Abstract
Background:
Bisphenol A (BPA) is an endocrine-disrupting chemical (EDC) that might be harmful to human health. Recently, there has been widespread usage of bisphenol chemicals (BPs), such as bisphenol AF (BPAF) and bisphenol S (BPS), as replacements for BPA. However, the potential biological actions, toxicity, and the molecular mechanism of these compounds are still poorly understood.
Objectives:
Our objective was to examine the estrogenic effects of BPA, BPAF, and BPS and the molecular mechanisms of action in the estrogen receptor alpha () complex.
Methods:
In vitro cell models were used to compare the estrogenic effects of BPA, BPAF, and BPS to estrogen. Microarray Assay for Real-Time Coregulator-Nuclear receptor Interaction (MARCoNI) analysis was used to identify coregulators of BPA, BPAF, and BPS, and molecular dynamic (MD) simulations were used to determine the compounds binding in the .
Results:
We demonstrated that BPA and BPAF have agonistic activity for both and , but BPS has -selective specificity. We concluded that coregulators were differentially recruited in the presence of BPA, BPAF, or BPS. Interestingly, BPS recruited more corepressors when compared to BPA and BPAF. From a series of MD analysis, we concluded that BPA, BPAF, and BPS can bind to the ER–ligand-binding domain with differing energetics and conformations. In addition, the binding surface of coregulator interactions on was characterized for the BPA, BPAF, and BPS complexes.
Conclusion:
These findings further our understanding of the molecular mechanisms of EDCs, such as BPs, in ER-mediated transcriptional activation, biological activity, and their effects on physiological functions in human health. https://doi.org/10.1289/EHP2505
Introduction
Estrogens, including the endogenous hormone estradiol (E2), are key regulators for growth, differentiation, and a variety of physiological functions. E2 acts through the two estrogen receptors (ERs), and (Lubahn et al. 1993; McDonnell and Norris 2002; Nilsson et al. 2001; Pettersson and Gustafsson 2001). These two receptors belong to the nuclear receptor (NR) superfamily of ligand-inducible transcription factors (TFs) (Hall and McDonnell 2005). Both exhibit distinct tissue-specific expression patterns and biological roles (Deroo and Korach 2006). Exposure to estrogen results in a dramatic increase in the amount of ER binding to chromatin (Taylor and Al-Azzawi 2000). When ERs bind to chromatin, their interaction with the RNA polymerase II holocomplex and the surrounding chromatin environment is modified by various coregulators (Lonard and O'Malley 2012). The major mechanisms of ER-mediated transcriptional gene regulation are a) the classical genomic mechanism, in which the ligand-bound ER interacts directly with DNA estrogen response elements (EREs) that are located in the regulatory regions of the ER target genes; b) the nonclassical genomic tethered mechanism, in which the ER interacts with other TFs, such as AP-1 or Sp1, to regulate gene expression; and c) the rapid nongenomic action mechanism, which activates intracellular signaling cascades involving p44/42 MAKP, Src, and Akt (Burns et al. 2011; Hall and McDonnell 2005; O'Lone et al. 2004).
In the past decade, more than 400 transcriptional coregulators of NRs have been identified and characterized (Dasgupta et al. 2014). Coregulators, which are either coactivators or corepressors, interact with NRs and other TFs to alter chromatin and facilitate gene activity (Dasgupta et al. 2014). Steroid receptor coactivators (SRCs), SRC-1, SRC-2, and SRC-3, were the first gene family to be identified and characterized as coactivators (Lonard and O'Malley B 2007). Each member of the SRC family can enhance the transcriptional activity of NRs by acting as a bridging molecule, which assists protein–protein interactions between NRs and multiple components that facilitate the assembly of the transcriptome complex at the target gene promoter (Dasgupta et al. 2014). Peroxisome proliferator-activated receptor gamma coactivator 1s (PGC1s), including and , are a family of coactivators that are key in energy metabolism (Knutti and Kralli 2001; Puigserver and Spiegelman 2003), but they also increase the transcriptional activity of many other NRs, including the ERs (Tcherepanova et al. 2000).
A large number of natural and synthetic chemicals are reported to disrupt normal functions of the endocrine system (Henley and Korach 2010). Bisphenol chemicals (BPs), including bisphenol A (BPA), bisphenol AF (BPAF), and bisphenol S (BPS), are a group of synthetic chemicals widely used in plastics, food packaging, and other products (Wetherill et al. 2007). BPA is an endocrine-disrupting chemical (EDC) that might be harmful to human health due to its ability to regulate the actions of ERs (Li et al. 2012; Rochester 2013; Welshons et al. 2006). Because of its use in a number of consumer products, concerns regarding the toxicological effects of BPA grew, and BPA is gradually being replaced with analogs such as BPAF and BPS (Liao et al. 2012a). BPAF is a fluorinated derivative of BPA used in many consumer products (Akahori et al. 2008). Humans are exposed to both BPA and BPAF, and effects on reproductive and developmental systems have been demonstrated in vitro (Bay et al. 2004; Bermudez et al. 2010). Recent studies have demonstrated that the estrogenic activity of BPS is similar to BPA (Grignard et al. 2012; Rochester and Bolden 2015). Thermal receipt paper is a potential source of occupational exposure to BPS (Thayer et al. 2015). Both BPA and BPS have been implicated in obesity and steatosis processes, but findings from a recent in vitro study suggest that they may act through different metabolic pathways (Héliès-Toussaint et al. 2014). However, overall, the molecular mechanisms attributed to the BPs are still poorly understood.
In this study, we performed functional analysis using several cell model systems to compare the agonistic effects of BPA, BPAF, and BPS on responses. Using Chromatin Immunoprecipitation (ChIP) analysis, we determined direct binding occurs in the presence of BPA, BPAF, and BPS to the ERE of an ER target gene promoter. We profiled the recruitment of coregulator-derived peptides in the ligand (BPA, BPAF, or BPS)– complexes using a peptide chip, Microarray Assay for Real-Time Coregulator-Nuclear receptor Interaction (MARCoNI) assay. Furthermore, we evaluated differences or similarities among E2, BPA, BPAF, and BPS in the ligand complex through computational analysis.
Materials and Methods
Chemicals
The 17 (E2) was purchased from Sigma-Aldrich, and ICI 182,780 (ICI) was purchased from Tocris Bioscience. BPA, BPAF, and BPS used in this study were provided by the Midwest Research Institute via a contract with the National Toxicology Program (NTP). All stock solutions were made in DMSO and kept at .
Plasmids
The expression vectors pcDNA3 was purchased from Invitrogen. An internal control plasmid for transfection efficiency, pRL-TK renilla luciferase (pRL-TK Luc), was purchased from Promega. The luciferase reporter plasmid, 3xERE Luc (synthetic vitellogenin ERE-TATA box sequence fused to a luciferase reporter gene), , and (full-length human ER expression plasmids) have been described previously. The following plasmids were gifts from D. McDonnell (Duke University): pcDNA/SRC-1, -2, -3, and , .
Cell Lines and Tissue Culture
The human endometrial adenocarcinoma stable cell lines Ishikawa/vector (Ishikawa/vec) and () have been described previously (Burns et al. 2011, 2014). The human breast cancer cell line MCF-7 and human hepatocellular cancer cell line HepG2 were purchased from American Type Culture Collection. The human ovarian cancer cell line BG-1 FR has been described previously (Li et al. 2014). The human bone osteosarcoma epithelial recombinant U2OS cells stably expressing enhanced green fluorescent protein (EGFP)–tagged human fused to the C-terminus of EGFP were purchased from Thermo Fisher Scientific. Ishikawa/vec and cells were maintained in phenol red–free Dulbecco’s Modified Essential Medium (DMEM):F12 medium (Invitrogen) supplemented with 10% Fetal Bovine Serum (FBS; Gemini Bio Products) and Geneticin® (G418; Invitrogen). HepG2 cells were maintained in phenol red–free Minimum Essential Medium (MEM; Invitrogen) supplemented with 10% FBS and L-glutamine (Invitrogen). MCF-7 and BG-1 FR cells were maintained in phenol red–free DMEM:F12 medium (Invitrogen) supplemented with 10% FBS and L-glutamine. Recombinant cells were maintained in phenol red–free DMEM supplemented with 10% FBS and L-glutamine. For cell-starving conditions, 5% or 10% charcoal/dextran stripped FBS (sFBS; Gemini Bio Products) was substituted for FBS in the medium.
Transient Transfection and Luciferase Reporter Analysis
Cells were seeded in 24 well plates with 10% sFBS medium overnight. Effectene transfection reagent (Qiagen) was used for transient transfection. For ER-negative HepG2 cells, a total of of DNA, including of or , of 3xERE Luc, and of pRL-TK Luc plasmids were transfected into the cells. For Ishikawa/vec, stable cells, and ER-positive cells (MCF-7 and BG-1 FR), a total of of DNA, including of 3xERE Luc and of pRL-TK Luc plasmids, were transfected. After 8 h, cells were changed to fresh 10% sFBS medium overnight and then treated with vehicle control (DMSO, final concentration ), E2, or BPAs (1, 10, 100, and ). For coregulator experiments in HepG2 cells, a total of of DNA, including of , of either SRC-1, SRC-2, SRC-3, , or , of 3xERE Luc, and of pRL-TK plasmids were transfected into the cells. After 8 h, cells were changed to fresh 10% sFBS medium overnight and then treated with vehicle control (DMSO, final concentration ), E2 (), or BPAs (100 and ) for 18 h.
Luciferase assays were performed using the Dual Luciferase Reporter Activity System (Promega). Transfection efficiency was normalized against the renilla luciferase. Fold changes were calculated relative to vehicle controls. All experiments were repeated at least three times. Data shown are the average of triplicate determinations in a representative experiment. Values were calculated relative to vehicle control and presented as (SEM).
RNA Extraction and Real-Time Polymerase Chain Reaction Analysis
Cells were seeded in dishes with 5% sFBS medium and starved for 2 d. After changing to fresh 5% sFBS medium, cells were treated with vehicle control (DMSO, final concentration ), E2, or BPs for 18 h. Total RNA was extracted using RNeasy Mini Kit (Qiagen). First-strand cDNA synthesis was performed using SuperScript reverse transcriptase according to the manufacturer’s protocol (Invitrogen). The mRNA levels were measured using SYBR™ green assays (Applied Biosystems). The sequences of primers used in quantitative real-time polymerase chain reaction (qPCR) and ChIP qPCR are shown in Table S1. Cycle threshold values were obtained using the ABI PRISM™ 7900 Sequence Detection System and analysis software (Applied Biosystems). Each sample was normalized against expression. Experiments were repeated three times, and results are presented as . Changes of endogenous gene expression by E2 were calculated as fold change relative to the vehicle control group in each cell line.
Chromatin Preparation and ChIP–Quantitative PCR Analysis
Cells were seeded in dish with 5% sFBS medium and starved for 2 d. After changing to fresh 5% sFBS medium, cells were treated with vehicle control (DMSO, final concentration ), E2 (), or BPs () for 40 min. Chromatin preparation and ChIP assay were carried out with , anti-p300, or anti-CBP antibody using the Chromatin Immunoprecipitation Assay Kit (Cat.# 17-295; Millipore/Upstate®). Purified chromatin DNA was used for qPCR analysis. The sequences of the ChIP primers used in real-time PCR were as follows: for amplicon (): the forward primer 5′-TGGCACATAGTAGGTCCAGC-3′, reverse primer 5′-GTTTCTGGCAGGCAGATTGG-3′; for amplicon (ERE): the forward primer 5′-TGTTGTGCCTCCAGCTCCTG-3′, reverse primer 5′-GGTTTCTGGCAGGCAGATT-3′. Relative recruitment was normalized to signals obtained from input DNA. Experiments were repeated three times, and results are presented as .
Cell Lysis Preparation and Microarray Assay for Real-Time Coregulator-Nuclear Receptor Interaction Assay
Recombinant U2OS cells stably expressing human fused to the C-terminus of EGFP were maintained in phenol red–free DMEM in the presence of L-Glutamine, 1% penicillin–streptomycin, G418, and 10% FBS at 37°C, 5% , 95% humidity incubator. Snap-frozen cell pellets were slowly thawed on ice and suspended in ice-cold lysis buffer (Perera et al. 2017) supplemented with Halt™ protease inhibitor cocktail (Pierce Biotechnology), using 150 uL per cells. The cell suspension was further snap-frozen, lysed, and centrifuged to obtain clear supernatant for the analyses later. Cell lysates were isolated from the full-length human U2OS cells. For MARCoNI, assay mixtures that contain cell lysates, of Alexa Fluor 488–conjugated GFP antibody (Invitrogen), and ligand (E2, BPA, BPAF or BPS, prediluted in DMSO, final concentration, 2%) were prepared on ice. The ligand-modulated coregulator interactions with the EGFP-tagged was assessed using a PamChip® plate that contained the 154 coregulator-derived binding peptides (Excel Table S1), including the LXXLL coactivator motif or the LXXXIXXXL corepressor motif from 66 different coregulators (PamGene International B.V.) (Beekmann et al. 2015; Koppen et al. 2009). The reaction was performed in a fully automated microarray processing platform at 20°C for 80 cycles (two cycles per minute). After removal of the unbound receptor by washing each array with in Tris-buffered Saline (TBS) buffer, images were obtained by the Charge-couple device (CCD) camera of the PamStation®96, and then were analyzed for quantification of ER binding using BioNavigator software (PamGene International B.V.). For data and statistical analysis, BioNavigator, Microsoft Excel (version 14.0.7,106.5,003; Microsoft Corporation) IBM SPSS Statistics (version 22; International Business Machines Corporation), and R (version 2.15.2; R Development Core Team), a language and environment for statistical computing, were used. Modulation index (MI) is the -transformed relative binding value, which is calculated by the compound’s binding value, relative to the vehicle control (DMSO) binding value. Positive interaction indicates that ligands increase motif interactions, and the cutoff is MI, ; the binding value, ; the relative binding value, ; p-value, . Negative interaction indicates that ligands decrease motifs interactions and the cutoff is MI, ; the binding value, ; the relative binding value, ; p-value, .
Molecular Modeling
Using the X-ray crystal structures of E2- (pdb ID 3UUD), BPA- (pdb ID 3UU7), and BPAF- (pdb ID 3UUA) bound to ER, starting structures of the ligand-binding domains of ER were created for molecular dynamics (MDs). Missing residues (462–464 in 3UUD, 462–466 in 3UU7, and 462–463 in 3UUA) were introduced using the program Modeller (version 9.14; Andrej Sali) (Sali and Blundell 1993) and missing side chains (305, 306, 397, 467–477, 492, 531, and 545 in 3UUD; 306, 330, 419, 467–472, 531–533, and 548 in 3UU7; and 306, 330, 334, 335, 337, 339, 342, 363, 413, 416, 417, 419, 423, 466–473, 477, 492, 523, 525, and 533 in 3UUA) were reconstructed using the program Coot (version 0.8.2; Paul Emsley) (Emsley et al. 2010). Initial configurations of E2–ER were directly constructed from the crystal structure 3UUD. Initial structures with the normal pose of the ligands BPA, BPAF, and BPS bound in agonist conformations were modeled using the crystal structure of 3UU7, and the alternate poses of BP compounds were based on the crystal structure of 3UUA. Starting conformations of the ligand-free ER came from 3UUD (in which ER was in the agonist conformation) and the antagonist conformation of ER found in BPC bound to ER (pdb ID 3UUC) after removing the ligands. Protons were added, followed by the addition of counter ions, and the resulting complex structures were then solvated in a box of water for each system. The amount of water molecules varied from 25,500 to 26,800 for these systems. Prior to equilibration, each system was subjected to the following steps: a) over a nanosecond of constrained MDs with the peptide and the ligand constrained to the original position with a force constant of , b) minimization, c) low-temperature, constant-pressure MD simulation to assure a reasonable starting density, d) minimization, e) stepwise heating MD at constant volume, and f) a constant-volume MD run for another 9 nanoseconds. All final unconstrained trajectories were calculated at 300 K under constant volume MD (300 ns total dynamics for ligand bound systems and 400 ns for ligand-free systems at 1 fs time step) using PMEMD from the Amber 14 program to accommodate long-range interactions (Case et al. 2014). The amino acid parameters were taken from the Amber SB14 force field. The charges of atomic positions of the ligands were derived using the ChelpG scheme with the 6–31g(d) basis, set at the B3LYP level using the program Gaussian 09.D01 (Gaussian Inc.). The other ligand force field parameters were generic parameters listed in the Amber force field. Additional variable length MD simulations were performed with the same protocol for each ligand in water to establish the solvation behavior and energies of each ligand. Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) calculations were performed for all the ligand-bound systems. Configurations saved from the final 250 ns of the dynamic trajectories were used for the MM/GBSA calculations. The Generalized Born model (Onufriev et al. 2004) () was used to calculate polar contributions of desolation. The dielectric constants of 80 and 1 were used for the solvent and solutes. Using the normal mode analysis, conformational entropy contributions to the binding energy were calculated using Amber 14.
Statistical Analysis
One-way and two-way analysis of variance with Dunnett’s Multiple Comparison test (**, *** or ****) were performed using GraphPad Prism (version 7.01; GraphPad Software, Inc.).
Results
Estrogenic Activation of and by BPA, BPAF, and BPS, Alterations in Expression of ER-Target Genes, and Direct Binding of to an ERE Located in the Promoter of ER-Regulated Genes
To evaluate the ERE-mediated transcriptional activity of when treated with BPA, BPAF, and BPS, promoter activation was performed using several cell systems. The chemical structures of compounds used in this study are shown in Figure 1A. The ER dependence of estrogenic activation was confirmed using human endometrial cancer Ishikawa cells stably expressing cells (Burns et al. 2011, 2014). As shown in Figure 1B, E2 induced ERE-mediated activation by fourfold. BPA-induced ERE-mediated activation by threefold at . BPAF induced ERE-mediated activation by threefold at , and matched the efficacy of E2 at . BPS only induced ERE-mediated activation by threefold at the highest concentration () in cells. ERE-mediated activation was weak or absent in Ishikawa/vector cells, suggesting that the activation by BPA, BPAF, and BPS are predominantly . To investigate the estrogenic activity in the different cell types through an endogenous receptor, human breast cancer MCF-7 ( positive) and ovarian cancer BG-1 FR ( and positive) cells (Burns et al. 2014; Li et al. 2014) were used in reporter assays. In MCF-7 cells, E2 induced ERE-mediated activation by fourfold. However, BPA, BPAF, and BPS induced the reporter activation weakly, even at the higher concentrations () (Figure 1C, left). In BG-1 FR cells, similar dose responses were observed after treatment with E2 or BPA and the two analogs (, Figure 1C, right). These data indicate all transcriptional activations with BPA, BPAF, and BPS occur in a concentration-dependent manner, and the sensitivity of responses to the individual compounds are cell type–specific.
Figure 1.
Bisphenol A (BPA), bisphenol AF (BPAF), and bisphenol S (BPS) activate estrogen receptor (ER) estrogen response element (ERE)–mediated transcriptional activity, alter expression of ER target genes, and directly bind to ERE sites in target genes. (A) The chemical structures of the compounds tested in this study. (B) Activation in Ishikawa cells stably expressing cells. (C) Activation of endogenous ER MCF-7 and BG-1 FR cells. (D). Co-transfection with or in HepG2 cells. Cells were transfected with 3xERE-luc and pRL-TK plasmids in MCF-7, BG-1 FR, or co-transfection with the or expression plasmid in HepG2 cells overnight. After changing to fresh starved medium, cells were treated with vehicle control, estradiol (E2), BPA, BPAF, and BPS for 18 h. The ER ERE-mediated activity was detected by the luciferase reporter assay as described in the materials and methods. Data shown are representative of triplicates as fold increases that were calculated relative to the vehicle , ***, **, * compared to vehicle control. (E) Changes of the ER target genes in MCF-7 cells. Cells were treated with vehicle control, E2, or BPA, BPAF, and BPS, with or without ICI 182,780 (ICI) for 18 h, respectively. Total RNA was extracted and used as a template for cDNA synthesis. Gene expression was measured by quantitative PCR (qPCR). Experiments were repeated three times, and results are presented as , **, ***, ****. (F) BPA-, BPAF-, and BPS-induced binding at an ERE site of the human WISP-2 gene promoter. Cells were treated with vehicle control, E2, and 100 or BPA, BPAF, or BPS for 40 min. ChIP assay was carried out with antibody using a ChIP Assay Kit as described in the materials and methods. Purified chromatin DNA was used for qPCR analysis by the special ChIP primers for the amplicons () or (ERE). Relative recruitments of were normalized to signals obtained from input DNA. Experiments were repeated three times, and results are presented as , **, ***, ****.
To compare the - and transcriptional activation, we performed reporter assays in human hepatocellular cancer HepG2 (ER negative) cells after transfecting with an or expression plasmid. For ERE-mediated activation, cells were highly responsive to E2, with up to 35-fold increases at (Figure 1D, left). BPA induced this activation by 25-fold at . BPAF induced activation by 17-fold at and 30-fold at . In contrast, BPS only weakly activated the ERE at (eightfold). With the highest concentration of , all three chemicals displayed strong activation of by greater than 30-fold. For , E2 induced ERE-mediated activation with a lower fivefold increase at (Figure 1D, right). BPA at and BPAF at had weaker activation when compared to E2 at the same concentrations. BPA at and BPAF at had strong activity for ERE-mediated activation. However, BPS did not have ERE-mediated activation in HepG2 cells. These results indicate that BPA and BPAF can activate ERE-mediated transcription via and , but BPS exhibited more ligand specificity for .
To investigate the ER-dependent response of BPA, BPAF, and BPS, we examined their effects on three well characterized ER target genes, GREB1 (gene regulation by estrogen in breast cancer 1), PGR (progesterone receptor), and WISP-2/CNN5 (WNT1-inducible signaling pathway protein 2) using real-time PCR analysis in MCF-7 cells (Figure 1E). All three chemicals, with varying degrees, significantly induced GREB1, PGR, and WISP-2/CNN5. In addition, the pure ER antagonist, ICI, blocked expression of all three ER target genes when cotreated with E2, BPA, BPAF, and BPS, indicating the ER receptor specificity of the activity response. These results indicate that BPA, BPAF, and BPS can alter transcriptional regulation directly on endogenous ER target genes.
To determine if directly interacts with ERE binding sites in the presence of BPA, BPAF, and BPS, we performed ChIP–qPCR analysis in MCF-7 cells. Promoter analysis showed that the upstream region of WISP-2/CNN5 harbors two AP-1 sites ( and ) and an ERE site () (Figure 1F, top). As expected, E2 increased enrichment significantly to both regions by 14-fold on the () and fivefold on the (ERE) regions (Figure 1F, bottom). BPA and BPAF significantly increased enrichments in both () and (ERE) regions. BPS weakly increased enrichments in the (ERE) region (Figure 1F, bottom). However, enrichment with BPS was not seen in the () region of the WISP-2 gene promoter. These data demonstrate directly binds differentially to EREs of ER target gene promoters when treated with BPA or BPAF.
Interaction Profiling and Comparison of BPA, BPAF, and BPS Coregulator-Derived Binding Peptides on
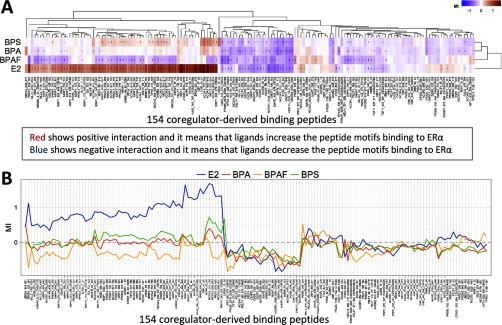
To further support the molecular basis of BPA actions, we investigated ligand-modulated coregulator recruitment using MARCoNI. A working model of MARCoNI is shown in Figure S1. The ligand (E2, BPA, BPAF, and BPS)-modulated interaction of with coregulators was quantified (Figure 2). In the heatmap, the red color depicts positive interactions, which suggests that the ligand increases peptide binding, and the blue color depicts negative interactions, which suggests that the ligand decreases peptide binding to (Figure 2A). As a parameter for modulation of the interaction of with each coregulator peptide motif on the array, MI was calculated (Figure 2B). The complete data set of the peptide array analysis for E2, BPA, BPAF, and BPS is summarized in Excel Table S2.
Figure 2.
Differential coregulators recruited by bisphenol A (BPA), bisphenol AF (BPAF), or bisphenol S (BPS) in the complex using Microarray Assay for Real-time Coregulator-Nuclear receptor Interaction (MARCoNI) analysis. (A) Heat map. The ligand-modulated coregulator of interaction with the enhanced green fluorescent protein (EGFP)–tagged was assessed using a PamChip® that contains 154 coregulator-derived motif peptides. Hierarchical clustering (Euclidean distance, average linkage) was applied to sort compounds and motifs, and visualize (dis)similarity by dendrograms. The red color shows positive interactions/increased bindings, and the blue color shows negative interactions/decreased bindings of the ligands. (B) Modulation Index (MI). binding was quantified using Bionavigator software. MI is the –transformed relative binding value, which is calculated by the compound’s binding value, relative to the vehicle control (DMSO) binding value. (C) Positive interactions. The cutoff for the binding peptides is MI, ; the binding value, ; the relative binding value, ; p-value, . (D) Negative interactions. The cut off for the binding peptides is MI, ; the binding value, ; the relative binding value, ; p-value, .
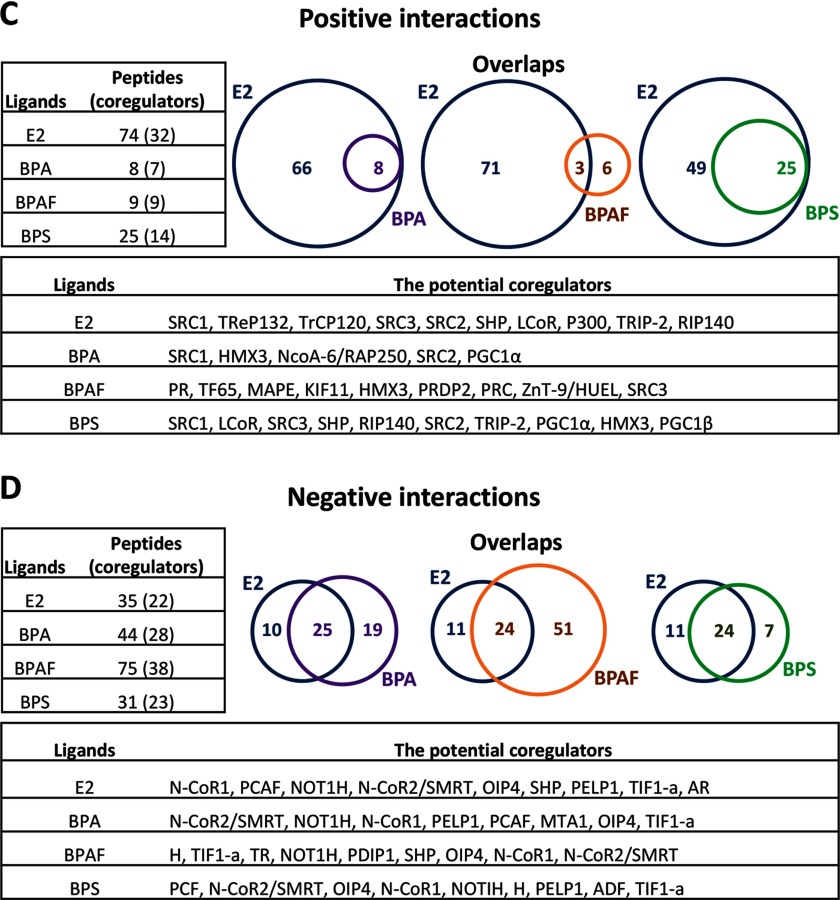
A summary of the positive interaction peptides in the complexes are shown in Figure 2C. E2 facilitated the binding of 74 peptides from 32 different coregulators. In contrast, BPA significantly enhanced the binding of 8 peptides from 7 different coregulators. BPAF significantly enhanced the binding of 9 peptides from 9 different coregulators, and BPS significantly enhanced the binding of 25 peptides from 14 different coregulators (Figure 2C, left). To further understand the differential effects of BPA, BPAF, and BPS on function, the recruitment of coregulator motifs, as induced by BPA, BPAF, and BPS, were compared to that observed with E2 stimulation of the . BPA- and BPS-bound peptides overlapped 100% with E2-modulated peptides. In comparison, only 33% of BPAF-bound peptides (3 out of 9) were also recruited by E2 (Figure 2C, right). Comparing the profiles of positive interactions of BPA, BPAF, and BPS to E2 on the complex show highly differential interactions (Tables S2–S5). In certain instances, the interactions were similar, but with a lower binding value compared to E2 (e.g., NCOA1_620_643 was 2.0 for BPA; 45.1 for E2) and with another portion of the same coactivator, NCOA1, no binding occurred (NCOA1_1421_1441) (Tables S2–S5). Analysis of the BPAF profile showed overall low binding values, but showed interactions to different factors not bound by E2 [e.g., peroxisome proliferator-activated receptor gamma, coactivator-related 1 (PPRC1)] (Tables S2–S5). The complexes exhibited more interactions than with BPA or BPAF, but these binding activities were generally lower than with E2 in the complexes, and in certain examples, selective to the BPS complex only (e.g., JHD2C, LCOR, and certain portions of MED1_591_614) (Tables S2–S5). In general, BPS had weaker interactions with the SRC family members than BPA or BPAF. The potential coregulators from positive interaction peptides in the ligand– complexes are summarized in Figure 2C, bottom. Interestingly, the unique peptides that were derived from PR, TF65, MAPE, KIF11, and PRDP2 were recruited only by BPAF.
A summary of the negative interaction peptides in the complexes are shown in Figure 2D. E2 was found to significantly modulate binding with 35 peptides from 22 different coregulators, BPA bound 44 peptides (28 coregulators), BPAF bound 75 peptides (38 coregulators), and BPS bound 31 peptides (23 coregulators) in the complexes (Figure 2CD, right). Comparing the profiles of the negative interactions showed that all three BPs overlapped with the E2 profile, but to varying degrees (Tables S6–S9). Fifty-seven percent of BPA-bound peptides (25 out of 44) overlapped with E2-modulated peptides, and 77% of BPS peptides (24 out of 31) overlapped with E2 peptides. However, only 32% of BPAF peptides (24 out of 75) recruited were the same as E2 (Figure 2D, right). Interestingly, all three BPs showed unique peptide interactions with factors not bound by E2 (e.g., BLIS1, CBP, GNAQ) (Tables S6–S9). With NCOR1 and NCOR2, there were interactions with E2 (NCOR1_2039_2061 and NCOR2_2330_2352), but at different sites in the coactivator (Tables S6–S9). NELFB interacted with E2, BPAF, and BPS, but not with the bound complex (Tables S6–S9). The potential coregulators (from negative interaction peptides) in the complexes are summarized in Figure 2D, bottom. Interestingly, the peptides that were derived from N-CoR1, N-CoR2/SMART, OIP4, and TIF1-a were recruited by all ligands. Thus, these results together demonstrate some of the first evidence that the coregulators in the complexes are differentially recruited in the presence of BPA, BPAF, or BPS.
Differential Recruitments of Coactivators SRCs and PGCs in Complexes
To evaluate the peptide screening results, we selected two well-characterized coactivator families, SRC and PGC1, for further experimentation. To determine the role of with coregulators in ERE-mediated transcriptional regulation using reporter assays, we co-transfected SRCs or PGC1s with in HepG2 cells. As expected, SRC-1, SRC-2, SRC-3, , and induced ERE-mediated activity when treated with E2 (Figure 3A). For SRCs, in the presence of 100 or BPA, SRC-1 significantly induced ERE-mediated activity at both concentrations, but SRC-2 had minimal effects at the lower concentration, and SRC-3 showed no effect at either concentration (Figure 3B). In contrast, BPAF at 100 or had further stimulation with SRC-3, but no additional effect was observed with SRC-1 or SRC-2 at either concentration. In the presence of BPS, all three SRCs significantly activated ERE-mediated transcription. However, there was no coactivation with either SRCs at . significantly induced ERE-mediated activity at high concentrations of BPA, and was active by BPA at the lower concentration with (Figure 3C). also significantly activated the ERE-mediated transcription in the presence of 100 or BPAF. , however, further induced activity only with BPAF. BPS showed a clear differential preference of activation with in the presence of BPS, but did not show any additional coactivator activity with BPS. These results confirmed the coactivator peptide profile experimentally detected by the MARCoNI assay as shown in Table 1. These data also demonstrate that BPA, BPAF, and BPS induce differential recruitments and interactions with coactivators such as SRCs and PGC1 in the different complexes.
Figure 3.
Differential recruitment of steroid receptor coactivators (SRCs) or peroxisome proliferator-activated receptor gamma coactivator 1 (PGC1) by bisphenol A (BPA), bisphenol AF (BPAF), or bisphenol S (BPS) in the complex in HepG2 cells. (A) Co-transfection with SRCs and PGC1s in the presence of estradiol (E2). (B) Co-transfection with SRCs in the presence of BPA, BPAF, or BPS. (C) Co-transfection with PGC1s in the presence of BPA,BPAF, or BPS. Cells were transfected with 3xERE-luc, pRL-TK, , and pcDNA/SRC1, pcDNA/SRC2, pcDNA/SRC3, , or plasmids overnight. After changing to fresh starved medium, cells were treated with the vehicle control, E2, 100 or BPA, BPAF, and BPS for 18 h. The ERE-mediated activity was detected by a luciferase reporter assay. Data are representative of triplicates as fold increase calculated relative to the vehicle , *** compared to vehicle (control).
Table 1.
Coactivator steroid receptor coactivators (SRCs) and peroxisome proliferator-activated receptor gamma coactivator 1 (PGC1) in the complex by Microarray Assay for Real-Time Coregulator-Nuclear receptor Interaction (MARCoNI) assay.
| Coactivators | Peptides | Peptide sequences | Motifs | Relative binding valuesa | |||
|---|---|---|---|---|---|---|---|
| E2 | BPA | BPAF | BPS | ||||
| SRC1 | NCOA1_737_759 | ASKKKESKDHQLLRYLLDKDEKD | LxxLL749 | 47.5 | 2.5 | — | 1.4 |
| NCOA1_620_643 | SDGDSKYSQTSHKLVQLLTTTAEQ | LxxLL633 | 45.1 | 2.0 | — | 3.6 | |
| NCOA1_1421_1441 | TSGPQTPQAQQKSLLQQLLTE | LxxLL1435 | 5.6 | — | — | 1.3 | |
| NCOA1_677_700 | PSSHSSLTERHKILHRLLQEGSPS | LxxLL690 | 5.5 | — | — | 1.6 | |
| SRC2 | NCOA2_628_651 | GQSRLHDSKGQTKLLQLLTTKSDQ | LxxLL641 | 12.8 | 1.3 | — | 1.9 |
| NCOA2_677_700 | STHGTSLKEKHKILHRLLQDSSSP | LxxLL690 | 9.7 | — | — | 1.7 | |
| NCOA2_733_755 | EPVSPKKKENALLRYLLDKDDTK | LxxLL745 | 8.6 | — | — | 1.4 | |
| SRC3 | NCOA3_609_631 | KKKGQGVIDKDSLGPLLLQALDG | LxxLL113 | 21.1 | — | — | 2.4 |
| NCOA3_609_631_C627S | QRGPLESKGHKKLLQLLTCSSDD | LxxLL621 | 13.1 | — | — | 1.7 | |
| NCOA3_725_747 | MHGSLLQEKHRILHKLLQNGNSP | LxxLL685 | 6.3 | — | — | 1.2 | |
| NCOA3_673_695 | QRGPLESKGHKKLLQLLTSSSDD | LxxLL621 | 5.8 | — | — | — | |
| NCOA3_104_123_N-KKK | HGSQNRPLLRNSLDDLLGPPSNA | LxxLL1041 | 2.5 | — | 1.5 | — | |
| PRGC1_130_155 | DGTPPPQEAEEPSLLKKLLLAPANTQ | LxxLL144 | 5.2 | 1.1 | — | 1.9 | |
| PRGC1_134_154 | PPQEAEEPSLLKKLLLAPANT | LxxLL144 | 5.8 | 0.9 | — | 1.5 | |
| PRGC2_146_166 | PAPEVDELSLLQKLLLATSYP | LxxLL156 | 7.1 | — | — | 1.5 | |
| PRGC2_338_358 | AEFSILRELLAQDVLCDVSKP | LxxLL343 | 6.0 | — | — | — | |
Note: —, no binding; BPA, bisphenol A; BPAF, bisphenol AF; BPS, bisphenol S; E2, estradiol.
Relative binding value shows each peptide binding relative to the control, 2% DMSO. (Cutoff: binding value, ; relative binding, ; p-value, .)
Simulating BPA-, BPAF-, and BPS-Bound by Molecular Dynamics
To investigate if any potential modifications to structure appear on the coregulator binding surface of in the presence of BPs at its natural ligand-binding site, we performed a series of MD simulations. In the X-ray crystal structure determined by Delfosse et al. in 2012, two distinct ligand conformations were identified for bisphenol compounds when bound to at the ligand-binding site (Delfosse et al. 2012). In that work, BPA was bound to in a conformation like the natural substrate, E2 (Figure S2A), and we refer to this mode of the ligand as the canonical conformation, or conformation 1. In this canonical mode of the ligand binding, one phenolic group of BPA interacted with H524 and M421, while the other phenolic oxygen interacted with E353 and R394 and helix 12 (h12) was in a similar conformation to (Figure S2B). In addition to the canonical conformation (Figure S2C), for , the ligand was found to be bound in an alternate conformation (Figure S2D), or conformation 2. Interactions of BPAF with ER residues in the canonical conformation resembled those of BPA, and the alternate conformation displayed a phenolic group interacting with T347 instead of H524 and M421 (Figure S2D). In the X-ray crystal structures of , even though BPAF conformations were different, h12 was observed in a similar conformation to BPA– or (i.e., was in the agonist form). Meanwhile, when another BPA analog, bisphenol C (BPC) was used in the crystal trials (Delfosse et al. 2012), only the alternate conformation of the ligand was observed in the X-ray structure of . Interestingly, in the case of , h12 was located elsewhere with a conformation labeled as an antagonist form of . It is worth noting that the used in the X-ray crystallographic study (Delfosse et al. 2012) was a mutant (Y537S) that enabled successful crystal trials. In contrast, we used the published X-ray crystal coordinates as our starting structures in the MD simulations with Y537 (wide-type ) in the simulated systems.
In the series of MD simulations reported here, three simulations were devoted to establishing a reference system for comparison. In the first, E2 was used to establish the structure dynamic of . Two additional MD simulations were performed without ligand presenting at the ligand-binding site to access the characteristics of tertiary structural elements of , with one having h12 in the agonist conformation, while the other was in the antagonist conformation. Both simulations may be essential, as h12 is implied to be a key player in the determination of the agonist or antagonist conformation. For each ligand, parallel simulations were performed by placing BPs into the two conformations described earlier. In all cases, helix h12 of was in an agonist conformation to match with the agonist effects described in the experimental setup. Furthermore, additional simulations were performed for E2, BPA, BPAF, and BPS ligands in a pure solvent environment (i.e., without present) so that the ligand behavior could be characterized for these ligands prior to their binding at the binding site of .
Convergence of Molecular Dynamic Simulations and Ligand Dynamics
MD simulations for ligand-bound systems were performed over 300 ns, and the ligand-free systems over 650 ns. Dynamic stabilities of the systems were analyzed using the root mean square deviations (RMSD) of the atomic positions of the heavy atoms of the receptor. RMSD values were calculated by superimposing the conformations at each time point with the starting reference conformation of each system while using only the protein heavy atoms for alignment. In general, a model system is said to be unstable if RMSD values continue to rise with time. The ligand-free system that started with the helix h12 in the agonist conformation showed relatively smaller deviations over the entire 650 ns segment of the simulation, indicating that the agonist conformation of is rather stable over a relatively lengthy period, even without the ligand present in the binding site. More dynamic, but still a stable conformation was observed in the RMSD of the ligand-free system with the h12 in an antagonist conformation. Comparison of the final structures with the corresponding starting configurations clearly demonstrate that once h12 is in the agonist conformation, the overall structural features of remain quite unaltered (Figure S4). Contrarily, significant changes in the structure were observed in the antagonist conformation (Figure S5). However, larger deviations in the backbone alignment of h4, a part of the coregulator binding surface, is noticeable in both cases.
Meanwhile, all ligand-bound systems showed relatively small RMSD values (), and the fluctuations of RMSD values (shown in Figure S6) are well within the acceptable range for a stable peptide system of this size with normal fluctuations in atomic positions. In general, this is a reasonable test to confirm that all ligand-bound systems are dynamic but stable during the trajectory calculations over 300 ns. showed relatively smaller RMSD values () over the 300-ns trajectory, and the final conformation (Figure S6) did not exhibit much divergence from the starting X-ray crystal conformation. Both and showed similar variations () to that found in , irrespective of ligand conformation. Meanwhile, displayed slightly more mobility in the canonical conformation of the ligand as opposed to the alternate conformation. Direct comparison of the two final configurations from each ligand-bound displayed minor changes in the tertiary structural features in ER (Figures S8–S10). Relatively larger deviations were observed among the BPS-bound systems (Figure S10), specifically h12.
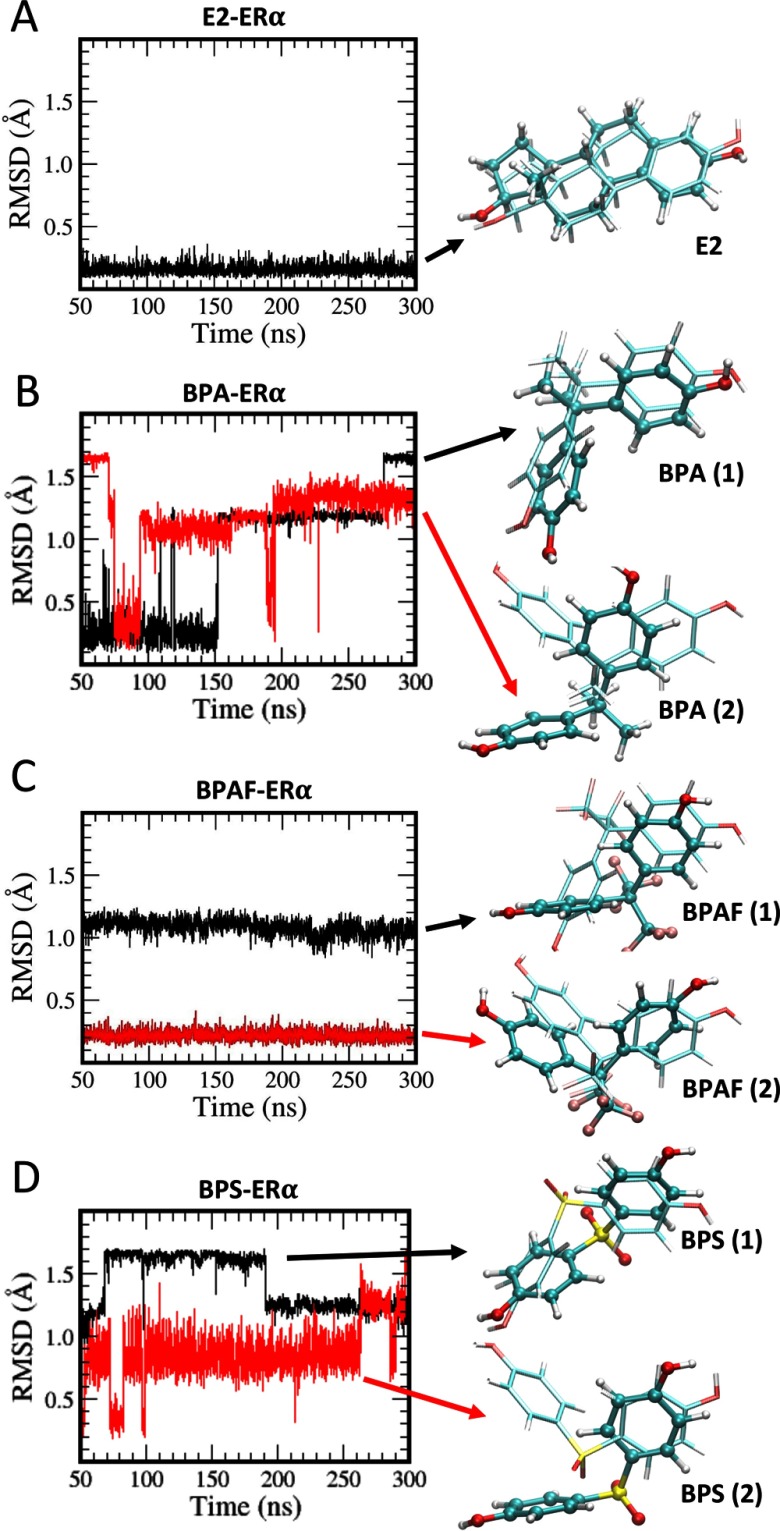
RMSD values calculated from the heavy atoms of the ligands may be an important parameter in analyzing the ligand dynamics, and these values for solvated ligands are given in Figure S11. E2 practically showed no deviations from the starting configuration for over 70 ns, but both BPA and BPS structures showed rapid variations due to faster ring-flipping dynamics, while BPAF conformations were rather stable over a long duration. Only three ring-flipping events were observed for BPAF during the longer 450-ns trajectory. Because we observed how ligand RMSDs correlate with the rigidity (or flexibility) of ligands in solution, one can use the RMSDs of ligands in complex with to obtain information on ligand dynamics at the ligand-binding site during a MD trajectory calculation. For E2, very little mobility was observed, as expected from a rigid ligand in its natural ligand conformation (Figure 4A). The positions of starting and final E2 conformations overlap quite well when structures were used in the alignment (Figure 4A, left).
Figure 4.
Root mean square deviations (RMSD) of ligands. (A) estradiol (E2), (B) bisphenol A (BPA), (C) bisphenol AF (BPAF), and (D) bisphenol S (BPS). The black curve represents the system starting with the ligand in its canonical conformation (conformation 1), and the red curve represents the system started with the ligand in its alternate conformation (conformation 2). In each case, the estrogen receptor () is in the agonist form. The final ligand structures are also shown (in dark ball and stick model) with their starting conformations (in lighter stick model).
Meanwhile, rearrangements in conformations are evident from the ligand heavy atom RMSDs in all three BPs, but with a lesser frequency when compared to the solution structures. BPA in the canonical conformation showed almost no variation in the positions except the ring flipping (Figure 4B), whereas the alternate conformation showed much larger heavy atom positional displacements. BPAF showed almost no ring flipping for both conformations, but showed a larger positional displacement during the early part of the simulation (during the first 50 ns). This final BPAF conformation was closer to the alternate conformation than the starting canonical conformation of the ligand (Figure 4C). BPS, in the canonical conformation, showed more ring flipping than displacement, but when started in the alternate conformation, there were positional displacements of BPS that brought the final conformation more towards the canonical conformation (Figure 4D).
Energetics of Ligand Binding
We observed the potential rearrangements of BPs within the binding site of during dynamics. Therefore, using the conformations selected from the final 250-ns production runs, we performed MM/GBSA calculations to obtain the binding free energies () of BPs to estimate how strongly different ligands bind to . The binding free energy results, along with their components, are summarized in Table 2. Enthalpic contributions to the binding free energy given by were the dominant component in the binding interaction. In all cases, the entropic contributions given by were unfavorable. However, all systems still showed favorable total binding free energies. Total ligand-binding free energies of all BPs were quite comparable to the endogenous E2. However, BPA seemed to show the weakest interactions with . Comparing the enthalpy contributions to the binding free energy , two conformations of BPA showed only difference, and that was further reduced to when the binding free energies () were compared. Similar energy behavior with slightly larger magnitudes was observed for both BPAF (about ) and BPS (about ), showing greater enthalpic contributions () to the binding free energy when compared with the values for configuration 1. However, BPAF and BPS configurations were favored only by 4.6 and when the binding free energies were compared. At this point, looking at the final conformations from dynamics and taking the free energies into consideration, we confirmed the fact that BPA and BPS favor conformation 1 that resemble the X-ray crystal conformation of BPA in , whereas BPAF prefers conformation 2 in the complex that differs from the former in its residue interactions.
Table 2.
Binding free energies () and their enthalphic () and conformational entropic () components (in kcal/mol) of ligands in predicted by MM/GBSA method.
| Energy | E2 | BPA (1) | BPA (2) | BPAF (1) | BPAF (2) | BPS (1) | BPS (2) |
|---|---|---|---|---|---|---|---|
Note: Using Generalized Born model, binding free energies () and their enthalpic () and conformational entropic () components were calculated using the equally spaced 1,000 configurations selected from the final 250 ns of the molecular dynamics trajectories.
Structures and Dynamics of Ligand Binding and a Close Inspection of the Coregulator Binding Surface
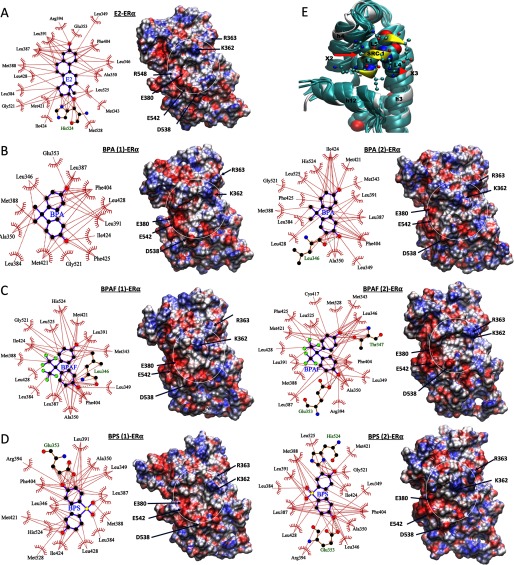
During the examination of ligand RMSDs, we concluded that there were rearrangements in ligand conformations. If the ligands undergo conformation changes, can they change the dynamics of interacting residues by searching for new interacting partners when accommodating changes in ligand conformations? Using the conformations of ligands and their ER interacting partners on the binding site in the X-ray crystal structures (Figure S2) as the reference, we systematically analyzed the ligand–receptor interactions during dynamics. Representative ligand conformations near the end of each trajectory are shown in Figure 5. In the conformation, the interactions between R394 and E353 of was enhanced during the simulation. The strengthening of the salt bridge between R394 and E353 may have weakened their interactions with the phenolic oxygen of E2 displayed in the X-ray crystal structure, but the phenolic oxygen (of E2) remained in the close proximity to these two residues during the entire time of the simulation. The other oxygen of E2 seemed to make more stable contacts with H524, G521, and L525 (Figure 5A), and these contacts remained intact during the entire simulation. It is noticeable (but not surprising, due to its bulkiness) that E2 displayed the largest number of interacting residues (18), and this number remained relatively un-changed (with residues T347 and M528 alternatively appearing in the coordinating shell). Between 13 and 17 residues were involved in direct interactions with BPA, BPAF, or BPS. Due to the flexibility and variable hydrophobicity of BPs, variations in residue participation in the stabilization of these compounds at the ligand-binding site were observed via close examination of the ligand interactions (Figure 5B–D). This variability of the bisphenol ligand interactions with may have led to the introduction of significant mobility into the assimilation of the coregulator binding surface formed by the helices h3, h4, and h12. In addition to the multiple BPAF configurations observed in the crystal structures (Delfosse et al. 2012), we found that the other two BPs were also capable of accommodating alternate binding conformations when they occupied the ligand-binding site of .
Figure 5.
Ligand interactions from the molecular dynamic (MD) structures. The ligand interactions with estrogen receptor (ER) that are found in the representative final structures from MD simulations of (A) estradiol (E2), (B) bisphenol A (BPA), (C) bisphenol AF (BPAF), and (D) bisphenol S (BPS). In B, C, and D, when the ligands were placed in the canonical conformation (conformation 1) at the beginning of the simulations, the systems are labeled as ligand (1)–, and when the ligands were placed in alternate conformations (conformation 2) in the beginning of the simulations, they are labeled as ligand (2)–. However, as noted from the figure, ligand conformations evolved during the dynamics. The corresponding electrostatic potential surface of is shown in left with the coregulator binding site of each system shown inside white circle. residues potentially interacting with coregulator residues are labeled. (E) Comparison of the structures near the coregulator binding surface from the MD simulations of ER with various ligands. The structures were superimposed based on the h3, h4, and h12 loops as the reference. All representative structures from various ligand-bound simulations are shown in teal, along with the starting X-ray structure of the E2-bound system (in white) with its steroid receptor coactivator 1 (SRC-1) peptide (shown in yellow). The 9-residue SRC-1 peptide from the X-ray crystal structure is also shown. The leucine residues of the motif are shown in the space-filling representation. The other residues from SRC-1 are represented in the ball and stick form.
Next, we focused on the variations in electrostatic structures of the coregulator binding surface defined by the helices h4, h12, and a part of h3. From the available X-ray crystal structures, the location of the cofactor binding surface was identified. Since we recognized the coregulator binding surface on , changes in structures and other associated properties of this surface during dynamics were used to characterize the coregulator binding. Information gathered from electrostatic potentials (ESP) mapped onto the surface of the molecule was a reasonable tool for predicting what type of interactions the molecule may have with an incoming molecule. Differentiation of coregulator binding due to variable ligand occupancy at the ligand-binding site can be carried out by comparing the ESP of the above surface. ESP surfaces calculated using the Amber charge distribution for the atoms in the amino acid residues of systems with different ligands at the ligand-binding site are given in Figure 5E (on the right side of each ligand-binding site conformation). In the figures showing electrostatic potential energy surfaces (Figures 5A–D, right), coregulator interaction areas bracketed by helices h3, h4, and h12 are circled in white. For (shown in Figure 5A), the basic residues K362 and R363 identified in blue depicted in the top part of the circle interacted with some coregulator residues. The acidic residues E380, D538, E542, and D545 (represented in red in the bottom of the circle) have also been shown to interact with coregulator residues. The middle region stayed charge neutral to facilitate the interaction with the coregulator binding motif. All three bisphenol compounds preserved general features of their electrostatic potential of this area suitable for cofactor binding (Figures 5B–D). However, the general electrostatic potential features of the coregulator interacting surfaces of the ER with various BPs were comparable to those of ; some changes in the fine details were observed in the ESP surface of the relevant area, especially when the same ligand occupied the ligand-binding site in two different conformations.
The three main helices, h3, h4, and h12, are presented in Figure 5E that define the cofactor binding surface. The embedded leucine residues in the motif of the SRC-1 peptide are shown using the space-filling model. The space for leucine binding is preserved in all systems in their representative structures, as in Figure 5E, and variations in the position of the backbone h4 and h12 can also be observed. Interactions of the residue with helix h4 were altered due to the mobility observed in that helix region. Also, the residues in the N-terminal side from of the SRC-1 peptide may potentially be in direct contact with h12 (specifically with the negatively charged residues marked in Figure 4), and even small variations in the position of h12 may lead to significant alteration in the binding strength of the coregulator peptide and . Therefore, taken together, the information from the electrostatic potential surfaces and the variations in the side chain locations of potentially cofactor interacting residues in helices h3, h4, and h12 supports the observed variations in binding affinity of various coregulators, even when the same ligand is present in the ligand-binding site.
Discussion
The ER ERE–Mediated Transcriptional Activity and Alteration of ER Target Genes by BPA, BPAF, and BPS
A recent review concluded BPA is a reproductive toxicant based on a series of studies reporting that BPA impacts female reproduction and potentially affects the male reproductive system in humans and animals (Peretz et al. 2014). As BPA substitutes are replacing BPA in manufacturing, these analogs, like BPAF and BPS, are now more prevalent in consumer products (Liao et al. 2012a; Liao et al. 2012b; Liao and Kannan 2014). Our studies concluded that BPA and BPAF can function as EDCs by acting as cell type–specific agonists () or antagonists () for mouse and (Li et al. 2012). In the comparison of twelve EDCs, including BPA and BPAF, we found that EDCs with similar chemical structures tended to have comparable mouse and ERE-mediated activities. However, similar chemical structures did not correlate with ligand-binding affinities of the EDCs to the ERs (Li et al. 2013). In the present study, we found that BPA and BPAF activate ERE-mediated transcription for human and , and the results were comparable due to the high homology between the DNA sequences of ERs in mouse and human. Interestingly, the estrogenic activity of BPS is more in the cell systems we used. Human WISP-2/CCN5 is an important regulator involved in the maintenance of a differentiated phenotype in breast tumor epithelial cells (Fritah et al. 2008). Promoter analysis showed that endogenous CBP and p21 were recruited in an E2-dependent manner to the chromosomal human WISP-2/CNN5 promoter, which contains an ERE site in MCF-7 cells (Fritah et al. 2006). Using an ERE site in the human WISP-2/CNN5 gene promoter as a tool, we found that BPA, BPAF, and BPS induced direct binding to the ERE site of this gene promoter. Using Ishikawa cells stably expressing mouse , we found that ER target genes, including GREB1, PGR, and WISP-2/CNN5, were increased by BPA and BPAF (Li et al. 2012, 2013). Here, we confirmed the effect of BPA, BPAF, and BPS on three ER target genes in MCF-7 cells, suggesting these target genes may be commonly upregulated by BPs.
Differential Coregulator Recruitment in the Complex
Previous studies showed that ER mediates transcription using a common cohort of coregulators through many target genes and recruitment was based on ER confirmation (DeVilbiss et al. 2013). Coregulators are intrinsically disordered proteins that become structured upon interaction with other proteins. This flexibility allows a single coregulator to harbor many TF binding motifs that cause intrinsically disordered regions of the protein (Millard et al. 2013). In this study, we used a peptide chip array that contains the most well-characterized 154 NR interaction surfaces from 66 coregulators. These included coactivators with highly conserved helical LXXLL (NR box) and corepressors with highly conserved helical LXXXIXXXL (CoRNR box) motifs, and were used to investigate the modulation of coregulators in the complex. Our findings indicated that coactivators such as SRCs and PGCs were differentially recruited in the presence of BPA, BPAF, and BPS. Interestingly, there are more corepressors, such as small heterodimer partner (SHP) and receptor-interacting protein 140 (RIP140), recruited by the complex; this may explain why BPS has weaker estrogenic activation than BPA and BPAF in our in vitro system. Of note, unlike other coregulators that are known either for the coactivating or co-repressing roles, RIP140 is capable of acting both as a coactivator and a co-repressor in the different biological systems (Nautiyal 2017). From our peptide studies, we noticed the differential bindings of nuclear receptor-interacting protein 1 (NRIP) peptides, which were deviated from RIP140 in the presence of BPA, BPAF, and BPS. Furthermore, a more recent study indicated that BPS had more than tenfold activity/toxicity in vivo compare to in vitro (Conley et al. 2016). Our new findings may also enhance our understanding of differential molecular mechanisms between the in vivo and in vitro studies and the biological activities in a ligand- and tissue-specific manner.
Dynamic Simulations of Bisphenol Chemicals Bound to
Binding interactions of several bisphenol compounds and the endogenous ligand E2 with were characterized using the solution structure evaluations by MD simulations. E2 was recognized as a ligand with relatively strong binding interactions and was used as a reference system. Among the bisphenol compounds studied, BPA showed weaker interactions with at the binding site, while BPAF and BPS showed comparable interaction strengths to E2 in their binding conformations. However, energetics and structures of bisphenol compounds display variable binding modes with differing strengths at the ligand-binding site of ER.
Ligand-free simulations are quite instructive in estimating stabilities brought into due to ligand binding. In addition, they can provide valuable information on intrinsic stabilities of tertiary structural motifs of . The major structural change that is said to be associated with the ligand-binding is the relocation of the h12 helix to create the coregulator binding surface. Our simulation, starting with the agonist form of , showed a rather stable structure around the coregulator binding site bracketed by helices h3, h4, and h12 over the 650-ns simulations, even in the absence of any ligand in the ligand-binding site. Once positioned, h12 in the agonist conformation seemed to be in a rather stable conformation compared to the other conformations for this helix in . However, some noticeable deviations of h4 can be seen in the backbone alignment during dynamics.
BPA, BPAF, and BPS show varying conformations in the ligand-binding site of ER. Dynamical trajectory calculations of bisphenol ligand-bound systems showed no major modifications in the coregulator-binding surface due to ligand mobility, even with a variable conformation space for ligands at the ligand-binding site. However, these minor alterations may be sufficient to introduce variability to the cofactor binding strengths specifically due to the repositioning of some key side chains in helices h4 and h12. Further studies are underway to characterize the antagonist features of BPs interacting with .
Conclusions
In summary, our data indicate that BPA, BPAF, and BPS have varying estrogenic activity for and , except BPS, which did not show activation of . We also conclude that coregulators are differentially recruited in the ligand– complex when bound with BPA, BPAF, and BPS, which may help explain their differential biological actions. This is the first report to investigate coregulator recruitment of the complex and the subsequent discovery that BPs stimulate formation of more corepressor complexes than E2. Molecular modeling suggests BPs can bind to the ER–ligand-binding domain with differing energetics and conformations. Furthermore, we characterized the surface of coregulator binding on in complexes with bisphenol compounds using molecular modeling results. Taken together, these findings provide an important basis for the understanding of the molecular mechanisms of BP activity in ER-mediated activation and the differential biological activities of these EDCs that are ubiquitous in our everyday products.
Supplemental Material
Supplemental Material
Acknowledgments
We thank B. Goodwin of the National Center for Advancing Translational Sciences (NCATS)/National Institutes of Health (NIH) for providing the U2OS cell lyses. We thank S. Fenton and L. Pedersen for critical review of this manuscript. Research support was provided by the Division Intramural Research of the NIEHS/NIH to K.S.K. through Z01 ES70065 and to L.P. through Z01 ES043010.
References
- Akahori Y, Nakai M, Yamasaki K, Takatsuki M, Shimohigashi Y, Ohtaki M. 2008. Relationship between the results of in vitro receptor binding assay to human estrogen receptor alpha and in vivo uterotrophic assay: comparative study with 65 selected chemicals. Toxicol In Vitro 22(1):225–231, PMID: 17904329, 10.1016/j.tiv.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Bay K, Andersson AM, Skakkebaek NE. 2004. Estradiol levels in prepubertal boys and girls–analytical challenges. Int J Androl 27(5):266–273, PMID: 15379966, 10.1111/j.1365-2605.2004.00487.x. [DOI] [PubMed] [Google Scholar]
- Beekmann K, de Haan LH, Actis-Goretta L, Houtman R, van Bladeren PJ, Rietjens IM. 2015. The effect of glucuronidation on isoflavone induced estrogen receptor and mediated coregulator interactions. J Steroid Biochem Mol Biol 154:245–253, PMID: 26361015, 10.1016/j.jsbmb.2015.09.002. [DOI] [PubMed] [Google Scholar]
- Bermudez DS, Gray LE Jr, Wilson VS. 2010. Modeling the interaction of binary and ternary mixtures of estradiol with bisphenol A and bisphenol AF in an in vitro estrogen-mediated transcriptional activation assay (T47D-KBluc). Toxicol Sci 116(2):477–487, PMID: 20498000, 10.1093/toxsci/kfq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KA, Li Y, Arao Y, Petrovich RM, Korach KS. 2011. Selective mutations in estrogen receptor alpha D-domain alters nuclear translocation and non-estrogen response element gene regulatory mechanisms. J Biol Chem 286(14):12640–12649, PMID: 21285458, 10.1074/jbc.M110.187773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KA, Li Y, Liu L, Korach KS. 2014. Research resource: Comparison of gene profiles from wild-type and hinge region mutants. Mol Endocrinol 28(8):1352–1361, PMID: 24947674, 10.1210/me.2014-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case DA, Babin V, Berryman JT, Betz RM, Cai Q, Cerutti DS, et al. 2014. Amber.14. San Francisco, CA:University of California, San Francisco. [Google Scholar]
- Conley JM, Hannas BR, Furr JR, Wilson VS, Gray LE Jr.. 2016. A demonstration of the uncertainty in predicting the estrogenic activity of individual chemicals and mixtures from an in vitro estrogen receptor transcriptional activation assay (T47D-KBluc) to the in vivo uterotrophic assay using oral exposure. Toxicol Sci 153(2):382–395, PMID: 27473340, 10.1093/toxsci/kfw134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S, Lonard DM, O'Malley BW. 2014. Nuclear receptor coactivators: master regulators of human health and disease. Annu Rev Med 65:279–292, PMID: 24111892, 10.1146/annurev-med-051812-145316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delfosse V, Grimaldi M, Pons JL, Boulahtouf A, le Maire A, Cavailles V, et al. 2012. Structural and mechanistic insights into bisphenols action provide guidelines for risk assessment and discovery of bisphenol A substitutes. Proc Natl Acad Sci USA 109(37):14930–14935, PMID: 22927406, 10.1073/pnas.1203574109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deroo BJ, Korach KS. 2006. Estrogen receptors and human disease. J Clin Invest 116(3):561–570, PMID: 16511588, 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVilbiss AW, Boyer ME, Bresnick EH. 2013. Establishing a hematopoietic genetic network through locus-specific integration of chromatin regulators. Proc Natl Acad Sci USA 110(36):E3398–E3407, PMID: 23959865, 10.1073/pnas.1302771110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66(Pt 4):486–501, PMID: 20383002, 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritah A, Redeuilh G, Sabbah M. 2006. Molecular cloning and characterization of the human WISP-2/CCN5 gene promoter reveal its upregulation by oestrogens. J Endocrinol 191(3):613–624, PMID: 17170219, 10.1677/joe.1.07009. [DOI] [PubMed] [Google Scholar]
- Fritah A, Saucier C, De Wever O, Bracke M, Bièche I, Lidereau R, et al. 2008. Role of WISP-2/CCN5 in the maintenance of a differentiated and noninvasive phenotype in human breast cancer cells. Mol Cell Biol 28(3):1114–1123, PMID: 18070926, 10.1128/MCB.01335-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grignard E, Lapenna S, Bremer S. 2012. Weak estrogenic transcriptional activities of bisphenol A and bisphenol S. Toxicol In Vitro 26(5):727–731, PMID: 22507746, 10.1016/j.tiv.2012.03.013. [DOI] [PubMed] [Google Scholar]
- Hall JM, McDonnell DP. 2005. Coregulators in nuclear estrogen receptor action: from concept to therapeutic targeting. Mol Interv 5(6):343–357, PMID: 16394250, 10.1124/mi.5.6.7. [DOI] [PubMed] [Google Scholar]
- Héliès-Toussaint C, Peyre L, Costanzo C, Chagnon MC, Rahmani R. 2014. Is bisphenol S a safe substitute for bisphenol A in terms of metabolic function? An in vitro study. Toxicol Appl Pharmacol 280(2):224–235, PMID: 25111128, 10.1016/j.taap.2014.07.025. [DOI] [PubMed] [Google Scholar]
- Henley DV, Korach KS. 2010. Physiological effects and mechanisms of action of endocrine disrupting chemicals that alter estrogen signaling. Hormones (Athens) 9(3):191–205, PMID: 20688617, 10.14310/horm.2002.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutti D, Kralli A. 2001. Pgc-1, a versatile coactivator. Trends Endocrinol Metab 12(8):360–365, PMID: 11551810, 10.1016/S1043-2760(01)00457-X. [DOI] [PubMed] [Google Scholar]
- Koppen A, Houtman R, Pijnenburg D, Jeninga EH, Ruijtenbeek R, Kalkhoven E. 2009. Nuclear receptor-coregulator interaction profiling identifies TRIP3 as a novel peroxisome proliferator-activated receptor gamma cofactor. Mol Cell Proteomics 8(10):2212–2226, PMID: 19596656, 10.1074/mcp.M900209-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Arao Y, Hall JM, Burkett S, Liu L, Gerrish K, et al. 2014. Research resource: STR DNA profile and gene expression comparisons of human BG-1 cells and a BG-1/MCF-7 clonal variant. Mol Endocrinol 28(12):2072–2081, PMID: 25321415, 10.1210/me.2014-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Burns KA, Arao Y, Luh CJ, Korach KS. 2012. Differential estrogenic actions of endocrine-disrupting chemicals bisphenol A, bisphenol AF, and zearalenone through estrogen receptor and in vitro. Environ Health Perspect 120(7):1029–1035, PMID: 22494775, 10.1289/ehp.1104689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Luh CJ, Burns KA, Arao Y, Jiang Z, Teng CT, et al. 2013. Endocrine-disrupting chemicals (EDCs): in vitro mechanism of estrogenic activation and differential effects on ER target genes. Environ Health Perspect 121(4):459–466, PMID: 23384675, 10.1289/ehp.1205951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C, Liu F, Kannan K. 2012a. Bisphenol S, a new bisphenol analogue, in paper products and currency bills and its association with bisphenol A residues. Environ Sci Technol 46(12):6515–6522, PMID: 22591511, 10.1021/es300876n. [DOI] [PubMed] [Google Scholar]
- Liao C, Liu F, Moon HB, Yamashita N, Yun S, Kannan K. 2012b. Bisphenol analogues in sediments from industrialized areas in the United States, Japan, and Korea: Spatial and temporal distributions. Environ Sci Technol 46(21):11558–11565, PMID: 23020513, 10.1021/es303191g. [DOI] [PubMed] [Google Scholar]
- Liao C, Kannan K. 2014. A survey of bisphenol A and other bisphenol analogues in foodstuffs from nine cities in China. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 31(2):319–329, PMID: 24262000, 10.1080/19440049.2013.868611. [DOI] [PubMed] [Google Scholar]
- Lonard DM, O'Malley BW. 2007. Nuclear receptor coregulators: Judges, juries, and executioners of cellular regulation. Mol Cell 27(5):691–700, PMID: 17803935, 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Lonard DM, O'Malley BW. 2012. Nuclear receptor coregulators: modulators of pathology and therapeutic targets. Nat Rev Endocrinol 8(10):598–604, PMID: 22733267, 10.1038/nrendo.2012.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubahn DB, Moyer JS, Golding TS, Couse JF, Korach KS, Smithies O. 1993. Alteration of reproductive function but not prenatal sexual development after insertional disruption of the mouse estrogen receptor gene. Proc Natl Acad Sci USA 90(23):11162–11166, 10.1073/pnas.90.23.11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell DP, Norris JD. 2002. Connections and regulation of the human estrogen receptor. Science 296(5573):1642–1644, PMID: 12040178, 10.1126/science.1071884. [DOI] [PubMed] [Google Scholar]
- Millard CJ, Watson PJ, Fairall L, Schwabe JW. 2013. An evolving understanding of nuclear receptor coregulator proteins. J Mol Endocrinol 51(3):T23–T36, PMID: 24203923, 10.1530/JME-13-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nautiyal J. 2017. Transcriptional coregulator RIP140: an essential regulator of physiology. J Mol Endocrinol 58(3):R147–R158, PMID: 28073818, 10.1530/JME-16-0156. [DOI] [PubMed] [Google Scholar]
- Nilsson S, Mäkelä S, Treuter E, Tujague M, Thomsen J, Andersson G, et al. 2001. Mechanisms of estrogen action. Physiol Rev 81(4):1535–1565, PMID: 11581496. [DOI] [PubMed] [Google Scholar]
- O'Lone R, Frith MC, Karlsson EK, Hansen U. 2004. Genomic targets of nuclear estrogen receptors. Mol Endocrinol 18(8):1859–1875, PMID: 15031323, 10.1210/me.2003-0044. [DOI] [PubMed] [Google Scholar]
- Onufriev A, Bashford D, Case DA. 2004. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins 55(2):383–394, PMID: 15048829, 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- Perera L, Li Y, Coons LA, Houtman R, van Beuningen R, Goodwin B, et al. 2017. Binding of bisphenol A, bisphenol AF, and bisphenol S on the androgen receptor: coregulator recruitment and stimulation of potential interaction sites. Toxicol In Vitro 44:287–302, PMID: 28751236, 10.1016/j.tiv.2017.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretz J, Vrooman L, Ricke WA, Hunt PA, Ehrlich S, Hauser R, et al. 2014. Bisphenol A and reproductive health: update of experimental and human evidence, 2007–2013. Environ Health Perspect 122(8):775–786, PMID: 24896072, 10.1289/ehp.1307728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson K, Gustafsson JA. 2001. Role of estrogen receptor beta in estrogen action. Annu Rev Physiol 63:165–192, PMID: 11181953, 10.1146/annurev.physiol.63.1.165. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Spiegelman BM. 2003. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev 24(1):78–90, PMID: 12588810, 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- Rochester JR. 2013. Bisphenol A and human health: a review of the literature. Reprod Toxicol 42:132–155, PMID: 23994667, 10.1016/j.reprotox.2013.08.008. [DOI] [PubMed] [Google Scholar]
- Rochester JR, Bolden AL. 2015. Bisphenol S and F: a systematic review and comparison of the hormonal activity of bisphenol A substitutes. Environ Health Perspect 123(7):643–650, PMID: 25775505, 10.1289/ehp.1408989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. 1993. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234(3):779–815, PMID: 8254673, 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Taylor AH, Al-Azzawi F. 2000. Immunolocalisation of oestrogen receptor beta in human tissues. J Mol Endocrinol 24(1):145–155, PMID: 10657006, 10.1677/jme.0.0240145. [DOI] [PubMed] [Google Scholar]
- Tcherepanova I, Puigserver P, Norris JD, Spiegelman BM, McDonnell DP. 2000. Modulation of estrogen receptor-alpha transcriptional activity by the coactivator PGC-1. J Biol Chem 275(21):16302–16308, PMID: 10748020, 10.1074/jbc.M001364200. [DOI] [PubMed] [Google Scholar]
- Thayer KA, Taylor KW, Garantziotis S, Schurman S, Kissling GE, Hunt D, et al. 2015. Bisphenol A, bisphenol S, and 4-hydroxyphenyl 4-isoprooxyphenylsulfone (BPSIP) in urine and blood of cashiers. Environ Health Perspect 24(4):437–44, PMID: 26309242, 10.1289/ehp.1409427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welshons WV, Nagel SC, vom Saal FS. 2006. Large effects from small exposures. III. Endocrine mechanisms mediating effects of bisphenol A at levels of human exposure. Endocrinology 147(6 suppl):S56–S69, PMID: 16690810, 10.1210/en.2005-1159. [DOI] [PubMed] [Google Scholar]
- Wetherill YB, Akingbemi BT, Kanno J, McLachlan JA, Nadal A, Sonnenschein C, et al. 2007. In vitro molecular mechanisms of bisphenol A action. Reprod Toxicol 24(2):178–198, PMID: 17628395, 10.1016/j.reprotox.2007.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



![Figure 1A shows chemical structures of 17 beta estradiol; ICI 182,780; Bisphenol A, Bisphenol AF, and Bisphenol S. Figure 1B comprises two plots respectively showing activation in Ishikawa cells stably expressing ER alpha cells and vector cells, plotting relative luc activity (fold change) (y-axis) across concentration (log[nanomolar]) (x-axis) for E2, BPA, BPAF, and BPS. Figures 1C and 1D plot the same for two plots showing activation of endogenous ER MCF-7 and BG-1 FR cells and HepG2 cells transfected with ER alpha and ER beta, respectively. Figure 1E comprises three bar graphs with standard error of the mean respectively plotting GREB1, PGR, and WISP-2/CNN5 (y-axis) treated with control, E2 10, BPA 100, BPAF 100, and BPS 100 (x-axis) for negative ICI and positive ICI. Figure 1F comprises two parts. The first part presents two bar graphs with standard error of the mean respectively plotting relative recruitment (percentage of input) (y-axis) treated with control and E2 10 (x-axis) for IgG and ER alpha under amplicon negative 699 virgule negative 422 (AP 1 plus ERE) and amplicon negative 593 virgule negative 422 (ERE). The second part presents the same data respectively plotting relative recruitment (percentage of input) (y-axis) treated with control, BPA 100, BPA 1000, BPAF 100, BPAF 1000, BPAS 100, and BPAS 1000 (x-axis).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/8a36/6014695/f818a29d4ecd/EHP2505_f1a.jpg)
![Figure 1A shows chemical structures of 17 beta estradiol; ICI 182,780; Bisphenol A, Bisphenol AF, and Bisphenol S. Figure 1B comprises two plots respectively showing activation in Ishikawa cells stably expressing ER alpha cells and vector cells, plotting relative luc activity (fold change) (y-axis) across concentration (log[nanomolar]) (x-axis) for E2, BPA, BPAF, and BPS. Figures 1C and 1D plot the same for two plots showing activation of endogenous ER MCF-7 and BG-1 FR cells and HepG2 cells transfected with ER alpha and ER beta, respectively. Figure 1E comprises three bar graphs with standard error of the mean respectively plotting GREB1, PGR, and WISP-2/CNN5 (y-axis) treated with control, E2 10, BPA 100, BPAF 100, and BPS 100 (x-axis) for negative ICI and positive ICI. Figure 1F comprises two parts. The first part presents two bar graphs with standard error of the mean respectively plotting relative recruitment (percentage of input) (y-axis) treated with control and E2 10 (x-axis) for IgG and ER alpha under amplicon negative 699 virgule negative 422 (AP 1 plus ERE) and amplicon negative 593 virgule negative 422 (ERE). The second part presents the same data respectively plotting relative recruitment (percentage of input) (y-axis) treated with control, BPA 100, BPA 1000, BPAF 100, BPAF 1000, BPAS 100, and BPAS 1000 (x-axis).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/8a36/6014695/e0994ddd46bf/EHP2505_f1b.jpg)
![Figure 3A shows two bar graphs with standard error of the mean plotting relative luc activity (fold change) (y-axis) for treated with VEC, ER alpha, SRC1, SRC1 plus ER alpha, SRC2, SRC2 plus ER alpha, SRC3, SRC3 plus ER alpha (x-axis) for controls and E2 10 for SRCs plus ER alpha and PGC1s plus ER alpha in the presence of E2, respectively. Figure 3b comprises nine line graphs plotting relative luc activity (fold change) (y-axis) across concentration (log[nanomolar]) (x-axis) in vec, ER alpha, SRC, SRC plus ER alpha for BPA: SRC1 plus ER alpha; BPA: SRC2 plus ER alpha; BPA: SRC3 plus ER alpha; BPAF: SRC1 plus ER alpha; BPAF: SRC2 plus ER alpha; BPAF: SRC3 plus ER alpha; BPS: SRC1 plus ER alpha; BPS: SRC2 plus ER alpha; and BPS: SRC3 plus ER alpha, respectively. Figure 3C comprises six line graphs plotting relative luc activity (fold change) (y-axis) across concentration (log[nanomolar]) (x-axis) in vec, ER alpha, PGC1, and PG1 plus ER alpha for BPA: PGC1 alpha plus ER alpha; BPA: PGC1 beta plus ER alpha; BPAF: PGC1 alpha plus ER alpha; BPAF: PGC1 beta plus ER alpha; BPS: PGC1 alpha plus ER alpha; and BPS: PGC1 beta plus ER alpha, respectively.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/8a36/6014695/a314eb9c469e/EHP2505_f3a.jpg)
![Figure 3A shows two bar graphs with standard error of the mean plotting relative luc activity (fold change) (y-axis) for treated with VEC, ER alpha, SRC1, SRC1 plus ER alpha, SRC2, SRC2 plus ER alpha, SRC3, SRC3 plus ER alpha (x-axis) for controls and E2 10 for SRCs plus ER alpha and PGC1s plus ER alpha in the presence of E2, respectively. Figure 3b comprises nine line graphs plotting relative luc activity (fold change) (y-axis) across concentration (log[nanomolar]) (x-axis) in vec, ER alpha, SRC, SRC plus ER alpha for BPA: SRC1 plus ER alpha; BPA: SRC2 plus ER alpha; BPA: SRC3 plus ER alpha; BPAF: SRC1 plus ER alpha; BPAF: SRC2 plus ER alpha; BPAF: SRC3 plus ER alpha; BPS: SRC1 plus ER alpha; BPS: SRC2 plus ER alpha; and BPS: SRC3 plus ER alpha, respectively. Figure 3C comprises six line graphs plotting relative luc activity (fold change) (y-axis) across concentration (log[nanomolar]) (x-axis) in vec, ER alpha, PGC1, and PG1 plus ER alpha for BPA: PGC1 alpha plus ER alpha; BPA: PGC1 beta plus ER alpha; BPAF: PGC1 alpha plus ER alpha; BPAF: PGC1 beta plus ER alpha; BPS: PGC1 alpha plus ER alpha; and BPS: PGC1 beta plus ER alpha, respectively.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/8a36/6014695/5952f11862b8/EHP2505_f3b.jpg)