Summary:
The Comparative Toxicogenomics Database (CTD; http://ctdbase.org) is a free resource that provides manually curated information on chemical, gene, phenotype, and disease relationships to advance understanding of the effect of environmental exposures on human health. Four core content areas are independently curated: chemical–gene interactions, chemical–disease and gene–disease associations, chemical–phenotype interactions, and environmental exposure data (e.g., effects of chemical stressors on humans). Since releasing exposure data in 2015, we have vastly increased our coverage of chemicals and disease/phenotype outcomes; greatly expanded access to exposure content; added search capability by stressors, cohorts, population demographics, and measured outcomes; and created user-specified displays of content. These enhancements aim to facilitate human studies by allowing comparisons among experimental parameters and across studies involving specified chemicals, populations, or outcomes. Integration of data among CTD’s four content areas and external data sets, such as Gene Ontology annotations and pathway information, links exposure data with over 1.8 million chemical–gene, chemical–disease and gene–disease interactions. Our analysis tools reveal direct and inferred relationships among the data and provide opportunities to generate predictive connections between environmental exposures and population-level health outcomes. https://doi.org/10.1289/EHP2873
Introduction
Since its release in 2004, the Comparative Toxicogenomics Database (CTD; http://ctdbase.org) has evolved into a premier resource that integrates manually curated data on toxicogenomic interactions among chemicals, genes, phenotypes, diseases, and pathways (Davis et al. 2009, 2011, 2015, 2017). Our recent integration of phenotype and exposure science content (Davis et al. 2016; Grondin et al. 2016) expand the available resources for data analysis and increase exponentially the number of toxicogenomic relationships available at CTD. Inclusion of exposure studies in CTD centralizes, standardizes, and organizes data obtained from multiple exposure-assessment methodologies across different life stages, which is critical for understanding and characterizing the exposome, the totality of an individual’s environmental exposures from the prenatal period onwards (Stingone et al. 2017; Wild 2005).
Using concepts derived from the Exposure Ontology (ExO) (Mattingly et al. 2012), CTD exposure data include statements connecting chemicals (i.e., stressors) and human populations (i.e., receptors) via exposure events and their resulting outcomes. Here we describe our newly updated web interface that allows querying and filtering of 33 unique fields of exposure data.
Technical Architecture
As previously documented (Davis et al. 2011), CTD’s database architecture comprises a curation database (containing manually curated data from the primary literature), a Third Party database (containing data extracted from external sources), and a Public Web Application (PWA) database (the basis for CTD’s public web site). Curation and Third Party databases are updated, integrated, and loaded on a monthly basis to the PWA database. All CTD data, including exposure data, are loaded into CTD’s PostgreSQL database management system. Load and validation processes are primarily Java-based and run in a Linux environment; the CTD web and curation applications utilize a J2EE-based Model-View-Controller architecture.
Although CTD’s initial exposure paradigm included curation of 54 data fields pertaining to five exposure categories, our initial release of exposure data was simplified to display nine and 13 data types on the Exposure Studies and Exposure Details pages, respectively (Grondin et al. 2016). The second phase of the Exposure project, which enables public access to the full exposure data set, necessitated significant expansion of the existing CTD database architecture, incorporation of new tables, and modification of existing ones. As well, the Exposure database load, validation processes, and web interface were enhanced to accommodate new search features to query exposure data directly, display additional key exposure data fields, and customize screen/report functionality. Finally, the hardware platforms on which all the web-based applications operate were upgraded to meet the demands of the new functionality.
Integration of CTD Content Areas
Four content areas are independently and manually curated in CTD, including chemical–gene interactions, chemical–disease and gene–disease interactions, chemical–phenotype interactions, and environmental exposure data. The first three content areas are curated across more than 570 species and include both in vitro and in vivo results. CTD’s exposure curation details the effects of chemical stressors on human receptors in vivo. Data from all four content areas are integrated with each other, as well as with external data sets, including Gene Ontology (GO) annotations and pathway information, to help elucidate connections across species and experimental parameters, and to link real-world examples of chemical exposures and corresponding outcomes with mechanistic knowledge. In this way, these integrated data sets support a link between exposome research and adverse outcome pathways (AOP), whereby AOP’s molecular initiating events (comparable to CTD’s chemical–gene interactions) and AOP’s key events (comparable to CTD’s chemical-induced phenotypes) in the disruption of networks result in an adverse outcome such as a disease end point (comparable to CTD’s chemical/gene–disease interactions) (Ankley et al. 2010; Escher et al. 2017; Vinken 2016). All data curated into CTD can be analyzed using CTD analysis tools Batch Query, Set Analyzer, MyGeneVenn, MyVenn, and VennViewer (Davis et al. 2015, 2017).
Accessing Exposure Data
Currently, CTD contains curated data for over 12,000 chemicals; 43,000 genes; and 6,900 diseases; each of these entities has its own unique page in CTD to easily access its associated data, comparable terms, GO annotations, pathways, and now exposure data. Exposure Studies and Exposure Details are new data tabs at the top of each chemical, gene, GO, and disease page in CTD. In our updated module, Exposure Studies tabs summarize 11 fields of basic information for each curated exposure paper, and Exposure Details tabs display up to 33 different data categories for each study, including measurements for detected chemicals and population demographics (e.g., age, race, sex, smoking status, geographical location, and time period of exposure event). All data link directly to the primary reference, and chemical, gene, disease, and phenotype terms hyperlink directly to their respective CTD pages. Webpage displays can be downloaded via CSV, Excel, XML, or TSV formats to a user’s desktop. Exposure data are now accessible via query pages. Below we discuss the two drop-down query pages (for Exposure Studies or Exposure Details) that are available under the “Search” menu on the main navigation bar at the top of every CTD page:
1. Exposure Studies
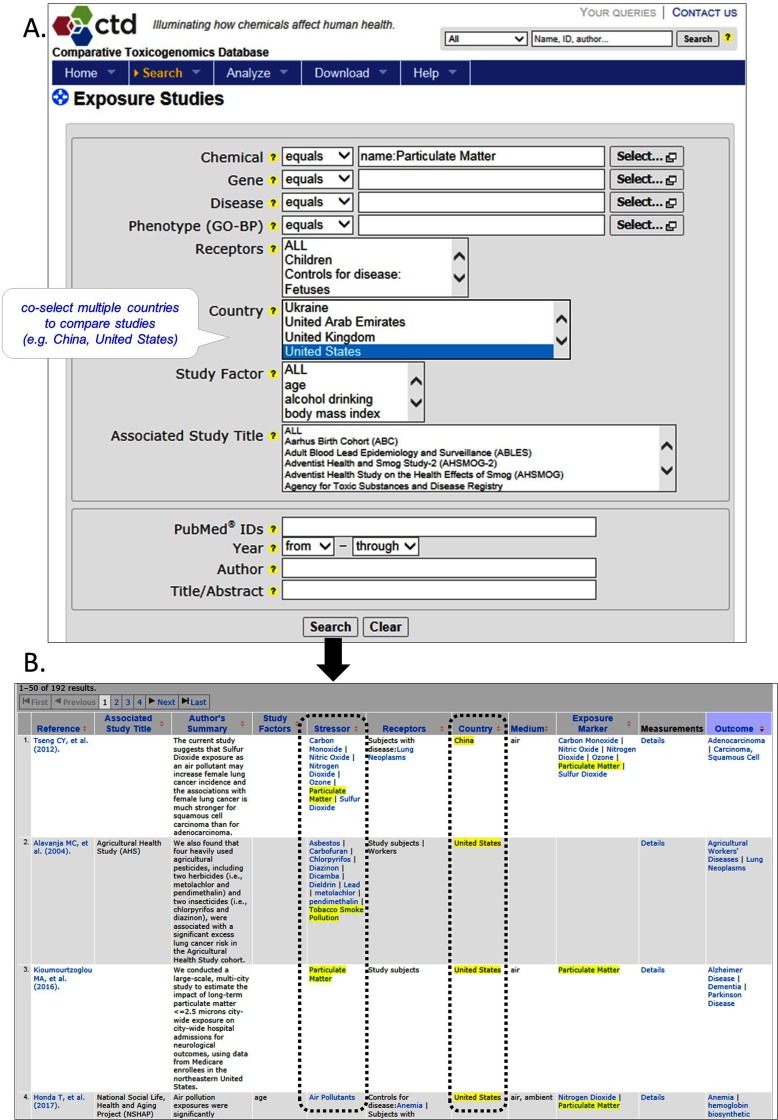
Users can query for summarized exposure data by selecting Exposure Studies from the “Search” tab (Figure 1A). Search terms from one or more of 11 data fields can be specified, allowing users to retrieve information restricted by chemical stressor, associated gene, disease or phenotype outcome, receptor population, country of study, influencing study factors (e.g., diet, genetics, body mass index, socioeconomic status, inter alia), associated study title, or reference attributes (e.g., PubMed accession identifier, author, year, title/abstract words). Returned results yield a summary view of all curated papers that meet the user-defined criteria (Figure 1B). Fields displayed include a link to the cited reference, the author’s summary statement, and data on stressors, receptors, influencing study factors, medium, exposure markers measured, outcome, and country. As of August, 2017, there were 1,731 exposure papers curated in CTD, comprising more than 97,000 exposure statements, which describe associations involving 955 unique chemicals, 325 genes, 359 diseases, and 268 phenotypic GO terms.
Figure 1.
Querying Exposure Studies in Comparative Toxicogenomics Database (CTD). Exposure studies can be queried in CTD from an Exposure Studies search page that allows specification of one or more of 12 query terms from different exposure categories (A). Here, the database is queried for studies conducted in China and/or the United States in which particulate matter is an exposure stressor by entering “particulate matter” in the chemical search field and co-selecting “China” and “United States” in the Country field. The bold black arrow connects the query form to the results page (B). Results are returned in summary format, with each curated exposure paper represented by one row of 11 data fields, including the primary reference, associated study title, author’s summary, study factors, stressor, receptor, country, medium, exposure marker, outcome, and a link to detailed measurements. Dashed rectangles correspond to Chemical Stressor and Country fields that were queried in part A. Query parameters are highlighted in yellow on results pages, along with any hierarchically related terms. A chemical may appear in both the Stressor and Exposure Marker fields if the chemical stressor was experimentally measured as a result of the exposure event, but the Exposure Marker may also be an unrelated entity whose concentration may have been affected by the exposure event, such as a metabolite or a gene. References, chemical, gene, disease, and phenotype-related GO-BP terms hyperlink to their individual CTD pages. Results can be downloaded via CSV, Excel, XML, or TSV formats. Image: ©2012–2017 MDI Biological Laboratory & North Carolina State University.
2. Exposure Details
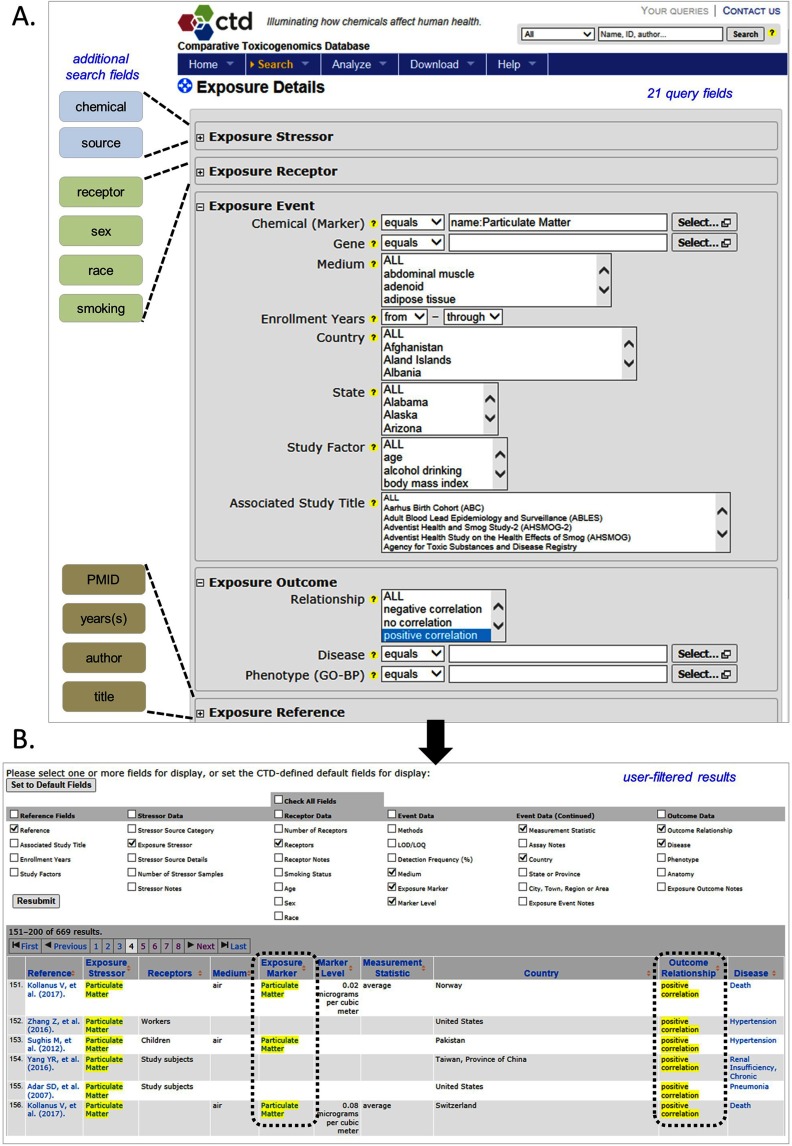
For more detailed queries, users can select the Exposure Details query page from the “Search” tab (Figure 2A). This query form is organized into five sections that can be opened or collapsed by toggle buttons that correspond to each of the four ExO concepts (Stressor, Receptor, Event, or Outcome), plus a reference section. Collectively, these five sections contain 21 different fields that can be specified in the query, either jointly or individually. The user-specified search terms are highlighted on the results page, in addition to 14 default fields (Figure 2B). To further filter these results, users can select any of 33 different exposure fields by checking or unchecking information boxes and clicking “Resubmit.” All results can be downloaded in a variety of formats at the bottom of the page.
Figure 2.
Accessing detailed exposure information in Comparative Toxicogenomics Database (CTD). Detailed exposure data in CTD can be accessed from an Exposure Details query page (A), which allows users to specify search terms in one or more of five categories (Exposure Stressor, Exposure Receptor, Exposure Event, Exposure Outcome, and Exposure Reference) that can be expanded or collapsed by clicking on the toggle boxes to the left of the headings. Additional search fields for each section are shown to the left of the toggle boxes, totaling 21 query fields. Here, a search for positive correlations between particulate matter and disease or phenotype outcomes can be done by entering “particulate matter” in the chemical (marker) field and “positive correlation” in the Exposure Outcome section. The bold black arrow connects the query form to the returned results (B). Users can specify up to 33 unique data fields to display on the results page, or display 14 default fields. The queried term(s) is highlighted in yellow, along with any hierarchically related terms (here, dashed rectangles correspond to the queried fields of Exposure Event Chemical Marker and Exposure Outcome Relationship). References, chemical, gene, disease, and phenotype-related GO-BP terms hyperlink to their individual CTD pages. Results can be downloaded via CSV, Excel, XML, or TSV formats. Image: ©2012–2017 MDI Biological Laboratory & North Carolina State University.
Application to Human Studies
The introduction of user-defined exposure queries, coupled with downloadable results, allows users to compare experimental parameters and outcomes within CTD-curated exposure studies across many different fields, such as life stage or geographic location. For example, a user interested in potential adverse outcomes of particulate matter might compare studies conducted in the United States to those conducted in China, where air pollution was noted as the single largest public health and environmental issue affecting Beijing (Zhang et al. 2011). To initiate this comparison, a researcher could perform an Exposure Studies query by selecting particulate matter as the “Chemical” and co-selecting China and the United States in the “Country” query field to yield summary views of 192 related exposure studies currently curated in CTD (Figure 1). Results can be sorted by clicking the column headings, including associated study title, influencing study factors, population demographics, medium, exposure markers measured, or observed outcomes. The entire data set can also be downloaded into an Excel spreadsheet for further analysis. Complementing these results, an Exposure Details query selecting particulate matter as the “Chemical Stressor” and co-selecting China and United States in the “Country” field returns all 3,599 statements that provide more detailed information from these 192 studies, such as the measured level and measurement statistics of the exposure markers and their correlated outcomes. These results can assist scientists addressing questions such as whether similar marker levels and outcomes are observed between the two countries, the limits of detection that were calculated for various methods of air-pollution monitoring, or which populations are at risk for adverse outcomes.
Integration of exposure data with CTD’s extensive knowledgebase of chemical–gene–disease content, phenotypes, GO terms, and pathways increases the number of potential relationships between chemical stressors and genes, diseases, and phenotypes, and promotes investigation beyond the curated exposure data to explore direct and inferred relationships that can be tested experimentally. For example, 42 unique genes that are associated with particulate matter were curated from exposure studies, in comparison with 7,055 genes that are associated with particulate matter in core CTD. CTD’s analytical tools can be used to search for common chemical, gene, disease, or pathway associations among these genes. Here, a Batch Query using particulate-matter–interacting genes as input reveals that Immune System, Signal Transduction, and Metabolism were the most common pathways affected, encompassing 14%, 13%, and 12% of the genes interacting with particulate matter, respectively. Similarly, particulate matter was associated with 90 unique disease outcomes curated from exposure studies, yet associated with 372 other diseases in all modules of CTD combined (including curated content from nonexposure papers in laboratory animals), which can be filtered by disease category, and explored by enriched pathways, GO terms, and diseases among genes in the inference network. Common disease categories can be visualized under the Diseases tab of the particulate matter chemical page, which identifies some of the top categories of curated particulate matter–disease associations as Respiratory Tract Diseases (15%) and Cardiovascular Diseases (10%).
Limitations
At CTD, exposure papers are manually triaged and curated from PubMed, and the sheer volume of exposure studies and related literature exceeds available CTD resources to curate every relevant publication. By prioritizing curation of exposure papers from high-impact journals, we aim to incorporate new, relevant exposure content as quickly as possible. As well, integration of new text-mining tools similar to those currently used successfully in conjunction with our CTD-inclusive curation (Davis et al. 2013) will automate and further inform our triage process. Finally, technological advances that will transition our curation from spreadsheets to our existing online curation tool are underway to help increase the efficiency of manual exposure curation. A second limitation remains the variability of data types and reporting practices among exposure studies (Grondin et al. 2016). Guidelines to assist researchers in the design and reporting of epidemiological studies are evolving, such as the STROBE-ME initiative (STrengthening Reporting of OBservational studies in Epidemiology-Molecular Epidemiology) (Gallo et al. 2011). Adoption of guidelines such as these by epidemiologists will contribute to broader transparency, consistency, and reuse of data by CTD and others in the exposure research community. Thirdly, the majority of the curated exposure studies to date have focused on the effects of single chemical compounds rather than chemical mixtures. We recognize the importance of including the effects of chemical mixtures in our curation paradigm to benefit both exposome research and complementary AOP networks, and we are working to incorporate data on chemicals as costressors, as well as chemical classes and chemical mixtures as stressors.
Conclusions
Since its inception, one of our goals for CTD’s exposure-curation initiative has been to establish a centralized resource of exposure data that will provide a more complete view of environmental exposures, identify gaps in knowledge, and help prioritize or refine future exposure studies. CTD centralizes exposure data by curating 33 unique content areas (when available) from exposure studies, using a combination of standardized and controlled vocabularies and making this information freely available. Collectively, CTD’s curated exposure content and integration with 1.8 million chemical–gene, chemical–disease, and gene–disease interactions and 80,500 chemical–phenotype relationships combined with our analytical tools, can be directly applied to facilitate these goals.
We invite feedback from the public to optimize future development of CTD’s exposure module, so that it can continue to evolve as an invaluable resource to the scientific community.
Acknowledgment
The authors would like to thank R. McMorran for system administration support. This project is supported by NIEHS grants ES014065, ES019604, ES023788, and ES025128.
References
- Ankley GT, Bennett RS, Erickson RJ, Hoff DJ, Hornung MW, Johnson RD, et al. 2010. Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environ Toxicol Chem 29(3):730–741, PMID: 20821501, 10.1002/etc.34. [DOI] [PubMed] [Google Scholar]
- Davis AP, Grondin CJ, Johnson RJ, Sciaky D, King BL, McMorran R, et al. 2017. The comparative toxicogenomics database: update 2017. Nucleic Acids Res 45(D1):D972–D978, PMID: 27651457, 10.1093/nar/gkw838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Grondin CJ, Lennon-Hopkins K, Saraceni-Richards C, Sciaky D, King BL, et al. 2015. The comparative toxicogenomics database's 10th year anniversary: update 2015. Nucleic Acids Res 43(Database issue):D914–D920, PMID: 25326323, 10.1093/nar/gku935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Murphy CG, Saraceni-Richards CA, Rosenstein MC, Wiegers TC, Mattingly CJ. 2009. Comparative toxicogenomics database: a knowledgebase and discovery tool for chemical-gene-disease networks. Nucleic Acids Res 37(Database issue):D786–D792, PMID: 18782832, 10.1093/nar/gkn580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Wiegers TC, Johnson RJ, Lay JM, Lennon-Hopkins K, Saraceni-Richards C, et al. 2013. Text mining effectively scores and ranks the literature for improving chemical-gene-disease curation at the comparative toxicogenomics database. PLoS One 8(4):e58201, PMID: 23613709, 10.1371/journal.pone.0058201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Wiegers TC, King BL, Wiegers J, Grondin CJ, Sciaky D, et al. 2016. Generating gene ontology-disease inferences to explore mechanisms of human disease at the comparative toxicogenomics database. PLoS One 11(5):e0155530, PMID: 27171405, 10.1371/journal.pone.0155530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Wiegers TC, Rosenstein MC, Murphy CG, Mattingly CJ. 2011. The curation paradigm and application tool used for manual curation of the scientific literature at the comparative toxicogenomics database. Database (Oxford) 2011:bar034, PMID: 21933848, 10.1093/database/bar034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escher BI, Hackermüller J, Polte T, Scholz S, Aigner A, Altenburger R, et al. 2017. From the exposome to mechanistic understanding of chemical-induced adverse effects. Environ Int 99:97–106, PMID: 27939949, 10.1016/j.envint.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo V, Egger M, McCormack V, Farmer PB, Ioannidis JP, Kirsch-Volders M, et al. 2011. STrengthening the reporting of observational studies in epidemiology–molecular epidemiology (STROBE-ME): an extension of the STROBE statement. PLoS Med 8(10):e1001117, PMID: 22039356, 10.1371/journal.pmed.1001117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin CJ, Davis AP, Wiegers TC, King BL, Wiegers JA, Reif DM, et al. 2016. Advancing exposure science through chemical data curation and integration in the comparative toxicogenomics database. Environ Health Perspect 124(10):1592–1599, PMID: 27170236, 10.1289/EHP174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly CJ, McKone TE, Callahan MA, Blake JA, Hubal EA. 2012. Providing the missing link: the exposure science ontology ExO. Environ Sci Technol 46(6):3046–3053, PMID: 22324457, 10.1021/es2033857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingone JA, Buck Louis GM, Nakayama SF, Vermeulen RC, Kwok RK, Cui Y, et al. 2017. Toward greater implementation of the exposome research paradigm within environmental epidemiology. Annu Rev Public Health 38:315–327, PMID: 28125387, 10.1146/annurev-publhealth-082516-012750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinken M. 2016. Adverse outcome pathways as tools to assess drug-induced toxicity. Methods Mol Biol 1425:325–337, PMID: 27311472, 10.1007/978-1-4939-3609-0_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild CP. 2005. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev 14(8):1847–1850, PMID: 16103423, 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- Zhang F, Li L, Krafft T, Lv J, Wang W, Pei D. 2011. Study on the association between ambient air pollution and daily cardiovascular and respiratory mortality in an urban district of Beijing. Int J Environ Res Public Health 8(6):2109–2123, PMID: 21776219, 10.3390/ijerph8062109. [DOI] [PMC free article] [PubMed] [Google Scholar]