric imaging) or acceptor photobleaching, the ability to measure the fluorescence lifetime of fluorescent proteins expressed in cells is independent of relative probe concentrations and intensities, as well as being independent of photo-bleaching and spectral bleed through11–13. FLIM is therefore a sensitive and accurate approach to measure FRET between two co-expressed molecules in biological specimens.
Slide preparation
HeLa cells were plated on glass coverslips and transfected using Lipofectamine to express fluorescent fusion proteins. 24 h later, cells were fixed with 4% paraformaldehyde, washed with PBS, quenched with 1 mg/ml sodium borohydride in PBS and mounted using Mowiol (Calbiochem).
For virion samples, a drop of 5 μg/ml fibronectin in PBS was added to coverslips, left for 1 h, removed and allowed to dry before addition of a suspension of sucrose purified HIV-1 virions (typically 20 μl at a concentration of an equivalent of 1 x 107 pg p24Gag protein/ml). After 1 h incubation at 37°C, virions were fixed with 4% paraformaldehyde, quenched, then permeabilized with 0.2% Triton before staining with labeled Fab fragments and mounting. Virions were produced by co-transfection of HEK293T cells with pNL4.3/ΔVif and expression vectors for fluorescent fusion proteins (A3G, CYPA or Vpr).
Cy3 labelled Fab fragment generation
To obtain RT specific Fab fragments, we first generated a polyclonal RT specific antibody in rabbits, with Lampire Biological Laboratories, using full length RT produced in E. coli. The RT expression plasmid was a kind gift from Stephen Hughes and purification was performed as reported14,15. Rabbit serum had titers, as determined by ELISA, of 4.84 x 104 and 3.62 x 105 at day 30 and 50 post injection respectively. Antigen specific antibody was purified from serum by first enriching IgG fractions using Melon Gel IgG purification resin (Pierce) and then binding to full length RT isolated from HEK293T cells and immobilized using the AminoLink Plus Immobilization Kit (Pierce). Bound antibody was eluted with low pH buffer, neutralized and subsequently used for Fab fragment preparation and purification using the Fab Micro Preparation Kit (Pierce). The same kit was used to produce Fab fragments from the anti-p24Gag antibody (mouse monoclonal, 24-28). All steps were carried out according to the manufacturer’s protocol and resulting products controlled by SDS-PAGE and Coomassie staining. Lastly, Fab fragments were covalently labeled with Cy3 using Cy3 Monoreactive Dye (GE Healthcare) and purified from excess dye by gel filtration (PD MiniTrap G25, GE Healthcare).
FRET-FLIM instrument
FLIM was used to measure FRET between protein pairs, which allows the determination of spatial protein interactions16. Time-domain FLIM was performed with a multi-photon microscope system as described previously16–18. The system is based on a modified Bio-Rad MRC 1024MP workstation, comprising a solid-state-pumped femtosecond Ti:Sapphire (Tsunami, Spectra-Physics) laser system, a focal scan-head and an inverted microscope (Nikon TE200). Enhanced detection of the scattered component of the emitted (fluorescence) photons was afforded by the use of fast response (Hamamatsu R7401-P) non-descanned detectors, developed in-house, situated in the re-imaged objective pupil plane. Fluorescence lifetime imaging capability was provided by time-correlated single photon counting (TCSPC) electronics (Becker & Hickl, SPC 700). A 40x objective was used throughout (Nikon, CFI60 Plan Fluor N.A. 1.3) and data collected at 500 ± 20 nm through a bandpass filter (Coherent Inc. 35-5040). Images were all acquired at 256 x 256 pixel resolution. Laser power was adjusted to give average photon counting rates of the order 104 – 105 photons s-1 (0.0001 to 0.001 photon counts per excitation event) to avoid pulse pile up that can lead to inaccurate lifetime quantification. Acquisition times up to 300 s at low excitation power were used to achieve sufficient photon statistics for monoexponential fitting, while avoiding either pulse pile-up or significant photobleaching. Excitation was at 890 nm. Widefield acceptor images were acquired using a CCD camera (Hammamatsu) at <100 ms exposure times.
FRET data analysis
Bulk measurements of FRET efficiency (i.e. intensity-based methods) cannot distinguish between an increase in FRET efficiency (i.e. coupling efficiency) and an increase in FRET population (concentration of FRET species) since the two parameters are not resolved. The data presented here were analyzed using a monoexponential decay model, and goodness of fit was confirmed by determination of chi-squared values close to 1. The FRET efficiency is related to the molecular separation of donor and acceptor and the fluorescence lifetime of the interacting fraction by:
Where R0 is the Förster radius, r the molecular separation, ηfret is the lifetime of the interacting fraction and τd the lifetime of the donor in the absence of acceptor. ηfret and τd can also be taken to be the lifetime of the interacting fraction and non-interacting fraction, respectively. All data were analysed using TRI2 software (developed by Paul Barber, King’s College London). Dot plot data presented here include individual measurements as well as the mean FRET efficiency from >8 cells (or >5 areas for virion FRET) per sample +/- SD. Lifetime images of example cells are presented using a pseudocolour scale whereby blue depicts normal GFP lifetime (no FRET) and red depicts lower GFP lifetime (areas of FRET).
Purification of A3G proteins
A3G sequences were subcloned into pVL1392 transfer vector as described above. Recombinant baculovirus stocks expressing wild type A3G or the R24A mutant with C-terminal 6xHis tags were prepared using the BaculoGold Baculovirus Expression System (BD Biosciences) on SF9 monolayer cells, according to the manufacturer’s protocol. High titer virus stocks were achieved by 5 rounds of viral amplification on Sf9 monolayer cells at 3 days each. Proteins were expressed in suspension Sf9 insect cells, cultured in Sf900 II medium (Gibco) supplemented with 1% Pen/Strep (Gibco), by infection with recombinant virus. Three days after infection cells were lysed in 50 mM Tris HCl pH 8.8, 0.5% NP40, 200 mM NaCl, 10% glycerol, 1mM DTT supplemented with 50 μg/ml RNase A (Sigma) and protease inhibitor (Roche), and incubated for 10 min on ice. The lysate was homogenized by two 10 s sonication steps before incubation for 1 h at 25°C with slow rocking for complete RNA digestion. Lysates were clarified by centrifugation at 1000 x g for 10 min at 4°C, and adjusted to 0.6 M NaCl, before addition of nickel-nitrilotriacetic acid-agarose (Novagen). Proteins were allowed to bind for 2 h at 4°C while rotating. The suspension was centrifuged at 500 x g for 10 min at 4°C, the supernatant discarded, and lysis buffer added to the beads. The suspension was loaded onto a poly-prep chromatography column (Bio-Rad) and washed 5 times with 5 ml of wash buffer (50 mM Tris HCl pH 8.8, 125 mM NaCl, 10% glycerol, 1 mM DTT, 1% NP40, 50 mM imidazole) before elution with 50 mM Tris HCl pH 8.8, 125 mM NaCl, 10% glycerol, 1 mM DTT, 1% NP40, 200 mM imidazole. To avoid protein precipitation off the column, elution was performed with 1.2 ml elution buffer into 5 ml buffer (50 mM Tris HCl pH 8.8, 125 mM NaCl, 10% glycerol, 1 mM DTT, 1% NP40). The solution was then loaded onto a DEAE-FF sepharose column (GE Healthcare) on an AKTA Purifier 10 (Pharmacia) and eluted with a gradient (0-100%) of buffer A (50 mM Tris HCl pH 8.8, 0.5% NP40, 50 mM NaCl, 10% glycerol, 1 mM DTT) and buffer B (50 mM Tris HCl pH 8.8, 0.5% NP40, 1 M NaCl, 10% glycerol, 1 mM DTT). Peak fractions were dialyzed into a suitable buffer, concentration determined by Bradford assay with BSA standards, and homogeneity judged by Coomassie staining (see Supplemental Figure 3 and 8).
UDG functional assay
The efficiency of hUGI-mediated inhibition of cellular UDG activity was evaluated in cell lysates. Cells were lysed by sonication in a buffer containing 10 mM HEPES pH 7.4, 1 mM EDTA, 1 mM DTT. A 34-base, single-stranded uracil-containing oligonucleotide (5′-attattattattattcugcggatttatttattta-3′)19 was 5’ end labelled with γ-P32-ATP and then incubated with lysate in reaction buffer (10 mM Tris [pH 7.4], 1 mM EDTA, 50 mM NaCl);. negative and positive controls comprised the addition of recombinant UDG or UGI (both NEB). Reactions were left for 15 min at 37°C and stopped by addition of 0.75 volumes of stop buffer (70% formamide, 0.4 N NaOH, 1× TBE). Samples were heated to 95°C for 15 min, resolved on 8% denaturing urea-polyacrylamide gels and exposure to a phosphorimaging screen for detection using a Typhoon FLA Biomolecular Imager.
In vitro primer extension assays
Reconstituted primer extension assays were based on a previous method20. The HIV-1 RNA template was created using a dsDNA template for in vitro transcription that had been created by PCR using pNL4.3 as the template and the following primers: T7 HIV-1 TAR fwd 5’-aatttaatacgactcactataggggtctctctggttagaccag-3’ and UL244 rev 5'-gtcctgcgtcgagagatct-3'20,21. ssRNA was synthesized using the MAXIScript T7 transcription kit (Ambion) according to the manufacturer’s protocol, passed through a G-50 column and further purified by denaturing PAGE. An 18 nt DNA oligonucleotide corresponding to the natural HIV-1 primer binding site (5'-gtccctgttcgggcgcca-3') was radiolabeled using polynucleotide kinase (NEB) and γ-P32-ATP (Perkin Elmer). This labeled primer and RNA template were annealed in 50 mM Tris HCl pH 8.0, 75 mM KCl at 65 °C for 5 min followed by gradual cooling to 37 °C, and annealed template/primer then diluted to 0.1 μM in reaction buffer (50 mM Tris HCl pH 8.0, 75 mM KCl, 7 mM MgCl2, 1 mM DTT. Reactions of 10 μl were prepared in this buffer with 10 nM template, 200 μM dNTPs and 1 U/μl RNAase Inhibitor (Promega). Reactions were preincubated for 10 min at 37 °C with indicated amounts of purified A3G (dialyzed against 50 mM Tris HCL pH 8.9, 150 mM NaCl, 10% glycerol, 0.1% NP40, 1 mM DTT) or dialysis buffer alone. Unless otherwise noted, the reactions were initiated by addition of 10 nM HIV-1 RT (Worthington), and incubated for the indicated amount of time at 37°C. Reactions were stopped with formamide buffer, boiled, and separated on a 6% urea sequencing gel. The gels were dried and exposed to phosphorimager screens for detection using a Typhoon FLA Biomolecular Imager.
Single molecule RNA binding assay
To quantitatively assess the RNA binding properties of A3G proteins an assay employing imaging of fluorescently labeled single RNA molecules was carried out as described22; Protein binding increases the fluorescence intensity of the bound ssRNA molecule via a phenomenon referred to as protein-induced fluorescence enhancement, or PIFE23,24. The two ssRNA substrates were purchased from Integrated DNA Technologies: The HIV-1 LTR polyA loop sequence plus linker (5’-biotin-aaaaaaaagccucaauaaagcuugccuugagu-Cy3Sp-3’) and an RNA molecule with low predicted secondary structure as used in19: (5’-biotin-auuauuauuauuauuauuauuccgcggauuuauuuauuua-Cy3Sp-3’). Briefly, substrates were tethered to a glass surfaces in parallel chambers of flow cells in T-125 buffer (10 mM Tris HCl pH 7.5, 3 mM MgCl2, 125 mM NaCl) containing an oxygen scavenging system (1 mg/ml of glucose oxidase (Sigma), 0.04 mg/ml of catalase (Sigma), 0.4% dextrose) to reduce photobleaching, and 1% β-mercaptoethanol (Acros) serving as a triplet state quencher. A3G and A3G-R24A were purified from insect cells and dialyzed against T-125 buffer + 10% glycerol (stocks were 2.5 μM and 5 μM, respectively). Proteins were added at concentrations between 100 nM and 400 nM and fluorescent intensities of several hundred individual RNA molecules were measured over 2 min. Analysis was performed using in house software, developed by John Marko, Northwestern University, which was previously described22 and is available upon request.
Bacterial mutator assay
This assay was performed as described2,25. Briefly, pTrc99A-based vectors encoding, wild type or mutant A3G proteins were transformed into E. coli strain KL16. Individual colonies were picked and grown to saturation in Luria broth containing 100 μg/ml ampicillin and 1 mM isopropyl β-D-1-thiogalactopyranoside. Cultures were diluted 1:10, 1:100 or left undiluted and spread onto agar plates containing 100 μg/ml rifampicin. The same cultures were also diluted 1:106, 1:107, 1:108 onto agar plates containing 100 mg/ml ampicillin and incubated overnight at 37°C. Mutation frequencies were recorded as the number of rifampicin-resistant colonies per 109 viable cells, which were enumerated using the ampicillin containing plates. The experiment was carried out at least twice. To average the repeat experiments, the average colony count for wild type A3G was set at 100 and all other scores were normalized against this value.
Data and code availability
The data supporting this study and custom software are available from the corresponding author upon reasonable request. There are no restrictions to data availability. Raw MiSeq® sequencing files analyzed in this study (presented in Fig 1g, 2d, 6e as well as Supplementary Fig 2, 4, 5 and 7) are also publicly available at the European Nucleotide Archive (ENA) under study accession number PRJEB22170. Individual accession codes for each sample are listed in Supplementary Figure 4b. Custom computer code used to analyze the raw MiSeq® reads is deposited in GitHub and publicly available (http://doi.org/10.5281/zenodo.1004571).
Supplementary Material
Acknowledgments
We would like to acknowledge the support and fruitful debate with members of the Malim laboratory, the insights of Jernej Ule and Rebecca Oakey on optimization of the sequencing protocol, and Michael Emerman and Stephen Hughes for the generous provision of reagents. We also thank Matt Arno at the King’s College London Genomic Center and Debbie Hughes at the University College London (UCL) Institute for Neurology Next Generation Sequencing Facility for help with MiSeq sequencing runs. The work was supported by the U.K. Medical Research Council (G1000196 and MR/M001199/1 to M.M.; and MR/K015664/1 to M.P.), the Wellcome Trust (106223/Z/14/Z to M.M.), the European Commission's Seventh Framework Programme [FP7/2007-2013] under grant agreement n°PIIF-GA-2012-329679 (to D.P.), King’s alumni community sponsored King’s Undergraduate Research Fellowships (to R.D.L), King’s School of Medicine Summer Studentship Award (to J.C), and the Department of Health via a National Institute for Health Research comprehensive Biomedical Research Centre award to Guy’s and St. Thomas’ NHS Foundation Trust in partnership with King’s College London and King’s College Hospital NHS Foundation Trust (guysbrc-2012-1).
Footnotes
Author Contributions:
D.P. co-wrote the manuscript and executed all experiments with the following exceptions: R.D.L. performed the co-immunoprecipitation shown in Figure 5a, M.P. carried out all the microscopy and FRET-FLIM experiments. A.E.S. wrote and ran the analysis software for analyzing raw FASTQ sequencing data. S.C. carried out the double alanine scan for the A3G-RT binding site mapping. A.M.B. and C.M.H carried out and analyzed the single molecule RNA binding assays (Supplementary Figure 3c). S.P., R.D.L. and J.C. contributed to reagent generation, in particular for Fig 5. J.M.M. contributed to the SPR experiments (Figure 3c) and performed the analysis. L.A and A.E.S contributed to the sequencing library design. D.P and M.H.M. conceived the experiments and co-wrote the manuscript. All authors cross-checked the manuscript.
Competing financial interest:
The authors have no conflict of financial interest.
Michael H. Malim: ORCID iD: 0000-0002-7699-2064
References
- 1.Doyle T, Goujon C, Malim MH. HIV-1 and interferons: who's interfering with whom? Nat Rev Microbiol. 2015;13:403–13. doi: 10.1038/nrmicro3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simon V, Bloch N, Landau NR. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol. 2015;16:546–53. doi: 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desimmie BA, et al. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J Mol Biol. 2014;426:1220–45. doi: 10.1016/j.jmb.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris RS, Dudley JP. APOBECs and virus restriction. Virology. 2015;479–480:131–45. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop KN, et al. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–6. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 6.Hultquist JF, et al. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol. 2011;85:11220–34. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–50. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 8.Apolonia L, et al. Promiscuous RNA binding ensures effective encapsidation of APOBEC3 proteins by HIV-1. PLoS Pathog. 2015;11:e1004609. doi: 10.1371/journal.ppat.1004609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo K, et al. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J Virol. 2004;78:11841–52. doi: 10.1128/JVI.78.21.11841-11852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soros VB, Yonemoto W, Greene WC. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA, and subsequently activated by RNase H. PLoS Pathog. 2007;3:e15. doi: 10.1371/journal.ppat.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9:1404–7. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 12.Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med. 2003;9:1398–403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 13.Yu X, et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–60. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 14.Gillick K, et al. Suppression of HIV-1 infection by APOBEC3 proteins in primary human CD4(+) T cells is associated with inhibition of processive reverse transcription as well as excessive cytidine deamination. J Virol. 2013;87:1508–17. doi: 10.1128/JVI.02587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phalora PK, Sherer NM, Wolinsky SM, Swanson CM, Malim MH. HIV-1 replication and APOBEC3 antiviral activity are not regulated by P bodies. J Virol. 2012;86:11712–24. doi: 10.1128/JVI.00595-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris RS, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–9. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 17.Mangeat B, et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, et al. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–8. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bishop KN, Holmes RK, Malim MH. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J Virol. 2006;80:8450–8. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4:e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes RK, Koning FA, Bishop KN, Malim MH. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J Biol Chem. 2007;282:2587–95. doi: 10.1074/jbc.M607298200. [DOI] [PubMed] [Google Scholar]
- 22.Iwatani Y, et al. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007;35:7096–108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schrofelbauer B, Yu Q, Zeitlin SG, Landau NR. Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J Virol. 2005;79:10978–87. doi: 10.1128/JVI.79.17.10978-10987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang B, Chen K, Zhang C, Huang S, Zhang H. Virion-associated uracil DNA glycosylase-2 and apurinic/apyrimidinic endonuclease are involved in the degradation of APOBEC3G-edited nascent HIV-1 DNA. J Biol Chem. 2007;282:11667–75. doi: 10.1074/jbc.M606864200. [DOI] [PubMed] [Google Scholar]
- 25.Kaiser SM, Emerman M. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase Apobec3G. J Virol. 2006;80:875–82. doi: 10.1128/JVI.80.2.875-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langlois MA, Neuberger MS. Human APOBEC3G can restrict retroviral infection in avian cells and acts independently of both UNG and SMUG1. J Virol. 2008;82:4660–4. doi: 10.1128/JVI.02469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mbisa JL, et al. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J Virol. 2007;81:7099–110. doi: 10.1128/JVI.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman EN, et al. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15:166–70. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 29.Mbisa JL, Bu W, Pathak VK. APOBEC3F and APOBEC3G inhibit HIV-1 DNA integration by different mechanisms. J Virol. 2010;84:5250–9. doi: 10.1128/JVI.02358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaurasiya KR, et al. Oligomerization transforms human APOBEC3G from an efficient enzyme to a slowly dissociating nucleic acid-binding protein. Nat Chem. 2014;6:28–33. doi: 10.1038/nchem.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, et al. The cellular antiviral protein APOBEC3G interacts with HIV-1 reverse transcriptase and inhibits its function during viral replication. J Virol. 2012 doi: 10.1128/JVI.06594-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu WS, Hughes SH. HIV-1 reverse transcription. Cold Spring Harb Perspect Med. 2012;2 doi: 10.1101/cshperspect.a006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herschhorn A, Hizi A. Retroviral reverse transcriptases. Cell Mol Life Sci. 2010;67:2717–47. doi: 10.1007/s00018-010-0346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gillick K, et al. The suppression of HIV-1 infection by APOBEC3 proteins in primary human CD4+ T cells is associated with the inhibition of processive reverse transcription as well as excessive cytidine deamination. submitted to J Virol. 2012 doi: 10.1128/JVI.02587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arts EJ, Li Z, Wainberg MA. Analysis of Primer Extension and the First Template Switch during Human Immunodeficiency Virus Reverse Transcription. J Biomed Sci. 1995;2:314–321. doi: 10.1007/BF02255218. [DOI] [PubMed] [Google Scholar]
- 36.Driscoll MD, Golinelli MP, Hughes SH. In vitro analysis of human immunodeficiency virus type 1 minus-strand strong-stop DNA synthesis and genomic RNA processing. J Virol. 2001;75:672–86. doi: 10.1128/JVI.75.2.672-686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harrison GP, Mayo MS, Hunter E, Lever AM. Pausing of reverse transcriptase on retroviral RNA templates is influenced by secondary structures both 5' and 3' of the catalytic site. Nucleic Acids Res. 1998;26:3433–42. doi: 10.1093/nar/26.14.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim EY, et al. Human APOBEC3 Induced Mutation of Human Immunodeficiency Virus Type-1 Contributes to Adaptation and Evolution in Natural Infection. PLoS Pathog. 2014;10:e1004281. doi: 10.1371/journal.ppat.1004281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu Q, et al. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–42. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 40.Schormann N, Ricciardi R, Chattopadhyay D. Uracil-DNA glycosylases-structural and functional perspectives on an essential family of DNA repair enzymes. Protein Sci. 2014;23:1667–85. doi: 10.1002/pro.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Visnes T, et al. Uracil in DNA and its processing by different DNA glycosylases. Philos Trans R Soc Lond B Biol Sci. 2009;364:563–8. doi: 10.1098/rstb.2008.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z, Mosbaugh DW. Uracil-DNA glycosylase inhibitor gene of bacteriophage PBS2 encodes a binding protein specific for uracil-DNA glycosylase. J Biol Chem. 1989;264:1163–71. [PubMed] [Google Scholar]
- 43.Mansky LM, Preveral S, Selig L, Benarous R, Benichou S. The interaction of vpr with uracil DNA glycosylase modulates the human immunodeficiency virus type 1 In vivo mutation rate. J Virol. 2000;74:7039–47. doi: 10.1128/jvi.74.15.7039-7047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willetts KE, et al. DNA repair enzyme uracil DNA glycosylase is specifically incorporated into human immunodeficiency virus type 1 viral particles through a Vpr-independent mechanism. J Virol. 1999;73:1682–8. doi: 10.1128/jvi.73.2.1682-1688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adolph MB, Webb J, Chelico L. Retroviral restriction factor APOBEC3G delays the initiation of DNA synthesis by HIV-1 reverse transcriptase. PLoS One. 2013;8:e64196. doi: 10.1371/journal.pone.0064196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voss TC, Demarco IA, Day RN. Quantitative imaging of protein interactions in the cell nucleus. Biotechniques. 2005;38:413–24. doi: 10.2144/05383RV01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burnett A, Spearman P. APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker. J Virol. 2007;81:5000–13. doi: 10.1128/JVI.02237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004;23:2451–8. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wedekind JE, et al. Nanostructures of APOBEC3G support a hierarchical assembly model of high molecular mass ribonucleoprotein particles from dimeric subunits. J Biol Chem. 2006;281:38122–6. doi: 10.1074/jbc.C600253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gallois-Montbrun S, et al. Comparison of cellular ribonucleoprotein complexes associated with the APOBEC3F and APOBEC3G antiviral proteins. J Virol. 2008;82:5636–42. doi: 10.1128/JVI.00287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huthoff H, Autore F, Gallois-Montbrun S, Fraternali F, Malim MH. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 2009;5:e1000330. doi: 10.1371/journal.ppat.1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao X, Li SX, Yang H, Chen XS. Crystal structures of APOBEC3G N-domain alone and its complex with DNA. Nat Commun. 2016;7:12193. doi: 10.1038/ncomms12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Menendez-Arias L, Sebastian-Martin A, Alvarez M. Viral reverse transcriptases. Virus Res. 2016 doi: 10.1016/j.virusres.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 54.Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 2010;7:754–74. doi: 10.4161/rna.7.6.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Basu VP, et al. Strand transfer events during HIV-1 reverse transcription. Virus Res. 2008;134:19–38. doi: 10.1016/j.virusres.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 56.Iwatani Y, Rosen AE, Guo J, Musier-Forsyth K, Levin JG. Efficient initiation of HIV-1 reverse transcription in vitro. Requirement for RNA sequences downstream of the primer binding site abrogated by nucleocapsid protein-dependent primer-template interactions. J Biol Chem. 2003;278:14185–95. doi: 10.1074/jbc.M211618200. [DOI] [PubMed] [Google Scholar]
- 57.Masuda T, et al. Fate of HIV-1 cDNA intermediates during reverse transcription is dictated by transcription initiation site of virus genomic RNA. Sci Rep. 2015;5:17680. doi: 10.1038/srep17680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kharytonchyk S, et al. Transcriptional start site heterogeneity modulates the structure and function of the HIV-1 genome. Proc Natl Acad Sci U S A. 2016;113:13378–13383. doi: 10.1073/pnas.1616627113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klarmann GJ, Schauber CA, Preston BD. Template-directed pausing of DNA synthesis by HIV-1 reverse transcriptase during polymerization of HIV-1 sequences in vitro. J Biol Chem. 1993;268:9793–802. [PubMed] [Google Scholar]
- 60.Altfeld M, Gale M., Jr Innate immunity against HIV-1 infection. Nat Immunol. 2015;16:554–62. doi: 10.1038/ni.3157. [DOI] [PubMed] [Google Scholar]
- 61.Ahn J, et al. HIV-1 Vpr loads uracil DNA glycosylase-2 onto DCAF1, a substrate recognition subunit of a cullin 4A-ring E3 ubiquitin ligase for proteasome-dependent degradation. J Biol Chem. 2010;285:37333–41. doi: 10.1074/jbc.M110.133181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steagall WK, Robek MD, Perry ST, Fuller FJ, Payne SL. Incorporation of uracil into viral DNA correlates with reduced replication of EIAV in macrophages. Virology. 1995;210:302–13. doi: 10.1006/viro.1995.1347. [DOI] [PubMed] [Google Scholar]
- 63.Kennedy EM, et al. Abundant non-canonical dUTP found in primary human macrophages drives its frequent incorporation by HIV-1 reverse transcriptase. J Biol Chem. 2011;286:25047–55. doi: 10.1074/jbc.M111.234047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan N, O'Day E, Wheeler LA, Engelman A, Lieberman J. HIV DNA is heavily uracilated, which protects it from autointegration. Proc Natl Acad Sci U S A. 2011;108:9244–9. doi: 10.1073/pnas.1102943108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hansen EC, et al. Diverse fates of uracilated HIV-1 DNA during infection of myeloid lineage cells. Elife. 2016;5 doi: 10.7554/eLife.18447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weil AF, et al. Uracil DNA glycosylase initiates degradation of HIV-1 cDNA containing misincorporated dUTP and prevents viral integration. Proc Natl Acad Sci U S A. 2013;110:E448–57. doi: 10.1073/pnas.1219702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen R, Wang H, Mansky LM. Roles of uracil-DNA glycosylase and dUTPase in virus replication. J Gen Virol. 2002;83:2339–45. doi: 10.1099/0022-1317-83-10-2339. [DOI] [PubMed] [Google Scholar]
- 68.Sire J, Querat G, Esnault C, Priet S. Uracil within DNA: an actor of antiviral immunity. Retrovirology. 2008;5:45. doi: 10.1186/1742-4690-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–53. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 70.Stremlau M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A. 2006;103:5514–9. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Malim MH, Bieniasz PD. HIV Restriction Factors and Mechanisms of Evasion. Cold Spring Harb Perspect Med. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Warren K, Warrilow D, Meredith L, Harrich D. Reverse Transcriptase and Cellular Factors: Regulators of HIV-1 Reverse Transcription. Viruses. 2009;1:873–94. doi: 10.3390/v1030873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salter JD, Bennett RP, Smith HC. The APOBEC Protein Family: United by Structure, Divergent in Function. Trends Biochem Sci. 2016;41:578–94. doi: 10.1016/j.tibs.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1.Phalora PK, Sherer NM, Wolinsky SM, Swanson CM, Malim MH. HIV-1 replication and APOBEC3 antiviral activity are not regulated by P bodies. J Virol. 2012;86:11712–24. doi: 10.1128/JVI.00595-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huthoff H, Autore F, Gallois-Montbrun S, Fraternali F, Malim MH. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 2009;5:e1000330. doi: 10.1371/journal.ppat.1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huthoff H, Malim MH. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J Virol. 2007;81:3807–15. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swanson CM, Sherer NM, Malim MH. SRp40 and SRp55 promote the translation of unspliced human immunodeficiency virus type 1 RNA. J Virol. 2010;84:6748–59. doi: 10.1128/JVI.02526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaiser SM, Emerman M. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase Apobec3G. J Virol. 2006;80:875–82. doi: 10.1128/JVI.80.2.875-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallois-Montbrun S, et al. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J Virol. 2007;81:2165–78. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Apolonia L, et al. Promiscuous RNA binding ensures effective encapsidation of APOBEC3 proteins by HIV-1. PLoS Pathog. 2015;11:e1004609. doi: 10.1371/journal.ppat.1004609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaddis NC, Chertova E, Sheehy AM, Henderson LE, Malim MH. Comprehensive investigation of the molecular defect in vif-deficient human immunodeficiency virus type 1 virions. J Virol. 2003;77:5810–20. doi: 10.1128/JVI.77.10.5810-5820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12:279–84. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 10.Voss TC, Demarco IA, Day RN. Quantitative imaging of protein interactions in the cell nucleus. Biotechniques. 2005;38:413–24. doi: 10.2144/05383RV01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Mills JD, Periasamy A. Protein localization in living cells and tissues using FRET and FLIM. Differentiation. 2003;71:528–41. doi: 10.1111/j.1432-0436.2003.07109007.x. [DOI] [PubMed] [Google Scholar]
- 12.Wallrabe H, Periasamy A. Imaging protein molecules using FRET and FLIM microscopy. Curr Opin Biotechnol. 2005;16:19–27. doi: 10.1016/j.copbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Becker W. Fluorescence lifetime imaging--techniques and applications. J Microsc. 2012;247:119–36. doi: 10.1111/j.1365-2818.2012.03618.x. [DOI] [PubMed] [Google Scholar]
- 14.Boyer PL, Clark PK, Hughes SH. HIV-1 and HIV-2 reverse transcriptases: different mechanisms of resistance to nucleoside reverse transcriptase inhibitors. J Virol. 2012;86:5885–94. doi: 10.1128/JVI.06597-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyer PL, Ding J, Arnold E, Hughes SH. Subunit specificity of mutations that confer resistance to nonnucleoside inhibitors in human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1994;38:1909–14. doi: 10.1128/aac.38.9.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peter M, et al. Multiphoton-FLIM quantification of the EGFP-mRFP1 FRET pair for localization of membrane receptor-kinase interactions. Biophys J. 2005;88:1224–37. doi: 10.1529/biophysj.104.050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsons M, et al. Spatially distinct binding of Cdc42 to PAK1 and N-WASP in breast carcinoma cells. Mol Cell Biol. 2005;25:1680–95. doi: 10.1128/MCB.25.5.1680-1695.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prag S, et al. Activated ezrin promotes cell migration through recruitment of the GEF Dbl to lipid rafts and preferential downstream activation of Cdc42. Mol Biol Cell. 2007;18:2935–48. doi: 10.1091/mbc.E06-11-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwatani Y, Takeuchi H, Strebel K, Levin JG. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J Virol. 2006;80:5992–6002. doi: 10.1128/JVI.02680-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwatani Y, et al. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007;35:7096–108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwatani Y, Rosen AE, Guo J, Musier-Forsyth K, Levin JG. Efficient initiation of HIV-1 reverse transcription in vitro. Requirement for RNA sequences downstream of the primer binding site abrogated by nucleocapsid protein-dependent primer-template interactions. J Biol Chem. 2003;278:14185–95. doi: 10.1074/jbc.M211618200. [DOI] [PubMed] [Google Scholar]
- 22.Bruns AM, et al. ATP hydrolysis enhances RNA recognition and antiviral signal transduction by the innate immune sensor, laboratory of genetics and physiology 2 (LGP2) J Biol Chem. 2013;288:938–46. doi: 10.1074/jbc.M112.424416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang H, Myong S. Protein induced fluorescence enhancement (PIFE) for probing protein-nucleic acid interactions. Chem Soc Rev. 2014;43:1221–9. doi: 10.1039/c3cs60201j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myong S, et al. Cytosolic viral sensor RIG-I is a 5'-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–4. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting this study and custom software are available from the corresponding author upon reasonable request. There are no restrictions to data availability. Raw MiSeq® sequencing files analyzed in this study (presented in Fig 1g, 2d, 6e as well as Supplementary Fig 2, 4, 5 and 7) are also publicly available at the European Nucleotide Archive (ENA) under study accession number PRJEB22170. Individual accession codes for each sample are listed in Supplementary Figure 4b. Custom computer code used to analyze the raw MiSeq® reads is deposited in GitHub and publicly available (http://doi.org/10.5281/zenodo.1004571).